Abstract
Glioblastoma multiforme (GBM) is the most common and malignant type of glioma, with a poor prognosis for patients. The survival time of patients varies greatly due to the complexity of the human genome, which harbors diverse oncogenic drivers. In order to identify the specific driving factors, 325 glioma samples from the Chinese Glioma Genome Atlas (CGGA) database were analyzed in the present study. The level of RELB proto-oncogene, NF-κβ subunit (RELB) expression increased with the pathological grade progression of the gliomas, and higher expression levels were present in the mesenchymal subtype and isocitrate dehydrogenase 1 (IDH1) wild-type gliomas. This RELB expression pattern was identified in the CGGA database and observed in three large independent databases. In patients with GBM from the CGGA database, a higher RELB expression level was associated with a shorter survival time, a mesenchymal subtype and IDH1 wild-type gliomas. Kaplan-Meier survival analysis, survival nomograms and Cox analysis demonstrated an independent prognostic value for RELB expression. Moreover, biological function analysis indicated the association of RELB with the ‘immune response’, ‘cell activation’ and the ‘apoptotic process’. In addition, RELB expression levels exhibited a negative correlation with the levels of microRNA (miR)-139-5p and miR-139-3p. The present study identified the pathological and biological roles of RELB in glioma and revealed its independent prognostic effect. These results suggested that RELB may be used as a prognostic biomarker and potential therapeutic target in glioma.
Keywords: glioma, gene expression, prognosis, RELB, glioblastoma
Introduction
Glioma is the most common and lethal type of intracranial tumor, accounting for ~46% of intracranial tumors (1). The World Health Organization (WHO) classifies glioma into four grades according to the density and polymorphism of the cancer cells, from grade I to IV as the malignancy increases (2). Patients with gliomas of the low and high grades have significantly different outcomes (3). Glioblastoma multiforme (GBM), defined as grade IV glioma, is the most lethal form among all of the grades. Despite receiving standard treatment including surgery, radiation and chemotherapy, patients with GBM have a median survival time of 14.4 months and a five-year survival rate of 10% (4,5). The survival time of patients varies greatly due to the complexity of the human genome, which harbors diverse oncogenic drivers (6,7). Therefore, identification of specific tumor-related molecular markers based on the pathogenesis and development of glioma may aid individual treatment and prognosis evaluation.
Nuclear factor κβ (NF-κβ) proteins are a family of transcription factors that play central roles in a wide range of biological processes, including cell survival and inflammatory and immune responses (8). The five mammalian family members, consisting of RELA proto-oncogene NF-κβ subunit (RelA), RELB proto-oncogene NF-κβ subunit (RelB), REL proto-oncogene NF-κβ subunit, NF-κβ subunit 1 and 2, share a conserved Rel homology domain that mediates dimerization and DNA binding (9). Previous studies have mainly focused on the canonical NF-κβ signaling pathway mediated by RelA-containing dimers and have demonstrated its important role in regulating cancer invasion and progression (10–12). The noncanonical NF-κβ signaling pathway, which has been more recently described, is mediated by RelB-containing dimers and regulates important biological processes, including B-cell survival and maturation, dendritic cell activation and lymphoid organogenesis (13). Although NF-κβ pathways have been extensively investigated, the specific roles of individual NF-κβ proteins in tumorigenesis are not well understood. A previous study suggested the correlation between RELB and breast cancer (14). However, the expression characteristics of RELB and its effect on the prognosis of patients with glioma studied using a high-throughput sequencing method of large clinical samples have not been reported, particularly in Chinese patients.
In the present study, the expression levels of RELB in glioma samples in the Chinese Glioma Genome Atlas (CGGA; www.cgga.org.cn) database, as well as its prognostic value, were investigated. Furthermore, the biological functions of RELB and related microRNAs (miRNAs/miRs) were analyzed. The current study provided a novel insight into the development of glioma and identified RELB as a prognostic biomarker and potential therapeutic target.
Materials and methods
Patients and samples
The CGGA database was used as the discovery set in the present study. The establishment and management of the dataset were described in a previous study (15). The CGGA RNA sequence dataset consisted of 325 samples, including 109 grade II samples, 72 grade III samples and 144 grade IV samples. Of the 144 GBM samples, 6 samples were lost to follow-up; therefore, 138 samples were included in the survival analysis. The patients with GBM were followed up every 3 months. A further three databases were used as validation sets, which included The Cancer Genome Atlas RNA sequencing database (TCGA; http://cancergenome.nih.gov), the GSE16011 mRNA microarray database (16) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16011) and the mRNA microarray data of the Repository for Molecular Brain Neoplasia Data (REMBRANDT; http://caintegrator-info.nci.nih.gov/rembrandt). The four databases were normalized.
Statistical analysis
Differences in variables between groups were evaluated using the student's t-test or the one-way analysis of variance (ANOVA) followed by the Holm-Sidak test. Kaplan-Meier survival curves were generated in order to estimate survival distributions, and the log-rank test was used to assess statistical significance between the groups. Univariate and further multivariate Cox regression analyses were performed to assess the prognostic value of RELB in patients. The HR and 95% CI were calculated. A nomogram was formulated based on the results of the multivariate Cox regression analysis. The ‘total points’ in the nomogram, which is the sum of the individual point value of each clinical factor, may be used to predict patient survival time. Receiver operating characteristic curves were constructed to determine the predictive effects of RELB expression for diagnosis. Gene ontology (GO) analysis of the RELB expression level-related genes was performed using the online Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov). Correlations between miRNAs and RELB were analyzed by Pearson's correlation in our CGGA miRNA microarray database. All statistical analyses were conducted using GraphPad Prism (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA), SPSS (version 16.0; SPSS, Inc., Chicago, IL, USA) or several packages of R statistical software (version 3.2.1, http://cran.r-project.org/src/base/R-3/R-3.2.1.tar.gz), such as ‘pheatmap’ (17), ‘circlize’ (18) and ‘rms’ (19). P<0.05 was considered to indicate a statistically significant difference.
Results
Analysis of RELB expression in patients with glioma
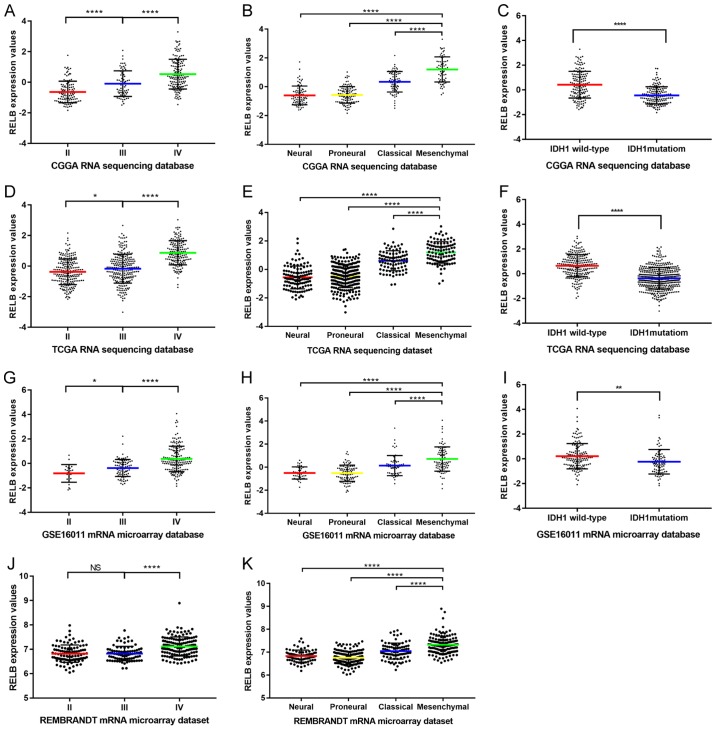
In total, 325 samples containing RNA sequencing data were collected from the CGGA. The clinical characteristics of the patients in the CGGA dataset are summarized in Table I. The clinical characteristics of the patients in the other three datasets are summarized in Tables II–IV. Gene expression characteristics of RELB in CGGA database were comprehensively analyzed. The results demonstrated that the levels of RELB expression increased with the pathological grade of gliomas. The highest expression level was identified in grade IV gliomas, and the lowest expression level was exhibited by grade II gliomas (Fig. 1A). Grade I gliomas were not included in the present study as patients with grade I gliomas have the lowest degree of malignancy and a good prognosis. Based on the TCGA subtype classification system (20), the mesenchymal subtype exhibited the highest expression level of RELB, while the neural subtype had the lowest (Fig. 1B). As the somatic mutations of isocitrate dehydrogenase 1 gene (IDH1) occurred in a majority of malignant gliomas and may be used as a prognosis indicator (7), the correlation between RELB expression levels and IDH1 mutations was investigated. RELB expression levels in patients harboring wild-type IDH1 were increased compared with patients with mutant IDH1 (Fig. 1C). The aforementioned expression characteristics of RELB were validated in TCGA (Fig. 1D-F), GSE16011 (Fig. 1G-I) and REMBRANDT (Fig. 1J and K) datasets. These results indicated that RELB expression was associated with the glioma grade and that patients with the mesenchymal subtype and wild-type IDH1 have a higher expression level of RELB compared with other subtypes and mutant IDH1.
Table I.
Clinical and molecular characteristics of patients in CGGA database.
| Variable | No. of cases (n, %) |
|---|---|
| Age | |
| Age ≥60 | 289 (89) |
| Age <60 | 36 (11) |
| Sex | |
| Male | 203 (62) |
| Female | 122 (38) |
| WHO grade | |
| II | 109 (34) |
| III | 72 (22) |
| IV | 144 (22) |
| TCGA subtype | |
| Neural | 81 (25) |
| Proneural | 102 (31) |
| Classical | 74 (23) |
| Mesenchymal | 68 (21) |
| IDH1 status | |
| Mutation | 167 (51) |
| Wild-type | 158 (49) |
| MGMT promoter status | |
| Methylated | 117 (36) |
| Unmethylated | 139 (43) |
| NA | 69 (21) |
| Radiotherapy | |
| Yes | 212 (65) |
| No | 84 (26) |
| NA | 29 (9) |
| Chemotherapy | |
| Yes | 158 (49) |
| No | 128 (39) |
| NA | 39 (12) |
CGGA, Chinese glioma genome atlas; WHO, World Health Organization; TCGA, The Cancer Genome Atlas; IDH1, isocitrate dehydrogenase1; MGMT, O6-methylguanine-DNA methyltransferase; NA, not available.
Table II.
Clinical and molecular characteristics of patients in the TCGA database.
| Variable | No. of cases |
|---|---|
| WHO grade | |
| II | 223 |
| III | 245 |
| IV | 168 |
| TCGA subtype | |
| Neural | 42 |
| Proneural | 448 |
| Classical | 168 |
| Mesenchymal | 41 |
| IDH1 status | |
| Mutation | 443 |
| Wild-type | 246 |
TCGA, The Cancer Genome Atlas; WHO, World Health Organization; IDH1, isocitrate dehydrogenase 1.
Table IV.
Clinical and molecular characteristics of patients in the Repository for Molecular Brain Neoplasia Data dataset.
| Variable | Case |
|---|---|
| WHO grade | |
| II | 99 |
| III | 84 |
| IV | 225 |
| TCGA subtype | |
| Neural | 47 |
| Proneural | 140 |
| Classical | 86 |
| Mesenchymal | 135 |
WHO, World Health Organization; TCGA, The Cancer Genome Atlas.
Figure 1.
RELB expression patterns in the CGGA database and other validation datasets. (A) The expression level of RELB in tissues from the CGGA database was positively correlated with tumor grade. (B) RELB expression was highest in mesenchymal subtype glioma samples from the CGGA database. (C) Patients with wild-type IDH1 had higher levels of RELB expression compared with those with mutant IDH1 in the CGGA database. (D) The expression level of RELB in tissues from TCGA database was positively correlated with tumor grade. (E) RELB expression was highest in mesenchymal subtype glioma samples from TCGA database. (F) Patients with wild-type IDH1 had higher levels of RELB expression compared with those with mutant IDH1 in TCGA database. (G) The expression level of RELB in tissues from the GSE16011 database was positively correlated with tumor grade. (H) RELB expression was highest in mesenchymal subtype glioma samples from the GSE16011 database. (I) Patients with wild-type IDH1 had higher levels of RELB expression compared with those with mutant IDH1 in the GSE16011 database. (J) The expression level of RELB from the REMBRANDT database was positively correlated with tumor grade. (K) RELB expression was highest in mesenchymal subtype glioma samples from the REMBRANDT database. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, as indicated. RELB, RELB proto-oncogene, NF-κβ subunit; CGGA, Chinese Glioma Genome Atlas; TCGA, The Cancer Genome Atlas; IDH1, isocitrate dehydrogenase 1; REMBRANDT, Repository for Molecular Brain Neoplasia Data.
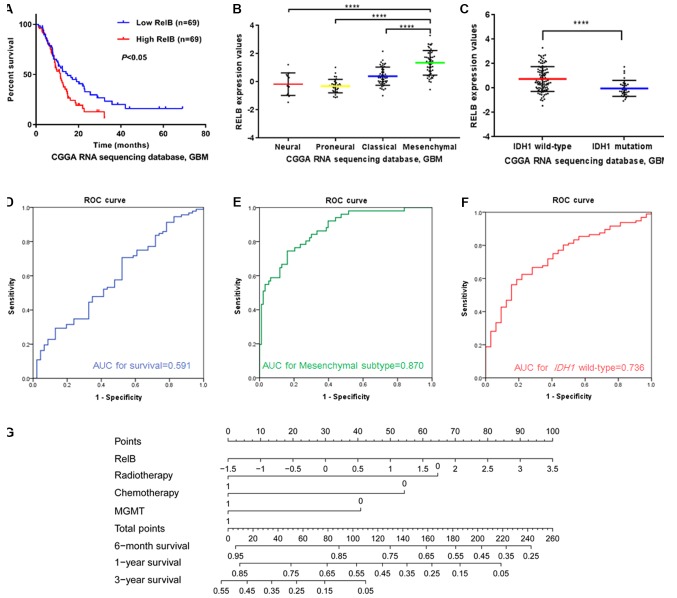
RELB is a predictive marker in patients with GBM
The expression characteristics of RELB in patients with GBM from the CGGA dataset were further analyzed. Of the 144 GBM samples, 6 samples were lost at follow-up; therefore, 138 samples were included in the survival analysis. Kaplan-Meier survival analysis was used to evaluate the relationship between RELB expression and the prognosis of patients. Half of the patients with relatively high RELB expression had a significantly shorter survival time than those with low RELB expression (P<0.05; Fig. 2A). Additionally, the characteristics of RELB expression in patients with GBM were the same as those concluded from the four independent databases. The expression levels of RELB were higher in the mesenchymal subtype (Fig. 2B) and IDH1 wild-type gliomas (Fig. 2C) compared with the corresponding control groups (P<0.01). The area under the curve for RELB expression levels as a predictor of one-year survival, mesenchymal subtype and IDH1 wild-type in the CGGA dataset was 0.591 (Fig. 2D), 0.870 (Fig. 2E) and 0.736 (Fig. 2F), respectively.
Figure 2.
Expression patterns and prognosis efficiency of the expression levels of RELB in patients with GBM from the CGGA. (A) Comparison of the overall survival time between the RELB high- and low-expression groups of patients with GBM. (B) Correlation of the RELB expression level and subtype classification. (C) Correlation of the RELB expression level and IDH1 mutation. (D) The predictive value of RELB expression for one-year survival. (E) The predictive value of the RELB expression level for the mesenchymal subtype. (F) The predictive value of RELB expression for patients with wild-type IDH1. (G) Nomograms for predicting the survival with risk score and clinical information of patients with GBM. The ‘point’ represents the impact of each clinical information on patients’ survival. The ‘total points’ is the sum of the individual points. ****P<0.0001. RELB, RELB proto-oncogene, NF-κβ subunit; GBM, glioblastoma multiforme; CGGA, Chinese Glioma Genome Atlas; IDH1, isocitrate dehydrogenase 1; ROC, receiver operating characteristic; AUC, area under the curve; MGMT, O6-methylguanine-DNA methyltransferase.
In order to further identify the prognostic value of RELB in patients with GBM, the Cox proportional hazards model was used (Table V). O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status is a prognostic and predictive factor for patients with GBM (4), and was therefore analyzed in the current study. The results of the univariate analysis demonstrated that age, RELB expression level, radiotherapy, chemotherapy and MGMT promoter methylation status affected the overall survival (OS) of patients (P<0.05), while sex was not a significant factor (P>0.05). Subsequently, multivariate Cox proportional hazards analysis of the aforementioned significant influencing factors was conducted. The expression of RELB was demonstrated to be an independent effective factor in the survival time of patients with GBM (P<0.05) and could be used independently to predict the prognosis of patients with GBM. In order to facilitate the utilization of RELB expression, different nomograms of survival time were plotted that incorporated the RELB expression level and the aforementioned clinical information (Fig. 2G). The results showed that RELB expression contributed the most risk points (range, 0–100), whereas the other clinical information had smaller contributions.
Table V.
Cox regression analysis of clinical parameters in patients with glioblastoma multiforme.
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sex (male vs. female) | 1.227 | 0.795–1.893 | 0.355 | – | – | – |
| Age (≥60 vs. <60) | 1.722 | 1.041–2.850 | 0.034a | 0.930 | 0.494–1.750 | 0.822 |
| RELB expression (high vs. low) | 1.265 | 1.043–1.533 | 0.017a | 1.332 | 1.005–1.766 | 0.046a |
| Radiotherapy (yes vs. no) | 0.412 | 0.259–0.653 | <0.001a | 0.404 | 0.243–0.674 | <0.001a |
| Chemotherapy (yes vs. no) | 0.336 | 0.214–0.528 | <0.001a | 0.466 | 0.282–0.768 | 0.003a |
| MGMT promoter status (methylated vs. unmethylated) | 0.564 | 0.364–0.872 | 0.010a | 0.572 | 0.352–0.931 | 0.024a |
P<0.05. HR, hazard ratio; CI, confidence interval; RELB, RELB proto-oncogene, NF-κβ subunit; MGMT, O6-methylguanine-DNA methyltransferase.
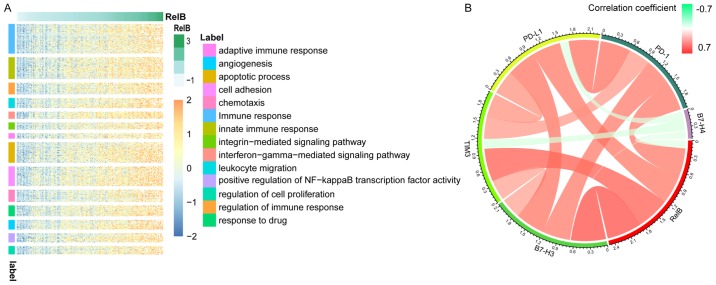
RELB-related biological processes in patients with GBM
In order to investigate the biological processes associated with RELB expression in patients with GBM, Pearson's correlation analysis was performed between RELB expression and other genes in the dataset. In total, 766 significant positively correlated genes (r>0.5) were identified and used for subsequent GO analysis using the DAVID website. The genes that positively correlated with RELB expression were involved in the ‘immune response’, ‘cell activation’, ‘apoptotic process’ and ‘cell adhesion’ (Fig. 3A). The association between RELB and immune checkpoint genes in glioma was analyzed and that PD-1, B7-H3, TIM3 and PD-L1 were positively correlated with RELB expression (Fig. 3B) (21).
Figure 3.
(A) Gene ontology analysis of RELB in the CGGA dataset. The selected genes were positively correlated with RELB expression. (r>0.5). (B) Correlation of RELB expression and immune checkpoint genes in glioma. RELB, RELB proto-oncogene, NF-κβ subunit; GBM, glioblastoma multiforme; CGGA, Chinese Glioma Genome Atlas.
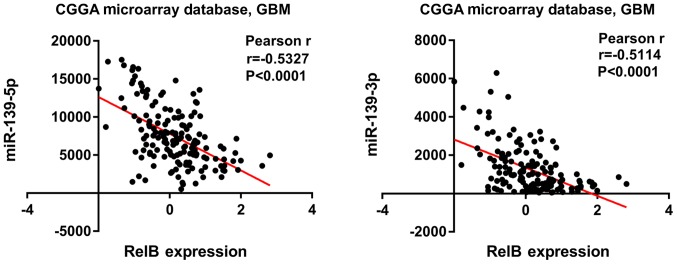
Correlation between RELB expression and miRNA levels in patients with GBM
In order to investigate the correlation between RELB and miRNA, 829 candidate miRNAs were analyzed in patients with GBM from the CGGA dataset. The results showed that an increased RELB expression was associated with the decreased expression of miR-139-5p and miR-139-3p (P<0.0001), which were the most negatively correlated with RELB expression (Fig. 4).
Figure 4.
Negative correlation between the expression levels of RELB and miR-139-5p/miR-139-3p. Increased RELB expression levels were associated with decreased expression levels of miR-139-5p and miR-139-3p. RELB, RELB proto-oncogene, nuclear factor κβ subunit; GBM, glioblastoma multiforme; miR, microRNA; CGGA, Chinese Glioma Genome Atlas.
Discussion
GBM is the most common and lethal type of brain tumor, in which cancer cells penetrate the adjacent normal tissues with no definite range. Current therapeutic approaches, including surgery, radiotherapy and chemotherapy, do not achieve satisfactory results (22). Genome differences in patients make a difference in prognosis. Therefore, it is necessary to identify effective and differential molecular markers in order to assist with the accurate prognosis of patients. An in-depth study of cancer development revealed that NF-κβ transcription factors participate in a wide variety of biological processes, including inflammation, apoptosis and proliferation (23). Therefore, clarifying the individual role of key NF-κβ subunits in cancer may aid the development of novel therapeutic agents.
The present study focused on RELB, a member of the alternative NF-κβ signaling pathway, which was initially identified as a regulator of the adaptive immune response (24). A previous study revealed that abnormal activity of RELB serves a role in the development of solid tumors and hematopoietic malignancies (25). In breast cancer, high RELB expression was demonstrated to confer more highly invasive phenotypes (26), and inhibition of RELB decreased proliferation (27). In prostate cancer, high RELB expression was observed to enhance cell growth and exert a radioprotective role in cancer cells (28,29). According to these studies, RELB exerts a tumor-supportive role in a several types of cancer. In glioma cells, RELB promoted cell survival and invasion (30,31). However, these studies were conducted in cell lines or animal models and not based on human clinical specimens. In the present study, the pathological and biological role of RELB in glioma was investigated in a large number of Chinese patients.
In the present study, RELB expression was identified to be upregulated in higher stage gliomas, mesenchymal subtypes and IDH1 wild-type gliomas in four independent databases, indicating the association of RELB expression levels with oncological biological processes. Based on RNA-sequencing analysis of patients with GBM, RELB may serve as an indicator of mesenchymal subtype and IDH1 wild-type gliomas. Moreover, high expression of RELB predicted a significantly shorter survival time for patients with glioma. The independent prognostic value of the RELB expression level was observed via multivariate analysis (P<0.05).
RELB is involved in regulating the biological activities of cancer cells. Cormier et al (32) reported that RELB activation was important for promoting the survival of multiple myeloma cells through the upregulation of anti-apoptotic proteins. Ge et al (33) revealed that RELB was associated with the levels of certain key regulators in endometrioid adenocarcinoma, and that high RELB expression levels may lead to endometrial cell tumorigenicity. In order to elucidate the role of RELB in the progression of glioma, biological functional annotation of RELB-related genes was performed in the present study. The GO analysis showed that RELB was mainly related to ‘apoptotic processes’ and ‘cell adhesion’ in patients with glioma. High expression of RELB may increase the adhesion, invasion and proliferation of cancer cells, inhibit apoptosis of cells and lead to the progression of glioma. In addition, the results also demonstrated an association between RELB and the immune response in patients. The findings of the present study are consistent with the results of previous studies (32,33). The NF-κβ family regulates a number of processes, ranging from the development and survival of lymphocytes and lymphoid organs to the control of immune responses and malignant transformation (34,35). RELB is involved in dendritic cell maturation and immune tolerance to inflammation (36,37). The GO term analysis results of the present study identified that RELB regulates the immune process in patients with glioma, including the immune response, the interferon-gamma-mediated signaling pathway and leukocyte migration. The expression levels of PD-1, PD-L1, TIM3 and B7-H3 were positively correlated with the expression level of RELB in the present study, suggesting that RELB may be associated with immune checkpoints (38).
miRNAs are small non-coding RNA molecules that regulate a large variety of biological processes in sequence-specific manners (39). miRNAs destabilize the target mRNAs or suppress their translation in order to regulate gene expression (40). Aberrantly expressed miRNAs were demonstrated to serve as oncogenes or tumor suppressors in cancer (41). The present study identified a negative correlation between the expression level of RELB and that of miR-139-3p and miR-139-5p. miR-139-3p and miR-139-5p are derived from pre-miR-139 and serve roles in the development of cancer. Sun et al (42) reported that ectopic expression of miR-139-5p significantly suppressed cell growth and metastasis through inhibition of cyclin D1 and matrix metalloproteinases in non-small cell lung cancer. Additionally, previous studies indicated that miR-139-5p has antitumor effects in several types of cancer (43,44). Previous studies revealed that miR-139-3p was involved in the carcinogenesis and development of various types of cancer (45,46). Huang et al (47) investigated the mechanism of miR-139-3p in the progression of cervical cancer and revealed that miR-139-3p inhibited cell proliferation and induced cell apoptosis through downregulation of NIN1 (RPN12) binding protein 1 homolog expression. As miR-139-5p and miR-139-3p have demonstrated anticancer effects, their downregulation may contribute to the progression of cancer. This is consistent with the results of the analysis of patients with glioma in the present study. Decreased expression of miR-139-5p and miR-139-3p may result in the upregulation of RELB, which, based on the GO term analysis results, may subsequently induce the proliferation, migration and progression of glioma cells and lead to a poor prognosis in patients. A limitation of the current study was that the effects of RELB were not demonstrated experimentally. Future studies are required to investigate the effects of the expression levels of RELB in vitro and in vivo. The RELB related pathway which take part in the progression of glioma was also interesting. It is a further investigation.
In summary, the present study identified the expression patterns and biological functions of RELB in patients with glioma. RELB expression levels are increased in patients with the mesenchymal subtype and wild-type IDH1, resulting in a shorter OS. RELB may therefore be used as an independent prognostic indicator in patients with glioma. In addition, the activity of RELB was found to be associated with the immune response, apoptosis and cell adhesion of cancer cells. Furthermore, the expression levels of RELB displayed a negative correlation with miR-139-5p and miR-139-3p. The results obtained in the current study suggested that RELB may be a novel and promising prognostic marker or therapeutic target for patients with glioma.
Table III.
Clinical and molecular characteristics of patients in GSE16011 database.
| Variable | Case |
|---|---|
| WHO grade | |
| II | 24 |
| III | 85 |
| IV | 159 |
| TCGA subtype | |
| Neural | 40 |
| Proneural | 97 |
| Classical | 58 |
| Mesenchymal | 89 |
| IDH1 status | |
| Mutation | 83 |
| Wild-type | 143 |
WHO, World Health Organization; TCGA, The Cancer Genome Atlas; IDH1, isocitrate dehydrogenase 1.
Acknowledgements
Not applicable.
Funding
The current study was supported by the National Nature Science Foundation of China (grant nos. 81802994, 81502495 and 81702460).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HH and FZ designed the experiments. FZ, KW and RH analyzed the data and contributed to the analytical tools. YZ and YL analyzed the data and reviewed the literature. FZ and KW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: Genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 4.Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang X, Jiang C, Kang C, Li X, Chen L, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375:263–273. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14:712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, Hu BQ, Yan W, Zhang W, Akers J, et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014;24:1765–1773. doi: 10.1101/gr.165126.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayden MS, Ghosh S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baud V, Collares D. Post-translational modifications of RelB NF-κB subunit and associated functions. Cells. 2016;5(pii):E22. doi: 10.3390/cells5020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raychaudhuri B, Han Y, Lu T, Vogelbaum MA. Aberrant constitutive activation of nuclear factor kappaB in glioblastoma multiforme drives invasive phenotype. J Neurooncol. 2007;85:39–47. doi: 10.1007/s11060-007-9390-7. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Guerra M, Colomer D. NF-kappaB as a therapeutic target in chronic lymphocytic leukemia. Expert Opin Ther Targets. 2010;14:275–288. doi: 10.1517/14728221003598930. [DOI] [PubMed] [Google Scholar]
- 13.Sun SC. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Belguise K, Kersual N, Kirsch KH, Mineva ND, Galtier F, Chalbos D, Sonenshein GE. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat Cell Biol. 2007;9:470–478. doi: 10.1038/ncb1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, Wang Z, Liu Y, Zhang C, Li M, Zhang W, Wang K, Cai J, Cheng W, Huang H, Jiang T. Genome-wide transcriptional analyses of Chinese patients reveal cell migration is attenuated in IDH1-mutant glioblastomas. Cancer Lett. 2015;357:566–574. doi: 10.1016/j.canlet.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, Zhang W, Chen D, Lv Y, Zheng J, Lilljebjörn H, Ran L, Bao Z, Soneson C, Sjögren HO, et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc Natl Acad Sci USA. 2014;111:3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Zhang C, Liu X, Wang Z, Sun L, Li G, Liang J, Hu H, Liu Y, Zhang W, Jiang T. Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology. 2016;5:e1196310. doi: 10.1080/2162402X.2016.1196310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 19.Zhang ZL, Zhao LJ, Chai L, Zhou SH, Wang F, Wei Y, Xu YP, Zhao P. Seven LncRNA-mRNA based risk score predicts the survival of head and neck squamous cell carcinoma. Sci Rep. 2017;7:309. doi: 10.1038/s41598-017-00252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 23.Park JY, Nam JH. Progestins in the fertility-sparing treatment and retreatment of patients with primary and recurrent endometrial cancer. Oncologist. 2015;20:270–278. doi: 10.1634/theoncologist.2013-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burkly L, Hession C, Ogata L, Reilly C, Marconi LA, Olson D, Tizard R, Cate R, Lo D. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 25.Baud V, Jacque E. The alternative NF-kB activation pathway and cancer: Friend or foe? Med Sci (Paris) 2008;24:1083–1088. doi: 10.1051/medsci/200824121083. (In French) [DOI] [PubMed] [Google Scholar]
- 26.Mineva ND, Wang X, Yang S, Ying H, Xiao ZX, Holick MF, Sonenshein GE. Inhibition of RelB by 1,25-dihydroxyvitamin D3 promotes sensitivity of breast cancer cells to radiation. J Cell Physiol. 2009;220:593–599. doi: 10.1002/jcp.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demicco EG, Kavanagh KT, Romieu-Mourez R, Wang X, Shin SR, Landesman-Bollag E, Seldin DC, Sonenshein GE. RelB/p52 NF-kappaB complexes rescue an early delay in mammary gland development in transgenic mice with targeted superrepressor IkappaB-alpha expression and promote carcinogenesis of the mammary gland. Mol Cell Biol. 2005;25:10136–10147. doi: 10.1128/MCB.25.22.10136-10147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Josson S, Fang F, Oberley TD, St Clair DK, Wan XS, Sun Y, Bakthavatchalu V, Muthuswamy A, St Clair WH. RelB enhances prostate cancer growth: Implications for the role of the nuclear factor-kappaB alternative pathway in tumorigenicity. Cancer Res. 2009;69:3267–3271. doi: 10.1158/0008-5472.CAN-08-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josson S, Xu Y, Fang F, Dhar SK, St Clair DK, St Clair WH. RelB regulates manganese superoxide dismutase gene and resistance to ionizing radiation of prostate cancer cells. Oncogene. 2006;25:1554–1559. doi: 10.1038/sj.onc.1209186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DW, Ramakrishnan D, Valenta J, Parney IF, Bayless KJ, Sitcheran R. The NF-κB RelB protein is an oncogenic driver of mesenchymal glioma. PLoS One. 2013;8:e57489. doi: 10.1371/journal.pone.0057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherry EM, Lee DW, Jung JU, Sitcheran R. Tumor necrosis factor-like weak inducer of apoptosis (TWEAK) promotes glioma cell invasion through induction of NF-κB-inducing kinase (NIK) and noncanonical NF-κB signaling. Mol Cancer. 2015;14:9. doi: 10.1186/s12943-014-0273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cormier F, Monjanel H, Fabre C, Billot K, Sapharikas E, Chereau F, Bordereaux D, Molina TJ, Avet-Loiseau H, Baud V. Frequent engagement of RelB activation is critical for cell survival in multiple myeloma. PLoS One. 2013;8:e59127. doi: 10.1371/journal.pone.0059127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge QL, Liu SH, Ai ZH, Tao MF, Ma L, Wen SY, Dai M, Liu F, Liu HS, Jiang RZ, et al. RelB/NF-κB links cell cycle transition and apoptosis to endometrioid adenocarcinoma tumorigenesis. Cell Death Dis. 2016;7:e2402. doi: 10.1038/cddis.2016.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 35.Baldwin AS. Regulation of cell death and autophagy by IKK and NF-κB: Critical mechanisms in immune function and cancer. Immunol Rev. 2012;246:327–345. doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- 36.Vogel CF, Wu D, Goth SR, Baek J, Lollies A, Domhardt R, Grindel A, Pessah IN. Aryl hydrocarbon receptor signaling regulates NF-κB RelB activation during dendritic-cell differentiation. Immunol Cell Biol. 2013;91:568–575. doi: 10.1038/icb.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu HC, Qiu T, Liu XH, Dong WC, Weng XD, Hu CH, Kuang YL, Gao RH, Dan C, Tao T. Tolerogenic dendritic cells generated by RelB silencing using shRNA prevent acute rejection. Cell Immunol. 2012;274:12–18. doi: 10.1016/j.cellimm.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Lu L, Bai Y, Wang Z. Elevated T cell activation score is associated with improved survival of breast cancer. Breast Cancer Res Treat. 2017;164:689–696. doi: 10.1007/s10549-017-4281-x. [DOI] [PubMed] [Google Scholar]
- 39.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 41.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C, Sang M, Li S, Sun X, Yang C, Xi Y, Wang L, Zhang F, Bi Y, Fu Y, Li D. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget. 2015;6:39756–39792. doi: 10.18632/oncotarget.5476. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Wong CC, Wong CM, Tung EK, Au SL, Lee JM, Poon RT, Man K, Ng IO. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–331. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Wang K, Jin J, Ma T, Zhai H. miR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J Cell Mol Med. 2017;21:3730–3740. doi: 10.1111/jcmm.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Duan B, Dong Y, He C, Zhou H, Sheng H, Gao H, Zhang X. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7:8046–8052. [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M, Wen TF, He LH, Li C, Zhu WJ, Trishul NM. A six-microRNA set as prognostic indicators for bile duct cancer. Int J Clin Exp Med. 2015;8:17261–17270. [PMC free article] [PubMed] [Google Scholar]
- 47.Huang P, Xi J, Liu S. miR-139-3p induces cell apoptosis and inhibits metastasis of cervical cancer by targeting NOB1. Biomed Pharmacother. 2016;83:850–856. doi: 10.1016/j.biopha.2016.07.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.