Abstract
Knowledge of etiology causes of diarrheal illness is essential for development and implementation of public health measures to prevent and control this disease syndrome. There are few published studies examining diarrhea in children aged <5 years in Iraq. This study aims to investigate the occurrences and epidemiology of selected bacterial (Salmonella spp. and Campylobacter spp.), viral (adenovirus, norovirus GI and GII, and astrovirus), and parasitic (Entamoeba spp. and Giardia spp.) agents in stool samples from 155 child diarrheal cases enrolled between March and August 2017, in a hospital-based cross-sectional study in Thi-Qar, southeastern Iraq. Using molecular techniques and sequence-based characterization, adenovirus was the most frequently detected enteropathogen (53/155 (34.2%)), followed by Salmonella spp. (23/155 (14.8%)), Entamoeba spp. (21/155 (13.5%)), and Campylobacter spp. (17/155 (10.9%)). Mixed infection with Salmonella spp. and Campylobacter spp. was evident, and the same was revealed between various enteric viruses, particularly adenovirus and norovirus. The most frequent co-infection pattern was between adenovirus and Campylobacter spp., in seven cases (7/155 (4.5%)). Whole-genome sequencing-derived typing data for Salmonella isolates (n = 23) revealed that sequence type 49 was the most prevalent in this sample set (15/23 (65.2%)). To the best of our knowledge, this study provides the first report on detection and identification of floR, blaCARB-2, and mphA antimicrobial resistance genes in Salmonella isolated from children in the Middle East region. Logistic regression analysis pointed to few enteropathogen-specific correlations between child age, household water source, and breastfeeding patterns in relation to the outcome of detection of individual enteropathogens. This study presents the first published molecular investigation of multiple enteropathogens among children <5 years of age in Iraq. Our data provide supporting evidence for planning of childhood diarrhea management programs. It is important to build on this study and develop future longitudinal case-control research in order to elaborate the epidemiology of enteropathogens in childhood diarrhea in Iraq.
Keywords: Iraq, adenovirus, Salmonella, Entamoeba, Campylobacter
1. Introduction
Diarrheal diseases accounted for 8% of all deaths in children under five years of age in 2016, and this translates to over 1300 young children dying each day, or approximately 480,000 children a year [1]. In Iraq, the impact of war, sanctions, and sectarian violence left a dysfunctional health system and an on-going public health emergency impacting vulnerable sections of the population, particularly children. Several viral, bacterial, and parasitic infections are among the most common causes of acute diarrheal cases in children [2]. Published studies on childhood diarrhea are lacking in Iraq and, therefore, the pathogen spectrum associated with diarrheal disease requires investigation.
Among enteric viruses, rotavirus is the most commonly identified cause of severe diarrhea among children in Iraq, as well as in many developing countries [3,4]. Adenoviruses are also implicated in several viral outbreaks and sporadic cases across all age groups, causing a broad spectrum of clinical symptoms and occurring throughout the year [5,6,7]. Within the adenovirus F subgenera, serotypes HAdV-40 and HAdV-41 are associated with significant outbreaks of disease in infants and children [7]. Other viruses commonly associated with acute gastroenteritis globally include noroviruses (NoVs) and human astroviruses (HAstVs) [8,9]. After enteric viruses, bacterial causes are ranked as the second most common cause of diarrhea in developing countries. Campylobacter is a potential etiological agent of bacterial enteritis both in children and adults, and it is second in prevalence to Salmonella and similar to Shigella in many countries [10]. Non-typhoidal Salmonella spp. are among the leading causes of gastroenteritis worldwide, with an increased incidence observed in children less than five years old [11,12]. Invasive cases of non-typhoidal Salmonella are frequently reported in infants and young children with a higher risk of secondary complications such as bacteremia and meningitis [13]. In addition, the recent increase of multidrug resistance (MDR) among non-typhoidal Salmonella species is a serious problem worldwide, due to the widespread use of traditional antibiotics in human and veterinary medicine, raising global public health concern [14]. Next to viral and bacterial causes, amebiasis and giardiasis are among the major intestinal parasitic infections causing childhood diarrhea in many developing countries [15], and are endemic throughout socio-economically deprived communities [16,17]. Given the multifactorial nature of diarrheal illnesses, it is suggested that enteric pathogen co-infections play an important role in gastroenteritis; however, research efforts often focus on a small range of species belonging to a few pathogen groups [13,14,15,16,17,18]. Thus, studies oriented at investigating the role of co-infections with enteric pathogens in cases of acute diarrhea are required.
In Iraq, the morbidity and mortality associated with diarrhea is high, particularly among children <5 years [4]. Elevated morbidity and mortality is predominantly due to serious challenges facing the delivery of basic public health and environmental sanitation services across Iraq, after decades of war and political instability. Previously, we investigated gastroenteritis caused by Salmonella infection among children aged below five years in Thi-Qar, southeastern Iraq [18]. Thi-Qar is one of the least developed and poorest governorates in Iraq, and it is important to investigate the spectrum of infectious causes of children diarrhea in such an unprivileged setting in Iraq. Hence, we transported aliquots of fecal samples from child diarrheal cases recruited in Thi-Qar (Iraq) to the Antimicrobial Resistance and Infectious Disease (AMRID) laboratory at Murdoch University (Australia). The present study is pilot in nature, and aims to conduct a comprehensive molecular screening survey of selected viral, bacterial, and parasitic agents. This molecular-based survey hopes to explore the coexistence between several infectious pathogens, along with their related clinical and epidemiological features among children with acute diarrhea in Thi-Qar.
2. Materials and Methods
2.1. Study Setting and Design
The study population consisted of children below five years of age presenting with acute diarrhea to the Enteric Diseases Clinic of two referral children hospitals in Thi-Qar, a regional governorate situated in southeastern Iraq, between March and August 2017. This survey is a follow-up from an initial hospital-based cross-sectional study that focused on culture-based screening and characterization of non-typhoidal Salmonella [18]. The initial study included 320 diarrhea cases of children below five years; details of case enrolment, stool specimen collection, and questionnaires administered to the child’s parent or guardian to gather information on basic socio-demographic information and potential risk factors for infection are presented in full details elsewhere [18].
For the present study, aliquots of fecal samples from half of the diarrhea cases (n = 320) enrolled in the primary study [18] were selected for further molecular screening of a panel of enteropathogens. The decision to select half of the cases was based on feasibility and cost-effectiveness. Random selection of the cases was done using the “select cases” tool in the Statistical Package for the Social Sciences software (SPSS for windows, version 15.0), with the option for selecting approximately 50% of the cases. Thus, in this study, 155 aliquots of stool specimens from children below five years presenting with acute diarrhea were selected for molecular screening of a panel of viral, bacterial, and parasitic enteropathogens.
2.2. Stool Samples Processing and DNA Extraction
The sampled stool specimens were kept at 4 °C at the hospital facility, and each sample was divided into two aliquots; one aliquot was placed in Amies transport media with charcoal (COPAN, Italy), labeled and transported under cold chain to the Microbiology Laboratory, University of Thi-Qar, for Salmonella detection using the culture-based method [18]. The second aliquot was stored in RNA later® solution (Ambion, USA) as per the manufacturer’s instructions and then shipped from Iraq to Australia. Molecular analysis was conducted at the AMRID Laboratory of Murdoch University. Genomic DNA was extracted from all fecal samples suspended in RNA later® solution using a Bioline Isolate Fecal DNA kit (ISOLATE II, Genomic DNA Kit, Bioline), according to the manufacturer’s recommended protocol. Purified DNA was stored at −20 °C until further analysis. The following panel of enteropathogens was screened (Table 1): (a) Salmonella spp. and Campylobacter spp. as targeted common bacterial enteric pathogens; (b) adenovirus, norovirus (GI and GII), and astrovirus as representative viral causes of diarrhea; (c) Entamoeba spp. and Giardia spp. as targeted parasitic causes. In this study, we use the term co-infection to denote cases where different classes of enteropathogens were detected together, for example, bacteria and virus from the same stool specimen. We use the term mixed infection to refer to cases where different agents from the same class were detected together, such as two bacterial species.
Table 1.
Primers and probes used for molecular screening of a panel of seven enteropathogens in diarrheal cases (n = 155) among children <5 years. F—forward; R—reverse; rRNA—ribosomal RNA.
| Target Pathogen | Gene | Sequence (5′ to 3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| Salmonella spp. |
invA F invA R |
TTGTTACGGCTATTTTGACCA CTGACTGCTACCTTGCTGATG |
521 | [20] |
| Campylobacter spp. | 16S rRNA F 16S rRNA R |
GGATGACACTTTTCGGAGC CATTGTAGCACGTGTGTC |
812 | [21] |
| Astrovirus | PreCAP1 F 82b R |
GGACTGCAAAGCAGCTTCGTG GTGAGCCACCAGCCATCCCT |
719 | [22] |
| Norovirus GI | G1SK F G1SK R |
CTGCCCGAATTYGTAAATGA CCAACCCARCCATTRTACA |
330 | [22] |
| Norovirus GII | COG2 F G2SK R |
CARGARBCNATGTTYAGRTGGATGAG CCRCCNGCATRHCCRTTRTACAT |
387 | [22] |
| Adenovirus | Ad1 F Ad2 R |
TTCCCCATGGCICAYAACAC CCCTGGTAKCCRATRTTGTA |
482 | [23] |
| Entamoeba spp. | E-1 F E-1 R |
TAAGATGCACGAGAGCGAAA GTACAAAGGGCAGGGACGTA |
439 | [24] |
| Giardia spp. | gdf F gdf R Probe |
GGGCAAGTCCGACAACGA GCACATCTCCTCCAGGAAGTAGAC TCATGCGCTTCTGCCAG BHQ2 |
261 | [25] |
2.3. Molecular Screening of Enteropathogens
2.3.1. Bacteria
In the present study, the randomly selected aliquots of fecal samples encompassed 23 out of the total 33 non-typhoidal Salmonella isolates identified in previous study [18]. We further characterized those 23 non-typhoidal Salmonella isolates using whole-genome sequencing (WGS). The WGS was utilized to validate the previous serotype identities of Salmonella isolates, to screen and match between antimicrobial resistance genes and the resistance phenotypes, and to screen for multilocus sequence types (MLST). For WGS, library preparation was performed using an Illumina NexTera® XT library preparation kit (Illumina) as per the manufacturer’s instructions. Sequencing was performed on an Illumina Nextseq platform using a mid-output 2 × 150 kit. Reads were de novo assembled using SPAdes 3.11.1 software [19]. Contig files were uploaded to the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/) to screen for MLST and serotypes, and to extract antimicrobial resistance gene data. From all transferred stool aliquots (n = 155), we screened for the S. enterica gene invA using conventional PCR as previously described by Swamy et al. [20]. Screening for Campylobacter was undertaken using the conventional PCR assay targeting the 16S ribosomal RNA (rRNA) gene according to protocol described by Barletta et al. [21].
2.3.2. Viruses
Reverse-transcription PCR (RT-PCR) assays were performed using SuperScript III One-Step Platinum@ Taq (Invitrogen, USA) for detection of three enteric viruses: human astrovirus, norovirus group 1 (GI), and norovirus group 2 (GII) [22], while adenovirus was detected by conventional PCR [23]. The specific primers used in this reaction are outlined in Table 1. Bands of the expected size from each assay were excised from 1.5% agarose gels and DNA was purified though filter tips. DNA sequencing was performed at the Australian Genome Research Facility (Perth, WA). The results of the sequencing were analyzed and edited using FinchTV (Version 1.4), then compared to the most similar sequence deposited in public databases on National Center for Biotechnology Information GenBank by applying the Basic Local Alignment Search Tool (BLASTn).
Two randomly chosen samples which were positive on the adenovirus screening PCR were analyzed using WGS, utilizing the same procedures described above. Initial screening for adenovirus genomes was performed using SPAdes, and generation of complete genomes was performed using Geneious V10.2.3 to map raw read data against a representative HAdV-41 genome (GenBank Accession KY316161). Annotation of adenovirus genomes was performed using Geneious V10.2.3. The adenovirus sequences were submitted to NCBI GenBank under the accession numbers MG925782 (MU22) and MG925783 (MU35).
2.3.3. Parasites
A nested PCR assay for the detection of Entamoeba species in stool aliquots was utilized according to the procedure previously described by Al-Areeqi et al. [24]. The presence of Giardia spp. in all samples was screened at the glutamate dehydrogenase (gdh) locus using a quantitative PCR (qPCR) [25].
2.4. Statistical Analysis
Descriptive data analysis was used to determine the frequency of enteropathogen occurrence and distribution over a range of variables related to study subjects. Statistical analyses were performed using univariable logistic regression analysis (STATA software package, version 11.0). Univariable logistic regression models were used to examine the correlation between demographic characteristics, household features, and breastfeeding patterns in relation to the binary outcome variable of pathogen detection (presence vs. absence of “a pathogen” of concern in diarrheal stool samples). The analysis examined the correlation between the predictor variables and each of adenovirus, Salmonella spp., Campylobacter spp., and Entamoeba spp. Those four enteropathogens were the most frequently detected in diarrheal stool samples. The analysis did not involve the other enteropathogens detected in less than 10% of the diarrheal samples, nor the various mixed and co-infection combinations detected in low number (minimum = 1, and maximum = 7) of samples.
2.5. Ethics and Consent Approval
The study protocol was approved by the Murdoch University Human Research Ethics Committee (Permit No. 2015/224). Permission to conduct the study was also obtained from the Ministry of Health, Iraq (Permit No.11/5/393) and the children’s hospitals in Thi-Qar Governorate (Permit No.1/4/26885). As the study subjects were children under the age of five, informed verbal consent was obtained from their caregivers (parents/guardians) before enrolment. Movement of samples from Iraq to Australia was granted by the Department of Agriculture (Australian Government), under quarantine import permit number 0000369563.
3. Results
In this study, we tested a total of 155 stool samples from children with acute diarrhea. Of all cases, the male:female ratio was 1.4:1 and 93 (60%) were under two years of age (Table 2). Descriptive information about demographic characteristics of the cases, their breastfeeding patterns in the first six months of age, and recorded household features, together with information about caregivers’ hygiene practices, is presented in Table 2.
Table 2.
Descriptive characteristics of diarrheal cases (n = 155) among children <5 years.
| Variables | Category | No. of Cases (%) |
|---|---|---|
| Gender | Male Female |
90 (58.1) 65 (41.9) |
| Age (years) | <2 years 2–5 years |
93 (60.0) 62 (40.0) |
| Mean age (months) ± SD | 22.7 (±12.7) | |
| Residence | Rural Urban |
110 (71.0) 45 (29.0) |
| Breastfeeding pattern in the first 6 months of age | Exclusively bottle-fed Exclusively breastfed Mix—breast- and bottle-fed |
60 (38.7) 64 (41.3) 31 (20.0) |
| Water source in household | Reverse osmosis water Municipal (pipe) water |
58 (37.4) 97 (62.6) |
| Domestic animals in the household | No Yes |
62 (40.0) 93 (60.0) |
| Caregiver hand washing before food preparation | No Sometimes Always |
51 (32.9) 86 (55.5) 18 (11.6) |
| Caregiver hand washing after cleaning child defecation | No Sometimes Always |
15 (9.7) 40 (25.8) 100 (64.5) |
| Caregiver hand washing before feeding the child | No Sometimes Always |
36 (23.2) 49 (31.6) 70 (45.2) |
Among all samples, adenovirus was the most frequently detected enteropathogen (53/155 (34.2%)), followed by Salmonella spp. (23/155 (14.8%)), Entamoeba spp. (21/155 (13.5%),) and Campylobacter spp. (17/155 (10.9%)) (Table 3). Those four etiologic agents accounted for 73.4% of the spectrum of enteropathogens detected in the study samples. Group I noroviruses were the least detected (5/155 (3.2%)) among the panel of enteropathogens screened for in this study (Table 3). WGS analysis of two adenovirus PCR positive samples using SPAdes de novo assembly of raw reads returned large contigs consistent with HAdV-41 adenovirus genomes. Raw read files were mapped to the HAdV-41 KY316161 to obtain an entire genomic sequence for each strain. BLASTn analysis of strains MU22 and MU35 demonstrated most homology to existing HAdV-41 strains, with 98.6% pairwise homology to Genbank accessions AB728839 and KY316161, and 98.56% homology to each other.
Table 3.
Frequency of isolation, co-infection, and mixed infection patterns of enteropathogens detected in diarrheal cases (n = 155) among children <5 years. CI—confidence interval.
| Enteropathogens | No. of Cases | % of Cases (95% CI) | Co-Infection (No. of Cases) | Mixed Infection (No. of Cases) |
|---|---|---|---|---|
| Salmonella spp. | 23 | 14.8 (9.6–21.4) | Adenovirus (5) Astrovirus (3) Giardia spp. + astrovirus (3) Giardia spp. (2) Norovirus GII (1) Norovirus GII + adenovirus (1) Astrovirus + adenovirus (1) |
Campylobacter spp. (3) |
| Campylobacter spp. | 17 | 10.9 (6.5–16.9) | Adenovirus (7) Entamoeba spp. + adenovirus (3) Norovirus GI (2) Norovirus GI + adenovirus (2) Norovirus GII (1) |
Salmonella spp. (3) |
| Astrovirus | 11 | 7.1 (3.6–12.3) |
Salmonella spp.+ Giardia spp. (3) Salmonella spp. (3) Giardia spp. (1) |
Adenovirus (2) Adenovirus + norovirus GII (1) |
| Adenovirus | 53 | 34.2 (26.7–42.2) |
Campylobacter spp. (7) Salmonella spp. (5) Entamoeba spp. (3) Entamoeba spp. + Campylobacter spp. (3) Salmonella spp. + Giardia spp. (3) Salmonella spp. + Campylobacter spp. (2) |
Norovirus GII (4) Norovirus GI (3) Astrovirus (2) Astrovirus + norovirus GII (1) |
| Norovirus GI | 5 | 3.2 (1.0–7.3) | Campylobacter spp. (2) | Adenovirus (3) |
| Norovirus GII | 10 | 6.4 (3.1–11.5) |
Salmonella spp. (1) Campylobacter spp. (1) |
Adenovirus (4) |
| Entamoeba spp. | 21 | 13.5 (8.5–19.9) | Adenovirus + Campylobacter spp. (3) Adenovirus (3) Astrovirus + adenovirus (1) |
None |
| Giardia spp. | 11 | 7.1 (3.6–12.3) | Astrovirus + Salmonella spp. (3) Adenovirus + Salmonella spp. (3) Salmonella spp. (2) Campylobacter spp. (1) Astrovirus (1) |
None |
Results presented in Table 3 highlight the diverse nature of pathogens among cases of acute diarrhea in Iraqi children. Mixed infection with the bacterial pathogens Salmonella spp. and Campylobacter spp. was evident, and the same was revealed between various enteric viruses, particularly adenoviruses and noroviruses (Table 3). Nevertheless, there was no mixed infection between the two parasitic agents Entamoeba spp. and Giardia spp. (Table 3). Moreover, co-infection with different classes of enteropathogens was common among the tested samples. Of interest, co-infection with adenovirus and Campylobacter spp. was detected in seven cases (7/155 (4.5%)). Co-infection with a bacterial, viral, and a parasitic etiologic agent all together was detected in nine cases (9/155 (5.8%)).
Table 4 summarizes some WGS-derived typing data for Salmonella isolates (n = 23). A total of four multilocus sequence types (STs) were characterized among the 23 isolates; of which, S. typhimurium ST49 was the most common (15/23 (65.2%)). All of the whole-genome sequenced Salmonella isolates harbored at least one tet gene, with the tetB gene having the highest frequency (n = 12), followed by tetA (n = 8) and tetG (n = 3). Five groups of streptomycin-resistance genes were detected among 17 out of the 23 Salmonella isolates, consisting of aadA7 (n = 5), strB (n = 4), strA (n = 3), aadA2 (n = 3), and aadA1 (n = 2). For aminoglycoside resistance genes, 11 of the 23 Salmonella isolates carried aph(3’)-Ic (n = 7) and aac(3)-Id (n = 4). The sul1 gene was identified in eight of the sulfonamide-resistant Salmonella isolates. Each of β-lactamase (blaCARB-2) and kanamycin (aph(3’)-Ia) resistance genes were found in four Salmonella isolates. Furthermore, trimethoprim (dfrA14), azithromycin (mphA), erythromycin (erm42), and florfenicol (floR) resistance genes were also detected in a few isolates (Table 4).
Table 4.
Whole-genome sequencing (WGS)-derived typing data and resistance phenotype among Salmonella (n = 23) isolated from diarrheal cases (n = 155) among children <5 years.
| Serovars | MLST | Resistance Genes | Resistance Phenotypes a |
|---|---|---|---|
| S. typhimurium | ST-49 | tetB | TET, S |
| S. typhimurium | ST-49 | tetB | TET, S |
| S. typhimurium | ST-49 | tetB | TET, ATH |
| S. typhimurium | ST-49 | tetB | TET, CTX |
| S. typhimurium | ST-49 | tetB | ATH, NA, CTX |
| S. typhimurium | ST-49 | tetB | TET, ATH, TS |
| S. typhimurium | ST-49 | tetB | TET, ATH, CTX |
| S. typhimurium | ST-49 | tetB | TET, ATH, CTX, CIP |
| S. typhimurium | ST-49 | tetB | NA, ATH, CTX, TS, S |
| S. typhimurium | ST-3020 | tetG, sul1, aadA2, blaCARB-2 | TET, ATH, CTX, CIP, TS, S |
| S. typhimurium | ST-49 | tetA, sul1, aadA7, aph(3’)-Ic, aac(3)-Id | TET, ATH, CTX, GM |
| S. typhimurium | ST-49 | tetB, strB, aph(3’)-Ic | TET, ATH, NA, S |
| S. typhimurium | ST-49 | tetB, strB, strA, aph(3’)-Ic | TET, ATH, CTX, CIP, TS |
| S. typhimurium | ST-3020 | tetG, aadA2, sul1, blaCARB-2, floR | TET, ATH, CIP, CRO, TS, NA, GM |
| S. typhimurium | ST-49 | tetB, aadA7, sul1, aph(3’)-Ia, blaCARB-2 | TET, TS, NA, AMP, CRO, ATH, CIP |
| S. hadar | ST-198 | tetA, aadA7, sul1, aph(3’)-Ic, aac(3)-Id | NA, ATH, CTX, TS, S |
| S. hadar | ST-198 | tetA, aadA7, sul, aph(3’)-Ia, aac(3)-Id | TET, CRO, CIP, TS, GM |
| S. hadar | ST-198 | tetG, aadA2, sul, aph(3’)-Ia, blaCARB-2, dfrA14, erm(42) | TET, ATH, CIP, NA, GM |
| S. hato | ST-52 | tetA, aadA1, sul1, aph(3’)-Ic | TET, CRO, S, GM |
| S. hato | ST-52 | tetA, strB, strA, aph(3’)-Ic, mphA | ATH, NA, CTX, TS |
| S. hato | ST-52 | tetA, strB, strA, aph(3’)-Ic, mphA | ATH, CRO, S, NA, TS |
| S. muenchen | ST-1825 | tetA, aadA1, sul1, dfrA14 | TET, ATH, CIP, S, TS, NA |
| S. muenchen | ST-1825 | tetA, aadA7, sul1, aph(3’)-Ia, aac(3)-Id | TET, ATH, CIP, S, NA, CTX |
a TET: tetracycline; ATH: azithromycin; S: streptomycin; TS: trimethoprim/sulfamethoxazole; CIP: ciprofloxacin; NA: nalidixic acid; CTX: cefotaxime; CRO: ceftriaxone; AMP: ampicillin; GM: gentamicin.
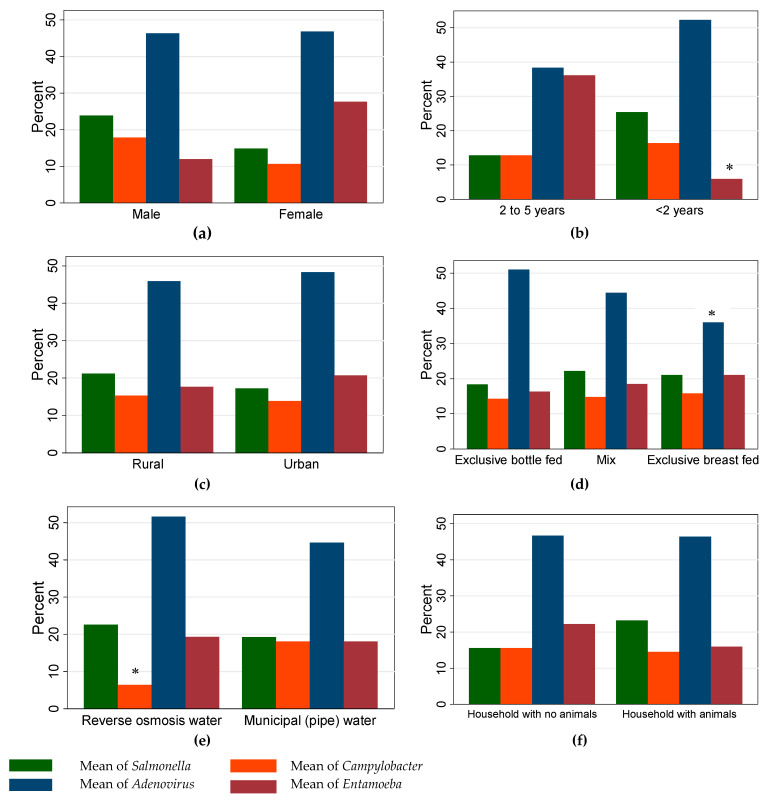
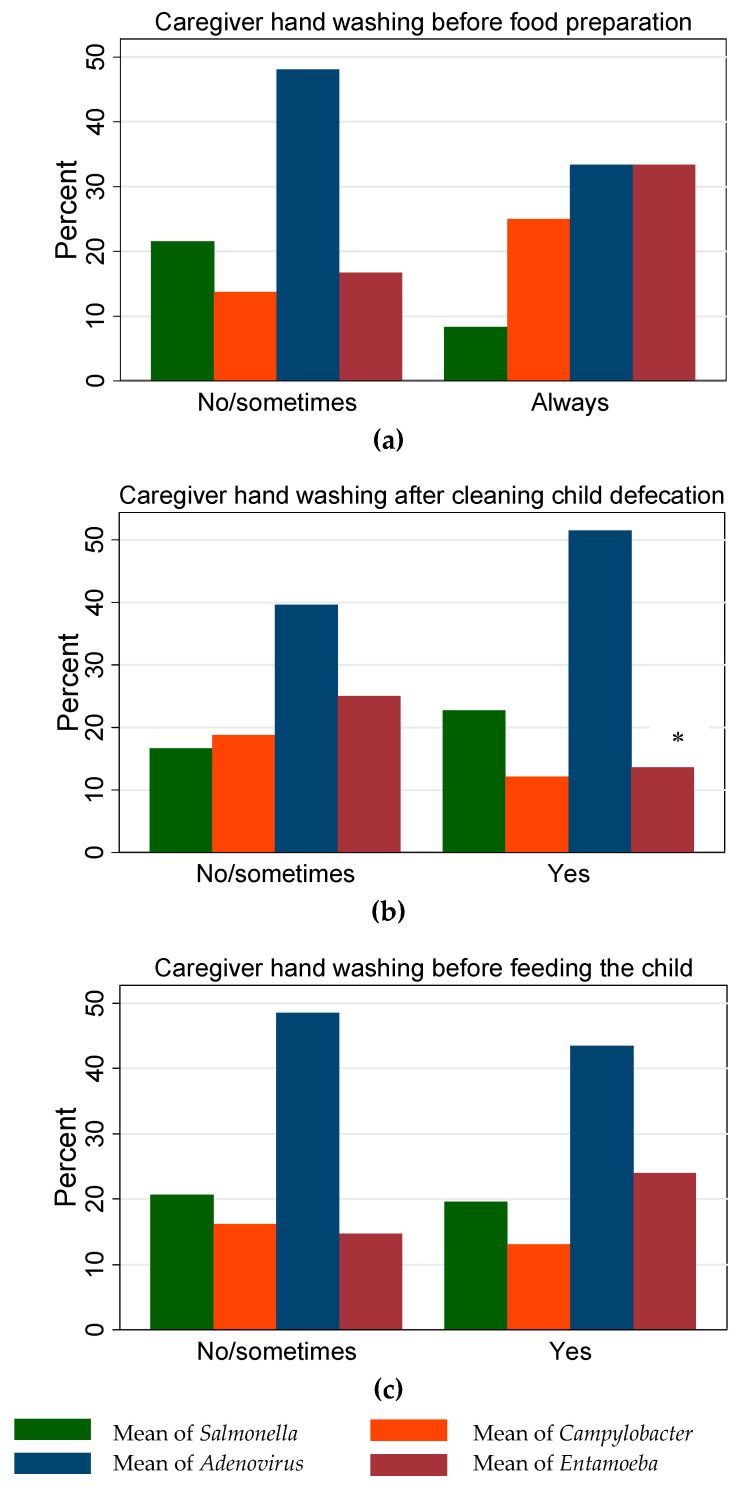
Few enteropathogen-specific associations were significant based on logistic regression analysis (Figure 1). Among the cases enrolled in this study, detection of Entamoeba spp. was less likely (p < 0.001) to occur among children younger than two years (odds ratio (OR) = 0.12, 95% confidence interval (CI) = 0.03–0.37). Lower odds (p = 0.051; OR= 0.46, 95% CI = 0.21–0.99) of adenovirus detection were associated with children exclusively breastfed compared to children exclusively bottle-fed. Among the present study subjects, the likelihood of PCR detection of Campylobacter spp. in children from households supplied by pipe water was 5.12 (95% CI = 1.12–23.27) times higher (p = 0.034) compared to children from households supplied with (purchased) reverse osmosis-treated water. Figure 2 shows the relationship between caregivers’ hygienic practices and occurrence of the four frequently detected enteropathogens in diarrheal cases. According to results from the logistic regression model, the odds of Entamoeba spp. detection in children belonging to caregivers who reported always washing hands after cleaning child defecations was three times lower (p = 0.030; OR = 0.34, 95% CI = 0.14–0.90) compared to children belonging to caregivers who did not (at all/not always) wash hands.
Figure 1.
Distribution (percentage of cases) of demographic characteristics (sex (a), age (b) and residence (c)), breastfeeding pattern in the first six months of age (d), and household features (water source (e) and domestic animals (f)) in relation to frequently (>10%) detected enteropathogens in diarrheal cases (n = 155) among children <5 years old. The symbol (*) denote bars of categories with statistical differences.
Figure 2.
Caregivers’ hygienic practices; (a) hand washing before food preparation; (b) hand washing after cleaning child defecation; (c) hand washing before feeding the child, in relation to percentage of enteropathogens cases (Figure 1) detected in diarrheal children (n = 155) <5 years old in Thi-Qar Governorate, Iraq. The symbol (*) denote bars of categories with statistical differences.
4. Discussion
The majority of enteric bacterial and viral pathogens are not routinely screened for in Iraqi hospitals, due to a lack of basic diagnostics and sufficiently trained personnel [26]. The war in the last decade destroyed substantial capacities of hospitals and public health laboratories in Iraq, and nearly two-thirds of its qualified medical personnel emigrated [27]. In general, published research on diarrheal illnesses in the Iraqi population is very limited, and has mainly focused on screening for single-pathogen infections or, at best, infections with pathogen groups [4,5,6,7,8,9,10,11,12,13,14,15,16,17,18]. In the present study, we report the first molecular epidemiological investigation describing the occurrence and co-existence of several enteropathogens in stool samples from diarrheal children <5 years of age in one of the least developed governorates in Iraq. Added to that, we profiled sequence types and genes conferring resistance to several antimicrobial groups among non-typhoidal Salmonella isolated for the diarrheal cases. This study demonstrates the value of WGS as a tool for comprehensive analysis of bacterial and viral pathogens commonly detected in diarrheal patients.
In the present study, human adenovirus (HAdV) was the most common enteropathogen, detected in 53 (34.2%) cases. A survey of patients under five years in Australia from 2007 to 2010 revealed that adenovirus was also the most common cause of gastroenteritis in the studied population, emphasizing the importance of this virus in childhood diarrhea in both developing and developed countries [8]. Detection rates in the present study are considerably higher than published research on HAdV in children with diarrhea from other Middle East and North African countries such as in Kuwait (4%) [28], Qatar (6.25%) [29], Saudi Arabia (8%) [16], and Egypt (20%) [30]. Also, our finding is higher compared to the published occurrence of HAdV in East Asia (9.8–20%) [31] and in Bangladesh (10.7%) [32]. The reasons for the higher HAdV detection rate are not clear. However, it is important to note that the pan-adenovirus PCR assay used in this study detects all adenovirus serotypes and not just the enteric serotypes F40 and F41 [33]. Nevertheless, WGS analysis of two of the PCR positive samples collected in this study allowed the extraction of complete genomes of HAdV-41 serotypes, providing evidence that this enteric serotype, as expected, is circulating in the study population in Iraq. Also worth highlighting is that our study design may have impacted our observed frequency of enteropathogens, including HAdV. Because the diarrheal cases were sampled in the warmer summer months, it is possible our observed frequencies are higher than for other times of year, as described elsewhere [5]. Hence, future case-control studies are needed to accurately predict the frequency of pathogens and potential changes due to seasonality.
Non-typhoidal Salmonella was detected in 14.8% of the stool samples from children with diarrhea, and was the second most frequently detected enteric pathogen in this study. This result is consistent with studies conducted previously in Iraq (15%) [34], and in other neighboring countries such as Kuwait (18%) [35] and Saudi Arabia (15.3%) [16]. Our results re-emphasize the importance of non-typhoidal Salmonella in the epidemiology of childhood bacterial diarrhea in Iraq. We previously demonstrated that a higher likelihood of positive isolation of non-typhoidal Salmonella from children diarrheal cases in Thi-Qar was associated with source of water and presence of domestic animals in the household, as well as with caregiver education level and hygienic practices [18].
Entamoeba spp. were found to be the third ranked among the seven enteropathogens screened for in the present research. This finding is consistent with previous surveillance data from Saudi Arabia [16], Oman [36], Yemen [24], and Libya [17], where Entamoeba spp. were commonly isolated from children diarrheal samples. Intestinal parasitic infection is a significant public health burden, especially in poor and socio-economically deprived communities [16], which is relevant to the situation in Thi-Qar where 37.8% of the population lives below the poverty line of United States dollar (USD) $2.5 per day [37]. Added to that, the proportion of the population in Thi-Qar using an improved sanitation facility is very low, with only 20.8% utilizing the public sewage system as the primary system, while 39.4% rely on a covered canal outside the house, and 30% primarily use a septic tank [37]. An alarming 54.8% of the population in Thi-Qar disposes of garbage in open areas [37]. A number of case-control and cohort studies on diarrhea in children demonstrated that unsafe water supply and poor sanitation are important risk factors associated with enteric parasite infections [16,17,18,19,20,21,22,23,24].
Very limited research was conducted on Campylobacter occurrence in childhood diarrhea in Iraq, as it is not screened for in pediatric hospitals, hampering our understanding of the role of Campylobacter spp. in diarrheal illness in this setting. Our results indicate positive PCR detection of Campylobacter spp. in 10.9% (17/155) of the screened stool samples from child diarrheal cases. Interestingly, recent findings from the Global Enteric Multicenter Study [38] indicate that the fraction of severe diarrheal cases in infants attributed to Campylobacter jejuni or Campylobacter coli ranged from 6% in Kenya to 12% in Bangladesh, which is comparable to the present study finding from Iraq. We also conclude, based on logistic regression analysis, that the likelihood of detection of Campylobacter spp. in children from households supplied by pipe water was higher compared to those supplied with reverse osmosis-treated water. This finding is in accordance with a cross-sectional study on Campylobacter infections among diarrheic children in Ethiopia, where the highest rates of infections were reported in children whose family did not use a protected water source [39]. In spite of growing evidence regarding the burden of Campylobacter-attributed diarrhea in developing countries, we know little about what, how, and where children contract infection [40]. Further research is urgently required to investigate the role of supplied household water and the role of domestic animals in the transmission of Campylobacter jejuni, especially in populations living with poor sanitary conditions, similar to those in Thi-Qar in south of Iraq.
The spectrum of co-existence of enteric pathogens and their role in diarrheal illnesses could be understood better by utilizing recent advances in diagnostic tools. The utilization of molecular tools in the present study shed light on the potential occurrence of mixed infection between the bacterial pathogens Salmonella spp. and Campylobacter spp., and the same was revealed between various enteric viruses (Table 3). Children can be exposed to multiple pathogens at home, the playground, and daycare [41]. The presence of mixed infections complicates diagnosis of a specific pathogen responsible for the disease and may result in an additive impact, leading to a more severe clinical disease [42]. Our results also point to an intriguing frequency (4.5%) of co-infection between adenovirus and Campylobacter spp. This co-infection pattern should be viewed in parallel with the results of the logistic regression modeling, as our results pointed to higher odds of adenovirus detection in children exclusively bottle-fed (compared to exclusively breastfed), as well as a higher likelihood of PCR detection of Campylobacter spp. in children from households supplied by pipe water (compared to reverse osmosis water). In settings where potable water may be limited or surfaces contaminated, cleaning feeding bottles adequately may be impossible, placing infants at a heightened risk of infectious disease. The role water plays in the epidemiology of adenoviruses and Campylobacter, as well as the potential health risks constituted by these pathogens in water environments, is widely recognized [43,44,45]. Adenoviruses are considered the only DNA viral pathogens in the enteric virus group. They are robust viruses which are non-enveloped with a double-stranded DNA (dsDNA) genome and are, thus, more resistant in the environment, including water sources, than other enteric viruses [44]. A recent multi-country study suggests that treatment of drinking water and improved sanitation reduced risk associated with Campylobacter infection [46]. The frequent co-infection that we report in the present study between adenoviruses and Campylobacter warrants a hypothesis that an interaction between hygiene and contaminated water might be a possible route of children co-exposure to both pathogens in Thi-Qar.
Our results demonstrate the usefulness of WGS-derived data in providing in-depth insight into non-typhoidal Salmonella isolated for children with diarrhea. To the best of our knowledge, this is the first published WGS-based characterization of Salmonella from clinical samples from a Middle-Eastern country. ST49 was the most frequent genotype, followed by ST198 and ST52. The standardization of data and the portable nature of the sequence-based typing allow this method to be used as a worldwide epidemiological tool to study source attribution of enteric pathogens. The three STs characterized in Salmonella isolates in our study were recently reported in human salmonellosis cases from neighboring Qatar [47], as well as in the United Kingdom [48]. In several studies, ST49, ST198, and ST52 were also frequently carried in cattle and poultry sources contaminated with Salmonella, which might have played an important role in human exposure to infection through food and environmental sources [47,48,49,50].
Analysis of WGS data also revealed that tetracycline, streptomycin, and aminoglycoside resistance genes were commonly harbored by Salmonella isolates characterized in this study (Table 4). The emergence and spread of antimicrobial resistance in Salmonella is a threat to human public health [14]. The high resistance rates to traditional antibiotics in the current study could be explained by the fact that many of these antibiotics in Iraq, as in other developing countries, are still indiscriminately prescribed in human medicine due to their low cost and wide availability [51]. Three tetracycline resistance genes (tetB, tetA, and tetG) were detected among the sequenced Salmonella isolates. A study in Iran also found a similar pattern, as the same three genes were the most commonly identified in tetracycline-resistant Salmonella from human stool samples [52]. Florfenicol is a chemosynthesis broad-spectrum antibiotic related to the chloramphenicol class and is mainly used in veterinary medicine [53]. In this study, WGS identified floR in 4.3% of S. enterica isolates, which is lower compared to findings from a study in Taiwan where floR was identified in 19% of the Salmonella isolates from children [54]. In the isolates characterized in the present study, blaCARB-2 and floR genes were all associated with S. typhimurium, with one exception of a strain of S. hadar that harbored the blaCARB-2 resistance gene. A similar finding was demonstrated by Randall et al. [55] who also found blaCARB-2 and floR genes to be linked with S. typhimurium isolated from humans and animals. Using WGS, Nair et al. [48] observed resistance to azithromycin among Salmonella serovars isolated from humans to be linked with the presence of mphA gene. This is consistent with our results, which also found two mphA genes to be associated with the azithromycin resistance profile. To the best of our knowledge, this is the first report of detection and identification of floR, blaCARB-2, and mphA in Salmonella isolated from children in the Middle East region.
Effective hand hygiene is essential to prevent the spread of microbes from person to person and reduce cross-contamination from hands to food [56]. In the present study sample, the likelihood of Entamoeba spp. detection in children belonging to caregivers who reported always washing hands after cleaning child defecations was significantly lower (compared with those belonging to caregivers who did not wash hands). However, our study data could not conclude a relationship between caregivers’ hygienic practices and occurrence of the other frequently detected bacterial and viral pathogens (Figure 2). It is possible that caregivers’ hygienic practice is a limited route of exposure, compared to other sanitary and environmental routes, and, hence, it did not reveal a tangible relationship among the present study samples. It is worth noting that it is not uncommon to experience difficulty in establishing statistical relationships between hygiene-related factors and infections that are multifactorial in nature, as is often the case for diarrheal illnesses. For instance, recently concluded randomized controlled trials that tested the efficacy of improvements in drinking water, sanitation, and hand washing (WSH) in low- and middle-income countries found no significant effects on gut markers of environmental enteric dysfunction, growth at 18 months of age, or diarrhea incidence in two out of three sites [57]. Lack of statistical significance of individual studies should not be taken as implying that the totality of evidence supports no effect. Hence, it is recommended that mothers should always be encouraged to wash their hands following the use of a toilet, cleaning the child’s bottom after defecation, and before feeding the child, as inadequate hand hygiene can transfer contamination to surfaces and foods in the home [58].
5. Conclusions
To the best of our knowledge, this study presents the first published molecular investigation of multiple enteropathogens among children <5 years of age in Iraq. Although this was not a case-control study, the frequency of detection of adenovirus, Salmonella, Campylobacter, and Entamoeba suggests that these organisms are important causes of diarrhea in this population. More information is needed about the sources, modes of transmission and risk factors of enteropathogens in Iraqi children in order to develop methods to control these infections. In future work, it is important to build on the present study and plan for longitudinal case-control research to investigate in depth the epidemiology of enteropathogens in childhood diarrhea, and to perform environmental, water source, and animal sampling. Phenotypic and genotypic characterization of Salmonella resistance to some clinically important antimicrobial emphasizes the need to carry out long-term monitoring. Overall, this work fills a gap in research on the frequency of a range of enteropathogens and could be used by public health authorities for informing diarrhea control programs among infants and children in Iraq.
Acknowledgments
The authors would like to thank the physicians, staffs, and technicians of the children hospitals for their assistance with sampling and processing. We would also like to thank the caregivers of children who participated in this study. We are grateful to the Department of Biology, College of Science, Thi-Qar University for their technical assistance.
Author Contributions
Conceptualization, S.A., M.O., and I.H.; formal analysis, A.H.; investigation, B.R. and T.L.; methodology, S.A., M.O., and I.H.; project administration, A.H.; resources, S.A., M.O., and I.H.; supervision, S.A., M.O., and I.H.; writing—original draft, A.H.; writing—review and editing, M.O. and I.H.
Funding
This research received no external funding. Ali Harb was supported by a PhD scholarship from the Higher Committee for Education Development in Iraq and Murdoch University in Western Australia.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.United Nations International Children’s Emergency Fund Diarrhoeal Diseases. UNICEF Data: Monitoring the Situation of Children and Women. [(accessed on 17 March 2018)]. Available online: https://data.unicef.org/topic/child-health/diarrhoeal-disease/
- 2.Kelly P. Infectious diarrhoea. Medicine. 2015;43:253–258. doi: 10.1016/j.mpmed.2015.02.005. [DOI] [Google Scholar]
- 3.Khoury H., Ogilvie I., El Khoury A.C., Duan Y., Goetghebeur M.M. Burden of rotavirus gastroenteritis in the Middle Eastern and North African pediatric population. BMC Infect. Dis. 2011;11:9. doi: 10.1186/1471-2334-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed S., Klena J., Albana A., Alhamdani F., Oskoff J., Soliman M., Heylen E., Teleb N., Husain T., Matthijnssens J. Characterization of human rotaviruses circulating in Iraq in 2008: Atypical G8 and high prevalence of P [6] strains. Infect. Genet. Evol. 2013;16:212–217. doi: 10.1016/j.meegid.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Cruz J.R., Caceres P., Cano F., Flores J., Bartlett A., Torun B. Adenovirus types 40 and 41 and rotaviruses associated with diarrhea in children from Guatemala. J. Clin. Microbiol. 1990;28:1780–1784. doi: 10.1128/jcm.28.8.1780-1784.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu H., Phan T.G., Nishimura S., Okitsu S., Maneekarn N., Ushijima H. An outbreak of adenovirus serotype 41 infection in infants and children with acute gastroenteritis in Maizuru City, Japan. Infect. Genet. Evol. 2007;7:279–284. doi: 10.1016/j.meegid.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Dey R.S., Ghosh S., Chawla-Sarkar M., Panchalingam S., Nataro J.P., Sur D., Manna B., Ramamurthy T. Circulation of a novel pattern of infections by enteric adenovirus serotype 41 among children below 5 years of age in Kolkata, India. J. Clin. Microbiol. 2011;49:500–505. doi: 10.1128/JCM.01834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher S., Van Hal S., Andresen D., McLaws M.L., Stark D., Harkness J., Ellis J. Gastrointestinal pathogen distribution in symptomatic children in Sydney, Australia. J. Epidemiol. Glob. Health. 2013;3:11. doi: 10.1016/j.jegh.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett S., Gunson R.N. The development of a multiplex real-time RT-PCR for the detection of adenovirus, astrovirus, rotavirus and sapovirus from stool samples. J. Virol. Methods. 2017;242:30–34. doi: 10.1016/j.jviromet.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry F. Textbook of Pediatric Infectious Diseases. 4th ed. W.B Saunders Company; Philadelphia, PA, USA: 1998. [Google Scholar]
- 11.Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 12.Kovanen S.M., Kivisto R.I., Rossi M., Schott T., Karkkainen U.M., Tuuminen T., Uksila J., Rautelin H., Hanninen M.L. Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland. J. Clin. Microbiol. 2014;52:4147–4154. doi: 10.1128/JCM.01959-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham S.M. Salmonellosis in children in developing and developed countries and populations. Curr. Opin. Infect. Dis. 2002;15:507–512. doi: 10.1097/00001432-200210000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Mukerji S., O’Dea M., Barton M., Kirkwood R., Lee T., Abraham S. Development and transmission of antimicrobial resistance among Gram-negative bacteria in animals and their public health impact. Essays Biochem. 2017;61:23–35. doi: 10.1042/EBC20160055. [DOI] [PubMed] [Google Scholar]
- 15.Cheun H.I., Cho S.H., Lee J.H., Lim Y.Y., Jeon J.H., Yu J.R., Kim T.S., Lee W.J., Cho S.H., Lee D.Y., et al. Infection status of hospitalized diarrhea patients with gastrointestinal protozoa, bacteria, and viruses in the Republic of Korea. Korean J. Parasitol. 2010;48:113. doi: 10.3347/kjp.2010.48.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegazi M.A., Patel T.A., El-Deek B.S. Prevalence and characters of Entamoeba histolytica infection in Saudi infants and children admitted with diarrhea at 2 main hospitals at South Jeddah: A re-emerging serious infection with unusual presentation. Braz. J. Infect. Dis. 2013;17:32–40. doi: 10.1016/j.bjid.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghenghesh K.S., Ghanghish K., BenDarif E.T., Shembesh K., Franka E. Prevalence of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium spp. in Libya: 2000–2015. Libyan J. Med. 2016;11:32088. doi: 10.3402/ljm.v11.32088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harb A., O’Dea M., Hanan Z.K., Abraham S., Habib I. Prevalence, risk factors and antimicrobial resistance of Salmonella diarrhoeal infection among children in Thi-Qar Governorate, Iraq. Epidemiol. Infect. 2017;145:3486–3496. doi: 10.1017/S0950268817002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swamy S.C., Barnhart H.M., Lee M.D., Dreesen D.W. Virulence determinants invA and spvC in Salmonellae isolated from poultry products, wastewater, and human sources. Appl. Environ. Microbiol. 1996;62:3768–3771. doi: 10.1128/aem.62.10.3768-3771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barletta F., Mercado E.H., Lluque A., Ruiz J., Cleary T.G., Ochoa T.J. Multiplex Real-Time PCR for Detection of Campylobacter, Salmonella, and Shigella. J. Clin. Microbiol. 2013;51:2822–2829. doi: 10.1128/JCM.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan H., Yagyu F., Okitsu S., Nishio O., Ushijima H. Detection of norovirus (GI, GII) Sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J. Virol. Methods. 2003;114:37–44. doi: 10.1016/j.jviromet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Yan H., Nguyen T.A., Phan T.G., Okitsu S., Li Y., Ushijima H. Development of RT-multiplex PCR assay for detection of adenovirus and group A and C rotaviruses in diarrheal fecal specimens from children in China. Kansenshogaku Zasshi. 2004;78:699–709. doi: 10.11150/kansenshogakuzasshi1970.78.699. [DOI] [PubMed] [Google Scholar]
- 24.Al-Areeqi M.A., Sady H., Al-Mekhlafi H.M., Anuar T.S., Al-Adhroey A.H., Atroosh W.M., Dawaki S., Elyana F.N., Nasr N.A., Ithoi I., et al. First molecular epidemiology of Entamoeba histolytica, E. dispar and E. moshkovskii infections in Yemen: Different species-specific associated risk factors. Trop. Med. Int. Health. 2017;22:493–504. doi: 10.1111/tmi.12848. [DOI] [PubMed] [Google Scholar]
- 25.Yang R., Jacobson C., Gardner G., Carmichael I., Campbell A.J.D., Ryan U. Development of a quantitative PCR (qPCR) for Giardia and analysis of the prevalence, cyst shedding and genotypes of Giardia present in sheep across four states in Australia. Exp. Parasitol. 2014;137:46–52. doi: 10.1016/j.exppara.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Guha-Sapir D., Burkle F.M. Health trends in Iraq with a focus on children: No cause for optimism. J. Trop. Pediatr. 2014;60:177–178. doi: 10.1093/tropej/fmu027. [DOI] [PubMed] [Google Scholar]
- 27.Burkle F., Garfield R. Civilian mortality after the 2003 invasion of Iraq. Lancet. 2013;381:877–879. doi: 10.1016/S0140-6736(12)62196-5. [DOI] [PubMed] [Google Scholar]
- 28.Sethi S.K., Khuffash F.A., Al-Nakib W. Microbial etiology of acute gastroenteritis in hospitalized children in Kuwait. Pediatr. Infect. Dis. J. 1989;8:593–597. doi: 10.1097/00006454-198909000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Al-Thani A., Baris M., Al-Lawati N., Al-Dhahry S. Characterising the aetiology of severe acute gastroenteritis among patients visiting a hospital in Qatar using real-time polymerase chain reaction. BMC Infect. Dis. 2013;13:329. doi: 10.1186/1471-2334-13-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Sayed Zaki M., Abo El Kheir N. Molecular study of astrovirus, adenovirus and norovirus in community acquired diarrhea in children: One Egyptian center study. Asian. Pac. J. Trop. Biomed. 2017;7:987–990. doi: 10.1016/j.apjtb.2017.10.003. [DOI] [Google Scholar]
- 31.Han H.J., Wen H.L., Zhao L., Liu J.W., Luo L.M., Zhou C.M., Qin X.R., Zhu Y.L., Liu M.M., Qi R., et al. Novel coronaviruses, astroviruses, adenoviruses and circoviruses in insectivorous bats from northern China. Zoonoses Public Health. 2017;64:636–646. doi: 10.1111/zph.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afrad M.H., Avzun T., Haque J., Haque W., Hossain M.E., Rahman A.R., Ahmed S., Faruque A.S.G., Rahman M.Z., Rahman M. Detection of enteric- and non-enteric adenoviruses in gastroenteritis patients, Bangladesh, 2012–2015. J. Med. Virol. 2018;90:677–684. doi: 10.1002/jmv.25008. [DOI] [PubMed] [Google Scholar]
- 33.Lekana-Douki S.E., Kombila-Koumavor C., Nkoghe D., Drosten C., Drexler J.F., Leroy E.M. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. Int. J. Infect. Dis. 2015;34:90–95. doi: 10.1016/j.ijid.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Alrajab W.J., Abdullah B.A., Shareef A.Y. Salmonella responsible for infantile gastroenteritis in Mosul, Iraq. J. Trop. Med. Hyg. 1988;91:315–318. [PubMed] [Google Scholar]
- 35.Sethi S.K., Khuffash F. Bacterial and viral causes of acute diarrhoea in children in Kuwait. J. Diarrhoeal. Dis. Res. 1989;7:85–88. [PubMed] [Google Scholar]
- 36.Prakash K.P. Epidemiology and Antimicrobial Resistance of Enteric Pathogens in Dhahira Region, Oman. Iran. J. Public Health. 2008;37:60–69. [Google Scholar]
- 37.Joint Analysis Unit Thi-Qar Governorate Profile. Iraq: United Nations. 2013. [(accessed on 5 October 2013)]. Available online: http://www.cybermanual.com/thi-qar-ir-joint-analysis-unit-jau.html?page=5.
- 38.Liu J., Platts-Mills J.A., Juma J., Kabir F., Nkeze J., Okoi C., Operario D.J., Uddin J., Ahmed S., Alonso P.L., et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: A reanalysis of the GEMS case-control study. Lancet. 2016;388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lengerh A., Moges F., Unakal C., Anagaw B. Prevalence, associated risk factors and antimicrobial susceptibility pattern of Campylobacter species among under five diarrheic children at Gondar University Hospital, Northwest Ethiopia. BMC Pediatr. 2013;13:82. doi: 10.1186/1471-2431-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnee A.E., Petri W.A. Campylobacter jejuni and associated immune mechanisms: Short-term effects and long-term implications for infants in low-income countries. Curr. Opin. Infect. Dis. 2017;30:322–328. doi: 10.1097/QCO.0000000000000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akihara S., Phan T.G., Nguyen T.A., Hansman G., Okitsu S., Ushijima H. Existence of multiple outbreaks of viral gastroenteritis among infants in a day care center in Japan. Arch. Virol. 2005;150:2061–2075. doi: 10.1007/s00705-005-0540-y. [DOI] [PubMed] [Google Scholar]
- 42.Zheng S., Yu F., Chen X., Cui D., Cheng Y., Xie G., Yang X., Han D., Wang Y., Zhang W., et al. Enteropathogens in children less than 5 years of age with acute diarrhea: A 5-year surveillance study in the Southeast Coast of China. BMC Infect. Dis. 2016;16:434. doi: 10.1186/s12879-016-1760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enriquez C.E., Hurst C.J., Gerba C.P. Survival of the enteric andenoviruses 40 and 41 in tap, sea, and waste water. Water Res. 1995;29:2548–2553. doi: 10.1016/0043-1354(95)00070-2. [DOI] [Google Scholar]
- 44.Van Heerden J., Ehlers M.M., Hiem A., Grabow W.O.K. Prevalence, quantification and typing of adenoviruses detected in river and treated drinking water in South Africa. J. Appl. Microbiol. 2005;99:234–242. doi: 10.1111/j.1365-2672.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 45.Bronowski C., Mustafa K., Goodhead I., James C.E., Nelson C., Lucaci A., Wigley P., Humphrey T.J., Williams N.J., Winstanley C. Campylobacter jejuni transcriptome changes during loss of culturability in water. PLoS ONE. 2017;12:e0188936. doi: 10.1371/journal.pone.0188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amour C., Gratz J., Mduma E., Svensen E., Rogawski E.T., McGrath M., Seidman J.C., McCormick B.J.J., Shrestha S., Samie A., et al. Epidemiology and Impact of Campylobacter Infection in Children in 8 Low-Resource Settings: Results From the MAL-ED Study. Clin. Infect. Dis. 2016;63:1171–1179. doi: 10.1093/cid/ciw542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang Y.C., Scaria J., Ibraham M., Doiphode S., Chang Y.F., Sultan A., Mohammed H.O. Distribution and factors associated with Salmonella enterica genotypes in a diverse population of humans and animals in Qatar using multi-locus sequence typing (MLST) J. Infect. Public. Health. 2016;9:315–323. doi: 10.1016/j.jiph.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 48.Nair S., Ashton P., Doumith M., Connell S., Painset A., Mwaigwisya S., Langridge G., de Pinna E., Godbole G., Day M. WGS for surveillance of antimicrobial resistance: A pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J. Antimicrob. Chemother. 2016;71:3400–3408. doi: 10.1093/jac/dkw318. [DOI] [PubMed] [Google Scholar]
- 49.Liu F., Kariyawasam S., Jayarao B.M., Barrangou R., Gerner-Smidt P., Ribot E.M., Knabel S.J., Dudley E.G. Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs) Appl. Environ. Microbiol. 2011;77:4520–4526. doi: 10.1128/AEM.00468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu B., Liu W., Zhu X., Yu S., Shi X. Diversity of Salmonella isolates using serotyping and multilocus sequence typing. Food Microbiol. 2011;28:1182–1189. doi: 10.1016/j.fm.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Jassim A.M. In-home drug storage and self-medication with antimicrobial drugs in Basrah, Iraq. Oman. Med. J. 2010;25:79–87. doi: 10.5001/omj.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tajbakhsh M., Hendriksen R.S., Nochi Z., Zali M.R., Aarestrup F.M., Garcia-Migura L. Antimicrobial resistance in Salmonella spp. recovered from patients admitted to six different hospitals in Tehran, Iran from 2007 to 2008. Folia Microbiol. 2012;57:91–97. doi: 10.1007/s12223-012-0099-4. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X., Yang J., Zhang B., Sun S., Chang W. Characterization of Integrons and Resistance Genes in Salmonella Isolates from Farm Animals in Shandong Province, China. Front. Microbiol. 2017;8:1300. doi: 10.3389/fmicb.2017.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang S., Chiu C., Chiou C., Yang Y. Multidrug-resistant Salmonella enterica serovar Panama carrying class 1 integrons is invasive in Taiwanese children. J. Formos. Med. Assoc. 2013;112:269–275. doi: 10.1016/j.jfma.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 55.Randall L.P., Cooles S.W., Osborn M.K., Piddock L.J.V., Woodward M.J. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004;53:208–216. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- 56.Robinson A.L., Lee H.J., Kwon J., Todd E., Rodriguez F.P., Ryu D. Adequate hand washing and glove use are necessary to reduce cross-contamination from hands with high bacterial loads. J. Food Prot. 2016;79:304–408. doi: 10.4315/0362-028X.JFP-15-342. [DOI] [PubMed] [Google Scholar]
- 57.Humphrey J.H., Jones A.D., Manges A., Mangwadu G., Maluccio J.A., Mbuya M.N.N., Moulton L.H., Ntozini R., Prendergast A.J., Stoltzfus R.J., et al. The sanitation hygiene infant nutrition efficacy (SHINE) trial: Rationale, design, and methods. Clin. Infect. Dis. 2015;61:S685–S702. doi: 10.1093/cid/civ844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jensen D.A., Danyluk M.D., Harris L.J., Schaffner D.W. Quantifying the effect of hand wash duration, soap use, ground beef debris, and drying methods on the removal of enterobacter aerogenes on hands. J. Food Prot. 2015;78:685–690. doi: 10.4315/0362-028X.JFP-14-245. [DOI] [PubMed] [Google Scholar]