Abstract
Nucleic acid nanoparticles (NANPs) have evolved as a new class of therapeutics with the potential to detect and treat diseases. Despite tremendous advancements in NANP development, their immunotoxicity, one of the major impediments in clinical translation of traditional therapeutic nucleic acids (TNAs), has never been fully characterized. Here, we describe the first systematically studied immunological recognition of 25 representative RNA and DNA NANPs selected to have different design principles and physicochemical properties. We discover that, unlike traditional TNAs, NANPs used without a delivery carrier are immunoquiescent. We show that interferons (IFNs) are the key cytokines triggered by NANPs after their internalization by phagocytic cells, which agrees with predictions based on the experiences with TNAs. However, in addition to type I IFNs, type III IFNs also serve as reliable biomarkers of NANPs, which is usually not characteristic of TNAs. We show that overall immunostimulation relies on NANP shapes, connectivities, and compositions. We demonstrate that, like with traditional TNAs, plasmacytoid dendritic cells serve as the primary interferon producers among all peripheral blood mononuclear cells treated with NANPs, and scavenger receptor-mediated uptake and endosomal Toll-like receptor signaling are essential for NANP immunorecognition. The TLR involvement, however, is different from that expected for traditional TNA recognition. Based on these results, we suggest that NANP technology may serve as a prototype of auxiliary molecular language for communication with the immune system and the modulation of immune responses.
Keywords: RNA and DNA nanoparticles, immunorecognition, interferons, TLR, pDC, human PBMC
Graphical Abstract
The diverse field of therapeutic nucleic acids (TNAs) includes a wide range of materials made of RNA, DNA, and their analogs. Because there is no universal nomenclature, we can classify all TNAs based on the timeline of their discovery and translation into (pre)clinical studies as traditional TNAs and nano-TNAs. Some well-known examples of traditional TNAs are aptamers, antisense oligonucleotides, miRNAs, and siRNAs.1–3 Nano-TNAs can be further subdivided into the nanocarrier (e.g., liposomes)4–6 formulated traditional TNAs and programmable nucleic acid nanoparticles (NANPs), in which nucleic acids are used as building blocks to assemble novel entities with defined properties.7–9
Traditional TNAs have a long history in both basic research and clinical use, which has enabled researchers to identify their structure–activity relationships and understand their toxicities, immunorecognition, and issues with systemic delivery.10 This massive research effort has created a solid background for the traditional TNAs with diverse types of chemical modifications, conjugation strategies, and nanocarrier formulations, all of which have been used to address major translational barriers for these materials.11,12
NANP technology is a relatively new field that has already given rise to a host of self-assembling nucleic acid nanoparticles that are increasingly viewed as promising biological materials for medical applications.7,8 Programmable self-assembling NANPs are amenable to chemical modifications, control over functionalization, and consistent batch-to-batch formulation. Additionally, NANPs’ tunable physicochemical properties can be predicted and regulated. However, despite rapid advancements in basic research, clinical translation of these materials is still in its infancy because no NANPs have reached clinical trials yet.
The ability to control how the immune system responds to exogenous stimuli (therapeutic proteins, antibodies, small molecules, and nucleic acids) has the potential to either avoid adverse effects or promote the desirable activation of immune responses that are beneficial to the host. For over a decade following the proposal of the pattern recognition and danger signal theories that explain the function of the immune system, the so-called pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) have been extensively tested for applied immunomodulation, a regulated way to adjust the immune responses.13 The chemical identity of such molecular patterns is diverse and, among other types of materials, includes proteins, peptides, oligonucleotides, lipids, lipoproteins, and polysaccharides. In the field of therapeutic nucleic acids, tremendous attention and efforts were directed toward CpG oligonucleotides. However, immediate recognition of CpG oligos by the immune cells often triggers a robust stimulation of the immune system, resulting in uncontrollable severe inflammatory responses. Attempts to deliver certain types of therapeutic nucleic acids (e.g., antisense oligonucleotides, aptamers, siRNAs, and miRNAs) using nanotechnology-based carriers to control undesirable immunostimulation also had limited success because, as was discovered, commonly used biodegradable nanocarriers are not immunologically inert and can stimulate undesirable activation of the immune cells.14 In some cases, the resulting immunostimulation led to the dramatic responses and even caused patient death.15 Some attempts have been made to use NANPs as carrier for delivery of traditional TNAs. Despite the success in demonstrating functionality of these materials in animal models,16–24 their potential immunotoxicity has never been thoroughly investigated. Based on the knowledge from the field of traditional TNAs and natural nucleic acid studies, some predictions were made regarding the anticipated immune recognition of DNA-based NANPs.25 While these predictions provide an opinion, their direct application to NANPs would be inaccurate because the true knowledge of NANPs immune recognition can only be obtained empirically. While some features of NANPs’ immune behavior may appear similar to that of traditional TNAs, unexpected drastic changes in immunorecognition may stem from NANPs’ unique shapes, structures, sizes, connectivities, and compositions.26,27 This notion is consistent with the general experience of nanotechnology, wherein the materials with known properties when assembled into nanosized structures often acquire new qualities characteristic to their particular nanoscale dimensions.28,29 This point is further emphasized by the fact that all traditional TNAs are categorized by the U.S. FDA and Drug Information Association as a separate class of therapeutic products due to their distinct features separating TNAs from small molecules and biologics.30 Using similar categorization principles, NANPs were placed into a separate class due to several properties distinguishing them from traditional TNAs.26 To verify such a categorization of NANPs as drug products, it is imperative to fill the gaps regarding their immunological properties. This task can be achieved by performing a systematic investigation of NANPs immune recognition in the context of their physicochemical properties. While several attempts have been made by our groups31,32 and others33–35 to detect cytokine response to NANPs, systematic investigation across various particle designs and mechanistic investigation have not been performed.
Previously, we reported that upon introduction to human immune cells, some types of NANPs stimulated the production of type I interferons (IFNs; proteins that help regulate the antiviral activity of the immune system), while other types of NANPs were quiescent.27,32,36 Here, we set out to identify critical structural attributes that characterize the interaction of NANPs with human peripheral blood mononuclear cells (PBMCs), and we attempt to distinguish the cellular and molecular mechanisms that drive this immunological recognition. The PBMC model is known to accurately recapitulate the cytokine storm related immunotoxicities in patients and has been shown to be more predictive and reliable than any traditional rodent and nonhuman primate models.37
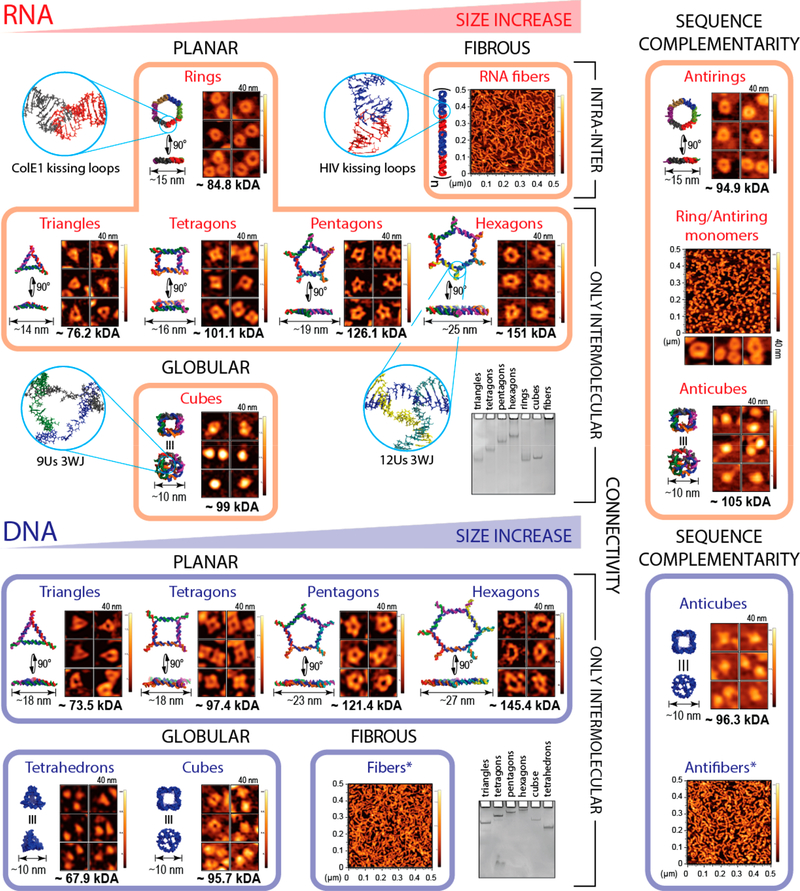
We constructed a library of 25 representative planar, globular, and fibrous RNA- or DNA-based NANPs of different sizes and molecular weights (Figure 1). NANPs with similar sizes, shapes, and compositions were further designed to have different connectivity, nucleic acid sequence complementarity, and number of single-stranded nucleotides (ssNTs) present in their structures (Figures S1 and S2). To ensure that the experimental set is representative of the field at large, we included NANPs developed by our groups and therapeutically valuable NANPs designed by others.38–40 We determined the cell types in PBMCs that preferentially internalize NANPs and studied a panel of 29 different cytokines produced by PBMCs in response to NANPs, with the goal of identifying the key biomarkers for the NANPs’ immune recognition. We further experimentally gained insight into molecular pathways triggering the immune responses and demonstrated that scavenger receptors drive the NANPs’ internalization through the endolysosomal pathway, where endosomal Toll-like receptors (TLRs) initiate the IFN response.
Figure 1.
NANPs used in this study. Representative NANPs organized to emphasize their differences in size, shape, composition, connectivity, and sequence complementarity: the key parameters contributing to the immunological recognition of NANPs. Energy-minimized 3D models of NANPs with corresponding atomic force microscopy images, relative electrophoretic mobility, estimated sizes, and molecular weights are included. The asterisk indicates that DNA fibers contain ssRNA components in their structure.
Assembly and Characterization of NANPs
We assembled and characterized under equivalent conditions 25 different NANPs, carefully chosen to address all possible links between their physicochemical properties and immunological responses (Figures 1 and S1). To study the effect of NANP sizes, various polygons (ranging from approximately 75 kDa to approximately 150 kDa) with the same connectivity27,41 were compared. The effect of 3D shapes was assessed by comparing globular cubes42,43 with planar rings31 (or polygons)27 and fibers.31 To understand the effect of chemical composition, RNA NANPs were compared with their DNA analogs. The effect of sequence complementarity was studied using anti-NANPs, assembled from NANPs’ reverse complement strands.36 We also studied the possible contributions from NANPs’ connectivity, defined by the nature of the hydrogen bonding (intramolecular and intermolecular) holding individual strands within a single NANP assembly (Figure S1). For example, in cubes, polygons, tetrahedrons, and DNA fibers, all strands are programmed to form only intermolecular hydrogen bonds. In contrast, RNA rings and RNA fibers are assembled from monomers, prefolded via intramolecular hydrogen bonds that expose long-range RNA–RNA interaction motifs (kissing loops)44 and initiate magnesium-dependent intermolecular assembly with their cognate partners.
All assembled NANPs were confirmed by electrophoretic mobility shift assay and visualized by atomic force microscopy (AFM) (Figures 1, S2, and S3). Their relative sizes and thermodynamic stabilities were assessed by dynamic light scattering and UV-melting experiments, respectively (Figures S2 and S3). Prior to all cell-culture experiments, NANPs were screened for endotoxin contamination because its presence may independently trigger the cytokine secretion. We summarized all measured parameters in Table S1.
NANPs Require a Delivery Carrier to Induce Interferon Response
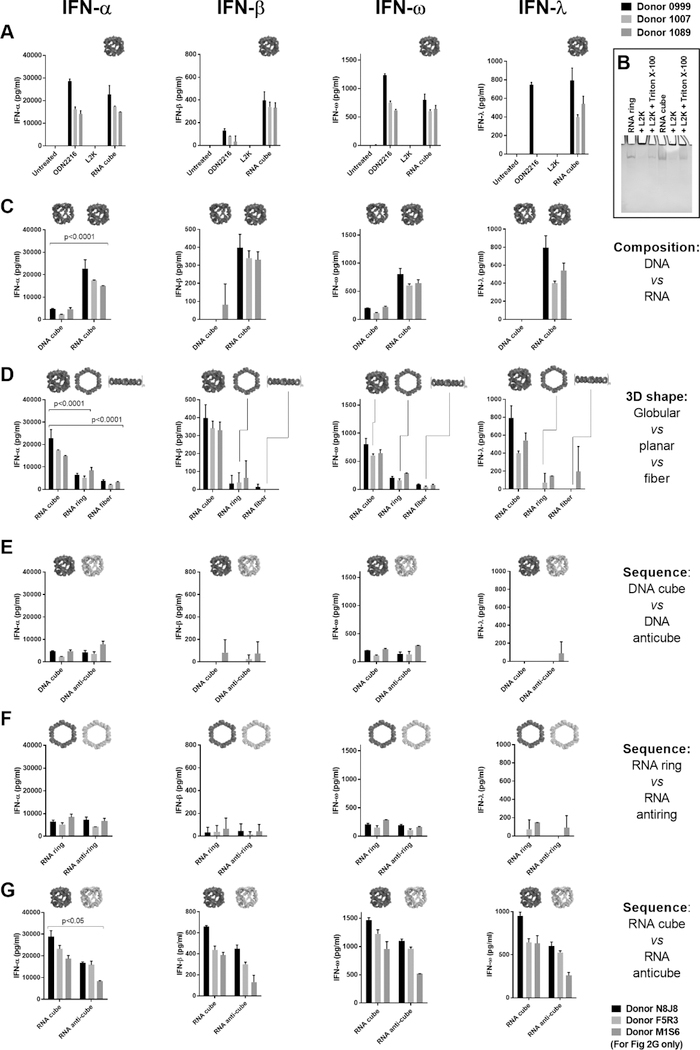
We assayed our library of NANPs for IFN induction in primary cultures of PBMCs obtained from healthy donors. NANPs that were added to the cell cultures without a delivery carrier (Lipofectamine 2000 (L2K)) were unable to stimulate any IFN response (Figures S4A,B), whereas those complexed with L2K before addition to PBMC cultures induced various levels of IFN secretion (Figure 2). L2K itself did not induce an IFN response (Figure 2A). A traditional CpG oligonucleotide (ODN 2216) specifically designed to stimulate IFN production45 exhibited its immunostimulatory properties regardless of the presence of L2K (Figure 2A). NANPs maintained their structural integrity after their release from complexes with L2K (Figures 2B and S5). These data suggest that, unlike traditional ODNs, NANPs’ complexation with L2K is critical for their recognition by PBMCs.
Figure 2.
Complexation with lipofectamine, material type, and 3D structure are critical for the NANP induction of interferon responses. Graphs show the production of IFNs by PBMCs after the delivery of NANPs with Lipofectamine 2000 (L2K). (A) Delivery of RNA cubes stimulates the production of all IFNs assayed. ODN 2216 is a positive control. (B) Ethidium bromide total staining native polyacrylamide gel electrophoresis (PAGE) results demonstrate complexation and retention of NANPs’ structural integrity upon interaction with L2K; NANPs’ complexation with L2K prevents them from entering the gel, while detergent treatment releases NANPs and restores their electrophoretic mobility. (C) RNA cubes induce greater IFN production by PBMCs than DNA cubes. (D) Comparison of RNA cubes, rings, and fibers shows that cubes are the most immunostimulatory. (E–G) Reversing the nucleic acid sequences for (E) DNA cubes and (F) RNA rings does not affect IFN induction. However, RNA cubes are stronger inducers of IFN than (G) anti-cubes. Corresponding 3D models of NANPs are shown in-line with IFN data. Each bar represents data from a single donor, showing a mean response and standard deviation (n = 3 donors). For panels A–F, donor numbers are 0999, 1007, and 1089; for panel G, donor numbers are N8J8, F5R3, and M1S6. Statistical analysis was performed by one-way ANOVA. The differences are significant for all INF in panels C, D, and G with the same p values as for INFα.
Based on our previous work,32 we expected RNA cubes complexed with L2K to stimulate high amounts of type I IFNs. Both lymphoid and myeloid cells typically produce these IFNs to provide the host with protection from viral and bacterial invasion. Indeed, RNA cubes stimulated all type I IFNs tested (IFN-α, IFN-β, and IFN-ω) at levels equivalent to or higher than the positive control (ODN 2216) (Figure 2A). RNA cubes also stimulated the induction of type III IFN (IFN-λ), a more recently characterized set of IFNs that also coordinate antiviral responses.46 RNA cubes did not induce IFN-γ, the singular type II IFN typically induced by lymphocytes (NK, NKT, and T cells) to orchestrate the adaptive immune response (Figure S6C). RNA cubes, therefore, induce strong and broad IFN responses only after complexation with the delivery carrier L2K. When electroporation was evaluated as a tool for NANPs’ delivery to the immune cells, no IFN responses were observed (data not shown), in agreement with previous reports.47,48 However, despite there being no changes in cell viability, electroporated PBMCs also lost the ability to respond to the TLR9 agonist ODN 2216, suggesting that electroporation adversely affects endosomal TLR signaling, which could also contribute to the recognition of NANPs. Based on these findings, we used L2K for all subsequent experiments.
To test if NANPs can stimulate additional cytokines, we chose RNA and DNA cubes as representative NANPs and assessed their ability to induce a panel of 29 cytokines and chemokines in both PBMCs and whole-blood cultures (Figure S7). In addition to the IFNs mentioned above, both RNA and DNA cubes stimulated IP-10 and MCP-2. This data is consistent with the IFN-dependent nature of these cytokines.49,50 Expression patterns of other tested cytokines varied between individual donors. Therefore, in all subsequent experiments, we focused on type I and III IFNs as major biomarkers of NANPs’ immune recognition. Because the CpG oligonucleotide ODN 2216 is a well-known stimulant of IFN production,45,51 we used it in all assays as a positive control.
NANP Structure and Composition Define Immunorecognition
Despite having nearly identical size and shape, RNA cubes induced higher amounts of type I and III IFNs than DNA cubes (Figure 2C), confirming our previous finding that RNA cubes are more potent immunostimulators than their DNA analogs.32 Notably, while DNA cubes induced the production of IFN-α and IFN-ω, only RNA cubes could stimulate significant quantities of IFN-β and the type III IFN-λ. Furthermore, globular RNA cubes were more immunostimulatory than any other tested RNA-based NANPs, including planar hexameric RNA rings and RNA fibers (Figure 2D). Both DNA- and RNA-based planar structures were more immunostimulatory than their respective fibers (Figures 2D and S8). Collectively, these data indicate that both composition and 3D shape of NANPs are critical factors that determine their immunorecognition.
Next, we examined whether the induction of IFNs was dependent on the sequences of the NANPs. In our library, several NANPs (e.g., DNA cubes, RNA rings, and RNA cubes) have corresponding anti-NANPs (DNA anti-cubes, RNA anti-rings, and RNA anti-cubes). In anti-NANPs, the sequences for each monomer are reverse complements of corresponding NANP strands (Figure S1). Thus, NANPs and anti-NANPs have nearly identical 3D shapes but completely different sequences. We also previously observed that combining single ring monomer with its reverse complement (here called “R/AR monomers”) resulted in the formation of mixed structures of rings and fibers. In current experiments, both DNA cubes and RNA rings induced similar IFN levels to their reverse complement analogs (Figure 2E,F), indicating that the composition (RNA versus DNA) and 3D shape of the NANPs were more-critical determinants of immunostimulatory potential than their sequences. RNA cubes, however, induced higher IFN levels than RNA anti-cubes (Figure 2G). This finding warranted the following investigation of the role of NANPs’ connectivity and size in immunorecognition.
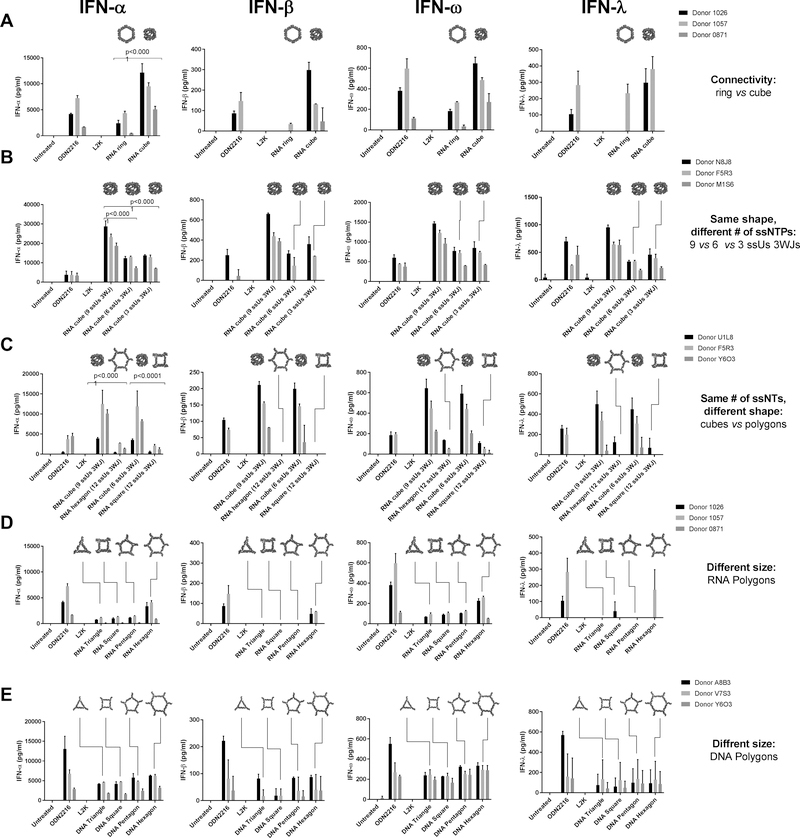
All six-stranded RNA and DNA cubes are assembled through a total of 120 intermolecular base-pairs per cube, and each contains eight (8) 3-way junctions (3WJs) at its corners (Figure S1). Each 3WJ is a flexible motif, made of 9 ssNTs—ssUs for RNA cubes and ssTs for DNA cubes.43 We hypothesized that the presence of these ssNTs may potentially contribute to the immune response. RNA rings, however, are also assembled from 6 prefolded monomers but have a total of 90 intramolecular and 42 intermolecular interactions and no ssNTs in their structure. Therefore, we compared RNA cubes and RNA rings, which are similar in size but have different connectivities (Figure S1). Consistent with previous results, RNA rings are less immunostimulatory than RNA cubes (Figures 2D and 3A), suggesting that connectivity may play a role in the IFN response.
Figure 3.
NANPs’ connectivity and size contribute to interferon induction in PBMCs. Bar graphs show the production of IFNs by PBMCs after the delivery of NANPs with L2K. (A) Comparison of NANPs with similar size but different connectivity. IFN stimulation by RNA cubes is stronger than that by RNA rings. (B) Comparison of NANPs with similar shape but different ssNTs. (C) Comparison of NANPs with the same number of ssNTs but different shape. When the number of ssUs is kept constant, 3D globular RNA NANPs (cubes) are more immunostimulatory than planar NANPs (polygons). (D) IFN induction by RNA polygons of different sizes. (E) IFN induction by DNA polygons of different sizes. Each bar shows a mean response and standard deviation from experimental duplicates (n = 3 donors). Schematics explaining connectivity can be viewed in Supplemental Figure S1. Statistical analysis was performed by one-way ANOVA. The differences are significant for all INF in panels A, B, and C with the same p values as for INFα.
To determine whether the content of the ssNTs influences the strength of the IFN response to globular NANPs, we analyzed RNA cubes with 3WJs containing 9, 6, and 3 ssUs (i.e., 72, 48, and 24 total ssUs per cube, respectively). All cubes, analyzed in blood cells from 11 different donors, showed higher immunostimulation than the assay positive control ODN 2216 (Figures 3B and S9). Interestingly, in the blood cells of some donors that show a higher response to ODN 2216, cubes with smaller 3WJs (3 or 6 ssUs) were found to be less immunostimulatory than cubes with 9-ssU 3WJs (Figure S9B, donors 1169, 0147, and 1202). No substantial difference between cubes with 3-, 6-, and 9-ssU 3WJs was observed in donor cells with lower response to the ODN 2216 (Figures 3B and S9B). These data suggest that the overall number of ssNTs may contribute to the immune response in a manner that depends on the interindividual variability in nucleic acid recognition as judged by the IFN response to ODN 2216.
We then compared NANPs that have the same connectivity and equal ssNT content but different shapes (Figure 3C). All planar RNA polygons are joined at their vertices by 3WJs composed of 12-ssNT linkers (ssUs)27 and have the same connectivity as globular cubes. Despite having a different number of ssUs per 3WJ (RNA cubes have 9 ssUs per 3WJ and RNA hexagons have 12), both RNA cubes and RNA hexagons carry a total of 72 ssUs in their structures. The results showed that RNA hexagons were less potent IFN inducers than RNA cubes. Likewise, RNA squares were less immunostimulatory than RNA cubes containing 6 ssUs per 3WJ even though both materials had a total of 48 ssUs per NANP (Figure 3C). Interestingly, introduction of the ssNTs’ regions into RNA rings52 did not make any difference in IFN induction by these particles (Figure S10). Collectively, these data suggest that the immunostimulatory effect of ssNTs is unique to the 3D RNA NANPs (i.e., RNA cubes). However, neither ssNTs alone nor the monomer oligonucleotide (data not shown) can fully explain the robust immunostimulatory behavior of these NANPs.
Finally, we considered the role of NANP size by testing three-, four-, five-, and six-sided RNA and DNA polygons. When tested at the equimolar concentrations in cells with the strongest response to ODN 2216, RNA hexagons induced more IFNs than any smaller RNA polygons (Figure S9D, donors 1026 and 1057). Testing these polygons at equal mass concentration did not provide any advantage (Figure S11), suggesting that larger size and higher number of 3WJs per NANP, but not the overall amount of RNA material, were responsible for the increased IFN response in donor cells with the highest response to ODN 2216. Interestingly, when the same polygons were assayed in donor cells with lower IFN response to ODN 2216, no difference between NANPs was observed (Figures 3D and S9D; donors 0871, 0673, 0639, and 0544). For analogous DNA polygons, no physiologically significant (i.e., at least 2-fold) difference was observed between NANPs of different sizes regardless of the magnitude of the cellular response to the ODN 2216 (Figures 3E and S9E). Testing DNA polygons at equal mass also had no advantage (Figure S12). Overall, consistent with our previous reports,27,32 when tested side-by-side in cell cultures or primary cells derived from the same donors, DNA NANPs were less immunostimulatory than RNA NANPs. These results suggest that connectivity, shape, and size contribute to IFN responses for NANPs made of RNA but not DNA.
PBMC Internalize NANPs via the Endolysosomal Pathway
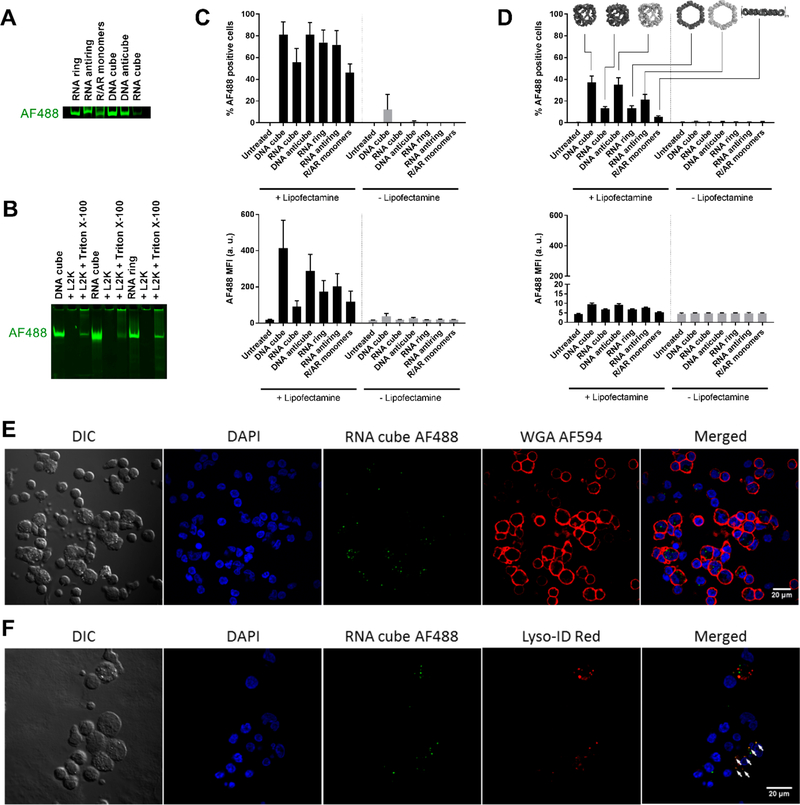
To determine which cells in PBMCs preferentially internalize NANPs, we labeled NANPs with Alexa Fluorophore 488 (AF488). Prior to testing in cells, we analyzed fluorescently labeled NANPs and found that all AF488-labeled NANPs, when compared at equimolar concentrations, had different levels of fluorescence (Figure 4A). This was likely due to different yields of covalently labeled monomers acquired from a commercial source. Therefore, the results of the uptake experiment could only be used to make qualitative comparisons (i.e., to confirm the NANPs uptake at different experimental conditions). Importantly, similar to the unlabeled NANPs, the structural integrity of AF488-labeled NANPs was retained after release from L2K (Figure 4B). PBMCs were exposed to AF488-labeled NANPs overnight, and cells with NANP-associated fluorescence were counted using flow cytometry. Lymphocytes and monocytes, the main cell types in PBMCs distinguishable by their forward and side scattering, were included in the analysis (Figure S13). Due to their overlapping scatter profiles, dendritic cells (DCs) could not be distinguished from the monocyte population in the PBMCs. Typically, immunophenotyping is used to discriminate between DCs and monocytes by staining the cells with antibodies specific to the surface markers of DCs but not monocytes. However, due to the small proportion of DCs (<1% of the monocytic fraction), our experiment did not include the phenotyping. NANP-associated fluorescence was predominantly detected on monocytes (Figure 4C), which are part of the mononuclear phagocyte system and play central roles in the internalization and clearance of various nanoparticles in the bloodstream.24 A smaller fraction was associated with lymphocytes (Figure 4D). In agreement with the IFN induction results (Figures 2, S3, and S4), presence of L2K was crucial for NANPs’ uptake. Without L2K, neither lymphocyte nor monocyte populations associated with the AF488-labeled NANPs (Figure 4C,D).
Figure 4.
Internalization of NANPs by PBMCs occuring through the endolysosomal pathway. All tested NANPs were assembled with AF488-labeled strands. (A) Fluorescence imaging of AF488-labeled NANPs analyzed by native PAGE. Due to variations in the labeling efficiencies of purchased monomers, various NANPs have different amounts of AF488 in their structures. (B) Labeled NANPs form complexes with L2K and retain their structural integrity upon detergent-mediated release. (C, D) AF488-labeled NANPs with L2K were delivered to PBMCs, which were then studied using flow cytometry. Each bar shows the mean response derived from three individual donors and a standard deviation. Samples in each individual donor were analyzed in duplicate. (C) Monocytes and (D) lymphocytes were assessed for both the percentage of cells that took up NANPs (upper plots) and the number of NANPs taken up as represented by geometric mean fluorescence intensity (MFI) (lower plots). NANP fluorescence was greatly associated with monocytes but minimally associated with lymphocytes. (E, F) PBMCs treated with AF488-labeled RNA cubes were visualized with confocal microscopy. (E) Intracellular localization of RNA cubes (green) was demonstrated using Alexa Fluor 594-labeled wheat germ agglutinin (WGA 594) to delineate cell membranes (red). (F) RNA cubes (green) were also shown to localize with the endolysosomal pathway (white arrows) using Lyso-ID Red as a stain for acidic vesicles (red). Shown is the representative image of several fields of view with a total count of at least 100 cells.
Next, we used RNA cubes, which were the most-potent inducers of IFNs, to determine intracellular localization in PBMCs. Following transfection, fixed PBMCs were stained with wheat-germ agglutinin, a lectin that binds to cell membranes. Confocal microscopy images show that NANP-associated fluorescence was mostly localized to the interior of cells (Figures 4E and S14 for the z-stacked image), which agrees with observations from flow cytometry that show NANPs were primarily associated with the phagocytic monocyte population. Labeled RNA cubes could also be seen within and on the surface of smaller cells with minimal cytoplasm that resemble lymphocytes, which was also in agreement with our earlier observations. PBMCs were subsequently stained with Lyso-ID Red, a photostable dye that localizes to acidic vesicles and labels the endolysosomal pathway. Confocal microscopy images show NANP-associated fluorescence colocalized with Lyso-ID Red staining (Figure 4F), confirming that the internalized cubes are taken up by the endolysosomal pathway after L2K-mediated delivery to PBMCs. Even though the levels of endosomal and lysosomal markers in the primary cells were lower than in the immortalized cancer cell lines, the current results are consistent with our previous observations for L2K-associated RNA–DNA hybrids, internalized by human cancer cells via endolysosomal pathway.24
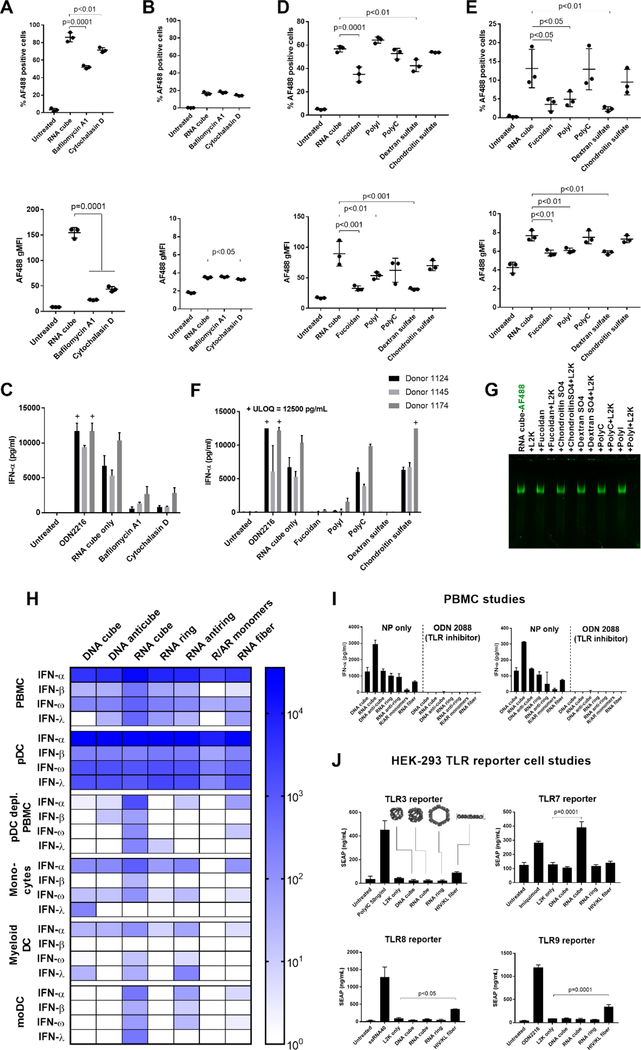
Using common inhibitors, we investigated the mechanism of endocytic uptake of the RNA cubes. First, we incubated labeled RNA cubes with PBMCs at 4 °C. This incubation significantly reduced their uptake by monocytes (Figure S15A), confirming the involvement of active energy-dependent processes in NANPs’ uptake. We then used bafilomycin A1, a vacuolar H+ ATPase inhibitor that blocks endosomal function, and cytochalasin D, an inhibitor of actin polymerization and phagocytosis. Both inhibitors significantly reduced RNA cube uptake by monocytes (Figure 5A) but not lymphocytes (Figure 5B), suggesting that phagocytosis and endosomal acidification were essential for the internalization of NANPs by monocytes. This finding was consistent with the phagocytic function of monocytes as opposed to the nonphagocytic lymphocytes. Both inhibitors also blocked the production of IFN-α that otherwise would be stimulated by RNA cube delivery (Figure 5C). The relevance of these in vitro data to the in vivo recognition of NANPs would involve a clinical study with cells derived from the patients treated with NANPs.
Figure 5.
Cellular and molecular mechanisms involved in the immune recognition of NANPs. The study involved 12 donors in total. After pretreatment with the inhibitors bafilomycin A1 and cytochalasin D, (A) monocytes and (B) lymphocytes were analyzed by flow cytometry for the percentage of cells that took up AF488-labeled RNA cubes (upper plots) and the number of particles taken up as represented by MFI (lower plots). A total of three donors (1083, 1094, and 1150) were tested, with each dot representing data from an individual donor. Phagocytosis and endosomal acidification are essential for NANP internalization by monocytes. (C) PBMCs from three donors treated with bafilomycin A1 and cytochalasin D did not produce an IFN-α response when exposed to unlabeled RNA cubes. (D, E) PBMCs were pretreated with several scavenger receptor inhibitors [fucoidan, polyinosinic acid (polyI), and dextran sulfate] and controls [polycytidylic acid (polyC) and chondroitin sulfate] before being exposed to AF488-labeled RNA cubes. (D) Monocytes and (E) lymphocytes were analyzed by flow cytometry. Fucoidan and dextran sulfate blocked the uptake of RNA cubes in monocytes (D), whereas fucoidan, polyI, and dextran sulfate blocked uptake in lymphocytes (E). A total of three donors (0794, 1155, and 1157) were tested, with each dot representing data from an individual donor. (F) PBMCs were treated with scavenger receptor inhibitors and then treated with unlabeled RNA cubes and assayed for IFN-α production. (G) Native PAGE showing that none of the inhibitors induced release of L2K complexation with AF488-labeled RNA cubes. “+ ULOQ” refers to the IFN levels above the assay’s upper limit of quantification. Statistical analysis was performed by one-way ANOVA with Dunnett’s post-test, comparing all groups to the RNA cube alone (single asterisk, p < 0.05; double asterisks, p < 0.01; triple asterisks, p < 0.001; quadruple asterisks, p = 0.0001). (H–J) Cells from major DC subsets [plasmacytoid DCs (pDCs), monocytes, and myeloid DCs] were purified from whole blood by negative selection, treated with NANPs, and assayed for IFN production (H). Each box represents an averaged value across separate groups of three donors. Plasmacytoid DCs were depleted from PBMCs (pDC-depleted PBMC) by positive selection, and the resulting cells were treated with NANPs. Purified monocytes were differentiated into monocyte-derived DCs, treated with NANPs, and tested for IFN induction. Complete data sets are presented in Figure S16. (I) NANPs were complexed with L2K and added to PBMCs either alone or with a pan-TLR inhibitor, ODN 2088. Production of IFN-α (left) and IFN-ω (right) was measured by multiplexed ELISA. (J) HEK-293 reporter cell lines overexpressing TLR3, TLR7, TLR8, or TLR9 were used to estimate the recognition of NANPs. Known agonists to the respective TLRs were used as positive controls: poly(I:C) for TLR3, Imiquimod for TLR7, ssRNA40 for TLR8, and ODN 2216 for TLR9. Only RNA cubes activated the SEAP reporter gene in the TLR7-over-expressing cell line. Activation of the reporter gene by RNA fibers was observed in TLR3-, TLR8-, and TLR9-over-expressing cells (possibly due to the expression of endogenous TLR3 in all cell lines). Each bar in panels H–J shows a mean response and a standard deviation (n = 3). Statistical analysis was performed by one-way ANOVA with Dunnett’s post-test, comparing all groups to the “L2K only” results.
Because scavenger receptors (SR) have been implicated in the cellular uptake and immunological sensing of therapeutic oligonucleotides53 and nucleic-acid-containing nanoparticles,54 we tested for the involvement of SR in NANPs’ uptake and processing. Both the percent of positive cells and the geometric mean of fluorescent intensity (gMFI) were assessed because they allow the assessment of different parameters. The percent of positive cells reflects the total number of cells that internalized the NANPs, while gMFI assesses the degree of NANPs’ uptake by an individual cell. A decrease in either or both of these parameters was interpreted as the inhibition of the NANPs’ uptake by cells. SR inhibitors fucoidan, dextran sulfate, and polyinosinic acid all inhibited the uptake of AF488-labeled RNA cubes by monocytes (Figure 5D). Interestingly, these inhibitors also blocked the association of NANPs with lymphocytes (Figure 5E), suggesting that SRs are involved in the association of NANPs with the cell membrane, even in the absence of bona fide endocytosis. In addition to NANPs’ internalization, the SR inhibitors also blocked the production of IFN-α in treated PBMCs (Figure 5F). Chondroitin sulfate and polycytidylic acid, which are structurally analogous to dextran sulfate and polyinosinic acid, respectively, do not bind to SRs and serve as respective negative controls. These inhibitors had no impact on either NANP uptake or IFN-α production (Figure 5D–F). Similar results were obtained with DNA cubes, RNA rings, and RNA fibers (Figure S15B). None of the inhibitors induced a detectable release of RNA cubes or rings from electrostatically driven complexation with L2K (Figures 5G and S15C), indicating that the reduction in IFN response was not due to decreased amounts of NANPs delivered into immune cells. It is important to note that complexation with L2K does not encapsulate NANPs; it only changes the net charge of the complex but does not eliminate NANPs polyanionic structures available for interaction with cell receptors. Our data suggest that L2K-delivered NANPs associate with the cell membrane via SR interactions and are subsequently taken up into the endolysosomal pathway via endocytosis. Because low levels of uptake and IFN are also observed in the presence of SR inhibitors, the data also suggested that some interactions of the NANP-L2K complex could occur through the L2K–cell membrane contact. Yet, the significant drop in both the uptake and IFN production suggest that SR-mediated contact with polyanionic NANPs was the major route of uptake.
pDCs Recognize NANPs and TLR7 Contributes to IFN Induction
Monocytes and dendritic cells play a key role in coordinating the innate and adaptive immune response. They are composed of circulating blood cells of the mononuclear phagocyte system, and they express multiple TLRs and other nucleic acid sensors that recognize various PAMPs, including foreign nucleic acids.55 In particular, plasmacytoid DCs (pDCs) are known as specialized producers of type I and III IFNs,56 while inflammatory monocytes also contribute to the IFN response.57 To determine which cells in PBMCs are responsible for IFN production upon uptake of NANPs, we isolated monocytes, myeloid DCs, and pDCs from PBMCs (Table S2), and generated monocyte-derived DCs. We then treated these purified immune cells with NANPs and determined the resulting IFN response. Purified pDCs responded strongly to all tested RNA- and DNA-based NANPs, producing large quantities of all IFNs assayed (Figure 5H). Interestingly, the response to RNA cubes did not dominate in pDCs as in PBMCs. Instead, all NANPs stimulated strong IFN induction in pDCs. This finding indicates that the structure–activity relationships observed are dependent on cellular interactions within the PBMC population. In addition, the depletion of pDCs from PBMCs (Figure S16D and Table S2) resulted in the near-total loss of IFN response to all NANP types, with a significantly lower IFN response stimulated by RNA cubes (Figure S17). This confirms that pDCs are the primary source of IFNs in the immune response to NANPs.
Purified monocytes produced low amounts of IFN-α and IFN-ω but no IFN-β or IFN-λ for all NANP types. However, among all NANP types, RNA cubes stimulated the greatest amount of type I IFN in this purified monocyte population (Figures 5H and S17). In myeloid DCs and monocyte-derived DCs, the responses were donor-dependent and cells isolated from two out of three donors did not produce IFN in response to NANPs, while cells obtained from responding donors showed the most robust response to RNA cubes. Therefore, monocytes and myeloid DCs may produce IFNs in response to RNA cubes in the absence of pDCs and may contribute to IFN induction within PBMC cultures.
Given the importance of TLRs in the detection of foreign nucleic acids by the immune system58,59 and NANPs’ uptake via the endocytic route, we investigated the involvement of these pattern recognition receptors in the IFN response to NANPs. This study did not involve the entire library of the NANPs; instead, we focused on those particles that demonstrated the most prominent IFN induction in earlier experiments. The oligonucleotide inhibitor, ODN 2088, inhibits all human endosomal TLRs (Figure S18A). Upon pretreatment with ODN 2088, IFN responses to all tested NANPs disappeared in PBMCs, suggesting the importance of TLR signaling in the IFN response to NANPs (Figure 5I). ODN 2088 also completely inhibited the IFN response of purified pDCs to DNA and RNA cubes (Figure S19). Despite broad claims of specificity to particular TLRs, other commercial oligonucleotide-based TLR inhibitors were similarly effective at inhibiting multiple human TLRs in our reporter-cell-based assay used to validate the inhibitors. The broadly inhibitory effects of these inhibitors mean that they are not suitable for investigating the involvement of specific TLRs in NANP-mediated type I IFN response (Table S3).
We then used HEK 293 cells over-expressing human TLR3, TLR7, TLR8, or TLR9 to evaluate the involvement of individual TLR signaling in the recognition of NANPs. These cells are engineered to contain a secreted embryonic alkaline phosphatase (SEAP) gene under control of the IFN-β promoter fused to NF-κB and AP-1 binding sites. RNA cubes specifically induced SEAP production in TLR7 over-expressing reporter cells, while RNA fibers induced SEAP production in TLR3, TLR8, and TLR9 cells (Figure 5J). However, as specified by the manufacturer, all HEK-293 cells also express endogenous TLR3. Therefore, it is possible that RNA fibers induced SEAP production in both TLR8 and TLR9 reporter cells through the involvement of endogenous TLR3.
Questions remain concerning the exact identity of the immune sensors that mediate the immune response to NANPs. While our data point firmly to the involvement of TLRs, we cannot completely rule out the contributions of other, non-endosomal nucleic acid sensors, such as the RIG-I/MDA5, cGAS-STING, and DHX9/DHX36 pathways that are also present in pDCs.56
Conclusions
The immunomodulatory effects of NANPs are largely unknown and must be defined for their successful translation into the clinic. Here, we show that immunological recognition of NANPs depends on multiple physicochemical parameters, including particle size, 3D structure, composition, and connectivity. By assembling RNA monomers into compact globular structures with intramolecular connectivity, the desirable immunostimulatory effects of NANPs, such as IFN induction by locally administered vaccine adjuvants, can be enhanced. Unwanted effects, such as IFN induction at cytokine storm levels by systemically administered nanoscaffolds carrying drugs, can be reduced by using DNA-based NANPs or by forming either planar or fibrous-shaped RNA materials with intra- and intermolecular connectivity. By adjusting the overall lengths of the single-stranded regions, further finetuning to control the magnitude of the desirable immunostimulation can be achieved for globular NANPs. Unlike traditional nucleic acids (e.g., CpG oligonucleotides), which activate the immune cells upon exposure, NANPs exhibit immunological activity only after complexation with delivery carriers, regardless of their size, shape, composition, and connectivity. Our mechanistic studies show that NANPs are associated with scavenger receptors and are subsequently taken up into the endolysosomal pathway via endocytosis, in which endosomal TLRs initiate the IFN response. Plasmacytoid dendritic cells are the primary source of IFNs in the immune response to NANPs. Our findings highlight the key parameters that inform the way NANPs interact with the immune system. These new insights improve the current understanding of NANPs’ immunostimulatory properties25 and pave the way to development of a new auxiliary molecular language that can be expressed through the script of rationally designed NANPs.
Such communication with the immune system will become instrumental, for example, in either achieving desirable activation of the immune cells that are beneficial for vaccines and cancer immunotherapies or reducing undesirable immunostimulation that commonly limits the translation of systemically delivered traditional nucleic-acid-based therapies.
Materials and Methods
All materials and methods are detailed in the Supporting Information.
NANP Synthesis and Characterization
All NANPs were assembled from individually combined monomers following different one-pot assembly protocols. For electrophoretic mobility shift assays, ethidium bromide total staining (or fluorescently labeled NANPs) nondenaturing native polyacrylamide gel electrophoresis (PAGE; 8%; 37.5:1) was used. All gels were visualized with a Bio-Rad ChemiDoc MP System.
AFM imaging was done on a MultiMode AFM Nanoscope IV system (Bruker Instruments, Santa Barbara, CA) in tapping mode. The images were collected with a 1.5 Hz scanning rate using a TESPA-300 probe from Bruker (resonance frequency of 320 kHz, spring constant of about 40 N/m). Images were processed by the FemtoScan Online software package (Advanced Technologies Center, Moscow, Russia).60,61
The average hydrodynamic radii for all NANPs were measured in a microcuvette (Starna Cells, Inc.) using a Zetasizer Nano-ZS (Malvern Instruments, Ltd.). All measurements were done at room temperature according to the instrumentation protocol.
To measure the melting temperature (Tm) of NANPs, temperature-dependent absorption measurements were recorded at 260 nm on an Agilent 8453 spectrophotometer coupled with a Agilent 89090 Peltier temperature controller.
For fluorescent labeling, one monomer strand (specified in the Supporting Information) from each NANP was purchased from Integrated DNA Technologies with AF488 covalently attached to the 3′ end. Fluorescently labeled NANPs were assembled via a one-pot protocol (as described above) from a mixture of monomers containing AF488-labeled strands and were characterized as described above.
Prior to immunological assays, all NANPs were tested for endotoxin by kinetic turbidity limulus amebocyte lysate (LAL) assay using a Pyros Kinetix Flex instrument (Associates of Cape Cod, East Falmouth, MA). Acceptance criteria was endotoxin levels of <0.05 EU/mL in 1 μM stocks of NANPs (Table S1).
Primary Human PBMC Isolation
Blood was collected from healthy volunteers under Institutional Review Board approved Frederick National Laboratory for Cancer Research Protocol OH9-C-N046. Each donor was anonymized and assigned a random number, which is denoted in the data. At least three donors were tested for each experiment. Blood was collected from more than 60 unique donors over the course of this study. Blood was drawn into tubes containing Li–heparin as an anticoagulant and processed within 2 h after collection. Whole blood was mixed 1:1 with phosphate-buffered saline and layered onto Ficoll-Paque Plus (GE Healthcare Biosciences, Pittsburgh, PA), then centrifuged at 900g with low acceleration and no brake. PBMCs at the buffy coat were collected, washed twice with 1× Hank’s balanced salt solution (HBSS), then resuspended in complete Roswell Park Memorial Institute (RPMI) medium (RPMI 1640 with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and penicillin–streptomycin). Live cells were enumerated and used in subsequent experiments.
Stimulation of PBMCs with DNA and RNA Nanoparticles for Cytokine Induction
PBMCs were seeded at 1.25 × 106 cells/mL in 96-well U-bottomed plates with 160 μL per well. Nanoparticles at 1 μM stock solution were complexed to L2K at a 5:1 v/v ratio. After a 30 min incubation at room temperature, complexed nanoparticles were made up to 50 nM in OptiMEM and added to PBMCs at 40 μL per well for a final stimulation concentration of 10 nM. After a 20 h incubation at 37 °C, the supernatants were collected and analyzed for cytokines by multiplexed enzyme-linked immunosorbent assay (ELISA) (Quansys Biosciences, Logan, UT). Samples producing IFN-α responses above the upper quantitation limit were reassayed using an all-subtype IFN-α ELISA (PBL Assay Science, Piscataway, NJ).
Purification and Culture of DC Subsets for Nanoparticle Stimulation
Plasmacytoid DCs and myeloid DCs were purified using negative selection kits per the manufacturer’s instructions (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Briefly, PBMCs were purified and incubated with a blocking reagent and biotinylated antibodies. Streptavidin-coated microbeads were then bound to undesired cells and retained in a magnetic column, while the unbound desired fraction was eluted from the column. Live cells were enumerated, phenotyped for appropriate surface markers (Figure S16), and used for nanoparticle stimulation as above. Monocytes were similarly purified by negative selection according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Purified monocytes were seeded at 20 000 cells per well, while all other dendritic cell subsets were seeded at 10 000 cells per well. To produce monocyte-derived dendritic cells, monocytes were cultured at 0.5 × 106 cells per milliliter in 6-well plates, supplemented with 100 ng/mL rhGM-CSF and 20 ng/mL rhIL-4 (Peprotech, Rocky Hill, NJ). Plasmacytoid DCs were depleted from PBMCs using a CD304 microbead kit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Briefly, CD304+ pDCs were bound to magnetic microbeads coated with anti-CD304 antibodies. Bound pDCs were retained in a magnetic column, while the flow-through was collected, phenotyped for the depletion of pDCs (Figure S16D), and used in subsequent experiments. Phenotyping was performed using the following antibodies: BDCA-2 APC, BDCA-4 APC, CD1c FITC, CD141 PE (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), and CD14 FITC (eBioscience, San Diego, CA).
Characterization of Nanoparticle Association with PBMCs by Flow Cytometry and Confocal Microscopy
AF488-labeled nanoparticles were delivered to PBMCs as described earlier. Lymphocytes and monocytes were distinguished based on their forward-scatter and side-scatter characteristics (Figure S13). For confocal microscopy samples, 1 × 106 PBMCs per well were seeded in 900 μL of complete RPMI medium in a 24-well plate. For each well, Lipofectamine-complexed and AF488-labeled RNA cubes were added to cells at a final concentration of 10 nM as described earlier. The next day, cells were removed and washed with 1× HBSS. For wheat-germ agglutinin Alexa Fluor 594 (WGA 594) (Molecular Probes, Eugene, OR) staining, cells were fixed for 15 min in 4% formaldehyde and then washed and stained with 1 μg/mL WGA 594 in HBSS for 10 min at room temperature. For the labeling of acidic vesicles, cells were stained with Lyso-ID Red (Enzo Life Sciences, Farmingdale, NY). Lyso-ID Red staining solution was prepared by diluting the assay buffer provided in HBSS and adding 1:1000 Lyso-ID Red stock and 10% FBS. Lyso-ID Red staining was performed for 30 min. After staining with either WGA 594 or Lyso-ID Red, cells were washed twice with HBSS, and slides were prepared using a CytoSpin centrifuge. Images were acquired on a Nikon A1R+ confocal microscope (Nikon Instruments, Melville, NY).
Inhibitor Studies
For all experiments involving inhibitors, PBMCs were preincubated with designated inhibitors for 2 h at 37 °C before the addition of nanoparticles. Internalization inhibitor concentrations used were as follows: bafilomycin A1, 100 nM; cytochalasin D, 5 μM; and all scavenger receptor inhibitors and controls, 50 μg/mL (all internalization inhibitors listed above were from Sigma-Aldrich, Saint Louis, MO). ODN 2088 was used at 10 μM (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Treated PBMCs were then tested for nanoparticle uptake or IFN induction.
To study the individual scavenger receptor inhibitors’ interactions with the assemblies, AF488-labeled NANPs (10 nM) (with and without L2K) were incubated with the various scavenger receptor inhibitors (50 μg/mL) for 30 min at room temperature. Samples were analyzed on nondenaturing native PAGE (8%, 37.5:1).
Reporter Cell Culture and Experiments
HEK-Blue human TLR reporter cells were purchased from Invivogen (San Diego, CA). All reporter lines were cultured according to the manufacturer’s instructions. Briefly, cells were cultured in Dulbecco’s modified Eagle medium with 10% FBS, penicillin–streptomycin, and 100 μg/mL normocin. The selection antibiotics zeocin (100 μg/mL) and blasticidin (10–30 μg/mL) were added for two passages after the cells were thawed. Then, the cells were cultured for two additional passages before experimental use. For the reporter cell assay, cells were plated in 96-well flat-bottomed plates at 40 000–80 000 cells per well. NANPs were added, and cells were incubated at 37 °C for 24 h. Supernatants were collected and assayed using QUANTI-Blue (Invivogen, San Diego, CA) and recombinant SEAP as a standard. Specific TLR agonists were used as follows: 1 μg/mL poly(I:C) (TLR3), 5 μg/mL Imiquimod (TLR7), 5 μg/mL ssRNA40/LyoVec (TLR8), and 100 μg/mL ODN 2216 (TLR9) (all from Invivogen, San Diego, CA).
Statistics
Statistical analysis was performed using one-way ANOVA conducted with GraphPad Prism Software. All column means were compared by Tukey’s multiple comparison test. Where specified, the Dunnett test was used to compare column means with a specific control column. A p value of less than 0.05 was considered to be statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported in part (E.H., A.S., and M.A.D.) by federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under award no. R01GM120487 (to K.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Alexander Lushnikov and Alexey Krasnoslobodtsev for performing AFM imaging of the nanoparticles at the Nanoimaging Core Facility at the University of Nebraska Medical Center, Morgan Chandler of the University of North Carolina at Charlotte for help with figures, Ai Lin Chun of Science Story Lab for helpful critique, and Barry Neun and Edward Cedrone of the Nanotechnology Characterization Lab for the excellent technical assistance. M.A.D. is grateful to Scott E. McNeil for continuous support.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.8b01283.
All NANPs sequences used in this project; detailed experimental methods; NANP design, shapes, and connectivity and their physicochemical characterizations; structural integrity after complexation with L2K; electroporation experiments; flow cytometry; confocal 3D microscopy receptor inhibition experiments; and assessment of the NANP-mediated induction of 29 different cytokines. (PDF)
REFERENCES
- (1).Grijalvo S; Alagia A; Jorge AF; Eritja R Genes 2018, 9, (2).74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sasaki S; Guo S Nucleic Acid Ther. 2018, 28 (1), 1–9. [DOI] [PubMed] [Google Scholar]
- (3).Nimjee SM; White RR; Becker RC; Sullenger BA Annu. Rev. Pharmacol. Toxicol 2017, 57, 61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kaczmarek JC; Kowalski PS; Anderson DG Genome Med. 2017, 9 (1), 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lieberman J Nat. Struct. Mol. Biol 2018, 25 (5), 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sun Y; Zhao Y; Zhao X; Lee RJ; Teng L; Zhou C Molecules 2017, 22, (10).1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Jasinski D; Haque F; Binzel DW; Guo P ACS Nano 2017111142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chen YJ; Groves B; Muscat RA; Seelig G Nat. Nanotechnol 2015, 10 (9), 748–60. [DOI] [PubMed] [Google Scholar]
- (9).Veneziano R; Ratanalert S; Zhang K; Zhang F; Yan H; Chiu W; Bathe M Science 2016, 352 (6293), 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Dobrovolskaia MA; McNeil SE Expert Opin. Biol. Ther 2015, 15 (7), 1023–48. [DOI] [PubMed] [Google Scholar]
- (11).Craig K; Abrams M; Amiji M Expert Opin Drug Deliv 2018, 3, 1–12. [DOI] [PubMed] [Google Scholar]
- (12).Freeley M; Long A Biochem. J 2013, 455 (2), 133–47. [DOI] [PubMed] [Google Scholar]
- (13).Kay E; Scotland RS; Whiteford JR Biofactors 2014, 40 (3), 284–94. [DOI] [PubMed] [Google Scholar]
- (14).Peer D Adv. Drug Delivery Rev 2012, 64 (15), 1738–48. [DOI] [PubMed] [Google Scholar]
- (15).Rupaimoole R; Slack FJ Nat. Rev. Drug Discovery 2017, 16 (3), 203–222. [DOI] [PubMed] [Google Scholar]
- (16).Binzel DW; Shu Y; Li H; Sun M; Zhang Q; Shu D; Guo B; Guo P Mol. Ther 2016, 24 (7), 1267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rychahou P; Haque F; Shu Y; Zaytseva Y; Weiss HL; Lee EY; Mustain W; Valentino J; Guo P; Evers BM ACS Nano 2015, 9 (2), 1108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lee TJ; Haque F; Vieweger M; Yoo JY; Kaur B; Guo P; Croce CM Methods Mol. Biol 2015, 1297, 137–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shu Y; Pi F; Sharma A; Rajabi M; Haque F; Shu D; Leggas M; Evers BM; Guo P Adv. Drug Delivery Rev 2014, 66, 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Feng L; Li SK; Liu H; Liu CY; LaSance K; Haque F; Shu D; Guo P Pharm. Res 2014, 31 (4), 1046–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Li S; Jiang Q; Liu S; Zhang Y; Tian Y; Song C; Wang J; Zou Y; Anderson GJ; Han JY; Chang Y; Liu Y; Zhang C; Chen L; Zhou G; Nie G; Yan H; Ding B; Zhao Y Nat. Biotechnol 2018, 36 (3), 258–264. [DOI] [PubMed] [Google Scholar]
- (22).Lee H; Lytton-Jean AKR; Chen Y; Love KT; Park AI; Karagiannis ED; Sehgal A; Querbes W; Zurenko CS; Jayaraman M; Peng CG; Charisse K; Borodovsky A; Manoharan M; Donahoe JS; Truelove J; Nahrendorf M; Langer R; Anderson DG Nat. Nanotechnol 2012, 7 (6), 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA Nano Lett. 2014, 14 (10), 5662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Afonin KA; Viard M; Martins AN; Lockett SJ; Maciag AE; Freed EO; Heldman E; Jaeger L; Blumenthal R; Shapiro BA Nat. Nanotechnol 2013, 8 (4), 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Surana S; Shenoy AR; Krishnan Y Nat. Nanotechnol 2015, 10 (9), 741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dobrovolskaia MA DNA and RNA Nanotechnology 2016, 3, (1). DOI: 10.1515/rnan-2016-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Johnson MB; Halman JR; Satterwhite E; Zakharov AV; Bui MN; Benkato K; Goldsworthy V; Kim T; Hong E; Dobrovolskaia MA; Khisamutdinov EF; Marriott I; Afonin KA Small 2017, 13, (42).1701255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).National Nanotechnology Initiative. What’s so special about the nanoscale?. https://www.nano.gov/nanotech-101/special (accessed).
- (29).Roduner E Chem. Soc. Rev 2006, 35 (7), 583–92. [DOI] [PubMed] [Google Scholar]
- (30).Kornbrust D; Cavagnaro J; Levin A; Foy J; Pavco P; Gamba-Vitalo C; Guimond A Nucleic Acid Ther. 2013, 23 (1), 21–8. [DOI] [PubMed] [Google Scholar]
- (31).Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L Nat. Protoc 2011, 6 (12), 2022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Afonin KA; Viard M; Kagiampakis I; Case CL; Dobrovolskaia MA; Hofmann J; Vrzak A; Kireeva M; Kasprzak WK; KewalRamani VN; Shapiro BA ACS Nano 2015, 9 (1), 251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Guo S; Li H; Ma M; Fu J; Dong Y; Guo P Mol. Ther.–Nucleic Acids 2017, 9, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Khisamutdinov EF; Li H; Jasinski DL; Chen J; Fu J; Guo P Nucleic Acids Res. 2014, 42 (15), 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Schuller VJ; Heidegger S; Sandholzer N; Nickels PC; Suhartha NA; Endres S; Bourquin C; Liedl T ACS Nano 2011, 5 (12), 9696–702. [DOI] [PubMed] [Google Scholar]
- (36).Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN; Lu JS; Miller J; Khisamutdinov EF; Shapiro BA; Dobrovolskaia MA; Afonin KA Nucleic Acids Res. 2017, 45 (4), 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).St Clair EW J. Clin Invest 2008, 118 (4), 1344–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Goodman RP; Berry RM; Turberfield AJ Chem. Commun. (Cambridge, U. K.) 2004, No. 12, 1372–3. [DOI] [PubMed] [Google Scholar]
- (39).Shu D; Shu Y; Haque F; Abdelmawla S; Guo P Nat. Nanotechnol 2011, 6 (10), 658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhang H; Endrizzi JA; Shu Y; Haque F; Sauter C; Shlyakhtenko LS; Lyubchenko YL; Guo P; Chi YI RNA 2013, 19 (9), 1226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Bui MN; Brittany Johnson M; Viard M; Satterwhite E; Martins AN; Li Z; Marriott I; Afonin KA; Khisamutdinov EF Nanomedicine 2017, 13 (3), 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L Nat. Nanotechnol 2010, 5 (9), 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Afonin KA; Kasprzak W; Bindewald E; Puppala PS; Diehl AR; Hall KT; Kim TJ; Zimmermann MT; Jernigan RL; Jaeger L; Shapiro BA Methods 2014, 67 (2), 256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chworos A; Severcan I; Koyfman AY; Weinkam P; Oroudjev E; Hansma HG; Jaeger L Science 2004, 306 (5704), 2068–2072. [DOI] [PubMed] [Google Scholar]
- (45).Reynolds A; Anderson EM; Vermeulen A; Fedorov Y; Robinson K; Leake D; Karpilow J; Marshall WS; Khvorova A RNA 2006, 12 (6), 988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Donnelly RP; Kotenko SV J. Interferon Cytokine Res 2010, 30 (8), 555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Sioud MJ Mol. Biol 2005, 348 (5), 1079–90. [DOI] [PubMed] [Google Scholar]
- (48).Jensen K; Anderson JA; Glass EJ Vet. Immunol. Immunopathol 2014, 158 (3–4), 224–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Van Damme J; Proost P; Put W; Arens S; Lenaerts J-P; Conings R; Opdenakker G; Heremans H; Billiau AJ Immunol. 1994, 152, 5495–5502. [PubMed] [Google Scholar]
- (50).Vanguri P; Farber JM J. Biol. Chem 1990, 265 (25), 15049–15057. [PubMed] [Google Scholar]
- (51).Kerkmann M; Rothenfusser S; Hornung V; Towarowski A; Wagner M; Sarris A; Giese T; Endres S; Hartmann GJ Immunol. 2003, 170 (9), 4465–74. [DOI] [PubMed] [Google Scholar]
- (52).Sajja S; Chandler M; Fedorov D; Kasprzak WK; Lushnikov A; Viard M; Shah A; Dang D; Dahl J; Worku B; Dobrovolskaia MA; Krasnoslobodtsev A; Shapiro BA; Afonin KA Langmuir 2018. DOI: 10.1021/acs.langmuir.8b00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Chelobanov BP; Laktionov PP; Vlasov VV Biochemistry (Moscow) 2006, 71 (6), 583–596. [DOI] [PubMed] [Google Scholar]
- (54).Choi CH; Hao L; Narayan SP; Auyeung E; Mirkin CA Proc. Natl. Acad. Sci. U. S. A 2013, 110 (19), 7625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Iwasaki A; Medzhitov R Nat. Immunol 2004, 5 (10), 987–95. [DOI] [PubMed] [Google Scholar]
- (56).Swiecki M; Colonna M Nat. Rev. Immunol 2015, 15 (8), 471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Gary-Gouy H; Lebon P; Dalloul AH J. Interferon Cytokine Res 2002, 22, 653–659. [DOI] [PubMed] [Google Scholar]
- (58).Sharma S; Fitzgerald KA PLoS Pathog. 2011, 7 (4), e1001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Sioud M Trends Mol. Med 2006, 12 (4), 167–76. [DOI] [PubMed] [Google Scholar]
- (60).Shlyakhtenko LS; Gall AA; Lyubchenko YL Methods Mol. Biol 2012, 931, 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Shlyakhtenko LS; Gall AA; Filonov A; Cerovac Z; Lushnikov A; Lyubchenko YL Ultramicroscopy 2003, 97 (1–4), 279–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.