Abstract
Osteosarcoma (OS) is the most common type of malignant primary bone neoplasm. Although the application of neoadjuvant chemotherapy has improved the 5-year survival rate of patients suffering from OS, prognosis remains poor. Therefore, it is important to elucidate the molecular mechanisms underlying the occurrence, progression and metastasis of OS. The RNA-binding protein Quaking (QKI) is a member of the STAR family of proteins, and can function as a tumor suppressor gene to suppress the occurrence and progression of a variety of tumors; however, the role of QKI in OS remains to be fully elucidated. In the present study, it was identified that the expression of QKI2 was downregulated in OS using western blot analysis. In addition, subsequent functional investigations, including MTT, Transwell invasion and migration assays, revealed that QKI2 inhibited the proliferation, invasion and migration of an OS cell line in vitro. By implementing a series of experimental techniques in molecular biology, including reverse transcription-quantitative polymerase chain reaction and a double fluorescence reporter assay, it was demonstrated that the expression of miR-20a was high and inhibited the expression of QKI2 in OS. In conclusion, it was revealed that aberrantly upregulated miR-20a inhibited the expression of QKI2 in OS by targeting QKI2 mRNA, subsequently promoting the proliferation, migration and invasion of OS cells.
Keywords: microRNA-20a, Quaking 2, osteosarcoma, progression
Introduction
Osteosarcoma (OS) is the most common type of malignant primary bone neoplasm of mesenchymal origin. This high-grade tumor has a high incidence within adolescents. The metaphyseal regions of the long bones are the most common regions for the development of OS; ~42% of cases of occurrence are in the femur, 19% in the tibia, 10% in the humerus, and ~8% in the skull and mandibula (1). Metastatic disease is the major cause of cancer-associated mortality; the lungs are the main sites of OS-related metastasis (2), followed by the bones. Unfortunately, the 5-year survival rate of patients suffering from OS has not improved in the last 20 years (3). Therefore, the development of more effective therapeutic targets is required for improving the survival rate of patients with OS. However, the molecular mechanism underlying the progression and metastasis of OS remains to be fully elucidated.
Quaking (QKI) is a member of the STAR family of proteins and may regulate mRNA expression at the transcription level (4–7). Increasing evidence shows that QKI may act as a tumor suppressor gene, suppressing the occurrence and progression of various types of tumor, including human astrocytic tumors, glioblastoma multiforme, oral squamous cell carcinoma, gastric cancer and colon cancer (8–15). However, studies investigating the effects and mechanisms underlying QKI in the progression and metastasis of OS are required. MicroRNA (miR)-20a, one of the most well-characterized oncomirs, is a member of the miR-17-92 cluster (16) and is upregulated in multiple types of human tumor, including gastric cancer, non-small cell lung cancer, multiple myeloma, colorectal cancer, prostate cancer and gallbladder carcinoma (17–23). At present, few studies investigating miR-20a in relation to OS have been performed. Namløs et al (24) identified that members of the miR-17-92 cluster, including miR-20a, are expressed at high levels in OS cell lines relative to normal bone. Evidence indicates that higher expression levels of miR-20a are significantly associated with systemic spread and the OS of patients (25). However, the corresponding mechanisms by which miR-20a functions in OS remain to be elucidated. In the present study, the expression levels and effects of QKI2 in OS were elucidated, and it was shown that miR-20a directly targets QKI2 in OS.
Materials and methods
OS samples and cell lines
Primary OS samples and normal bone samples were collected from 10 patients (6 males and 4 females) via biopsy at Harbin Medical University Cancer Hospital (Harbin, China) between June 2014 and December 2016. The mean age of the patients was 19.3 years (range, 9–67 years). All patients provided written informed consent, and the study was approved by the Ethics Committee of Harbin Medical University. The 143B human osteosarcoma cell line, and human SV40-transfected hFOB 1.19 osteoblasts and 293TN cells were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The 143B cells were cultured in EMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 0.015 mg/ml 5-bromo-2′-deoxyuridine (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 atmosphere. The hFOB 1.19 cells were cultured in D-MEM/F-12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 0.3 mg/ml G418 (Gibco; Thermo Fisher Scientific, Inc.) and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 34°C in a 5% CO2 atmosphere. The 293TN cells were cultured in EMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 atmosphere.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from samples and cells by applying TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix kit was purchased from Beijing Transgen Biotech Co., Ltd. (Beijing, China). Then total RNA was converted into cDNA through RT according to the manufacturer's protocol. Following RT, qPCR was performed using the TransScript® Tip Green qPCR SuperMix kit according to the manufacturer's protocol (Transgen Biotech Co., Ltd.) with an ABI 7900HT Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following forward and reverse primers were used for RT-qPCR: miR-20a, forward 5′-TAAAGTGCTTATAGTGCAG-3′ and reverse 5′-TGCGTGTCGTGGAGTC-3′ (LNA); U6, forward 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse 5′-ACGCTTCACGAATTTGCGTGTC-3′. The PCR reaction mixtures contained 2× TransScript® Tip Green qPCR SuperMix (10 µl), 4 µM primers (2 µl), 1 µl cDNA and 7 µl ddH2O in a total volume of 20 µl. The PCR thermocycling conditions were as follows: 94°C for 30 sec, and 40 cycles of 5 sec at 94°C and 30 sec at 60°C. The comparative 2−ΔΔCq method (26) was used for relative quantification and statistical analysis.
Cell transfection
To establish 143B cells stably overexpressing QKI2, pWPXL lentiviral vectors were used (Addgene, Inc., Cambridge, MA, USA). To construct the plasmid overexpressing QKI2, the coding sequence of QKI2 was subcloned into the pWPXL lentiviral vector. The pWPXL lentiviral plasmid overexpressing QKI2 and the packaging mix (pSPAX2 and pMD.2G) were cotransfected into 293TN cells. After 48 h, the virus-containing supernatant was harvested for the infection of 143B cells. The miR-20a mimic (5′-UAAAGUGCUUAUAGUGCAGGUAG-3′), inhibitor (5′-CUACCUGCACUAUAAGCACACUUUA-3′), miR-negative control (miR-NC, scrambled sequence; 5′-UCACAACCUCCUAGAAAGAGUAGA-3′), small interfering (si)RNA-QKI2 (5′-GAAUUCAAGAACGGUCUUAAUU-3′) and siRNA negative control (5′-AAUUUCUUCACUUCUUCAACUGCUC-3) were obtained from Invitrogen; Thermo Fisher Scientific, Inc. and were transfected into 143B cells by applying Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Cell growth assay
To determine the effect of miR-20a and QKI2 on the proliferative ability of the 143B cells, an MTT assay was performed. The 143B cells (2×103 cells/well) were seeded in 96-well plates and incubated in complete growth medium for 1, 2 and 3 days. Subsequently, 10 µl MTT solution was mixed into the samples and then incubated for 4 h. Following removal of the culture medium, 150 µl DMSO was added to samples. The cell proliferative capacity was determined at a wavelength of 570 nm.
Transwell migration and invasion assays
The transfected 143B cells (2×104) in 200 µl of FBS-free EMEM were seeded in the upper part of a Transwell chamber (Costar; Corning Incorporated, Corning, NY, USA) with or without Matrigel. Subsequently, the lower part of the Transwell chamber was filled with complete growth medium. The 143B cells were then incubated and allowed to migrate or invade, respectively for 24 h; the 143B cells that migrated or invaded into the lower part were stained with 1% crystal violet for 30 min. Images of the migrated or invaded 143B cells were captured under a light microscope and counted using ImageJ 2.0 software (National Institutes of Health, Bethesda, MD, USA).
Western blotting
The transfected 143B cells were lysed using RIPA buffer, and the total protein was isolated and quantified by applying a BCA protein assay kit (Beyotime Institute of Biotechnology, Haimen, China). The proteins (30 µg per lane) were resolved via 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto PVDF membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Membranes were blocked with 5% non-fat dried milk in 1×TBS and 0.1% Tween-20 (TBST; Sigma-Aldrich; Merck KGa) for 3 h at room temperature. The PVDF membranes were incubated with rabbit polyclonal anti-human QKI (1:5,000; catalog no. ab78518; Abcam, Cambridge, MA, USA) and mouse monoclonal anti-human β-tubulin (1:5,000; catalog no. ab7751; Abcam) antibodies overnight at 4°C. The membranes were then washed with TBST, and horseradish peroxidase-conjugated secondary goat anti-rabbit IgG (1:5,000; catalog no. ab6721; Abcam) or goat anti-mouse IgG (1:5,000; catalog no. ab6789; Abcam) were applied for 60 min at room temperature. The protein expression of QKI2 was assessed using an ECL kit (Beyotime Institute of Biotechnology).
miR-20a target prediction
The potential target genes of miR-20a were predicted using miRanda (http://www.microrna.org/microrna/; August 2010 release) and TargetScan 7.2 (http://www.TargetScan.org). These databases search the presence of conserved seven-mer and six-mer sites on the 3′-UTRs of messenger RNA that match the seed region of miR-20a.
Dual-luciferase reporter assay
The wild-type (WT) and mutant (MUT) 3′-UTR fragment of QKI2 were synthesized directly and separately inserted into the pmiR-report plasmids. The 293TN cells were co-transfected with a pmirGlo Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI, USA) containing the WT or MUT 3′-UTR fragment of QKI2, and miR-20a mimic. The luciferase activities of Firefly and Renilla were measured using a Dual-Luciferase Reporter assay system (Promega Corporation).
Statistical analysis
Statistical analyses were performed using SPSS version 17 (SPSS, Inc., Chicago, IL, USA). All statistical data are reported as the mean ± standard deviation. Significant differences between groups were assessed using Student's t-test (two-tailed). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of miR-20a in OS
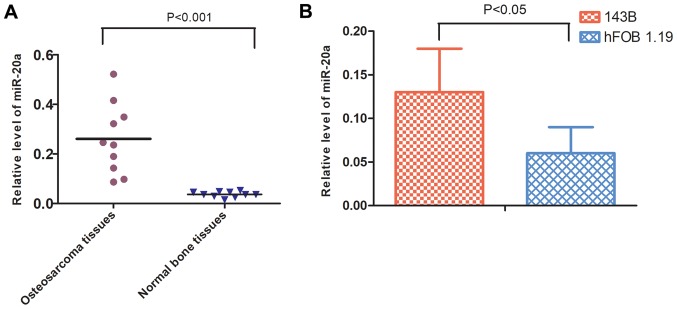
The expression levels of miR-20a in 10 primary OS and normal bone biopsy samples were initially quantified. The results of the RT-qPCR analysis showed that the expression levels of miR-20a were higher in OS tissues compared with those in the control group (Fig. 1A). Similarly, the expression levels of miR-20a were higher in 143B cells compared with those in hFOB 1.19 cells, also demonstrated by RT-qPCR analysis (Fig. 1B).
Figure 1.
miR-20a is overexpressed in OS tissues and 143B cells. (A) Relative expression levels of miR-20a were significantly increased in OS tissues compared with those in normal bone tissues (P<0.001). (B) Relative expression levels of miR-20a were increased in 143B cells compared with those in hFOB 1.19 cells (P<0.05). OS, osteosarcoma; miR, microRNA.
miR-20a promotes OS cell proliferation, migration and invasion
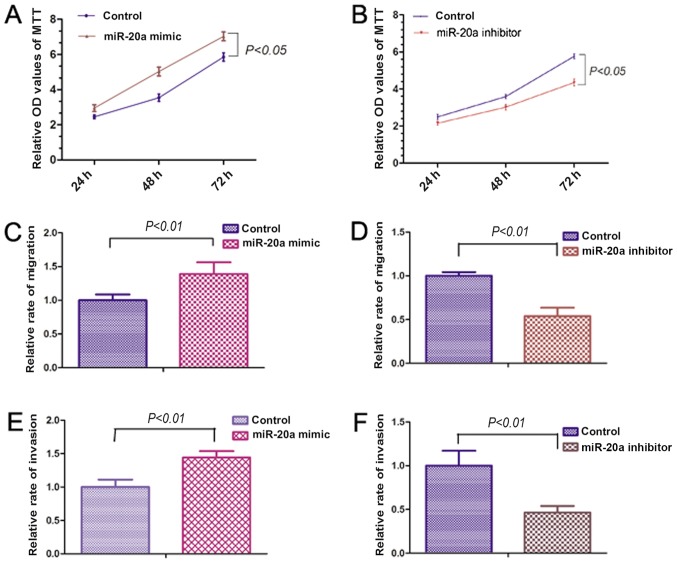
In order to examine the effects of miR-20a on OS cell proliferation, migration and invasion, 143B cells were transfected with miR-20a mimic or inhibitor. The MTT assay showed that the miR-20a mimic significantly increased the proliferation of 143B cells (Fig. 2A). However, miR-20a inhibitor significantly reduced the proliferative capacity of the 143B cells (Fig. 2B). Migration and invasion assays were performed to investigate the effects of miR-20a on the migration and invasive abilities of OS cells. The miR-20a mimic significantly promoted the migration and invasive abilities of 143B cells. By contrast, miR-20a inhibitor reduced the migration and invasive abilities of the 143B cells (Fig. 2C-F). The above results indicate that miR-20a may be involved in the proliferation, migration and invasion of OS cells.
Figure 2.
miR-20a promotes 143B cell proliferation, migration and invasion. (A) miR-20a mimic promoted the proliferative ability of 143B cells (P<0.05). (B) miR-20a inhibitor reduced the proliferative ability of 143B cells (P<0.05). (C) miR-20a mimic promoted the migration ability of 143B cells (P<0.01). (D) miR-20a inhibitor reduced the migration ability of 143B cells (P<0.01). (E) miR-20a mimic promoted the invasive ability of 143B cells (P<0.01). (F) miR-20a inhibitor reduced the invasive ability of 143B cells (P<0.01). miR, microRNA; OD, optical density.
QKI2 is a direct target of miR-20a
Using prediction tools, including TargetScan, QKI2 was hypothesized to be a direct target of miR-20a. In the present study, no difference in the mRNA expression of QKI2 was observed between the osteosarcoma tissue and normal bone tissue, or between the 143B cells and hFOB 1.19 cells, determined using RT-qPCR analysis. The protein expression of QK12 was decreased significantly in the OS tissue and 143B cells, as determined using western blot analysis. In order to further examine the association between QKI2 and miR-20a in OS, the miR-20a mimic and inhibitor were used for the transfection of 143B cells, following which the protein expression levels of QKI2 in the transfected 143B cells were detected using western blot analysis. The miR-20a mimic reduced the protein expression of QKI2 in 143B cells compared with that in the control group. However, the miR-20a inhibitor increased the protein expression levels of QKI2 (Fig. 3A). These data supported the hypothesis that QKI2 acts as a direct target of miR-20a. Additionally, in order to confirm that miR-20a can directly target QKI2 mRNA, a double fluorescence reporter assay was performed. The results of this assay showed that the miR-20a mimic significantly reduced the luciferase activity of the 293TN cells co-transfected with the plasmid containing the WT 3′-UTR fragment of QKI2, whereas the miR-20a mimic had no inhibitory effect when with the MUT QKI2 3′-UTR fragment (Fig. 3B and C). Taken together, the above findings show that miR-20a inhibited the protein expression of QKI2 by targeting the QKI2 3′-UTR in OS.
Figure 3.
miR-20a inhibits the protein expression level of QKI2 by directly targeting QKI2 mRNA in osteosarcoma cells. (A) miR-20a mimic reduced the protein expression of QKI2 in 143B cells compared with those in the control group, as determined by western blot analysis. (B) Sequences of interaction between QKI2 and miR-20a. (C) miR-20a mimic significantly decreased the luciferase activity in 293TN cells co-transfected with the WT 3′-UTR fragment of the QKI2 plasmid, as shown by a double fluorescence reporter assay (P<0.01). miR, microRNA; QKI, Quaking; WT, wild-type; MUT, mutant.
QKI2 inhibits OS cell proliferation, migration and invasion
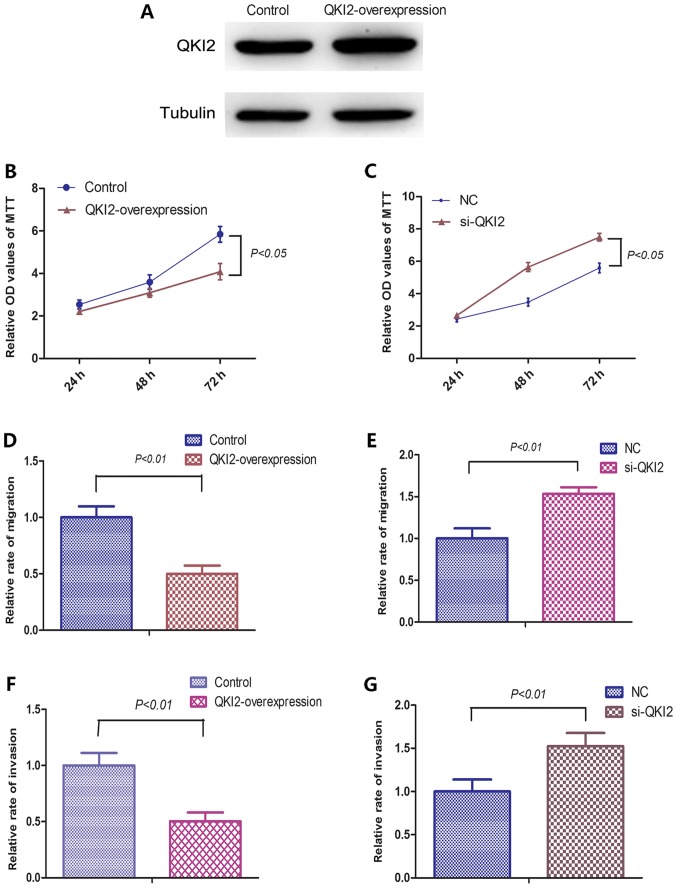
In order to further investigate the roles of QKI2 on OS cells, 143B cells stably overexpressing QKI2 were established using pWPXL lentiviral vectors. The subsequent western blot analysis showed that the protein expression levels of QKI2 were overexpressed in the stable cells (Fig. 4A). siRNAs against QKI2 were transfected into 143B cells to inhibit the expression of QKI2. An MTT assay, and Transwell migration and invasion assays were performed to examine the effects of QKI2 on OS cell proliferation, migration and invasive abilities. As shown in Figs. 4A-G and 5, the overexpression of QKI2 inhibited the proliferative, migration and invasive abilities of the 143B cells, whereas si-QKI2 evidently promoted the proliferation, migration and invasion of 143B cells. Taken together, the above results suggested that QKI2 acts as a tumor suppressor against the proliferation, migration and invasion of OS cells.
Figure 4.
QKI2 represses the proliferation, migration and invasion of 143B cells. (A) QKI2 protein was overexpressed in stable cells. (B) Overexpression of QKI2 reduces the proliferative ability of 143B cells (P<0.05). (C) si-QKI2 enhanced the proliferative ability of 143B cells (P<0.05). (D) Overexpression of QKI2 reduced the migration ability of 143B cells (P<0.01). (E) si-QKI2 enhanced the migration ability of 143B cells (P<0.01). (F) Overexpression of QKI2 reduced the invasive ability of 143B cells (P<0.01). (G) si-QKI2 enhanced the invasive ability of 143B cells (P<0.01). miR, microRNA; QKI, Quaking; si-, small interfering RNA; NC, negative control; OD, optical density.
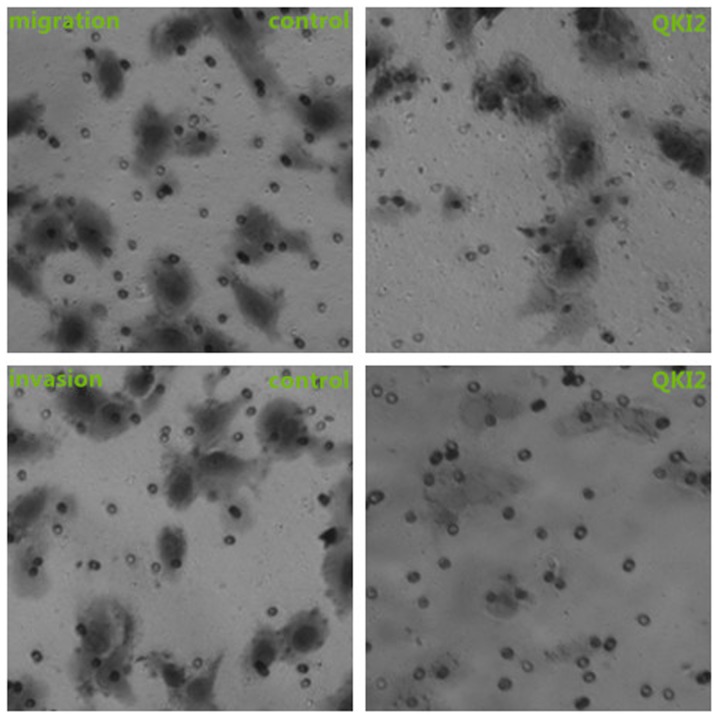
Figure 5.
Overexpression of QKI2 represses the migration and invasion ability of 143B cells (×100 magnification). QKI, Quaking.
Discussion
OS is the most common type of high-grade bone neoplasm during adolescence. Although neoadjuvant chemotherapy is administered, patients who develop relapse of disease and/or metastatic disease have a severely poor prognosis (27). To improve the 5-year survival rate of patients suffering from OS, further investigation of the cellular, molecular and signal transduction mechanisms underlying the initiation and progression of OS is required to develop novel and specific molecular targeted therapeutic strategies. miRNAs are a class of small, non-coding, single-stranded RNAs that are ~22 nucleotides in length and post-transcriptionally regulate the expression of protein-coding genes by inhibiting mRNA translation and/or directly cleaving target mRNA (28–33). In previous years, miRNAs have been reported to act as important regulators in cells (34). An increasing number of studies have shown that dysregulated miRNAs in cancer act as tumor suppressor genes or oncogenes (35,36). Marginal dysregulation in miRNA expression levels can be sufficient to affect the translation of protein-encoding genes. Consequently, one issue influencing the analysis of miRNA expression in relation to OS is that the majority of patients are treated with chemotherapeutic drugs prior to surgery; as a result, the nucleic acids required for analysis are degraded. In the present study, miRNAs were extracted from primary OS biopsy samples collected from untreated patients.
Several studies have indicated that miR-20a is a member of the oncogenic miRNAs and that it is overexpressed in multiple types of human tumor. One previous study demonstrated that the expression levels of miR-20a in gastric cancer samples were upregulated (8.9-fold) compared with those in normal adjacent samples (17). Zhang et al reported that the expression level of plasma miR-20a was significantly upregulated in early-stage non-small cell lung cancer samples when compared with control samples (18). miR-20a dysregulates the dynamic balance of colonic epithelial cells by interfering with the transforming growth factor-β-mediated regulation of Myc/p21, which is necessary for the development of colorectal cancer (21). In the cancer stem cells of glioma, the expression of miR-20a was reported to be upregulated and the invasive ability of the cells significantly promoted by targeting tissue inhibitor of metalloproteinase 2 (37). Another study showed that the overexpression of miR-20a was associated with the metastases and prognosis of patients suffering from OS (25). The present study revealed that the expression of miR-20a was upregulated in OS tissues and OS cells. To the best of our knowledge, the present study is the first to show that miR-20a can enhance the proliferative, invasive and migration abilities of OS cells.
The RNA-binding protein QKI is a member of the STAR family of proteins, and can act as a tumor suppressor gene in multiple types of human tumor. A previous study showed that QKI-5 reduces the proliferative ability of clear cell renal cell carcinoma by post-transcriptionally regulating the Ras-mitogen-activated protein kinase signaling pathway (38). de Miguel et al showed that QKI, as a tumor suppressor, is associated with the prognosis of lung carcinoma by targeting extended synaptotagmin-2 (39); the roles of QKI in OS remain to be fully elucidated. In the present study, target genes regulated by miR-20a were we screened using online databases miRanda and TargetScan. It was shown that QKI2 may interact with miR-20a, whereas other QKI members may not interact with miR-20a. Therefore, QKI2 was selected for investigation. To the best of our knowledge, the present study is among the first to demonstrate that QKI2 can inhibit OS cell proliferation, invasion and migration. These results demonstrated that miR-20a can directly target QKI2 mRNA in OS.
In conclusion, the present study first identified that miR-20a can inhibit the expression levels of QKI2 in OS by targeting QKI2 mRNA, which subsequently promotes the proliferation, invasion and migration of OS cells. The pathological role of the ‘miR-20a/QKI2’ axis in regulating the metastasis of OS was demonstrated. These results not only improve our understanding of the mechanism underlying the metastasis of OS, but also provide a novel strategy and target for OS treatment. However, at present, there are no agents available targeting miR-20a or QKI2 in the clinical setting; further investigation is required to determine these findings clinically.
The main purpose of the present study was to investigate pathological role of miR-20a/QKI2 in the invasion and migration of OS. The 143B cell line is a metastatic subline of HOS and has the ability to metastasize, therefore the 143B cell line was used in the present study. However, results obtained only in the 143B cell line are not fully convincing. In future investigations, experiments using other OS cell lines, including human OS U2OS, Saos-2, HOS and MG-63 cell lines, and in vivo experiments are to be performed.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data used and analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
YW conceived and designed the experiments; HY, YL and ZP performed the experiments and analyzed the data; YW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was performed in compliance with the Helsinki Declaration and approved by the Institutional Review Board of the First Affiliated Hospital of Harbin Medical University (IRB no. 2016-029). The data collection and analyses were performed without disclosing patient identities.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longhi A, Errani C, De Paolis M, Mercuri M, Bacci G. Primary bone osteosarcoma in the pediatric age: State of the art. Cancer Treat Rev. 2006;32:423–436. doi: 10.1016/j.ctrv.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Friebele JC, Peck J, Pan X, Abdel-Rasoul M, Mayerson JL. Osteosarcoma: A meta-analysis and review of the literature. Am J Orthop (Belle Mead NJ) 2015;44:547–553. [PubMed] [Google Scholar]
- 4.Kondo T, Furuta T, Mitsunaga K, Ebersole TA, Shichiri M, Wu J, Artzt K, Yamamura K, Abe K. Genomic organization and expression analysis of the mouse qkI locus. Mamm Genome. 1999;10:662–669. doi: 10.1007/s003359901068. [DOI] [PubMed] [Google Scholar]
- 5.Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Lu Z, Ku L, Chen Y, Wang H, Feng Y. Tyrosine phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. EMBO J. 2003;22:1801–1810. doi: 10.1093/emboj/cdg171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu F, Jin L, Yang G, Ji L, Wang F, Lu Z. Post-transcriptional repression of FOXO1 by QKI results in low levels of FOXO1 expression in breast cancer cells. Oncol Rep. 2014;31:1459–1465. doi: 10.3892/or.2013.2957. [DOI] [PubMed] [Google Scholar]
- 8.Ichimura K, Mungall AJ, Fiegler H, Pearson DM, Dunham I, Carter NP, Collins VP. Small regions of overlapping deletions on 6q26 in human astrocytic tumours identified using chromosome 6 tile path array-CGH. Oncogene. 2006;25:1261–1271. doi: 10.1038/sj.onc.1209156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li ZZ, Kondo T, Murata T, Ebersole TA, Nishi T, Tada K, Ushio Y, Yamamura K, Abe K. Expression of Hqk encoding a KH RNA binding protein is altered in human glioma. Jpn J Cancer Res. 2002;93:167–177. doi: 10.1111/j.1349-7006.2002.tb01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulholland PJ, Fiegler H, Mazzanti C, Gorman P, Sasieni P, Adams J, Jones TA, Babbage JW, Vatcheva R, Ichimura K, et al. Genomic profiling identifies discrete deletions associated with translocations in glioblastoma multiforme. Cell Cycle. 2006;5:783–791. doi: 10.4161/cc.5.7.2631. [DOI] [PubMed] [Google Scholar]
- 11.Yin D, Ogawa S, Kawamata N, Tunici P, Finocchiaro G, Eoli M, Ruckert C, Huynh T, Liu G, Kato M, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res. 2009;7:665–677. doi: 10.1158/1541-7786.MCR-08-0270. [DOI] [PubMed] [Google Scholar]
- 12.Fu X, Feng Y. QKI-5 suppresses cyclin D1 expression and proliferation of oral squamous cell carcinoma cells via MAPK signalling pathway. Int J Oral Maxillofac Surg. 2015;44:562–567. doi: 10.1016/j.ijom.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Bian Y, Wang L, Lu H, Yang G, Zhang Z, Fu H, Lu X, Wei M, Sun J, Zhao Q, et al. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem Biophys Res Commun. 2012;422:187–193. doi: 10.1016/j.bbrc.2012.04.138. [DOI] [PubMed] [Google Scholar]
- 14.Yang G, Fu H, Zhang J, Lu X, Yu F, Jin L, Bai L, Huang B, Shen L, Feng Y, et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology. 2010;138:231–240.e1-5. doi: 10.1053/j.gastro.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J, Ying H, Hu B, Hurt J, Farny N, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26:1459–1472. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jafarzadeh-Samani Z, Sohrabi S, Shirmohammadi K, Effatpanah H, Yadegarazari R, Saidijam M. Evaluation of miR-22 and miR-20a as diagnostic biomarkers for gastric cancer. Chin Clin Oncol. 2017;6:16. doi: 10.21037/cco.2017.03.01. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Mao F, Shen T, Luo Q, Ding Z, Qian L, Huang J. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol Lett. 2017;13:669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babu KR, Muckenthaler MU. miR-20a regulates expression of the iron exporter ferroportin in lung cancer. J Mol Med (Berl) 2016;94:347–359. doi: 10.1007/s00109-015-1362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng J, Thakur A, Zhang S, Dong Y, Wang X, Yuan R, Zhang K, Guo X. Expressions of miR-181a and miR-20a in RPMI8226 cell line and their potential as biomarkers for multiple myeloma. Tumour Biol. 2015;36:8545–8552. doi: 10.1007/s13277-015-3600-2. [DOI] [PubMed] [Google Scholar]
- 21.Sokolova V, Fiorino A, Zoni E, Crippa E, Reid JF, Gariboldi M, Pierotti MA. The effects of miR-20a on p21: Two mechanisms blocking growth arrest in TGF-β-responsive colon carcinoma. J Cell Physiol. 2015;230:3105–3114. doi: 10.1002/jcp.25051. [DOI] [PubMed] [Google Scholar]
- 22.Pesta M, Klecka J, Kulda V, Topolcan O, Hora M, Eret V, Ludvikova M, Babjuk M, Novak K, Stolz J, Holubec L. Importance of miR-20a expression in prostate cancer tissue. Anticancer Res. 2010;30:3579–3583. [PubMed] [Google Scholar]
- 23.Chang Y, Liu C, Yang J, Liu G, Feng F, Tang J, Hu L, Li L, Jiang F, Chen C, et al. MiR-20a triggers metastasis of gallbladder carcinoma. J Hepatol. 2013;59:518–527. doi: 10.1016/j.jhep.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arabi L, Gsponer JR, Smida J, Nathrath M, Perrina V, Jundt G, Ruiz C, Quagliata L, Baumhoer D. Upregulation of the miR-17-92 cluster and its two paraloga in osteosarcoma - reasons and consequences. Genes Cancer. 2014;5:56–63. doi: 10.18632/genesandcancer.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Wu PK, Chen WM, Chen CF, Lee OK, Haung CK, Chen TH. Primary osteogenic sarcoma with pulmonary metastasis: Clinical results and prognostic factors in 91 patients. Jpn J Clin Oncol. 2009;39:514–522. doi: 10.1093/jjco/hyp057. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Liu HP, Deng JE, Zhou M. Systematic analysis of genomic organization and heterogeneities of miRNA cluster in vertebrates. Mol Biol Rep. 2012;39:5143–5149. doi: 10.1007/s11033-011-1310-4. [DOI] [PubMed] [Google Scholar]
- 32.Wang QH, Zhou M, Sun J, Ning SW, Li Y, Chen L, Zheng Y, Li X, Lv SL, Li X. Systematic analysis of human microRNA divergence based on evolutionary emergence. FEBS Lett. 2011;585:240–248. doi: 10.1016/j.febslet.2010.11.053. [DOI] [PubMed] [Google Scholar]
- 33.Zhou M, Wang Q, Sun J, Li X, Xu L, Yang H, Shi H, Ning S, Chen L, Li Y, et al. In silico detection and characteristics of novel microRNA genes in the Equus caballus genome using an integrated ab initio and comparative genomic approach. Genomics. 2009;94:125–131. doi: 10.1016/j.ygeno.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passetti F, Ferreira CG, Costa FF. The impact of microRNAs and alternative splicing in pharmacogenomics. Pharmacogenomics J. 2009;9:1–13. doi: 10.1038/tpj.2008.14. [DOI] [PubMed] [Google Scholar]
- 36.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Wang B, Shi Y, Xu C, Xiao HL, Ma LN, Xu SL, Yang L, Wang QL, Dang WQ, et al. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34:1407–1419. doi: 10.1038/onc.2014.75. [DOI] [PubMed] [Google Scholar]
- 38.Zhang RL, Yang JP, Peng LX, Zheng LS, Xie P, Wang MY, Cao Y, Zhang ZL, Zhou FJ, Qian CN, Bao YX. RNA-binding protein QKI-5 inhibits the proliferation of clear cell renal cell carcinoma via post-transcriptional stabilization of RASA1 mRNA. Cell Cycle. 2016;15:3094–3104. doi: 10.1080/15384101.2016.1235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Miguel FJ, Pajares MJ, Martínez-Terroba E, Ajona D, Morales X, Sharma RD, Pardo FJ, Rouzaut A, Rubio A, Montuenga LM, Pio R. A large-scale analysis of alternative splicing reveals a key role of QKI in lung cancer. Mol Oncol. 2016;10:1437–1449. doi: 10.1016/j.molonc.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and analyzed in the present study are available from the corresponding author on reasonable request.