Abstract
RNA-binding proteins serve an essential role in post-transcriptional gene regulation. Cytoplasmic activation/proliferation-associated protein-1 (caprin-1) is an RNA-binding protein that participates in the regulation of cell cycle control-associated genes. Caprin-1 acts alone or in combination with other RNA-binding proteins, such as RasGAP SH3-domain-binding protein 1 and fragile X mental retardation protein. In the tumorigenesis process, caprin-1 primarily functions by activating cell proliferation and upregulating the expression of immune checkpoint proteins. Through the formation of stress granules, caprin-1 is also involved in the process by which tumor cells adapt to adverse conditions, which contributes to radiation and chemotherapy resistance. Given its role in various clinical malignancies, caprin-1 holds the potential to be used as a biomarker and a target for the development of novel therapeutics. The present review describes this newly identified putative oncogenic protein and its possible impact on tumorigenesis.
Keywords: cytoplasmic activation/proliferation-associated protein-1, tumorigenesis, RNA-binding proteins, epigenetics
1. Introduction
RNA-binding proteins (RBPs) can directly interact with RNA or can be part of ribonucleoprotein complexes without direct contact with RNA (1). The presence of RNA-binding domains (RBDs) in proteins and AU-enriched regions in target RNAs is essential for the synergistic function of RBPs on their targets, which include mRNA, tRNA, rRNA, microRNA (miRNA) and non-coding RNA (ncRNA). Transient or stable interactions of RBPs with RNAs are crucial for RNA regulatory processes. RBPs are involved in post-transcriptional gene regulation (PTGR) primarily through the formation of functional units termed ribonucleoprotein (RNP) complexes. mRNA-binding proteins and corresponding messenger RNPs are directly responsible for mRNA maturation and regulation; however, diverse ncRNA-targeted RBPs are also involved in the process of PTGR (2,3). RBPs can combine with miRNAs to form miRNPs, which are also involved in the regulation of mRNA translation and stability. According to an extensive classification of 1,542 human RBPs conducted by Gerstberger et al (1), approximately half of these RBPs can be grouped on the basis of their mRNA targets, and others may interact with different types of RNAs. Furthermore, approximately one-third of RBPs can bind to DNA and RNA (4). The diverse binding spectrum of RBPs indicates their diverse spectrum of function. Recent studies have demonstrated that RBPs often associate with a group of mRNAs encoding proteins with similar functions, forming an RNA operon (5). The different types of RBPs combine with the corresponding mRNAs at different times and in different positions inside cells, thereby meeting the need of the cell in a time- and location-dependent manner to adjust the regulation of target molecules. A particular mRNA can be bound by numerous different RBPs. RBPs can also act as a binding platform for other factors and enzymes involved in mRNA regulation. RBPs are considered the most important regulators of PTGR. In addition to sustaining cellular metabolism, coordinating maturation, transportation, stability and degradation of all classes of RNAs through PTGR, RBPs serve a critical role in maintaining genome integrity (6,7) and responding to a variety of cellular stresses, thus ensuring cellular homeostasis (8,9). Considering the multifaceted effects of RBPs, dysfunctional RBPs can initiate various pathological changes, including neurodegenerative disorders, cardiovascular diseases and certain types of cancer (10–12).
Cancer is a complex and heterogeneous disease, and is classically considered to be caused by genetic alterations that result in the activation of oncogenic signaling pathways and/or loss of tumor suppressor mechanisms (13). As RBPs serve a pivotal role in PTGR, it is not surprising that abnormal changes to RBPs can cause alterations of cancer-associated signaling pathways. Furthermore, as RBPs regulate multiple targets in various PTGR steps, small changes in their expression and/or activity can induce a large-scale alteration of downstream regulatory networks, potentially initiating cancer development (13). A wide range of mechanisms underlie RBP alteration-induced oncogenesis, including changes to alternative splicing and polyadenylation, and changes in RNA stability, subcellular localization and translation (13). In order for tumor cells to achieve survival, proliferation, metastasis and resistance to anticancer therapeutics, they must make adaptive changes to the gene expression. Regulating transcribed mRNA or PTGR is the most effective and rapid mechanism for doing so, and it has a pivotal role in tumorigenesis. Tumor cells can change the expression of target mRNAs and feedback regulators (miRNA and ncRNA) through the regulation of RBPs. The abnormal expression of RBPs has been detected in numerous types of tumor, and different RBPs act at different steps of mRNA metabolism. For example, Sam68 participates in alternative splicing, producing a variety of tumor-promoting mRNA variants (14); eukaryotic translation initiation factor 4E is involved in directing ribosomes to the 5′-cap of mRNAs and enhances the expression of specific mRNAs that regulate certain tumorigenesis-associated processes, such as proliferation [c-Myc and cyclin-dependent kinase (CDK)2], metastasis (matrix metalloproteinase 9) and angiogenesis (vascular endothelial growth factor) (15), while embryonic lethal abnormal visual-like RNA binding protein 1 regulates the stability and translation efficiency of tumor-related mRNAs (16). Cytoplasmic activation/proliferation-associated protein-1 (caprin-1) is an RBP that is essential for cell proliferation. As caprin-1 is closely associated with control of the cell cycle, alteration of caprin-1 is involved in oncogenesis, which has been demonstrated in multiple experimental cancer studies (17–20). It is important for the study of cancer and its therapeutic development to fully understand the biological function of caprin-1 and the association between its alteration and oncogenesis.
2. Caprin-1 and RBPs
Caprin-1 is a ubiquitously expressed and highly conserved cytoplasmic phosphoprotein. The gene is located on the long arm of human chromosome 11 (11p13), encoding a 709-amino acid protein, with a molecular weight of 116 kDa. Caprin-1 is part of the conserved caprin family, which consists of two members, caprin-1 and caprin-2, both of which contain two highly conserved regions, homologous region-1 and −2 (21). High expression of caprin-1 was initially identified in dividing cells of the thymus, and was also known to be upregulated in activated T or B lymphocytes, and hematopoietic progenitors. Caprin-1 expression has been reported to be low in slowly dividing cells, such as those of the kidney or muscles, but high levels have been detected in the brain (21). Caprin-1 is considered to be an RBP, as it possesses RNA binding characteristics, i.e., the arginine-glycine-glycine (RGG) motif and the RG enrichment region (18). Using RNA-sequencing technology, 6,064 mRNAs (>1,000 reads) were identified as caprin-1 binding targets in the CD3 and CD28 antibody-activated human T lymphoma cell line (Jurkat). Table I presents a partial list of caprin-1 target mRNAs divided into six categories according to their biological function: Cell structure, RNA metabolism, RNA translation, signaling transduction, ubiquitylation, and growth factors and growth factor receptors (Wang et al, unpublished results). Caprin-1 can affect cell survival and growth through selectively binding a variety of mRNAs that are involved in cell growth, differentiation and migration, including c-Myc and cyclin-D2 (20,22–25). c-Myc serves a central role in the transition from the G1 to the S phase of the cell cycle, and cyclin D2 functions as a regulatory subunit of the kinases CDK4 or CDK6, whose activity is required for the G1 to the S phase transition (26,27). Cells without caprin-1 expression exhibit delays in the transition from the G1 to the S phase of the cell cycle (17). Therefore, it is speculated that caprin-1 may serve an important role in tumorigenesis.
Table I.
Caprin-1-targeted mRNAs in activated Jurkat cellsa.
| Function | Gene | Gene ID | Fold enrichment |
|---|---|---|---|
| Cytoskeleton | Myosin heavy chain | MYH9 | 10.60 |
| Talin-1 | TLN1 | 14.97 | |
| Spectrin β chain | SPTBN2 | 11.95 | |
| Filamin-B | FLNB | 9.87 | |
| Receptors | Interleukin-17 receptor A | IL17RA | 16.84 |
| Tetraspanin-27 | CD82 | 10.82 | |
| Interlekin-27-α | IL27RA | 9.04 | |
| Fibroblast growth factor receptor 4 | FGFR4 | 7.75 | |
| Signaling | T-cell activation NFKB-like protein | NP_640332 | 15.56 |
| TNFR1-associated DEATH domain protein | TRADD | 11.07 | |
| Phospholipase C-β3 | PLCB3 | 10.81 | |
| Protein phosphatase 1 regulatory 14B p21ras, PI3-kinase, MAPK | PPP1R114B | 9.48 | |
| RNA translation/metabolism | Eukaryotic translation factor 4γ1 | eIF4-G1 | 10.83 |
| Pre-mRNA processing splicing factor 8 | PRPFS | 8.89 | |
| 40S ribosomal protein S19-binding protein 1 | RPS19BP1 | 8.69 | |
| Polyadenylate-binding protein 1 G3BP1, FXR1, FXR2 | PABPC1 | 5.02 | |
| Growth factor-associated | Wnt-8b | WNT8B | 15.56 |
| Fibroblast growth factor-binding protein 3 | FGFBP3 | 12.21 | |
| Insulin-like growth factor II receptor | IGF2R | 8.45 |
Caprin-1 immunoprecipitation + RNA-sequencing from Jurkat cells stimulated with anti-CD3 and anti-CD28 antibodies. ID, identifier.
3. Caprin-1 and cancer
Caprin-1 selectively binds to c-Myc and cyclin D2 mRNAs, which accelerates cell progression through the G1 phase into the S phase, enhances cell viability and promotes cell growth, indicating that it may serve an important role in tumorigenesis (17). This hypothesis is supported by increasing experimental and clinical evidence. Caprin-1 and associated regulatory factors (including miRNAs) are abnormally expressed in tumor tissues and tumor cell lines. For instance, miR-1 (24) and miR-223 (22) were revealed to be downregulated in renal carcinoma cells and breast cancer cells, respectively, resulting in a significant increase in the expression level of caprin-1. The overexpression of caprin-1 may contribute to the growth and invasion of tumor cells. Abnormal expression of caprin-1 was also observed in patients with osteosarcoma and in osteosarcoma cell lines (23). Increased expression of caprin-1 can promote tumor growth and lung metastasis from primary tumors, and shorten survival time (23). Tylophorine directly binds with caprin-1 and accelerates degradation of the RNP formed by caprin-1 and RasGAP SH3-domain-binding protein (G3BP), downregulates c-Myc and cyclins D1/D2, and ultimately inhibits the growth of the tumor (20). Our recent studies revealed the high expression of caprin-1 in clinical specimens from glioblastoma patients. The degree of caprin-1 expression was associated with clinical classification (28). Recently, a study by Casey et al (29) that focused on c-Myc, indirectly identified caprin-1 to be associated with certain immune checkpoint proteins. c-Myc has been demonstrated to directly bind to the promoters of two immune checkpoint protein genes, the innate immune regulator CD47 and the adaptive immune checkpoint protein programmed death-ligand 1 (PD-L1), thus regulating their expression on the surface of tumor cells.
4. Caprin-1 may function as a complex with other RBPs
In biological microenvironments, protein molecules do not act alone; they function through intracellular networks formed by interactions with other proteins. According to published experimental results, caprin-1 can directly bind with G3BP1 (18), fragile X mental retardation protein (FMRP) (30) and Japanese encephalitis virus core protein (31). G3BP is a classic RBP, containing one nuclear transport factor 2-like function domain (NTF2), one acidic amino acid region, one RBD and one C-terminal RGG/G-enriched sequence. The direct combination of G3BP1 and caprin-1 is formed through the NTF2 domain of G3BP1 interacting with amino acids 352–380 of caprin-1 (18). The RBD region and RGG/G-enriched sequence can selectively bind mRNA. FMRP is another important RBP, and its deficiency can induce fragile X syndrome (32). FMRP contains two Agenet-like functional regions, two heterogeneous nuclear ribonucleoprotein K homology functional regions and one RGG region. The FMRP 427–442 amino acid sequence can interact with the caprin-1 231–245 amino acid sequence (30), while the Agenet-like region, KH functional region and RGG region have the mRNA binding function. The existence of the caprin-1/G3BP1/FMRP-containing RNP was further confirmed by the recent crystallographic study by Wu et al (33). Crystal structures of a fragment of caprin-1 (residues 132–251) revealed that tight homodimerization can be formed through the combination of the HR1 regions of two molecules of caprin-1, creating a large and highly negatively charged concave surface, which acts as a protein-binding groove. This dimeric caprin-1 structure can be used as a scaffold to further combine with G3BP and FMRP to form a macromolecule polymer platform for more complex functions. Theoretically, two molecules of caprin-1 can combine with two molecules of G3BP1 and two molecules of FMRP. The resulting macromolecule polymer does not affect the integrity of the RNA-binding region of each component or their ability to bind mRNAs. Further precise research on the caprin-1/G3BP1/FMRP complex may have great potential to aid in developing novel and more effective anticancer drugs.
5. Caprin-1 may initiate carcinogenesis via stress granules
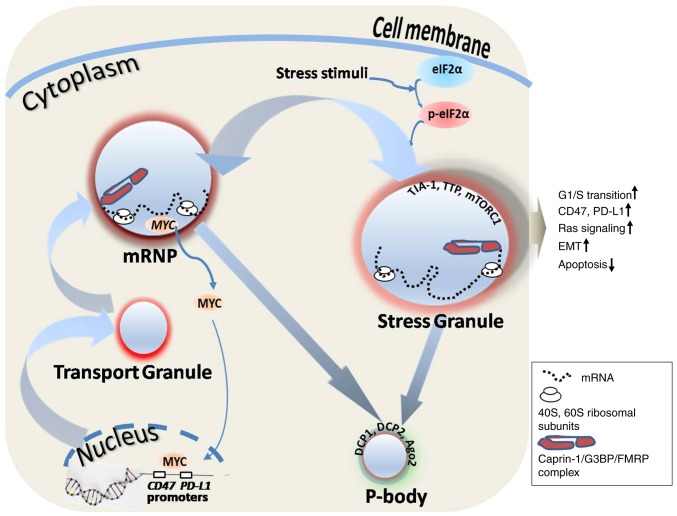
With increasing experimental evidence, the importance of SG in cancer has been awarded unprecedented attention. Our previous study demonstrated the presence of caprin-1 in RNA transport granules and stress granules (SGs) (18). These results indicated that caprin-1 may be involved in carcinogenesis through the formation of SGs. As illustrated in Fig. 1, there are numerous types of RNA-containing granule in the cell cytoplasm, including RNA transport granules, SGs and processing bodies (P-bodies). RNA transport granules are responsible for transporting mRNA to specific subcellular structures. For example, actin and microtubules are located at the leading edge of the cell membrane in the direction of fibroblast and tumor cell movement, which is closely associated with the invasive growth and metastasis of tumor cells. Caprin-1 is present in RNA transport granules (18), which are located at tubulin-enriched sites, such as mid-bodies, actin-enriched podosomes (34,35), the leading edge membrane of moving T lymphocytes (Wang et al, unpublished data) and migrating fibroblasts (36). Caprin-1 and G3BP1 can combine with FMRP and coexist in RNA transport granules. P-bodies are the site of RNA degradation; they contain multiple molecules that are responsible for mRNA degradation, surveillance, translation inhibition and RNA-mediated gene suppression. P-bodies also include marker proteins decapping mRNA, RNA degradation enzyme 1 and RNA degradation enzyme 2, GW182 (another RBP), argonaute 2, RNA-induced silencing complex and miRNAs. SGs are formed in response to cells undergoing different stress stimuli and are a conserved mechanism to reduce stress-associated damage and promote cell survival. SGs contain translationally stalled mRNAs, translation initiation factors, 40S and 60S ribosomal subunits, translation of suspended enzymes, specific RBPs and signaling molecules (37,38). The majority of classic SGs are produced by stress-induced phosphorylation of eukaryotic initiation factor 2α (eIF2α). Caprin-1, G3BP, T-cell-restricted intracellular antigen-1 (TIA-1), TIA-1-related protein, tristetraprolin (TTP) and FMRP can all induce the biogenesis of SGs. Caprin-1 combines with G3BP1 and/or TIA-1 and coexists in the classic SGs. However, high expression of caprin-1, G3BP, TIA-1, TTP or FMRP alone can induce the formation of SGs, even when there is no stress stimulus signal (18,39–43). SGs function to preserve RNAs against harmful conditions; they also serve a decisive role in facilitating further storage, translation reprogramming or degradation of untranslated mRNAs (38,44,45). The association between RNA granules and caprin-1-associated tumorigenesis is illustrated in Fig. 1.
Figure 1.
RNA granules and caprin-1-associated tumorigenesis. Caprin-1 mRNA is produced in the nucleus and transported into mRNP or the SG through the transport granule, and finally degraded in the P-body. Caprin-1-associated tumorigenesis takes place primarily through affecting SG-mediated tumorigenic characteristics and the MYC-mediated immune checkpoint. eIF2α can be activated by various stress stimuli and induces SG formation, thereby leading to tumorigenic characteristics, such as increased transition from the G1 to the S phase of the cell cycle, increased Ras signaling pathway, EMT and decreased apoptosis. Caprin-1 can also indirectly enhance the expression of immune checkpoint proteins, such as CD47 and PD-L1, through MYC, which is one of the regulatory targets of caprin-1. Caprin-1, cytoplasmic activation/proliferation-associated protein-1; mRNP, mRNA protein complexes; SG, stress granule; P-body, processing body; eIF2α, eukaryotic translation initiation factor 2α; EMT, epithelial to mesenchymal transition; PD-L1, programmed death-ligand 1; TIA-1, T-cell-restricted intracellular antigen-1; TTP, tristetraprolin; mTORC1, mammalian target of rapamycin complex 1; DCP, decapping enzyme; Ago2, argonaute 2; G3BP, GTPase-activating protein (Src homology 3 domain) binding protein; FMRP, fragile X mental retardation 1.
SG formation is part of the integrated stress response, which is a process that receives information from different ‘stress sensors’ and acts with cells to adapt to stress stimuli (46). These sensors are usually kinases that can phosphorylate serine/threonine residues of eIF2α. The tumor microenvironment is characterized by tissue hypoxia, high levels of reactive oxygen species and insufficient nutrients. These conditions can activate the cell stress response and promote SG formation, inducing tumor cells to adapt to the environment quickly and to alter metabolic pathways, thus maintaining their rapid growth. The existence of this mechanism has been validated in various clinical investigations. The presence of SGs has been verified in numerous types of human cancer, including glioblastoma, hepatocellular carcinoma and osteosarcoma (47–50). In addition, radiation therapy and chemotherapy can stimulate tumor cells to form SGs, which can result in resistance to treatment (46). A variety of approved chemotherapeutic agents have been indicated to induce SG, resulting in reduced therapeutic efficacy (46,51). Therefore, it can be hypothesized that blocking SG production may effectively kill tumor cells. Experimental evidence supports the hypothesis that the inhibition of SG formation can enhance cell mortality and inhibit tumor growth (52).
In addition to roles in cell mortality and tumorigenesis, SGs are also associated with tumor metastasis. In tumor xenograft animal experiments, SG-producing osteosarcoma cells were more resistant to treatment and were able to migrate to the lungs with greater ease, while inhibiting SG formation via G3BP1 limited tumor invasion and prevented metastasis in vivo (49). Dissemination of tumor cells can enhance metastasis. Bone marrow-derived tumor cells from patients with breast cancer have been revealed to contain SGs, and these SGs contribute to cell survival of disseminated tumor cells (45).
6. Clinical perspectives on caprin-1
As demonstrated in Table II, caprin-1 has been reported to be involved in almost all types of human malignancy, although the majority of the evidence obtained thus far comes from in vitro settings. Given its pivotal role in the PTGR of cell cycle control-associated genes, caprin-1 and/or caprin-1-containing complex act as an amplifier to drive tumorigenesis (17,18). The clinical significance of caprin-1 will be elucidated along with the progress of the corresponding investigations.
Table II.
Experimental evidence of caprin-1-associated human malignancies.
| Type of cancer | Type of evidence | (Refs.) |
|---|---|---|
| Hepatocellular | Tissue microarray and IHC of specimens from 239 patients | (53) |
| Human HepG2 and Hep3B cell lines | (20,47) | |
| Osteosarcoma | Tissue microarray and IHC of specimens from 59 patients | (23) |
| Human SaOS-2 and U2OS xenograft model | (23,49) | |
| Breast | PET imaging study of SaOS-2/Caprin-1 vs. 143B model | (54) |
| Melanoma | Human MCF-7, T-47D and MDA-MB-231 vs. MCF-10A | (20,22,47) |
| Human BT-474 and MDA-MB-453 ×enograft model | (45) | |
| Proteomic analysis of human BLM melanoma cells | (55) | |
| Glioblastoma | Human C6 and U87MG glioma cell lines | (48) |
| Prostate | Human PC-3 and LnCaP cell lines | (20,47) |
| Cervical | Human HeLa cell line | (20,47) |
| Gastric | Tissue microarray and IHC of specimens from 262 patients | (56) |
| Human NUGC-3, HGC-27 and MGC80-3 cell lines | (20,56) | |
| Pancreatic | Human H1299 ×enograft model | (57) |
| Colon | Human PANC-1, BxPC3, S2-013, SUIT-2, COLO357, HPAF, MIA-Paca2, Capan2 and AsPC-1 cell lines | (20,58) |
| Leukemia | Human DLD-1, SW480 and HCT116 ×enograft model | (20,59) |
| Human U-937 and human leukemic Jurkat T cells | (20) |
IHC, immunohistochemistry; PET, positron emission tomography.
Although being able to use caprin-1 as a therapeutic target in clinical practice may be a distant prospect, there are a number of established associations between caprin-1 and clinical cancer science: i) Caprin-1 acts as a biomarker for the clinical diagnosis of certain malignancies (53,54). Considering the close association between the expression level of caprin-1 and tumorigenesis, high expression levels of caprin-1 can be used as an indicator of certain malignancies. Caprin-1 mRNA and/or protein levels can be determined using molecular biology and liquid biopsy techniques in tissue or peripheral blood samples (28,55). ii) The degree of caprin-1 expression can be used to predict the prognosis of malignancies (23). iii) Caprin-1 and complexes can be used as a target for the development of cancer therapeutic drugs, such as specific short hairpin RNAs to knockdown caprin-1 expression, or specific peptides to block the formation of caprin-1/G3BP1/FMRP complexes (20,56–59).
7. Conclusions
Caprin-1 forms part of RNP complexes and directly interacts with mRNAs, and is therefore defined as an RBP. Caprin-1 acts alone or combines with other RBPs to participate in the regulation of cell cycle control-associated gene expression. Caprin-1 selectively interacts with c-Myc and cyclin D1/D2 mRNAs activating cell proliferation and upregulating the expression of immune checkpoint proteins CD47 and PD-L1. Through the formation of SGs, caprin-1 is also involved in the process of tumor cell adaption to adverse conditions, such as radiation and chemotherapy. Given its role in various clinical malignancies, caprin-1 holds the potential to be used as a biomarker and target for the development of novel therapeutics.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- caprin-1
cytoplasmic activation/proliferation-associated protein-1
- eIF2α
eukaryotic initiation factor 2α
- FMRP
fragile X mental retardation protein
- G3BP1
RasGAP-SH3-domain binding protein 1
- miRNA
microRNA
- ncRNA
non-coding RNA
- NTF2
nuclear transport factor 2
- PD-L1
programmed death-ligand 1
- PTGR
post-transcriptional gene regulation
- RBD
RNA-binding domain
- RBP
RNA-binding protein
- RNP
ribonucleoprotein
- SGs
stress granules
- TIA-1
T-cell-restricted intracellular antigen-1
Funding
This study was funded by the Key Projects of Precision Medicine (grant no. 2016JZ01) and the Education Department of Hubei Province (grant no. Q20162102).
Availability of data and material
The datasets used/analyzed in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
ZSY, HQ, HG, JL and BW collected data. LJD and BW contributed to the writing of the manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv Exp Med Biol. 2014;825:1–55. doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori T, Ngouv H, Hayashida M, Akutsu T, Nacher JC. ncRNA-disease association prediction based on sequence information and tripartite network. BMC Syst Biol. 2018;12(Suppl 1):S37. doi: 10.1186/s12918-018-0527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol. 2014;15:749–760. doi: 10.1038/nrm3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keene JD. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 6.Nishida K, Kuwano Y, Nishikawa T, Masuda K, Rokutan K. RNA binding proteins and genome integrity. Int J Mol Sci. 2017;18:E1341. doi: 10.3390/ijms18071341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kai M. Roles of RNA-binding proteins in DNA damage response. Int J Mol Sci. 2016;17:310. doi: 10.3390/ijms17030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey R, Dezi V, Pizzinga M, Willis AE. Post-transcriptional control of gene expression following stress: The role of RNA-binding proteins. Biochem Soc Trans. 2017;45:1007–1014. doi: 10.1042/BST20160364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheinberger J, Shav-Tal Y. mRNPs meet stress granules. FEBS Lett. 2017;591:2534–2542. doi: 10.1002/1873-3468.12765. [DOI] [PubMed] [Google Scholar]
- 10.Maziuk B, Balance HI, Wolozin B. Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front Mol Neurosci. 2017;10:89. doi: 10.3389/fnmol.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geuens T, Bouhy D, Timmerman V. The hnRNP family: Insights into their role in health and disease. Hum Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wurth L, Gebauer F. RNA-binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta. 2015;1849:881–886. doi: 10.1016/j.bbagrm.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: Old players and new actors. Trends Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Paronetto MP, Cappellari M, Busa R, Pedrotti S, Vitali R, Comstock C, Hyslop T, Knudsen KE, Sette C. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70:229–239. doi: 10.1158/0008-5472.CAN-09-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16:4914–4920. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, David MD, Schrader JW. Absence of caprin-1 results in defects in cellular proliferation. J Immunol. 2005;175:4274–4282. doi: 10.4049/jimmunol.175.7.4274. [DOI] [PubMed] [Google Scholar]
- 18.Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, Schrader JW. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reich J, Papoulas O. Caprin controls follicle stem cell fate in the Drosophila ovary. PLoS One. 2012;7:e35365. doi: 10.1371/journal.pone.0035365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu YQ, Yang CW, Lee YZ, Yang RB, Lee CH, Hsu HY, Chang CC, Lee SJ. Targeting a ribonucleoprotein complex containing the caprin-1 protein and the c-Myc mRNA suppresses tumor growth in mice: An identification of a novel oncotarget. Oncotarget. 2015;6:2148–2163. doi: 10.18632/oncotarget.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grill B, Wilson GM, Zhang KX, Wang B, Doyonnas R, Quadroni M, Schrader JW. Activation/division of lymphocytes results in increased levels of cytoplasmic activation/proliferation-associated protein-1: Prototype of a new family of proteins. J Immunol. 2004;172:2389–2400. doi: 10.4049/jimmunol.172.4.2389. [DOI] [PubMed] [Google Scholar]
- 22.Gong B, Hu H, Chen J, Cao S, Yu J, Xue J, Chen F, Cai Y, He H, Zhang L. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother. 2013;67:629–636. doi: 10.1016/j.biopha.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Sabile AA, Arlt MJ, Muff R, Husmann K, Hess D, Bertz J, Langsam B, Aemisegger C, Ziegler U, Born W, Fuchs B. Caprin-1, a novel Cyr61-interacting protein, promotes osteosarcoma tumor growth and lung metastasis in mice. Biochim Biophys Acta. 2013;1832:1173–1182. doi: 10.1016/j.bbadis.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Xiao H, Zeng J, Li H, Chen K, Yu G, Hu J, Tang K, Zhou H, Huang Q, Li A, et al. MiR-1 downregulation correlates with poor survival in clear cell renal cell carcinoma where it interferes with cell cycle regulation and metastasis. Oncotarget. 2015;6:13201–13215. doi: 10.18632/oncotarget.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumura I, Tanaka H, Kanakura Y. E2F1 and c-Myc in cell growth and death. Cell Cycle. 2003;2:333–338. doi: 10.4161/cc.2.4.428. [DOI] [PubMed] [Google Scholar]
- 27.Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Gui H, Tang XJ, Yang ZS, Zou DD, Lu JT, Yan LD, Dai LJ, Luo J, Wang B. Expression and tumor-Promoting effects of caprin-1 in human glioma. Glioma. 2018;1:136–141. doi: 10.4103/glioma.glioma_29_18. [DOI] [Google Scholar]
- 29.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Fatimy R, Tremblay S, Dury AY, Solomon S, De Koninck P, Schrader JW, Khandjian EW. Fragile X mental retardation protein interacts with the RNA-binding protein caprin1 in neuronal ribonucleoprotein complexes [corrected] PLoS One. 2012;7:e39338. doi: 10.1371/annotation/05374d07-34cf-483f-80f4-ec87374cbeb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, Mori Y, Kamitani W, Matsuura Y. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J Virol. 2013;87:489–502. doi: 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Zhu J, Huang X, Du Z. Crystal structure of a dimerization domain of human Caprin-1: Insights into the assembly of an evolutionarily conserved ribonucleoprotein complex consisting of Caprin-1, FMRP and G3BP1. Acta Crystallogr D Struct Biol. 2016;72:718–727. doi: 10.1107/S2059798316004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci. 2005;118:2425–2433. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carson JH, Barbarese E. Systems analysis of RNA trafficking in neural cells. Biol Cell. 2005;97:51–62. doi: 10.1042/BC20040083. [DOI] [PubMed] [Google Scholar]
- 36.Copsey AC, Cooper S, Parker R, Lineham E, Lapworth C, Jallad D, Sweet S, Morley SJ. The helicase, DDX3X, interacts with poly(A)-binding protein 1 (PABP1) and caprin-1 at the leading edge of migrating fibroblasts and is required for efficient cell spreading. Biochem J. 2017;474:3109–3120. doi: 10.1042/BCJ20170354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: The good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.e04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by fragile X mental retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 43.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchan JR, Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta N, Badeaux M, Liu Y, Naxerova K, Sgroi D, Munn LL, Jain RK, Garkavtsev I. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc Natl Acad Sci USA. 2017;114:1033–1038. doi: 10.1073/pnas.1525387114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adjibade P, St-Sauveur VG, Quevillon Huberdeau M, Fournier MJ, Savard A, Coudert L, Khandjian EW, Mazroui R. Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells. Oncotarget. 2015;6:43927–43943. doi: 10.18632/oncotarget.5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilas-Boas Fde A, da Silva AM, de Sousa LP, Lima KM, Vago JP, Bittencourt LF, Dantas AE, Gomes DA, Vilela MC, Teixeira MM, Barcelos LS. Impairment of stress granule assembly via inhibition of the eIF2alpha phosphorylation sensitizes glioma cells to chemotherapeutic agents. J Neurooncol. 2016;127:253–260. doi: 10.1007/s11060-015-2043-3. [DOI] [PubMed] [Google Scholar]
- 49.Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, Zhang F, Ng T, Delattre O, Evdokimova V, et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leprivier G, Rotblat B, Khan D, Jan E, Sorensen PH. Stress-mediated translational control in cancer cells. Biochim Biophys Acta. 2015;1849:845–860. doi: 10.1016/j.bbagrm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Szaflarski W, Fay MM, Kedersha N, Zabel M, Anderson P, Ivanov P. Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget. 2016;7:30307–30322. doi: 10.18632/oncotarget.8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fournier MJ, Coudert L, Mellaoui S, Adjibade P, Gareau C, Côté MF, Sonenberg N, Gaudreault RC, Mazroui R. Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation. Mol Cell Biol. 2013;33:2285–2301. doi: 10.1128/MCB.01517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan N, Dai L, Liu X, Pan G, Chen H, Huang J, Xu Q. Upregulation of caprin1 expression is associated with poor prognosis in hepatocellular carcinoma. Pathol Res Pract. 2017;213:1563–1567. doi: 10.1016/j.prp.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Campanile C, Arlt MJ, Kramer SD, Honer M, Gvozdenovic A, Brennecke P, Fischer CR, Sabile AA, Müller A, Ametamey SM, et al. Characterization of different osteosarcoma phenotypes by PET imaging in preclinical animal models. J Nucl Med. 2013;54:1362–1368. doi: 10.2967/jnumed.112.115527. [DOI] [PubMed] [Google Scholar]
- 55.Kedracka-Krok S, Jankowska U, Elas M, Sowa U, Swakon J, Cierniak A, Olko P, Romanowska-Dixon B, Urbanska K. Proteomic analysis of proton beam irradiated human melanoma cells. PLoS One. 2014;9:e84621. doi: 10.1371/journal.pone.0084621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Min L, Ruan Y, Shen Z, Jia D, Wang X, Zhao J, Sun Y, Gu J. Overexpression of Ras-GTPase-activating protein SH3 domain-binding protein 1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2015;67:677–688. doi: 10.1111/his.12695. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, Zhang SH, He HW, Zhang CX, Yu DK, Shao RG. Downregulation of G3BPs inhibits the growth, migration and invasion of human lung carcinoma H1299 cells by suppressing the Src/FAK-associated signaling pathway. Cancer Gene Ther. 2013;20:622–629. doi: 10.1038/cgt.2013.62. [DOI] [PubMed] [Google Scholar]
- 58.Taniuchi K, Nishimori I, Hollingsworth MA. The N-terminal domain of G3BP enhances cell motility and invasion by posttranscriptional regulation of BART. Mol Cancer Res. 2011;9:856–866. doi: 10.1158/1541-7786.MCR-10-0574. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H, Zhang S, He H, Zhao W, Chen J, Shao RG. GAP161 targets and downregulates G3BP to suppress cell growth and potentiate cisplaitin-mediated cytotoxicity to colon carcinoma HCT116 cells. Cancer Sci. 2012;103:1848–1856. doi: 10.1111/j.1349-7006.2012.02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used/analyzed in the present study are available from the corresponding author upon reasonable request.