Abstract
Chronic lymphocytic leukemia (CLL), a clonal expansion of CD5+ B cells, is the most common form of adult leukemia; however, the molecular mechanisms underlying its pathogenesis remain undetermined. It has been previously suggested that numerous biological factors, including cytokines, may be involved in the proliferation of malignant cells. For example, interleukin (IL)-4, IL-2, interferon-γ and tumor necrosis factor serve roles as inhibitors of cellular apoptosis; whereas IL-5 and IL-10 are inducers of cellular apoptosis. In the present study, the results demonstrated that the phosphorylation and activation of signal transducer and activator of transcription 6 (STAT6) was induced by IL-4 in a time-dependent manner. Notably, the expression level of microRNA (miR)-155 was increased in MEC-1 cells following treatment with IL-4; however, this effect was attenuated following STAT6 knockdown via RNA interference. In addition, STAT6 knockdown promoted cell apoptosis, which was partly attenuated by treatment with IL-4. Inhibition of miR-155 expression significantly increased cell apoptosis despite the presence of IL-4. The results of the present study suggested that treatment with IL-4 enhanced the expression of miR-155, which regulated CLL cell survival via the enhanced phosphorylation of STAT6.
Keywords: chronic lymphocytic leukemia, interleukin-4, signal transducer and activator of transcription 6, microRNA-155
Introduction
Chronic lymphocytic leukemia (CLL) is an incurable disease with extremely variable progression (1,2). CLL is characterized by a disruption of the homeostasis between the proliferation and apoptosis of leukemia cells and the accumulation of neoplastic B lymphocytes that co-express cluster of differentiation (CD)5 and CD19 antigens (3–6). However, the mechanism underlying the development of CLL remains unclear. The role of microRNA-155 (miR-155) in tumor immunity is receiving increasing attention (7).
Numerous T helper (Th)1 and Th2 microRNA (miRNA) molecules associated with inflammation, including miRNA-146, miR-155 and miRNA-324-5p, have been revealed to be conserved in mouse and human macrophages and are induced by lipopolysaccharide (LPS) and interleukin (IL)-4 (8). The signaling pathways associated with genome-wide miRNA expression levels and cellular functions regulated by miRNAs during alternative macrophage activation remain largely undetermined. The expression levels of miRNAs are useful for the prediction of the clinical manifestation of CLL (9). Structurally, miRNA are short RNA molecules measuring 19–25 nucleotides in length, processed from hairpin loop structures (pre-miRNA, 60–110 nucleotides in length), which regulate the expression of protein-coding genes via complementary binding with targeted messenger RNA (10). Previously, several chromosomal abnormalities, including 11q-, 13q-, 17p- and trisomy 12, and other molecular aberrations, including the degradation or downregulation of miRNA-15a and miRNA-16-1, and the overexpression of anti-apoptotic genes, have been identified in patients with CLL (11–14). Furthermore, a unique miRNA signature was revealed to be differentially expressed in patients with various immunoglobulin heavy variable 4-38-2-like (IgVH) and ζ chain of T cell receptor associated protein kinase 70 (ZAP-70) kinase statuses, and are composed of the most frequently deregulated miRNA molecules in different hematological malignancies, including miRNA-15/16, the miRNA-29 family and miR-155 (15,16).
As signal transducer and activator of transcription 6 (STAT6) and miRNA are associated with tumor growth (17), the present study aimed to determine the underlying pathways associated with these factors, and to explore the association between STAT6 and miR-155 in CLL.
Materials and methods
Cell culture
The human CLL MEC-1 cell line was obtained from the American Tissue Culture Collection (Manassas, VA, USA) and maintained at 37°C in 5% CO2. Cells were subsequently cultured in Iscove's modified Dulbecco's medium (HyClone: GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences).
Transfection of cells
MEC-1 cells (2×105 cells/well) were seeded in 24-well plates and incubated overnight. Subsequently, cells were transfected with the human miR-155 and negative control (NC) miRNA (both from Shanghai GenePharma Co., Ltd., Shanghai, China) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 48 h, according to the manufacturer's protocol. The concentration of all miRNA molecules transfected was 100 nM. The sequences of the miRNAs used for transfection were as follows: miR-155 mimic sense, UUAAUGCUAAUCGUGAUAGGGGU; and antisense, CCCUAUCACGAUUAGCAUUAAUU; NC mimic sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. The sequences of the miR-155 inhibitors used for transfection were as follows: anti-miR-155; 5′-ACCCCUAUCACGAUUAGCAUUAA-3′; and a anti-miR-155 NC 5′-CAGUACUUUUGUGUAGUACAA-3′. The miR-1555i and respective NC were purchased from Shanghai GenePharma Co., Ltd.
Knockdown of human STAT6 via RNA interference
STAT6 expression was knocked down via transfection with a small interfering (si)RNA complementary to STAT6 mRNA (5′-AAGCAGGAAGAACUCAAGUUUTT-3′ and 5′-AAACUUGAGUUCUUCCUGCUUTT-3′), as described previously (18). pGC-siSTAT6 (0.8 µg) was transfected into MEC-1 cells in 24-well plates (2×105 cells per well) using Lipofectamine® 2000 (2 µl; Invitrogen; Thermo Fisher Scientific, Inc.) for 4 h according to the manufacturer's protocol, and the medium was subsequently replaced with regular growth medium. Cells were used in subsequent experiments at 24 h following transfection.
Co-treatment and co-culture experiments
Recombinant human IL-4 (rIL-4) was added to the MEC-1 cell medium to investigate the effects of extracellular IL-4 on phosphorylated (p)-STAT6 and miR-155 levels in MEC-1 cells. The optimum concentration of rIL-4 used was 10 ng/ml. Following co-culture of the cells with rIL-4 for 0, 2, 5, 10, 15 and 20 h, levels of p-STAT6 and miR-155 were determined via western blot analysis and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays, respectively.
Western blot analysis
MEC-1 cells were treated with radioimmunoprecipitation assay lysis buffer containing 1% phenylmethylsulfonyl fluoride (Shanghai Shenergy Biocolor BioScience Technology Company, Shanghai, China) to extract total protein. For cytoplasmic and nuclear extracts, cells were washed with PBS and were lysed in NE-PER extraction reagent (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The protein concentrations of the samples were determined using a bicinchoninic acid assay (Shanghai Shenergy Biocolor BioScience & Technology Company). A total of 30 µg protein/lane was separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Following blocking with 5% skim milk in TBS with 0.1% Tween-20 (TBST) for at least 1 h at room temperature, the membranes were subsequently probed with primary antibodies at 4°C overnight. Following washing with TBST, a goat anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (1:3,000; cat no. TA140003; OriGene Technologies, Beijing, China) was added to the membranes for at least 1 h at room temperature. The bands were visualized by using an enhanced chemiluminescent western blot kit (EMD Millipore). The GAPDH antibody (1:1,000; cat no. sc-47724) was obtained from Santa Cruz Biotechnology, Inc., (Dallas, TX, USA), and anti-caspase-3 (1:1,000; cat no. ab32351) and p-STAT6 antibodies (1:1,000; cat no. ab28829) were obtained from Abcam (Cambridge, MA, USA). The anti-caspase-3 antibody bound to pro-caspase and cleaved forms of the protein. Western blot analysis results were analyzed using the Image J software 1.48 (National Institutes of Health, Bethesda, MD, USA) and SPSS version 20.0 (IBM Corp., Armonk, NY, USA).
RT-qPCR
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was used to extract total RNA from the MEC-1 cells. The procedure of RT and qPCR analyses was performed as described previously (19). Primers were purchased from BioSune Biotechnology Co., Ltd., (Shanghai, China), and the sequences are listed in Table I.
Table I.
Sequences of primers used for the reverse transcription quantitative polymerase chain reaction.
| Gene name | Sequence |
|---|---|
| miR-155 | Forward 5′-TTAATGCTAATCGTGA-3′ |
| Reverse 5′-TTTGGCACTAGCACATT-3′ | |
| U6 | Forward 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse 5′-AACGCTTCACGAATTTGCGT-3′ |
Assessment of cell apoptosis
MEC-1 cells (5×103 cells/well) were seeded into 96-well plates and then transfected with siSTAT6, miR-155 and anti-miR-155 (16). anti-miR-155 cells were subsequently incubated for 20 h in the presence and absence of rIL-4 (10 ng/ml). Staining with an Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) apoptosis detection kit (cat. no. FAK011.100; Neobioscience, Shenzhen, China) was performed to investigate the effect of rIL-4 on the levels of apoptotic and necrotic MEC-1 cells. MEC-1 cells (1×106) were incubated with Annexin V-FITC and PI (10 µl) for 10 min in the dark at room temperature. Cells were immediately analyzed using a flow cytometer (CytExpert version 1.1, Beckman Coulter, Inc., Brea, CA, USA). Necrotic cells were identified via positive staining of Annexin V-FITC and PI, whereas apoptotic cells were identified via positive staining of Annexin V-FITC and negative staining of PI.
Statistical analysis
All statistical analyses were performed using the SPSS version 20.0 (IBM Corp.) for Windows. Results are presented as mean ± standard deviation of at least 3 independent experiments. Differences between two groups were compared using Student's t-test. Multiple group comparisons were performed using one-way analysis of variance with a Student-Newman-Keuls post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
IL-4 induces activation of STAT6 at various time intervals
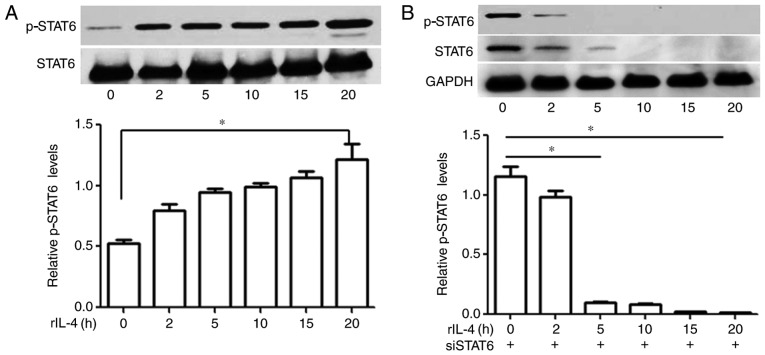
Expression levels of p-STAT6 in MEC-1 cells were investigated via western blot analysis, and the presence of p-STAT6 was determined by the visualization of a single band at ~100 kDa position. The results revealed that MEC-1 cells that were not treated with IL-4 did not exhibit marked levels of p-STAT6; whereas increased levels of p-STAT6 were demonstrated in MEC-1 cells treated with 10 ng/ml IL-4 in a time-dependent manner (Fig. 1A).
Figure 1.
Effects of rIL-4 on STAT6 phosphorylation in MEC-1 cells following transfection with siSTAT6. Untransfected MEC-1 cells and MEC-1 cells transfected with siSTAT6 were co-cultured with 10 ng/ml rIL-4 for different time intervals. rIL-4 was revealed to promote STAT6 phosphorylation in a time-dependent manner. The results revealed that siSTAT6 significantly attenuated the effects of rIL-4 on MEC-1 cells. (A) Levels of p-STAT6 in MEC-1 cells were increased following treatment with rIL-4 in a time-dependent manner. (B) Effects of rIL-4 on p-STAT6 levels were attenuated in MEC-1 cells following transfection with siSTAT6 in a time-dependent manner. *P<0.05. Data are presented as the mean ± standard deviation of at least 3 independent experiments. si, small interfering; STAT6, signal transducer and activator of transcription 6; p, phosphorylated; rIL-4, recombinant interleukin 4.
STAT6 knockdown in MEC-1 cells suppresses IL-4-induced phosphorylation of STAT6
Levels of p-STAT6 in the MEC-1 cells transfected with siSTAT6 were determined via western blot analysis at various time intervals. Following incubation with rIL-4 for 20 h, levels of p-STAT6 in the siSTAT6-transfected groups were almost undetectable. The results demonstrated that IL-4-induced phosphorylation of STAT6 was significantly attenuated following transfection with siSTAT6 (P>0.05; Fig. 1B).
Treatment with IL-4 upregulates miR-155 levels in MEC-1 cells, and transfection with siSTAT6 attenuates this effect
To additionally investigate the molecular mechanisms underlying the STAT6 signaling pathway, MEC-1 cells were treated with rIL-4 for 20 h. As presented in Fig. 2, treatment with rIL-4 increased miR-155 expression in the MEC-1 cells (P<0.05). Furthermore, this effect was revealed to be significantly attenuated in cells treated with rIL-4 and also transfected with siSTAT6 (P<0.05; Fig. 2). MEC-1 cells transfected with siSTAT6 alone, exhibited significantly decreased expression levels of miR-155 compared with MEC-1 cells not treated with anything (P<0.05; Fig. 2). These results suggested that rIL-4 promoted the expression of miR-155 via STAT6 phosphorylation.
Figure 2.
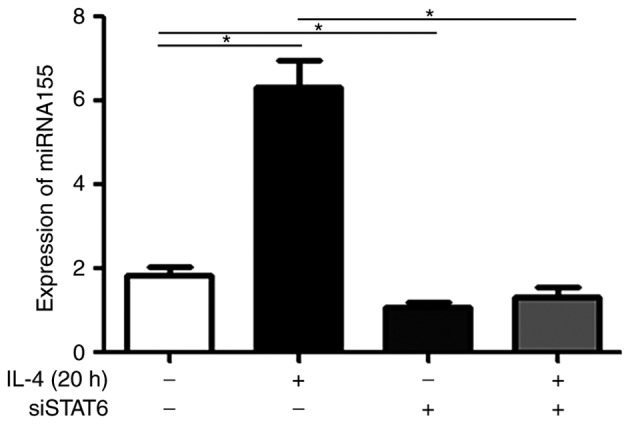
Effects of rIL-4 on miR-155 expression in MEC-1 cells. MEC-1 cells and MEC-1 cells transfected with siSTAT6 were incubated in the presence or absence of 10 ng/ml rIL-4 for 20 h. Expression levels of miR-155 were determined by reverse transcription-quantitative polymerase chain reaction. *P<0.05. Data are presented as the mean ± standard deviation of at least 3 independent experiments. miR/miRNA, microRNA; si, small interfering; STAT6, signal transducer and activator of transcription 6; rIL-4, recombinant interleukin 4.
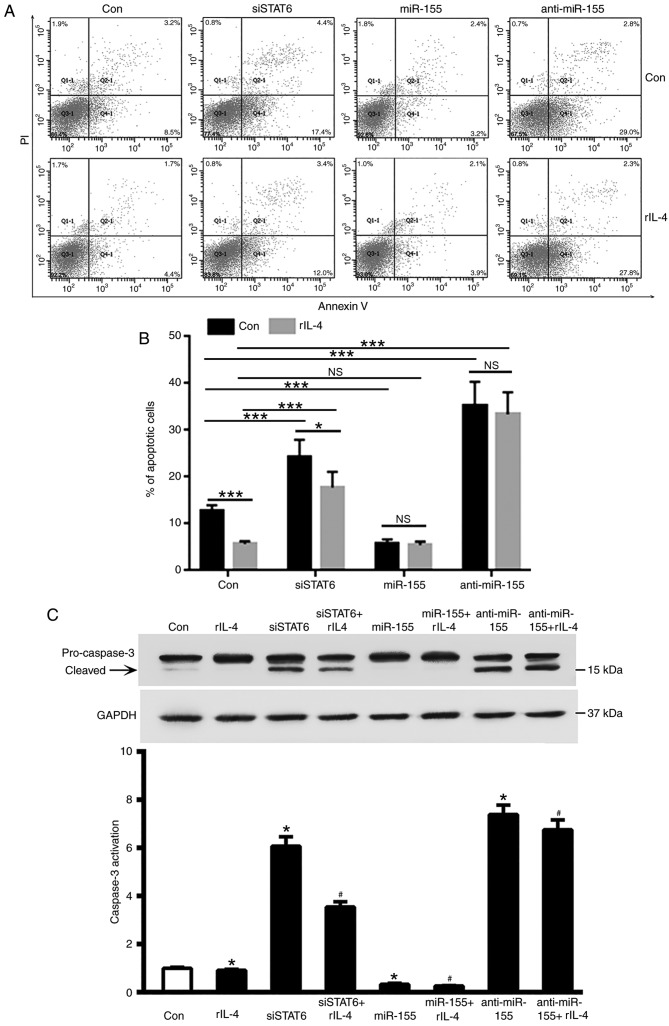
miR-155 inhibits MEC-1 cell apoptosis
As STAT6 was revealed to markedly upregulate miR-155 expression in MEC-1 cells, the roles of STAT6 and miR-155 in the apoptosis of MEC-1 cells were investigated. As demonstrated in Fig. 3A and B, inhibition of STAT6 in MEC-1 cells promoted cell apoptosis, and this was partly attenuated by treatment with rIL-4. Overexpression of miR-155 was revealed to significantly decrease MEC-1 cell apoptosis (P<0.001, Fig. 3B); whereas overexpression of anti-miR-155 enhanced cells apoptosis despite the presence of rIL-4 (P<0.001, Fig. 3B). It is therefore reasonable to suggest that treatment with rIL-4 may inhibit apoptosis in CLL cells. Transfection with siSTAT6 partly attenuated the anti-apoptosis effects of rIL-4 on MEC-1 cells (P<0.05; Fig. 3B). miR-155-treatment provided modest protection against apoptosis in CLL cells despite the presence of rIL-4 (P<0.001, Fig. 3B). In addition, knockdown of STAT6 increased caspase3 activation, which was attenuated by rIL-4 treatment (Fig. 3C).
Figure 3.
Effects of miR-155 on the apoptosis of MEC-1 cells. (A) MEC-1 cells were transfected with siSTAT6, miR-155 and anti-miR-155, and subsequently treated with rIL-4 for 20 h. Apoptosis and necrosis levels of MEC-1 cells were determined using flow cytometry. (B) Quantification of the flow cytometry apoptosis data. Columns indicate the mean apoptosis rate of three repeats. Error bars represent the standard deviation. *P<0.05 and ***P<0.001. (C) Caspase-3 activation, measured by relative levels of cleaved protein, was determined by western blot analysis. *P<0.05 vs. control group; #P<0.05 vs. rIL-4 treated group. Data are presented as the mean ± standard deviation of at least 3 independent experiments, and the exact values were as follows: Con Group=1±0.0153 (n=5); IL-4 group=0.9146±0.01809 (n=5); siSTAT6 group=6.069±0.1752 (n=5); siSTAT6+IL-4 group=3.543±0.09789 (n=5); miR-155 group=0.3274±0.01877 (n=5); miR-155+IL-4 group=0.2609±0.008641 (n=5); anti-miR-155 group=7.39±0.1753 (n=5); and anti-miR-155 + IL-4 group=6.753±0.1842 (n=5). miR, microRNA; si, small interfering; STAT6, signal transducer and activator of transcription 6; rIL-4, recombinant interleukin 4; PI, propidium iodide; Con, control; NS, not significant.
Discussion
Unique miRNA signatures are differentially expressed in patients with various IgVH and ZAP-70 kinase statuses and are composed of the most frequently deregulated miRNAs in different hematological malignancies, including miR-15/16, the miR-29 family and miR-155 (13,14). Increased expression levels of miR-155 in leukemia cells are associated with a more aggressive disease phenotype in patients with CLL (20). miR-155 is an important regulator of post-transcriptional gene expression in B cells (21). miR-155 has been previously demonstrated to be conserved in both mouse and human macrophages and is induced by LPS and IL-4 (22). A previous study revealed that the phosphorylation and activation of STAT6 in CLL cells may be rapidly induced by IL-4 in vitro (23). In the present study, the results demonstrated that a novel signaling pathway, the p-STAT6-mediated extracellular IL-4 upregulation of miR-155, is involved in CLL pathology.
IL-4-driven alternative macrophage activation and proliferation are characteristic features of anti-helminthic immune responses, which primarily occur during inflammatory responses (23,24). The signaling pathways associated with genome-wide miRNA expression and cellular functions regulated by miRNAs during alternative macrophage activation remain largely unknown. Numerous studies have investigated the roles of IL-4-regulated miRNAs in human and mouse alternative macrophage activation (25–27). Furthermore, ~1,000 miRNAs are present in the human genome; however, little is known about the transcriptional regulation of miRNAs (28,29). Considering that miRNA expression levels are deregulated in CLL, the present study aimed to determine whether STAT6 affected the transcription levels of miRNAs in CLL cells (30,31). Our previous study indicated that following treatment with rIL-4, levels of p-STAT6 in MEC-1 cells increased in a time-dependent manner (17). Based on these data, the present study aimed to determine whether miR-155 was involved in the development of CLL. The results demonstrated that pretreatment with rIL-4 promoted the expression of miR-155; however, this effect was attenuated following transfection with siSTAT6. These results suggested that rIL-4 induced the expression of miR-155 via STAT6 phosphorylation.
The effects of miR-155 were determined to additionally investigate the function of miR-155 in CLL pathogenesis. The results revealed that miR-155 decreased apoptosis levels in MEC-1 cells, which suggested that the upregulation of miR-155 may be associated with the pathogenesis of CLL.
In conclusion, the results of the present study demonstrated that IL-4 induced miR-155 expression and STAT6 phosphorylation. STAT6 knockdown was revealed to suppress IL-4-induced miR-155 expression. Overexpression of miR-155 was demonstrated to decrease the apoptotic rate of MEC-1 cells; whereas overexpression of anti-miR-155 was demonstrated to enhance MEC-1 cell apoptosis. The results of the present study suggested that miR-155 expression was induced by IL-4, which subsequently enhanced p-STAT6 levels to regulate CLL cell survival. The results of the present study have provided an improved understanding regarding the molecular mechanism of CLL pathogenesis and identified a novel therapeutic target for the treatment of patients with CLL.
Acknowledgements
Not applicable.
Funding
The present study was partly supported by the National Natural Science Foundation (grant nos. 81500124 and 81600121), Natural Science Foundations of Shandong Province (grant nos. Y2007C053, 2009ZRB14176 and ZR2016HQ46), Technology Development Projects of Shandong Province (grant nos. 2007GG10 and 2010GSF10250), Major Research Projects of Shandong Province (grant nos. 2016GSF201029 and 2017GSF218007), Program of Shandong Medical Leading Talent and Taishan Scholar Foundation of Shandong Province.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.
Authors' contributions
NC and XW conceived and designed the study. NC, LF, KL, XL and PL performed the experiments. NC and XL wrote the paper. NC, PL and XW reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lu K, Wang X. Therapeutic advancement of chronic lymphocytic leukemia. J Hematol Oncol. 2012;5:55. doi: 10.1186/1756-8722-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bilous N, Abramenko I, Saenko V, Chumak A, Dyagil I, Martina Z, Kryachok I. Clinical relevance of TP53 polymorphic genetic variations in chronic lymphocytic leukemia. Leuk Res. 2017;58:1–8. doi: 10.1016/j.leukres.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Nasr R, Marçais A, Hermine O, Bazarbachi A. Overview of targeted therapies for adult T-cell leukemia/lymphoma. Methods Mol Biol. 2017;1582:197–216. doi: 10.1007/978-1-4939-6872-5_15. [DOI] [PubMed] [Google Scholar]
- 4.Jin UH, Cheng Y, Zhou B, Safe S. Bardoxolone methyl and a related triterpenoid downregulate cMyc expression in leukemia cells. Mol Pharmacol. 2017;91:438–450. doi: 10.1124/mol.116.106245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quintanilla-Martinez L, Preffer F, Rubin D, Ferry JA, Harris NL. CD20+ T-cell lymphoma: Neoplastic transformation of a normal T-cell subset. Am J Clin Pathol. 1994;102:483–489. doi: 10.1093/ajcp/102.4.483. [DOI] [PubMed] [Google Scholar]
- 6.Moens L, Verbinnen B, Covens K, Wuyts G, Johnson M, Roalfe L, Goldblatt D, Meyts I, Bossuyt X. Anti-pneumococcal capsular polysaccharide antibody response and CD5 B lymphocyte subsets. Infect Immun. 2015;83:2889–2896. doi: 10.1128/IAI.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balatti V, Tomasello L, Rassenti LZ, Veneziano D, Nigita G, Wang HY, Thorson JA, Kipps TJ, Pekarsky Y, Croce CM. miR-125a and miR-34a expression predicts Richter syndrome in chronic lymphocytic leukemia patients. Blood. 2018;132:2179–2182. doi: 10.1182/blood-2018-04-845115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papageorgiou SG, Kontos CK, Diamantopoulos MA, Bouchla A, Glezou E, Bazani E, Pappa V, Scorilas A. MicroRNA-155-5p overexpression in peripheral blood mononuclear cells of chronic lymphocytic leukemia patients is a novel, independent molecular biomarker of poor prognosis. Dis Markers. 2017;2017:2046545. doi: 10.1155/2017/2046545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zitzer NC, Snyder K, Meng X, Taylor PA, Efebera YA, Devine SM, Blazar BR, Garzon R, Ranganathan P. MicroRNA-155 modulates acute graft-versus-host disease by impacting T cell expansion, migration, and effector function. J Immunol. 2018;200:4170–4179. doi: 10.4049/jimmunol.1701465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou B, Zhu H, Luo H, Gao S, Dai X, Li Y, Zuo X. MicroRNA-202-3p regulates scleroderma fibrosis by targeting matrix metalloproteinase 1. Biomed Pharmacother. 2017;87:412–418. doi: 10.1016/j.biopha.2016.12.080. [DOI] [PubMed] [Google Scholar]
- 11.Oscier D, Else M, Matutes E, Morilla R, Strefford JC, Catovsky D. The morphology of CLL revisited: The clinical significance of prolymphocytes and correlations with prognostic/molecular markers in the LRF CLL4 trial. Br J Haematol. 2016;174:767–775. doi: 10.1111/bjh.14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriston C, Bödör C, Szenthe K, Bánáti F, Bánkuti B, Csernus B, Reiniger L, Csomor J, Matolcsy A, Barna G. Low CD23 expression correlates with high CD38 expression and the presence of trisomy 12 in CLL. Hematol Oncol. 2017;35:58–63. doi: 10.1002/hon.2244. [DOI] [PubMed] [Google Scholar]
- 13.Underbayev C, Kasar S, Ruezinsky W, Degheidy H, Schneider JS, Marti G, Bauer SR, Fraidenraich D, Lightfoote MM, Parashar V, et al. Role of mir-15a/16-1 in early B cell development in a mouse model of chronic lymphocytic leukemia. Oncotarget. 2016;7:60986–60999. doi: 10.18632/oncotarget.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong C, Ji M, Ji C. microRNAs and their potential target genes in leukemia pathogenesis. Cancer Biol Ther. 2009;8:200–205. doi: 10.4161/cbt.8.3.7333. [DOI] [PubMed] [Google Scholar]
- 15.Smolej L, Vroblova V, Motyckova M, Jankovicova K, Schmitzova D, Krejsek J, Maly J. Quantification of ZAP-70 expression in chronic lymphocytic leukemia: T/B-cell ratio of mean fluorescence intensity provides stronger prognostic value than percentage of positive cells. Neoplasma. 2011;58:140–145. doi: 10.4149/neo_2011_02_140. [DOI] [PubMed] [Google Scholar]
- 16.Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, Ikuo M, Ghosh AK, Lerner S, Rassenti LZ, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891–1899. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan C, Chen Y, Kong W, Fu L, Liu Y, Yao Q, Yuan Y. PVT1-derived miR-1207-5p promotes breast cancer cell growth by targeting STAT6. Cancer Sci. 2017;108:868–876. doi: 10.1111/cas.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen N, Lu K, Li P, Lv X, Wang X. Overexpression of IL-9 induced by STAT6 activation promotes the pathogenesis of chronic lymphocytic leukemia. Int J Clin Exp Pathol. 2014;7:2319–2323. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N, Lv X, Li P, Lu K, Wang X. Role of high expression of IL-9 in prognosis of CLL. Int J Clin Exp Pathol. 2014;7:716–721. [PMC free article] [PubMed] [Google Scholar]
- 20.Kneitz C, Goller M, Seggewiss R, Yaman A, Serfling E, Tony HP. STAT6 and the regulation of CD23 expression in B-chronic lymphocytic leukemia. Leuk Res. 2000;24:331–337. doi: 10.1016/S0145-2126(99)00191-5. [DOI] [PubMed] [Google Scholar]
- 21.Jeon JI, Ko SH, Kim YJ, Choi SM, Kang KK, Kim H, Yoon HJ, Kim JM. The flavone eupatilin inhibits eotaxin expression in an NF-κB-dependent and STAT6-independent manner. Scand J Immunol. 2015;81:166–176. doi: 10.1111/sji.12263. [DOI] [PubMed] [Google Scholar]
- 22.Eigsti RL, Sudan B, Wilson ME, Graff JW. Regulation of activation-associated microRNA accumulation rates during monocyte-to-macrophage differentiation. J Biol Chem. 2014;289:28433–28447. doi: 10.1074/jbc.M114.599316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rückerl D, Jenkins SJ, Laqtom NN, Gallagher IJ, Sutherland TE, Duncan S, Buck AH, Allen JE. Induction of IL-4Rα-dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4-driven murine macrophage proliferation in vivo. Blood. 2012;120:2307–2316. doi: 10.1182/blood-2012-02-408252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautherot I, Burdin N, Seguin D, Aujame L, Sodoyer R. Cloning of interleukin-4 delta2 splice variant (IL-4delta2) in chimpanzee and cynomolgus macaque: phylogenetic analysis of delta2 splice variant appearance, and implications for the study of IL-4-driven immune processes. Immunogenetics. 2002;54:635–644. doi: 10.1007/s00251-002-0510-4. [DOI] [PubMed] [Google Scholar]
- 25.Wei Y, Schober A. MicroRNA regulation of macrophages in human pathologies. Cell Mol Life Sci. 2016;73:3473–3495. doi: 10.1007/s00018-016-2254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czimmerer Z, Varga T, Kiss M, Vázquez CO, Doan-Xuan QM, Rückerl D, Tattikota SG, Yan X, Nagy ZS, Daniel B, et al. The IL-4/STAT6 signaling axis establishes a conserved microRNA signature in human and mouse macrophages regulating cell survival via miR-342-3p. Genome Med. 2016;8:63. doi: 10.1186/s13073-016-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying W, Tseng A, Chang RC, Morin A, Brehm T, Triff K, Nair V, Zhuang G, Song H, Kanameni S, et al. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J Clin Invest. 2015;125:4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedländer MR, Lizano E, Houben AJ, Bezdan D, Báñez-Coronel M, Kudla G, Mateu-Huertas E, Kagerbauer B, González J, Chen KC, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014;15:R57. doi: 10.1186/gb-2014-15-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava AK, Sablok G, Hackenberg M, Deshpande U, Suprasanna P. Thiourea priming enhances salt tolerance through co-ordinated regulation of microRNAs and hormones in Brassica juncea. Sci Rep. 2017;7:45490. doi: 10.1038/srep45490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pallasch CP, Patz M, Park YJ, Hagist S, Eggle D, Claus R, Debey-Pascher S, Schulz A, Frenzel LP, Claasen J, et al. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood. 2009;114:3255–3264. doi: 10.1182/blood-2009-06-229898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abroun S, Saki N, Ahmadvand M, Asghari F, Salari F, Rahim F. STATs: An old story, yet mesmerizing. Cell J. 2015;17:395–411. doi: 10.22074/cellj.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.