Abstract
Triptolide (TPL) is an active extract from a Chinese herb, which has been used for centuries in China. TPL exhibits numerous bioactivities and pharmacological effects, including antitumor, anti-inflammatory and immunosuppressive activities. However, previous studies have further revealed a multi-target toxicity of TPL, including reproductive toxicity, hepatotoxicity and renal cytotoxicity. To validate the clinical benefit and reduce the risk of TPL application, studies have investigated the combination of TPL with other reagents to allow lower doses and decrease toxicity. The present study reported that TPL and the insulin-like growth factor-1 receptor (IGF1R) inhibitor AG1024synergistically inhibited cell proliferation and induced apoptosis in triple-negative breast cancer cells. Overexpression of B-cell lymphoma 2 partially reversed the TPL and AG1024-induced increase in apoptosis. A similar synergistic effect was observed with a combination of AG1024 and cisplatin, a DNA damage inducer, in MDA-MB-231 cells. These results suggested that inhibition of IGF1R may sensitize triple-negative breast cancer cells to DNA damage inducers. Using publicly available data from The Cancer Genome Atlas, an amplification and gain of copy number of IGF1R was observed in 38% of triple-negative breast tumors (n=82), 26% of estrogen receptor (ER)-negative tumors (n=174) and 10% of ER-positive tumors (n=594). Similarly, a higher alteration frequency of IGF1R was identified in basal-like breast tumors compared with luminal A/B-like breast tumors. Overexpressed proteins associated with these alterations were revealed to be significantly enriched in multiple oncogenic signaling pathways, key transcription factor networks and DNA repair pathways. In summary, the present study suggested that inhibition of IGFR signaling and induction of DNA damage may exhibit synergistic effects for the treatment of triple-negative and ER-negative breast cancer.
Keywords: triptolide, AG1024, insulin-like growth factor 1 receptor, triple-negative breast cancer, DNA damage, B-cell lymphoma 2
Introduction
Triptolide (TPL) is an active extract obtained from the Chinese herb Tripterygium wilfordii that has been used for centuries to treat autoimmune diseases in China (1). Numerous studies have reported that TPL has antitumor, anti-inflammatory and immunosuppressive activities. The antitumor activity of TPL has been investigated in various types of cancer cells in vitro and in vivo (2). Furthermore, recent studies have revealed that TPL enhances the sensitivity of cancer cells to chemotherapy by inducing apoptosis (3,4). To the best of our knowledge, among all the cancer types that have been investigated, breast cancer exhibits the highest sensitivity to TPL (5). Breast cancer is a common type of cancer in females. A number of distinct molecular subtypes of breast cancer exist, including luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-positive, basal-like and normal-like (6,7). In total, ~70% of cases of triple-negative breast cancer exhibit a basal-like subtype (8). The treatment of breast cancer, particularly triple-negative breast cancer, is a challenge due to the lack of targets, including for hormone receptors and amplified HER2 expression levels (9).
The associated molecular mechanisms by which TPL inhibits breast cancer cells have been investigated. n In breast cancer, TPL induces p53-dependent and lysosomal-mediated apoptosis, and inhibits numerous oncogenic signaling pathways, including the Wnt/β-catenin, the protein kinase B (Akt) and the focal adhesion kinase-signaling pathway (3,10,11). Notably, TPL inhibits key transcriptional factors, including MYC and estrogen receptor (ER), which suppresses associated target networks (12,13).
Although TPL exhibits a number of bioactivities and pharmacological effects, the clinical application of TPL has been restricted due to a number studies reporting multi-target toxicity of the extract, including reproductive toxicity, hepatotoxicity and renal cytotoxicity (14,15). Numerous studies have attempted to identify a combination of TPL with other regents, which support the function of TPL at a low dose and reduce the toxicity (16–18). It has been reported that inhibition of ER-negative breast cancer cell growth requires a high dose of TPL, which may induce toxicity (19). The present study investigated a new strategy to enhance TPL sensitivity in triple-negative breast cancer cells. Increased expression and activation of insulin-like growth factor-1 receptor (IGF-1R) and its associated downstream signaling components has been reported in clinical breast cancer samples, and has been associated with disease progression and recurrence (20). Furthermore, overexpression of IGF1R has been demonstrated to be associated with radio- or tamoxifen-resistant breast cancer cells. The IGF1R inhibitor AG1024 reduces this resistance by inhibition of IGF1R signaling (21). The present study reported that AG1024 synergistically enhanced apoptosis induced by low doses of TPL and cisplatin in the triple-negative breast cancer cells MDA-MB-231. In addition, high amplification of IGF1R in triple-negative tumors may serve as a potential target of a combination of AG1024 and DNA damage reagents.
Materials and methods
Chemicals, antibodies and plasmids
Triptolide (TPL), AG1024 and cisplatin were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Cell Counting Kit-8 (CCK-8) was obtained from Dojindo Molecular Technologies, Inc. (Kumamoto, Japan). Antibodies against B-cell lymphoma (Bcl-)2 (sc-7382; 1:1,000) and β-actin (sc-8432, 1:2,500) were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). The anti-phospho-H2A.X antibody (07–164, 1:1,000) was purchased from EMD Millipore (Billerica, MA, USA). Cleaved caspase-3, caspase-3 and IGF1R antibodies were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). The expression plasmid 3336 pcDNA3 Bcl-2 and the empty vector 336 pcDNA3 were kindly provided by Dr. Stanley Korsmeyer of the Dana-Farber Cancer Institute, Inc (cat. no. 8768; Addgene, Boston, MA, USA) (22).
Cell culture
Human breast carcinoma cell lines MDAMB231 and MDA-MB-436 were obtained from American Type Cell Culture Collection (Manassas, VA, USA). Cells were grown in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare, Logan, UT, USA), 100 U/ml penicillin and 100 U/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a humidified 5% CO2 atmosphere. Cells from <20 passages were used for experiments.
Cell viability assay
MDAMB231 and MDA-MB-436 cells were seeded in 96-well plates at a density of 4×103 cells/well, followed by treatment with DMSO or increasing doses of TPL (5 to 80 nM) and AG1024 (1 to 40 µM) for 3 days at 37°C. Cells were then incubated with 10 µl CCK-8 reagent for 1 h and the optical density value was measured at 450 nm, according to the manufacturer's protocol.
Cell cycle analysis
Cell cycle analysis was performed to evaluate the number of cells in the sub-G1 phase. MDA-MB-231 cells were seeded at 60% confluence and allowed to attach overnight. Cells were then treated with various concentrations of drugs (10 nM TPL, 10 µM AG1024, or 1 µM cisplatin) for 96 h at 37°C, trypsinized, washed with PBS and fixed in 70% ethanol at −20°C overnight. Prior to analysis, cells were washed with PBS, suspended in cold propidium iodide (PI; Sigma-Aldrich; Merck KGaA) solution (50 µl/ml in PBS) and incubated at room temperature in the dark for 30 min. Flow cytometry was then performed using a flow cytometer (BD FACSAria system) and analyzed using BD FACSDiva software version 5.0 (BD Biosciences, USA).
Transient transfection assay
A reverse transfection protocol was used to knockdown IGF1R by small interfering RNA (siRNA). 25 nM SMARTpool siRNAs (siIGF1R, L-003012-00-0005; and non-targeting siRNA;,D-001810-0X; GE Healthcare Dharmacon, Inc., Lafayette, CO. USA) was transfected into MDA-MB-231 cells using RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Transfected cells were plated in 96-well plates at a density of 2×103 cells/well for proliferation assays at various time points (day 0–5) or plated for protein collection 72 h after transfection.
Following treatment of the MDA-MB-231 cells with drugs (10 nM TPL and 10 µM AG1024) for 2 days at 37°C, cells were transfected with the expression plasmid 3336 pcDNA3 Bcl-2 (2 µg) or empty vector (2 µg) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) Cells were harvested 48 h after transfection for western blot analysis and cell cycle analysis.
Western blotting
Cells were harvested and lysed in radioimmunoprecipitation assay buffer (Invitrogen; Thermo Fisher Scientific, Inc.). Lysates were centrifuged at 10,000 × g for 15 min at 4°C and supernatants were collected as whole cell extracts. The protein concentration was determined using a Pierce™ Coomassie Plus (Bradford) assay kit (Thermo Fisher Scientific, Inc.). Equivalent amounts (20 µg) of protein were separated by SDS-PAGE (4–12%) and transferred to a PVDF membrane. Following blocking with 5% milk at room temperature for 1 h, the membranes were incubated with the corresponding primary antibodies for 2 h and horseradish peroxidase-conjugated secondary antibodies at room temperature (Santa Cruz Biotechnology, Inc.; cat. no. sc-2005 and sc-2004, 1:8,000) for 1 h. Protein bands were detected using an ECL reagent (Thermo Fisher Scientific, Inc.). ImageJ software (version 1.41; National Institutes of Health, Bethesda, MD, USA) was used to quantify the blots.
Bioinformatics analysis
The amplification of IGF1R in different subtypes of breast tumor was analyzed using data regarding breast invasive carcinoma from The Cancer Genome Atlas (TCGA) (23,24) with the online cBioPortal for Cancer Genomics platform (http://www.cbioportal.org) (25). The ER status determined by immunohistochemistry was selected for the analysis of the association between IGF1R expression and ER status. A list of the overexpressed proteins associated with genomic alterations of IGF1R in TCGA cohort was downloaded using cBioPortal. Gene ontology and pathway analysis of the overexpressed proteins were performed using the ToppGene online tool (26).
Statistical analysis
Experiments were replicated a minimum of three times. Data are presented as the mean ± standard error of the mean. GraphPad Prism software, version 7 (GraphPad Software, Inc., La Jolla, CA, USA) was used to conduct statistical analyses; unpaired t-test and one-way ANOVA (followed by Dunnett's test) were used to compare the experimental groups with the control group. P<0.05 was considered to indicate a statistically significant difference.
Results
TPL and AG1024 synergistically inhibit the proliferation of triple-negative breast cancer cells
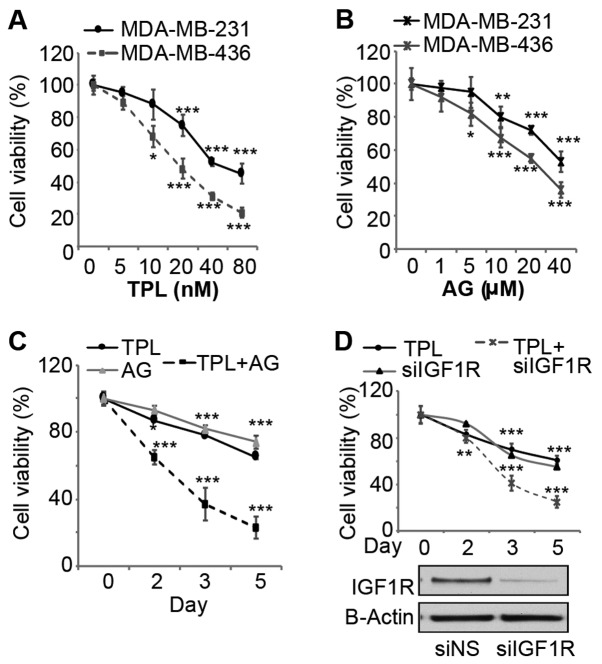
To evaluate effects of TPL and AG1024 on triple-negative breast cancer cell growth, MDA-MB-231 and MDA-MB-436 cells were treated with various doses of the two drugs and the viability was measured. After 3 days of treatment, TPL and AG1024 inhibited cell viability in a dose dependent-manner for the two cell lines (Fig. 1A and B). The sensitivity of MDA-MB-436 cells to TPL and AG1024 was higher compared with MDA-MB-231 cells, potentially due to the presence of a mutant BRCA1 (27). To evaluate synergistic effects, a combination of low doses (10 nM TPL and 10 mM AG1024) at which TPL or AG1024 alone cannot markedly inhibit cell growth was selected for cell proliferation assays with MDA-MB-231 cells. Notably, this combination of TPL and AG1024 inhibited cell viability by ~60% after 3 days of treatment (Fig. 1C). TPL and AG1024 at a low dose (10 nM TPL and 10 mM AG1024) alone only inhibited cell growth by ~20% (Fig. 1A and B). After 5 days, a combined treatment inhibited cell growth by ~80%. To specifically inhibit IGF1R, cells were transfected with siIGF1R and the transfection efficiency was confirmed by western blot analysis (Fig. 1D). siIGF1R alone inhibited cell viability by ~30% by day 3, while a combination of TPL and siIGF1R inhibited cell viability by ~60% (Fig. 1D). These results are consistent with the results of TPL and AG1024-treatment, which suggested that the synergistic effect was specific to the inhibition of IGF1R rather than other targets of AG1024.
Figure 1.
AG1024 enhances TPL-induced inhibition of the viability of triple-negative breast cancer cells. (A) TPL and (B) AG inhibited cell viability in a dose-dependent manner in MDA-MB-231 and MDA-MB-436 cells. Cells were treated with TPL or AG for 3 days and cell viability was determined by Cell Counting Kit-8. All values of cell viability were relative to the untreated cells. (C) A combination of TPL (10 nM) and AG (10 µM) inhibited cell viability to a greater extent than single drug treatment for 5 days. (D) A combination of TPL (10 nM) and siIGF1R inhibited cell viability to a greater extent compared with single treatment. Knockdown of IGF1R was confirmed by western blotting and b-actin was used as the loading control. Data are presented as the mean ± standard error of the mean (n=3). *P<0.05, **P<0.01 and ***P<0.001 vs. the control or single drug groups. AG, AG1024; si, small interfering; siNS, none specific siRNA; TPL, triptolide; IGF1R, insulin-like growth factor 1 receptor.
TPL and AG1024 synergistically induce apoptosis of triple-negative breast cancer cells
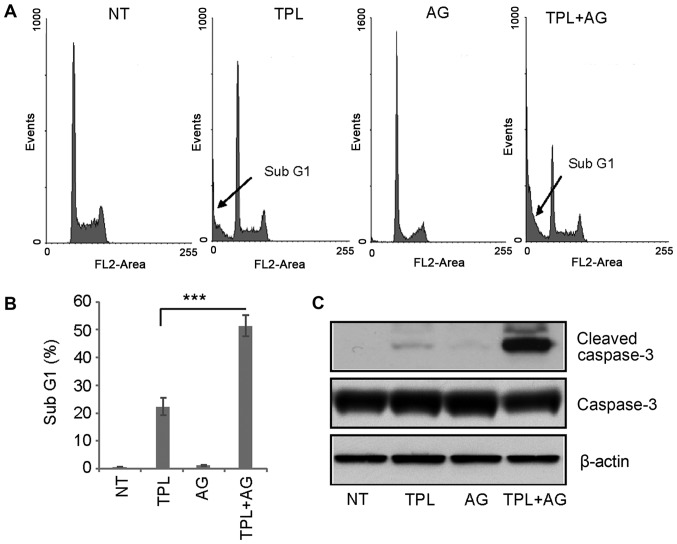
TPL has been reported to induce apoptosis of MDA-MB-231 cells (2). Therefore, flow cytometry was used in the present study to evaluate the effects of a combination of drugs on apoptosis. The sub-G1 cell population is understood to represent apoptotic cells. Following 4 days of treatment, 10 nM TPL increased the percentage of cells in the sub-G1 population to ~20%, while 10 µM AG1024 did not induce apoptosis (Fig. 2A and B). However, after 4 days of treatment with TPL and AG1024, the percentage of cells in the sub-G1 cell population was ~50% (Fig. 2A and B). These results were confirmed by western blotting. Notably, a high expression of cleaved caspase-3 was detected in TPL and AG1024-treated cells, while only a small amount of cleaved caspase-3 was detected in TPL-treated cells (Fig. 2C). In summary, these results suggested a synergistic effect of TPL and AG1024 in MDA-MB-231 cells.
Figure 2.
AG enhances TPL-induced apoptosis of MDA-MB-231 cells. (A) Cell cycle analysis was used to detected cells in the sub-G1 population. MDA-MB-231 cells were treated with AG and/or TPL for 4 days prior to analysis. (B) Quantification of the percentage of cells in the sub-G1 population. (C) Cleaved caspase-3 was detected by western blot analysis of all treated MDA-MB-231 cells. Data are presented as the mean ± standard error of the mean (n=3). ***P<0.001. NT, DMSO treatment; TPL, triptolide; AG, AG1024.
Overexpression of Bcl-2 partially inhibits TPL and AG1024-induced apoptosis
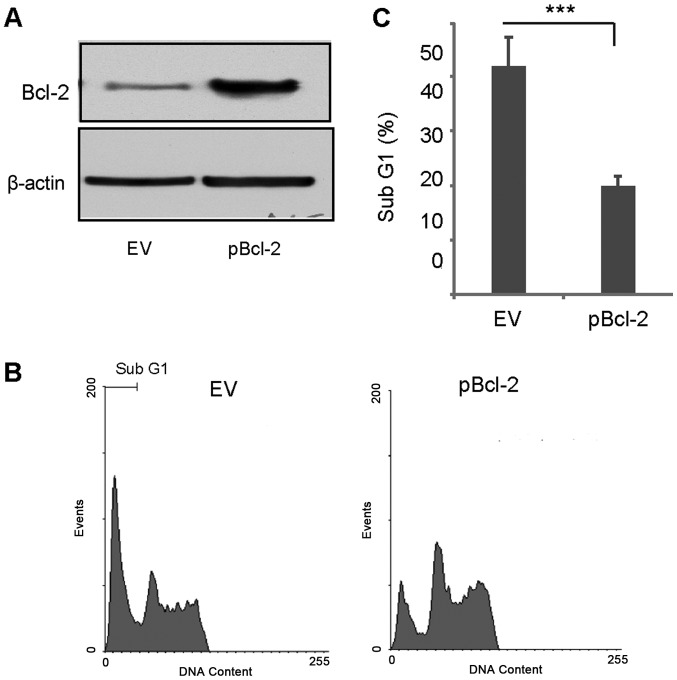
It has been reported that TPL induces caspase-mediated Bcl-2 cleavage, which contributes to TPL-induced apoptosis in MDA-MB-231 cells (28). Therefore, the present study transiently overexpressed Bcl-2 and treated the transfected cells with a combination of TPL and AG1024. Bcl-2 was successfully overexpressed in MDA-MB-231 cells (Fig. 3A). After 3 days of treatment with TPL and AG1024, the percentage of apoptotic cells in the Bcl-2 overexpressing group was significantly lower compared with the control group (Fig. 3B and C). This suggested cleavage of Bcl-2 may serve an important role in TPL and AG1024-induced apoptosis.
Figure 3.
Bcl-2 overexpression partially inhibited AG and TPL-induced apoptosis. (A) Bcl-2 was successfully overexpressed in the transfected cells. (B) Cells were treated with AG and TPL for 2 days and then transfected with the Bcl-2 overexpression plasmid. The percentage of cells in the sub-G1 population was detected in the EV-group and the pBcl-2 transfected group 2 days after transfection. (C) Quantified results of the percentage of cells in the sub-G1 population. Data are presented as the mean ± standard error of the mean (n=3). ***P<0.001. EV, empty vector; pBcl-2, Bcl-2 expression plasmid; TPL, triptolide; AG, AG1024; Bcl-2, B-cell lymphoma 2.
Synergy between AG1024 and DNA damage-inducing reagents enhances apoptosis in MDA-MB-231 cells
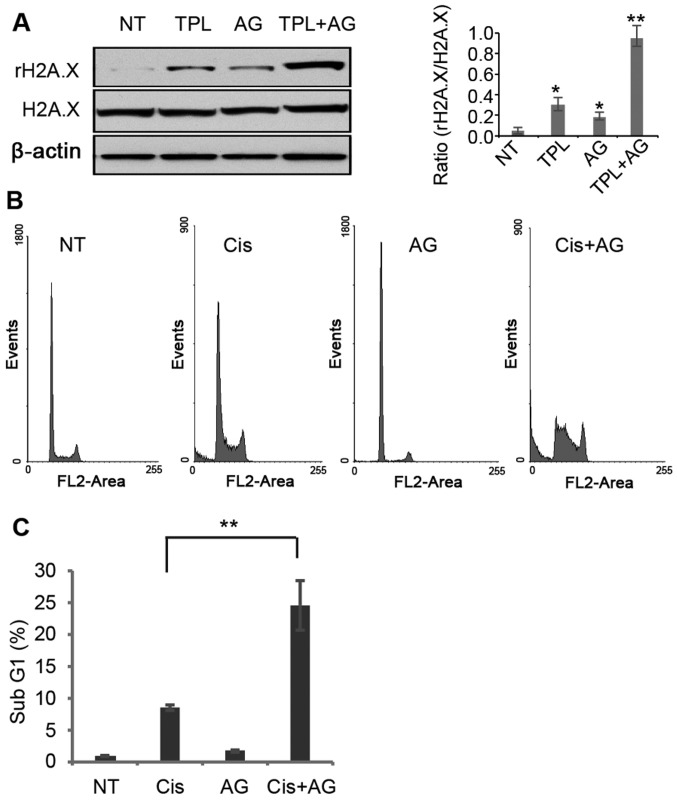
TPL has been reported to induce DNA damage in cancer cells (29). In MDA-MB-231 cells, TPL and AG1024 induced DNA damage, evidenced by a higher phosphorylation level of the DNA damage marker H2A.X (Fig. 4A). Notably, treatment with TPL and AG1024 increased H2A.X phosphorylation to a greater extent compared with treatment with TPL or AG1024 alone (Fig. 4A). It is hypothesized that AG1024 served a synergistic role with other DNA damage reagents. The DNA damage inducer cisplatin was selected to test this hypothesis. A low dose of cisplatin (1 µM) resulted in 8.3% of apoptotic cells, while a combined treatment of cisplatin (1 µM) and AG1024 (10 µM) significantly increased the number of apoptotic cells compared with cisplatin alone (Fig. 4B and C). These results suggest that a combined use of AG1024 and DNA damage reagents may provide a new approach for breast cancer treatment.
Figure 4.
Synergistic effect of AG and DNA damage-inducing reagents. (A) AG1024 enhanced TPL-induced DNA damage. Phosphorylation of H2A.X was detected by western blotting following treatment of cells with TPL and/or AG for 5 days. Values are presented as the mean ± standard error of the mean (n=3). *P<0.05 and **P<0.01 vs. the NT group. (B) AG1024 enhanced cis-induced apoptosis of MDA-MB-231 cells. Cells were treated with Cis and/or AG for 4 days and then cell cycle analysis was used to detect the percentage of cells in the sub-G1 population. (C) Quantified results of the percentage of cells in the sub-G1 population. Data are presented as the mean ± standard error of the mean (n=3). **P<0.01. Cis, cisplatin; AG, AG1024; TPL, triptolide; NT, DMSO treatment.
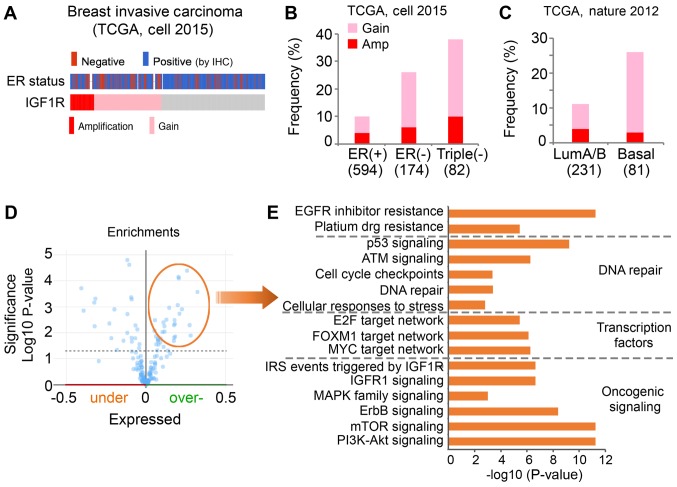
High level of IGF1R in ER-negative breast cancer
A high expression level of IGF1R has been reported in triple-negative MDA-MB-231 cells (30). Therefore, it was hypothesized that inhibition of IGF1R may increase the sensitivity of MDA-MB-231 cells to TPL and cisplatin, due to a higher activity of IGF1R signaling. The present study evaluated whether increased IGF1R expression was observed in patients with ER-negative breast cancer compared with patients with ER-positive breast cancer in TCGA clinical cohort (Fig. 5). Notably, genomic alteration of the IGF1R gene was observed in 18% of cases in the breast invasive TCGA cohort (Fig. 5A). In addition, a higher frequency of IGF1R amplification (Amp) and copy number gain (Gain) was observed in patients with ER-negative breast cancer compared with ER-positive breast cancer.
Figure 5.
A higher frequency of Amp and Gain of IGF1R is present in ER-negative breast tumors compared with ER-positive breast tumors. (A) Genomic alteration of IGF1R and ER status of breast invasive carcinoma cases of TCGA cohort. Gray represents individual tumors without an IGF1R alteration. (B) The frequency of IGF1R alteration in differential subtypes of tumors was analyzed in TCGA (cell, 2015) cohort. (C) The frequency of IGF1R alteration in differential subtypes of tumors was analyzed in TCGA (Nature, 2012) cohort. (D) Overexpressed proteins associated with genomic alterations of IGF1R in the Cell 2015 TCGA cohort were identified. (E) Pathway analysis was performed with the significantly overexpressed genes using the ToppGene online tool. The x-axis indicates the significance of the enrichment of the pathway. The y-axis presents the pathway term. TCGA, The Cancer Genome Atlas; ER, estrogen receptor; IGF1R, insulin-like growth factor 1 receptor; IHC, immunohistochemistry; Gain, copy number gain; Amp, amplification; LumA/B, Luminal A/B-like A/B; EGFR, epidermal growth factor receptor; E2F, E2 factor; FOXM1, forkhead box M1; IRS, insulin receptor substrate; MAPK, mitogen-activated protein kinase pathway; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase.
To further evaluate, the present study investigated the frequency of IGF1R alterations, including Amp and Gain, in patients with ER-positive (n=594), ER-negative (n=174) and triple-negative (n=82) cancer. IGF1R alteration was observed in 10% of the ER-positive group (4% Amp, 6% Gain), 26% of the ER-negative group (6% Amp, 20% Gain) and 38% of the triple-negative group (10% Amp, 28% Gain; Fig. 5B). It has been reported that basal-like breast cancers are more aggressive compared with luminal A/B-like tumors, and the majority of basal-like subtypes are typically negative for ER, PR and HER2 (24). Similarly, the present study observed genomic alteration of IGF1R in 26% of basal-like tumors (3% Amp, 23% Gain) and only 11% of luminal A/B-like tumors (4% Amp, 7% Gain; Fig. 5C). In summary, these data suggested IGF1R was highly amplified and expressed in ER-negative and basal-like breast cancer.
To understand the pathways associated with genomic alterations of IGF1R in clinical samples, the present study downloaded a list of overexpressed proteins that are positively associated with an alteration of IGF1R (Amp/Gain) using the cBioPortal (Fig. 5D). Pathway analysis was performed with these proteins using the ToppGene online tool. The overexpressed proteins associated with IGF1R alteration were identified to be significantly enriched in multiple oncogenic signaling pathways (Fig. 5E). As expected, proteins were revealed to be significantly enriched in IGF1R signaling. Furthermore, enrichment of phosphatidylinositol 3-kinase (PI3K)/Akt and mammalian target of rapamycin signaling activation was observed. Networks of key transcription factors, including E2 factor, forkhead box M1 and MYC, were revealed to be activated in the cases with IGF1R alteration. In addition, the proteins associated with IGF1R were enriched in DNA repair pathways. Certain proteins were identified to be enriched in the platinum drug resistance pathway, which may explain the synergistic effects of cisplatin and AG1024 observed in the present study.
Discussion
TPL has been reported to exert potent anticancer effects via multiple molecular targets and signaling pathways. However, the side effects of TPL limit its clinical application for the treatment of cancer (2). The present study demonstrated that the IGF1R inhibitor AG1024 increased the sensitivity of triple-negative breast cancer cells to TPL. Therefore, it was suggested that inhibition of IGF1R signaling and induction of DNA damage may have synergistic effects in the treatment of ER-negative or triple-negative breast cancer. Notably, the current study identified a higher amplification of IGF1R in ER-negative and basal-like breast cancer.
A previous study reported that the ER-positive breast cancer cells (MCF-7) were more sensitive to TPL treatment due to repression of the ER pathway by TPL, and that TPL-induced apoptosis of MDA-MB-231 cells was ER-independent (19). In addition, it has been reported that MDA-MB-231 cells possess a high IGF1R expression (30), which indicates IGFR1 may serve a role in cell survival. Consistent with previous studies, the present study observed that inhibition of IGF1R signaling by AG1024 inhibited cell viability. A combined use of TPL and AG1024 at low doses demonstrated a high potency compared with single drug application. This may be due to a higher level of IGFR1 signaling in ER-negative breast cancer cells (30). It has been reported that suppression of the IGF system with AG1024 augments cisplatin-induced DNA damage (30). Consistent with these findings, the present study identified that AG1024 enhanced cisplatin-induced apoptosis of MDA-MB-231 cells. Similarly, TPL has been revealed to induce DNA damage in cancer cells (31). These results suggested a possible synergistic function of IGF1R signaling inhibition and inducement of DNA damage.
Previously, it has been reported that patients with IGF1R-postive/ER-negative breast cancer have a worse prognosis compared with patients with IGF1R-negative/ER-negative breast cancer (32). However, the association of IGF1R signaling with ER status and the aggressiveness of breast cancer remains controversial. A previous study reported the differing effects of IGF1R expression on the prognosis of patients with ER-positive vs. triple-negative invasive ductal breast carcinoma (IDC) (33). IGF1R expression in ER-positive IDC is strongly associated with a favorable disease-free survival (DFS) time; however, IGF1R expression is associated with a shorter DFS for patients with TN-IDC. The aforementioned studies were all performed based on immunohistochemical staining of IGF1R to determine its protein expression. To the best of our knowledge, the present study was the first to identify a higher Amp and Gain of IGF1R in ER-negative breast cancer compared with ER-positive cancer. Furthermore, a difference in the amplification frequency of IGF1R was revealed between luminal A/B-like and basal-like breast tumors.
IGF1R signaling activates several downstream signaling pathways, including the PI3K/Akt signaling pathway and mitogen-activated protein kinase pathway (34,35). In the enrichment analysis performed in the current study, a number of oncogenic signaling pathways were identified to be associated with alterations of IGF1R, which may contribute to the IGF1R-driven aggressiveness. The EGFR inhibitor resistance pathway was also associated with the alteration of IGF1R. This observation was consistent with a previous study in which AG1024 has been demonstrated to enhance the apoptotic effects of EGFR inhibitor in human breast cancer cells (36). Therefore, a high alteration of IGF1R in ER-negative tumors may indicate a greater extent of activated oncogenic signaling compared with other tumors. Inhibition of IGF1R signaling indirectly targets multiple key pathways (37). The synergistic effect of TPL and AG1024 may be due to shared targets, including the PI3K/Akt signaling pathway and MYC. DNA repair pathways were revealed to be associated with the alteration of IGF1R, which suggests tumors with an IGF1R alteration may demonstrate a higher resistance to chemo- or radiotherapy. Therefore, a combination of IGF1R inhibitor and DNA damage reagents, including TPL and cisplatin, may serve as an effective strategy for the treatment of ER-negative or basal-like breast cancer.
In conclusion, the present identified a synergistic role of TPL and AG1024 in breast cancer cells. These results suggested that a potential new treatment strategy for ER-negative breast cancer may involve a combination of TPL and AG1024 at low doses, which may reduce the toxicity of the two drugs. Furthermore, a similar synergistic effect was revealed for AG1024 and cisplatin. Notably, a high amplification of IGF1R was identified in aggressive subtypes of breast cancer, including ER-negative and basal-like breast cancer. In summary, the results of the present study suggested that a combination of DNA damage reagents and inhibitors of IGF1R signaling may be a promising approach for the treatment of cancer types with activated IGF1R signaling.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HW designed the study, performed the experiments and analysis, visualized the data, wrote the manuscript and provides supervision. TS and RB performed the experiments and data analysis. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Corson TW, Crews CM. Molecular understanding and modern application of traditional medicines: Triumphs and trials. Cell. 2007;130:769–774. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng C, Zhu H, Song H, Wang Z, Huang G, Li D, Ma Z, Ma J, Qin Q, Sun X, Ma J. Targets and molecular mechanisms of triptolide in cancer therapy. Chin J Cancer Res. 2014;26:622–626. doi: 10.3978/j.issn.1000-9604.2014.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiong J, Su T, Qu Z, Yang Q, Wang Y, Li J, Zhou S. Triptolide has anticancer and chemosensitization effects by down-regulating Akt activation through the MDM2/REST pathway in human breast cancer. Oncotarget. 2016;7:23933–23946. doi: 10.18632/oncotarget.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng X, Shi W, Zhao C, Zhang D, Liang P, Wang G, Lu L. Triptolide sensitizes human breast cancer cells to tumor necrosis factor-α-induced apoptosis by inhibiting activation of the nuclear factor-κB pathway. Mol Med Rep. 2016;13:3257–3264. doi: 10.3892/mmr.2016.4931. [DOI] [PubMed] [Google Scholar]
- 5.Ateba SB, Ngeu ST, Mvondo MA, Tchoumtchoua J, Awounfack CF, Krenn L, Njamen D. Natural terpenoids against female breast cancer: A 5-year recent research. Curr Med Chem. 2018;25:3162–3213. doi: 10.2174/0929867325666180214110932. [DOI] [PubMed] [Google Scholar]
- 6.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims AH, Howell A, Howell SJ, Clarke RB. Origins of breast cancer subtypes and therapeutic implications. Nat Clin Pract Oncol. 2007;4:516–525. doi: 10.1038/ncponc0908. [DOI] [PubMed] [Google Scholar]
- 8.Peddi PF, Ellis MJ, Ma C. Molecular basis of triple negative breast cancer and implications for therapy. Int J Breast Cancer. 2012;2012:217185. doi: 10.1155/2012/217185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 10.Shao H, Ma J, Guo T, Hu R. Triptolide induces apoptosis of breast cancer cells via a mechanism associated with the Wnt/β-catenin signaling pathway. Exp Ther Med. 2014;8:505–508. doi: 10.3892/etm.2014.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan BJ, Tan BH, Chiu GN. Effect of triptolide on focal adhesion kinase and survival in MCF-7 breast cancer cells. Oncol Rep. 2011;26:1315–1321. doi: 10.3892/or.2011.1406. [DOI] [PubMed] [Google Scholar]
- 12.Yang A, Qin S, Schulte BA, Ethier SP, Tew KD, Wang GY. MYC inhibition depletes cancer stem-like cells in triple-negative breast cancer. Cancer Res. 2017;77:6641–6650. doi: 10.1158/0008-5472.CAN-16-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Wang J, Cheng J, Wang L. Antiestrogenic Activity of Triptolide in Human Breast Cancer Cells MDA-MB-231 and Immature Female Mouse. Drug Dev Res. 2017;78:164–169. doi: 10.1002/ddr.21387. [DOI] [PubMed] [Google Scholar]
- 14.Xi C, Peng S, Wu Z, Zhou Q, Zhou J. Toxicity of triptolide and the molecular mechanisms involved. Biomed Pharmacother. 2017;90:531–541. doi: 10.1016/j.biopha.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Li XJ, Jiang ZZ, Zhang LY. Triptolide: Progress on research in pharmacodynamics and toxicology. J Ethnopharmacol. 2014;155:67–79. doi: 10.1016/j.jep.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H, Shi P, Deng M, Jiang Z, Li Y, Kannappan V, Wang W, Li P, Xu B. Low dose triptolide reverses chemoresistance in adult acute lymphoblastic leukemia cells via reactive oxygen species generation and DNA damage response disruption. Oncotarget. 2016;7:85515–85528. doi: 10.18632/oncotarget.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SH, Kang JG, Kim CS, Ihm SH, Choi MG, Yoo HJ, Lee SJ. Synergistic cytotoxicity of BIIB021 with triptolide through suppression of PI3K/Akt/mTOR and NF-κB signal pathways in thyroid carcinoma cells. Biomed Pharmacother. 2016;83:22–32. doi: 10.1016/j.biopha.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Qiao Z, He M, He MU, Li W, Wang X, Wang Y, Kuai Q, Li C, Ren S, Yu Q. Synergistic antitumor activity of gemcitabine combined with triptolide in pancreatic cancer cells. Oncol Lett. 2016;11:3527–3533. doi: 10.3892/ol.2016.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Pan GF, Jiang ZZ, Yang J, Sun LX, Zhang LY. Triptolide inhibits human breast cancer MDA-MB-231 cell growth via downregulation of the ERα-mediated signaling pathway. Acta Pharmacol Sin. 2015;36:606–613. doi: 10.1038/aps.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiró G, Adrover E, Sánchez-Tejada L, Lerma E, Planelles M, Sánchez-Payá J, Aranda FI, Giner D, Gutiérrez-Aviñó FJ. Increased insulin-like growth factor-1 receptor mRNA expression predicts poor survival in immunophenotypes of early breast carcinoma. Mod Pathol. 2011;24:201–208. doi: 10.1038/modpathol.2010.191. [DOI] [PubMed] [Google Scholar]
- 21.Knowlden JM, Hutcheson IR, Barrow D, Gee JM, Nicholson RI. Insulin-like growth factor-I receptor signaling in tamoxifen-resistant breast cancer: a supporting role to the epidermal growth factor receptor. Endocrinology. 2005;146:4609–4618. doi: 10.1210/en.2005-0247. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19:8469–8478. doi: 10.1128/MCB.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. TCGA research network, perou cm. comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;90:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37(Web Server issue) 2009:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, van den Ouweland A, Merajver SD, Ethier SP, Schutte M. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res 1: 2006;66:41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 28.Wan CK, Wang C, Cheung HY, Yang M, Fong WF. Triptolide induces Bcl-2 cleavage and mitochondria dependent apoptosis in p53-deficient HL-60 cells. Cancer Lett. 2006;241:31–41. doi: 10.1016/j.canlet.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Chueh FS, Chen YL, Hsu SC, Yang JS, Hsueh SC, Ji BC, Lu HF, Chung JG. Triptolide induced DNA damage in A375.S2 human malignant melanoma cells is mediated via reduction of DNA repair genes. Oncol Rep. 2013;29:613–618. doi: 10.3892/or.2012.2170. [DOI] [PubMed] [Google Scholar]
- 30.Davison Z, de Blacquière GE, Westley BR, May FEB. Insulin-like Growth Factor-Dependent Proliferation and Survival of Triple-Negative Breast Cancer Cells: Implications for Therapy. Neoplasia. 2011;13:504–515. doi: 10.1593/neo.101590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeon JH, Kim SK, Kim HJ, Chang J, Ahn CM, Chang YS. Insulin-like growth factor-1 attenuates cisplatin-induced gammaH2AX formation and DNA double-strand breaks repair pathway in non-small cell lung cancer. Cancer Lett. 2008;272:232–241. doi: 10.1016/j.canlet.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Railo MJ, von Smitten K, Pekonen F. The prognostic value of insulin-like growth factor-I in breast cancer patients. Results of a follow-up study on 126 patients. Eur J Cancer 30A. 1994:307–311. doi: 10.1016/0959-8049(94)90247-X. [DOI] [PubMed] [Google Scholar]
- 33.Hartog H, Horlings HM, van der Vegt B, Kreike B, Ajouaou A, van de Vijver MJ, Marike Boezen H, de Bock GH, van der Graaf WT, Wesseling J. Divergent effects of insulin-like growth factor-1 receptor expression on prognosis of estrogen receptor positive versus triple negative invasive ductal breast carcinoma. Breast Cancer Res Treat. 2011;129:725–736. doi: 10.1007/s10549-010-1256-6. [DOI] [PubMed] [Google Scholar]
- 34.Martin JL, Baxter RC. Expression of insulin-like growth factor binding protein-2 by MCF-7 breast cancer cells is regulated through the phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway. Endocrinology. 2007;148:2532–2541. doi: 10.1210/en.2006-1335. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Moerkens M, Ramaiahgari S, de Bont H, Price L, Meerman J, van de Water B. Elevated insulin-like growth factor 1 receptor signaling induces antiestrogen resistance through the MAPK/ERK and PI3K/Akt signaling routes. Breast Cancer Res. 2011;13:R52. doi: 10.1186/bcr2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones HE, Goddard L, Gee JM, Hiscox S, Rubini M, Barrow D, Knowlden JM, Williams S, Wakeling AE, Nicholson RI. Insulin-like growth factor-I receptor signalling and acquired resistance to gefitinib (ZD1839; Iressa) in human breast and prostate cancer cells. Endocr Relat Cancer. 2004;11:793–814. doi: 10.1677/erc.1.00799. [DOI] [PubMed] [Google Scholar]
- 37.Riedemann J, Macaulay VM. IGF1R signalling and its inhibition. Endocr Relat Cancer. 2006;13(Suppl 1):S33–S43. doi: 10.1677/erc.1.01280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.