Abstract
The mosquitoes Aedes aegypti (L.) and Ae. albopictus Skuse are the major vectors of dengue, Zika, yellow fever, and chikungunya viruses worldwide. Wolbachia, an endosymbiotic bacterium present in many insects, is being utilized in novel vector control strategies to manipulate mosquito life history and vector competence to curb virus transmission. Earlier studies have found that Wolbachia is commonly detected in Ae. albopictus but rarely detected in Ae. aegypti. In this study, we used a two‐step PCR assay to detect Wolbachia in wild‐collected samples of Ae. aegypti. The PCR products were sequenced to validate amplicons and identify Wolbachia strains. A loop‐mediated isothermal amplification (LAMP) assay was developed and used for detecting Wolbachia in selected mosquito specimens as well. We found Wolbachia in 85/148 (57.4%) wild Ae. aegypti specimens from various cities in New Mexico, and in 2/46 (4.3%) from St. Augustine, Florida. Wolbachia was not detected in 94 samples of Ae. aegypti from Deer Park, Harris County, Texas. Wolbachia detected in Ae. aegypti from both New Mexico and Florida was the wAlbB strain of Wolbachia pipientis. A Wolbachia‐positive colony of Ae. aegypti was established from pupae collected in Las Cruces, New Mexico, in 2018. The infected females of this strain transmitted Wolbachia to their progeny when crossed with males of Rockefeller strain of Ae. aegypti, which does not carry Wolbachia. In contrast, none of the progeny of Las Cruces males mated to Rockefeller females were infected with Wolbachia.
Keywords: Aedes aegypti, Aedes albopictus, Florida, New Mexico, Texas, wAlbB, Wolbachia
1. INTRODUCTION
Wolbachia are obligate intracellular bacteria found in a wide range of terrestrial arthropods and nematodes (Werren, Baldo, & Clark, 2008). The bacterium was discovered in the reproductive tissues (testes and ovaries) of the mosquito Culex pipiens L. by Hertig and Wolbach in 1924 (Hertig & Wolbach, 1924) and was formally described as Wolbachia pipientis by Hertig in 1936 (Hertig, 1936). About 60%–70% of all insect species harbor Wolbachia, including some mosquito species (Hilgenboecker, Hammerstein, Schlattmann, Telschow, & Werren, 2008). Wolbachia can be a powerful reproductive manipulator, inducing cytoplasmic incompatibility (CI), parthenogenesis, feminization of males, and male killing in various host species (Werren et al., 2008). These properties have been exploited for the development of Wolbachia as a novel strategy for vector mosquito control. Wolbachia‐induced CI favors the reproductive success and spread of colonized females in populations, which can be used to drive desirable traits, including resistance to infection with vector‐borne pathogens, into a population. On the other hand, infected mosquito males can cause CI in a population with the presence of different Wolbachia strains or no infection, which can be used for sterile insect technique (SIT) to decrease vector populations (Flores & O'Neill, 2018).
Aedes (Stegomyia) aegypti (L.) and Ae. (Stegomyia) albopictus Skuse are the major vectors for the transmission of several arthropod‐borne viruses (arboviruses) among humans, particularly dengue, Zika, yellow fever, and chikungunya viruses. Wolbachia are commonly found in Ae. albopictus (de Albuquerque, Magalhaes, & Ayres, 2011; Joanne et al., 2015; Kitrayapong, Baimai, & O'Neill, 2002), but until recently Ae. aegypti was thought not to carry this bacterium (Gloria‐Soria, Chiodo, & Powell, 2018; Kitrayapong et al., 2002; Kittayapong, Baisley, Baimai, & O'Neill, 2000). However, Wolbachia sequences were found in wild Ae. aegypti in a few recent investigations. Wolbachia 16S ribosomal RNA gene sequencing reads (operational taxonomic units, OTUs) were detected in Ae. aegypti larvae collected from Jacksonville, Florida (Coon, Brown, & Strand, 2016), as well as in two Ae. aegypti adult pools collected from Thailand (Thongsripong et al., 2017). More recently, Wolbachia 16S reads were detected in a few individuals of Ae. aegypti collected from Houston, Texas, though the regular PCR to amplify other Wolbachia genes was not successful (Hegde et al., 2018).
Establishing the prevalence of Wolbachia in Ae. aegypti is critical to public health, because over the past decade, Ae. aegypti transinfected with Wolbachia have been generated with the goal of blocking transmission of dengue virus (Bian, Xu, Lu, Xie, & Xi, 2010; Bull & Turelli, 2013; Hoffmann et al., 2011; McMeniman et al., 2009; O'Neill, 2018; Walker et al., 2011). This approach was initially aimed at shortening mosquito life span below the extrinsic incubation period of the virus (McMeniman et al., 2009), but in the course of these experiments it was discovered that transinfection of Ae. aegypti with Wolbachia strain wMelPop also blocks dengue and chikungunya virus infections of the mosquito (Moreira et al., 2009). A successful large field trial in Australia showed a stable establishment and slow but steady spread of released Ae. aegypti transinfected with wMel in the study area (Schmidt et al., 2017). However, if a population of Ae. aegypti were to harbor an autochthonous strain of Wolbachia, then this native strain would have a high potential to prevent invasion of a virus‐blocking strain that exhibits incompatibility with the native strain (Hoffmann, Ross, & Rasic, 2015). This effect was demonstrated in a study of Ae. albopictus, wherein the wMel transinfected line produced complete bidirectional incompatibility with a wild‐type line carrying wAlbA and wAlbB, with 0% hatch rate from crossing between females of either strain with males of the other strain (Blagrove, Arias‐Goeta, Failloux, & Sinkins, 2012). On the other hand, complete CI could favor SIT for population reduction.
During a project to map the distribution of Ae. aegypti and Ae. albopictus in New Mexico in 2017 and characterize their microbiota, we unexpectedly detected Wolbachia 16S rRNA gene amplicon in wild‐caught Ae. aegypti in Las Cruces, New Mexico. A more comprehensive survey was then conducted using a two‐step PCR assay, which revealed a 57.4% prevalence of Wolbachia infection in 148 specimens of Ae. aegypti collected from eight cities across New Mexico. Wolbachia was also detected in two of 46 specimens of Ae. aegypti from St. Augustine, Florida (4.3% prevalence), but not detected in 94 specimens of Ae. aegypti from Deer Park, Harris County, Texas. A Wolbachia‐infected Ae. aegypti strain was established from wild pupae collected from Las Cruces. The cross of the strain with the Wolbachia‐uninfected Rockefeller strain demonstrated maternal transmission of Wolbachia to progeny.
2. MATERIALS AND METHODS
2.1. Mosquito collections and species identification
Aedes aegypti and Ae. albopictus mosquitoes were collected using gravid and sentinel traps in New Mexico in 2017 by the SouthWest Aedes Research and Mapping (SWARM) project team, in Florida in 2016 by the Anastasia Mosquito Control District, and in Texas in 2018 by the Harris County Public Health Mosquito and Vector Control Division. The location details of the samples are presented in Table 1 and Figure 1. Although Ae. aegypti and Ae. albopictus were commonly collected in the same trap in Texas, they were never collected in the same trap in New Mexico or Florida. Mosquitoes were sorted and identified as Ae. aegypti or Ae. albopictus based on morphology. The species identity was confirmed by a species‐diagnostic PCR assay that amplifies species‐specific fragments of internal transcribed spacer 1 (ITS1) of ribosomal DNA, as described by (Higa, Toma, Tsuda, & Miyagi, 2010). The primers used in this study are listed in Table S1. The PCR was conducted using 2× PCR master mix (MCLAB) with ~20 ng DNA, 0.2 µM primers, and cycling parameters as: 35 cycles of denaturing at 94°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 30 s with extra 5 min in the last cycle for final extension.
Table 1.
Mosquito collections in Florida, New Mexico, and Texas
| Species | Location | Coordinates | Collection time |
|---|---|---|---|
| Aedes aegypti | St. Augustine, FL | 29.895, −81.313 | July, 2016 |
| Aedes albopictus | St. Augustine, FL | 29.890, −81.332 | July, 2016 |
| Aedes aegypti | Deer Park, TX | 29.693, −95.115 | May, 2018 |
| Aedes albopictus | Deer Park, TX | 29.693, −95.115 | May, 2018 |
| Aedes aegypti | 8 cities, NM | See Table 2 | May–November, 2017 |
| Aedes albopictus | 2 cities, NM | See Table 4 | May–November, 2017 |
Figure 1.
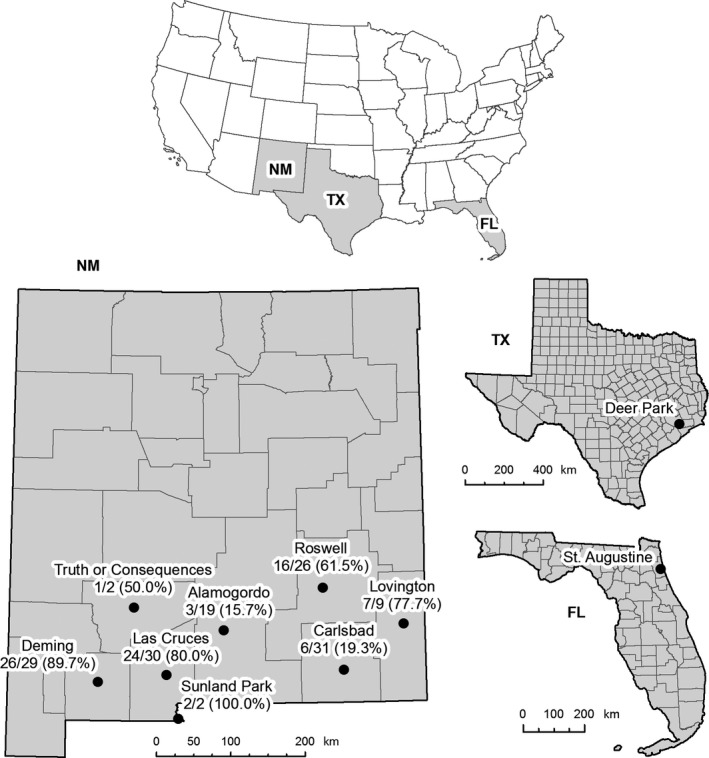
Maps of the sites where Aedes aegypti mosquitoes were sampled in this study. No. of Wolbachia positive/no. of tested (%) was displayed in sampling sites
2.2. DNA isolation, bacterial 16S rDNA PCR, cloning, and sequencing
Mosquito specimens from traps were desiccated in most cases. For each mosquito specimen, the abdomen was separated from the thorax by pulling gently with tweezers that were cleaned with 75% ethanol between samples. Abdomens were used for detecting associated microbiota. Metagenomic DNA was isolated individually from each abdomen using DNAzol following the manufacturer's protocol (Thermo Fisher Scientific). Briefly, one abdomen was homogenized in 100 µl DNAzol and centrifuged at 12,000 g for 10 min. Supernatant was transferred to a new tube, 50 µl ethanol was added, and the tube was centrifuged at 12,000 g for 10 min for DNA precipitation. The DNA pellet was dissolved in 30 µl H2O. A bacterial 16S rDNA fragment covering V1 to V3 was amplified from individual DNA using primers 27F and 519R (Table S1), as we previously reported (Wang, Gilbreath, Kukutla, Yan, & Xu, 2011). PCR was run using 2× PCR master mix with 0.2 µM primers and cycling parameters: 35 cycles of denaturing at 94°C for 15 s, annealing at 52°C for 15 s, and extension at 72°C for 30 s with extra 5 min in the last cycle for final extension. PCR products were purified and cloned into the plasmid pMiniT 2.0 using a NEB PCR cloning kit (New England Biolabs) following manufacturer's instructions. Colony PCR was conducted to amplify the inserts using SP6 forward and T7 reverse primers. The PCR products with a size of ~500 bp were sent for Sanger sequencing at a commercial provider (MCLAB).
2.3. Wolbachia PCR assays and sequencing
The PCR assays, using primer sets for the Wolbachia gatB and ftsZ gene from Baldo et al. (2006) (Table S1), were used for detecting Wolbachia in mosquitoes. PCR was run using 2× PCR master mix (MCLAB) with a primer concentration of 0.2 µM and the following cycling parameters: 35 cycles of denaturing at 94°C for 15 s, annealing at a temperature optimal for the amplicon (Table S1) for 15 s, and extension at 72°C for 30 s with an extra 5 min in the last cycle for final extension. For specimens that showed a faint band or no visible band after a Wolbachia target PCR, a second‐round PCR was conducted. The first‐round PCR product was diluted 100 times with H2O, and 1 µl was used as template for the second PCR. In 85 Wolbachia‐positive specimens from NM, 8 showed a clear band in the first PCR and the remaining specimens showed a faint or no band in the first PCR but a clear band in the second PCR.
To validate the Wolbachia detection in Ae. aegypti, we designed strain‐specific primers to amplify fragments from two Wolbachia genes encoding phosphoesterase (PE) and diaminopimelate epimerase (DE) based on the draft genomes of wAlbB (Mavingui et al., 2012) and wAlbA (GenBank accession NWVK00000000.1). The sequences of the two genes have distinctive interstrain differences, enabling the design of strain‐specific primers (Table S1). A subset of specimens that were positive from the first or second PCR were subjected to the validation PCR and sequencing. The products were sequenced at MCLAB, and the sequences were deposited in GenBank; the accession numbers are presented in Table S2.
2.4. Loop‐mediated isothermal amplification (LAMP) assay
Loop‐mediated isothermal amplification (LAMP) was developed as an additional assay for the detection of Wolbachia in mosquitoes. Oligonucleotides for LAMP were designed using Primer Explorer V5 software available on the website (http://primerexplorer.jp/lampv5e/index.html). The sequences of oligos for the 16S rRNA gene are listed in Table S1. The LAMP reactions were conducted using a NEB LAMP kit with Bst 3.0 (M0374, NEB). The reaction mixture, consisting of 1× isothermal amplification buffer II, 6 mM MgSO4, 1.4 mM of each of the deoxynucleotide triphosphates (dNTPs), 1.6 µM Forward Inner Primer/Backward Inner Primer, 0.4 µM F3/B3 primers, 0.8 µM Loop Forward/Backward, 8 U of Bst 3.0 DNA polymerase, and 1 µl genomic DNA in a total volume of 25 µl, was incubated at 65°C for 60 min in a T100 Thermal Cycler (Bio‐Rad). The amplified products (10 µl) were run on 1.5% agarose gel and visualized under ultraviolet light. For all the tests, a positive control (DNA from a female Wolbachia‐infected Ae. albopictus), a negative control (DNA from females of Ae. aegypti Rockefeller strain), and a no template control (nuclease‐free water) were used.
2.5. Maternal transmission of Wolbachia in Las Cruces strain
In August 2017, a colony of Ae. aegypti was established from pupae (n = 8) collected from a larval habitat in a residential area in Las Cruces. The eclosed adults were confirmed to carry Wolbachia by PCR and LAMP assays as described above. Unfortunately, the colony was lost in January 2018. In September 2018, a new colony was initiated from the pupae (n = 77) collected from the same location. Again, the eclosed adults tested positive for Wolbachia. The strain was named the LC (Las Cruces, NM) strain. To test whether Wolbachia can be transmitted to the progeny maternally, crosses between LC and Rockefeller (Rock) strains were conducted, which included a cross of virgin females (LC) × males (Rock), and a cross of virgin females (Rock) × males (LC). Rockefeller strain of Ae. aegypti is known not to carry Wolbachia (0% infected out of the 10 individuals screened with the LAMP assay). Each cross included 50 females and 30 males of the respective strains. All cages were maintained at 28°C, 72% RH and fed on 20% sucrose till day 3. On day 4, females were blood fed on the forearm of a human volunteer. Eggs were collected at day 3 postfeeding and reared to adults. The progeny adults from the respective crosses were screened for Wolbachia by the LAMP assay.
3. RESULTS
3.1. Prevalence of Wolbachia in Ae. aegypti and Ae. albopictus populations in New Mexico
Aedes aegypti and Ae. albopictus occur in the state of New Mexico (Hahn et al., 2017, 2016). In 2017, we conducted a survey to map the distribution and characterize the microbiomes of both species in New Mexico. To profile bacteriomes associated with wild Ae. aegypti collected from Las Cruces, a bacterial 16S ribosomal RNA gene fragment was amplified by PCR using primers 27F and 519R. The PCR products were cloned and sequenced to identify bacterial taxa, and a total of 80 sequences were obtained from eight individuals. Unexpectedly, 10 sequences of Wolbachia 16S amplicons were identified in six of eight individuals. It should be noted that no Wolbachia‐specific 16S PCR had been conducted in the laboratory previously, so the Wolbachia clones could not be derived from PCR product contamination in the laboratory working area. We then conducted a large survey by screening 148 specimens collected from eight cities across New Mexico, which largely span the distribution of Ae. aegypti in the state (Figure 1), by amplifying Wolbachia gatB or ftsZ (Table 2). Selected positive PCR products were validated by sequencing. Out of 148 specimens tested, 85 were Wolbachia‐positive, and the overall prevalence was 57.4% (Tables 2 and 3). Aedes albopictus is less common in New Mexico and is only present on the eastern part of the state. A total of 13 Ae. albopictus specimens from Roswell and Clovis were available for Wolbachia screening, 11 were positive for both wAlbA and wAlbB strains, and one was positive for wAlbB only. The overall Wolbachia prevalence in Ae. albopictus in New Mexico was 92.3% (Tables 4 and 5). It should be noted that no Ae. aegypti and Ae. albopictus were caught in the same trap. So Wolbachia detected in Ae. aegypti specimens could not possibly be derived from a cross contamination from Ae. albopictus specimens.
Table 2.
Wolbachia infection in Aedes aegypti populations in New Mexico from May to November 2017
| City (n) | Infection rate (%) | Coordinates of collection sites |
|---|---|---|
| Alamogordo (19) | 3 (15.8) | 32.861, −105.979; 32.918, −105.936 |
| Carlsbad (31) | 6 (19.4) | 32.356, −104.248; 32.440, −104.240; 32.427, −104.223 |
| Deming (29) | 26 (89.7) | 32.251, −107.763; 32.245, −107.761; 32.262, −107.745 |
| Las Cruces (30) | 24 (80.0) | 32.296, −106.732; 32.357, −106.769; 32.396, −106.816 |
| Lovington (9) | 7 (77.8) | 21.491, −103.364 |
| Sunland Park (2) | 2 (100) | 31.816, −106.603 |
| Roswell (26) | 16 (61.5) | 33.378, −104.513; 33.416, −104.529 |
| Truth or Consequences (2) | 1 (50.0) | 33.120, −107.272; 33.203, −107.228 |
| Total (148) | 85 (57.4) |
Table 3.
Wolbachia infection in Aedes aegypti popuations in New Mexico and Florida
| Wolbachia strain | ||||
|---|---|---|---|---|
| Specimens (n) | A & B (%) | A (%) | B (%) | Total no. (%) |
| Male (51), NM | 0 | 0 | 28 (54.9) | 28 (54.9) |
| Female (97), NM | 0 | 0 | 57 (58.8) | 57 (58.8) |
| Male (18), FL | 0 | 0 | 1 (5.5) | 1 (5.5) |
| Female (28), FL | 0 | 0 | 1 (3.6) | 1 (3.6) |
Table 4.
Wolbachia infection in Aedes albopictus populations in New Mexico from May to November 2017
| City (n) | Infection rate (%) | Coordinates of collection sites |
|---|---|---|
| Clovis (12) | 11 (91.7) | 34.406, −103.192; 34.424, −103.182; 34.414, −103.196; 34.399, −103.200; 34.404, −103.201 |
| Roswell (1) | 1 (100) | 33.389, −104.530 |
| Total (13) | 12 (92.3) |
Table 5.
Wolbachia strain distribution in Aedes albopictus in Texas, Florida, and New Mexico
| Wolbachia strain | ||||
|---|---|---|---|---|
| Specimens (n) | A & B (%) | A (%) | B (%) | Total no. (%) |
| Male (19), TX | 6 (31.6) | 1 (5.3) | 9 (47.4) | 16 (84.2) |
| Female (13), TX | 9 (69.2) | 1 (7.7) | 0 | 10 (76.9) |
| Male (20), FL | 20 (100) | 0 | 0 | 20 (100) |
| Female (18), FL | 14 (77.8) | 0 | 1 (5.6) | 15 (83.3) |
| Male (2), NM | 0 | 0 | 1 (50.0) | 1 (50.0) |
| Female (11), NM | 11 (100) | 0 | 0 | 11 (100) |
We also amplified DNA fragments of two additional Wolbachia genes, encoding PE and DE. The amplicons of Wolbachia 16S rDNA, gatB, ftsZ, PE, and DE were confirmed by sequencing. In Ae. aegypti specimens, only wAlbB sequences were detected. Representative sequences were deposited in GenBank (Table S2).
In August 2017, a local Ae. aegypti colony was derived from pupae collected in Las Cruces, NM. Wolbachia was detected in two of four females and two of four males that eclosed from the pupae. The infection persisted in F1 as well; 14/33 females (42.4%) and 14 of 14 males (100%) of F1 adults examined were positive for Wolbachia. Unfortunately, the colony was lost in January 2018 because of a failure of egg hatching at F3 generation. In September 2018, a new colony of Ae. aegypti was initiated from the pupae collected from the same location in Las Cruces. The strain, named LC, was Wolbachia‐positive, with a prevalence of 84.6% of the 13 individuals screened.
3.2. Prevalence of Wolbachia in Ae. aegypti and Ae. albopictus in Florida
Coon et al. (2016) previously reported the presence of Wolbachia in the larvae of Ae. aegypti collected from Jacksonville, Florida. We therefore examined the specimens of Ae. aegypti collected in July 2016 from St. Augustine, Florida, which is approximately 50 miles south of Jacksonville. Among 46 specimens screened, one male and one female were wAlbB‐positive, with a prevalence of 4.3% (Table 3). The wAlbA strain was not detected in these specimens. As expected, Wolbachia infection occurred at a high prevalence in Ae. albopictus. Among the 38 specimens tested, 35 were coinfected with wAlbA and wAlbB, and one carried only wAlbB. The overall Wolbachia prevalence in Ae. albopictus was 92.1% (Table 5). Ae. aegypti and Ae. albopictus were sampled at different sites in St. Augustine (Table 1).
3.3. Prevalence of Wolbachia in Ae. aegypti and Ae. albopictus in southeastern Texas
In 2018, we screened 94 Ae. aegypti and 32 Ae. albopictus collected in a neighborhood in Deer Park, Harris County, Texas (Table 1). In some traps, both Ae. aegypti and Ae. albopictus specimens were caught. No Wolbachia was detected in 94 Ae. aegypti specimens. In 32 Ae. albopictus specimens, 26 (81.3%) were Wolbachia‐positive. Among them, 15 carried both wAlbA and wAlbB, two carried wAlbA only, and nine carried wAlbB only (Table 5).
3.4. LAMP assays
The LAMP assay is a sensitive method for detecting target DNA with low abundance in a sample (Notomi et al., 2000). Goncalves et al. (2014) developed a LAMP assay for Wolbachia detection in insects (Goncalves Dda et al., 2014). Recently, Bhadra et al. (2018) reported a specific and sensitive assay that combines LAMP and oligonucleotide strand displacement (OSD) for detecting both species identity and Wolbachia. In our study, the Wolbachia density appears to be quite low in most infected specimens of Ae. aegypti. To corroborate the results from the Wolbachia PCR assay, we developed a LAMP assay to detect Wolbachia. The LAMP assay was sensitive and detected target Wolbachia DNA in infected Ae. aegypti directly. As shown in Figure 2, Wolbachia‐positive samples yield a ladder of bands between 200 bp–1 kb, and a ~100 bp band. Wolbachia‐negative samples show an accumulation of oligos around 50 bp. The infected Ae. albopictus specimen could be detected at 100 times dilution of template, but not at 500 times dilution, while the infected Ae. aegypti specimen could not be detected at 20 times dilution. Figure 2 shows a representative result of LAMP assay on the Wolbachia‐positive and Wolbachia‐negative specimens from New Mexico, Florida, and Texas.
Figure 2.
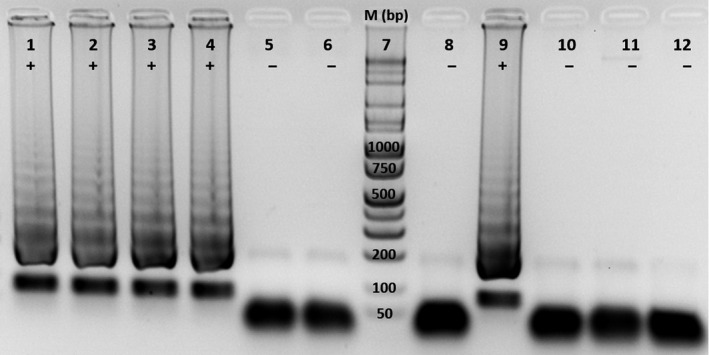
LAMP detection of Wolbachia 16S rDNA. 1,2 = Aedes aegypti (NM); 3,4 = Ae. aegypti (FL); 5,6 = Ae. aegypti (TX); 7 = Marker; 8 = Ae. aegypti (NM‐1, 1:20 dilution); 9 = Aedes albopictus (NM, 1:100 dilution); 10 = Ae. albopictus (NM, 1:500 dilution); 11 = Ae. aegypti Rockefeller; 12 = No template control. +: Wolbachia positive; −: Wolbachia negative
3.5. Maternal transmission of Wolbachia occurs in Las Cruces strain
Wolbachia are known to be transmitted vertically from female to the offspring (Werren et al., 2008). To test the occurrence of vertical transmission of Wolbachia in LC strain, crosses were set up between the adults of LC strain and Rockefeller strain (Rock). The F2 generation of LC strain was used for the experiment. From the LC females and males used for the crosses, five specimens from each sex were randomly selected and examined by the LAMP assay. They were all Wolbachia‐positive. The crosses between females (LC) × males (Rock) and females (Rock) × males (LC) both yielded viable progenies. The egg hatch rate was significantly higher in F/LC × M/Rock (98/463, 21.2%) than in M/LC × F/Rock (39/475, 8.2%), (chi‐square, p < 0.01). The progenies were screened for the presence of Wolbachia by the LAMP assay. As shown in Figure 3, the progenies of F/LC × M/Rock were Wolbachia‐positive (10/10, 100%), while the progenies of M/LC × F/Rock were Wolbachia‐negative (10/10, 100%). The results clearly demonstrated the maternal transmission of Wolbachia in LC Ae. aegypti.
Figure 3.
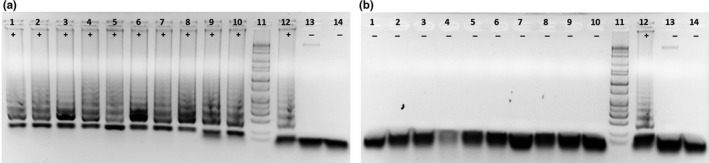
Maternal transmission of Wolbachia in LC strain. Wolbachia detection in the progeny of the respective crosses. (a) 1–10 = Progeny of the cross between females (LC) and males (Rock), 11 = Marker, 12 = Aedes albopictus (NM, 1:20 dilution), 13 = Aedes aegypti Rockefeller, 14 = No template control. +: Wolbachia positive, −: Wolbachia negative. (b) 1–10 = Progeny of the cross between females (Rock) and males (LC), 11 = Marker, 12 = Aedes albopictus (NM, 1:20 dilution), 13 = Aedes aegypti Rockefeller, 14 = No template control. +: Wolbachia positive, −: Wolbachia negative
4. DISCUSSION
Wolbachia is commonly associated with wild Ae. albopictus around the world. However, Wolbachia has not been detected in wild Ae. aegypti in previous surveys until recently. In the study by Coon et al. (2016), two Wolbachia 16S rDNA OTUs were detected in a pool of 30 larvae of Ae. aegypti that were collected from one of five larval sites in Jacksonville, Florida, in June 2014. The Wolbachia OTUs were detected in another collection from the same location in July 2014, and both wAlbA and wAlbB were detected based on the sequence comparison of several Wolbachia genes. The prevalence of Wolbachia in the Ae. aegypti population was not investigated in that study (Coon et al., 2016). Similarly, Wolbachia 16S OTUs were detected in two pools of 10 and 25 specimens of Ae. aegypti, respectively, collected in the suburb and urban areas of Thailand (Thongsripong et al., 2017). In a study by Hegde et al. (2018), a small number of Wolbachia 16S rDNA reads were found in a few Ae. aegypti individuals collected from Houston, Texas, but the results were not validated by PCR using several Wolbachia genes. Our survey of Florida mosquitoes was consistent with previous results, detecting a low prevalence (4.3%) of Wolbachia in Ae. aegypti from St. Augustine, Florida (Table 3). And, no Wolbachia was detected in 94 Ae. aegypti specimens from Deer Park, Texas.
In contrast, the screening of wild populations of Ae. aegypti from eight cities in New Mexico revealed a high prevalence (15.8%–100%, average of 57.4%) of Wolbachia, a level unprecedented for this species. The infection was validated by sequencing the PCR amplicons from the ftsZ, gatB, DE, and PE genes (Table S2). These sequences were identical to the sequences of respective genes in the genome of wAblB strain (Mavingui et al., 2012), indicating the Wolbachia detected in the Ae. aegypti samples belongs to the wAlbB strain. There were concerns about potential cross contamination from specimens of Wolbachia‐infected Ae. albopictus to uninfected specimens of Ae. aegypti. In our study, in the samples collected from New Mexico and Florida, no specimens of Ae. aegypti and Ae. albopictus were caught in same traps. In the samples collected from Texas, some specimens of Ae. aegypti and Ae. albopictus were caught in same traps, but no Ae. aegypti specimens were Wolbachia‐positive. So cross contamination was not a concern in our study.
A local Ae. aegypti strain LC was established from the pupae collected in the fall of 2018. The LC strain was Wolbachia‐positive. LC females were able to pass Wolbachia infection to their progeny when crossed with males of Wolbachia‐free Rockefeller strain (Figure 3), while the infected LC males did not produce Wolbachia‐infected offspring when crossed with Rockefeller females, demonstrating maternal transmission. Taken together, the molecular detection of Wolbachia in NM Ae. aegypti populations and establishment of Wolbachia‐positive colonies in both 2017 and 2018 indicated a persistent transmission of Wolbachia with the wild Ae. aegypti populations. Furthermore, the maternal transmission of Wolbachia to the progeny in the crosses of infected LC and uninfected Rockefeller strains ultimately provided satisfactory evidence that the presence of Wolbachia sequences in Ae. aegypti reflects authentic infection rather than environmental contamination or Wolbachia sequences derived from commensal or parasitic species, such as nematodes, within the mosquitoes.
Recently, Gloria‐Soria et al. (2018) reported screening for Wolbachia in 2,663 specimens of Ae. aegypti from 27 countries, including 60 specimens from Las Cruces, New Mexico, and no Wolbachia was detected in these samples. Their screen was conducted on DNA pools of up to 20 individuals. In our survey, we screened individual mosquitoes, and Wolbachia density in Ae. aegypti was substantially lower than in Ae. albopictus. Moreover, most positive specimens were identified by two rounds of PCR. We tested the sensitivity of our PCR assay using a DNA pool comprising 19 Wolbachia‐negative individuals and one positive individual for both Ae. aegypti and Ae. albopictus. Wolbachia could be detected in the pool containing DNA from the single positive Ae. albopictus specimen, but not in the pool containing DNA from the single positive Ae. aegypti specimen (data not shown). Wolbachia titer shows striking variation in infected individuals of Ae. albopictus (Ahantarig, Trinachartvanit, & Kittayapong, 2008; Calvitti, Marini, Desiderio, Puggioli, & Moretti, 2015). For example, in wild‐caught Ae. albopictus in North and Central Italy, 30.8%–50.0% of infected male Ae. albopictus had a low titer of wAlbA, which was not detectable by a standard PCR, but detectable by a quantitative PCR assay, while in the remaining males having higher Wolbachia densities it was detectable by a standard PCR (Calvitti et al., 2015). Similarly, Wolbachia load also varies substantially in Ae. aegypti into which the bacterium has been artificially transinfected (Ant, Herd, Geoghegan, Hoffmann, & Sinkins, 2018). The Wolbachia load was quite low in the Ae. aegypti samples in our study. Therefore, we hypothesize that assay sensitivity explains the different results between our study and that of Gloria‐Soria et al. (2018).
The conspicuous variation in the prevalence of Wolbachia infection in different populations of Ae. aegypti among the three states and eight cities within New Mexico raises several compelling questions. Chief among them are “what factors contribute to the low density of Wolbachia in Ae. aegypti relative to Ae. albopictus?” and “what makes the New Mexico populations more susceptible/hospitable to Wolbachia than other populations?”
Wolbachia density variation is common in natural populations of different insect hosts (Ahantarig et al., 2008; Unckless, Boelio, Herren, & Jaenike, 2009); however, it is not clear whether the variation is driven by genetic or environmental variation or both. A recent study revealed an amplification of a genome region that harbors a cluster of eight genes, called Octomom, which is responsible for the over‐replication and virulence of wMelPop in Drosophila melanogaster. The copy number of Octomom correlates with Wolbachia titers (Chrostek & Teixeira, 2015). Environmental factors such as temperature may play a role as well. A recent study in D. melanogaster demonstrated that Wolbachia abundance was higher when host flies developed at lower temperature (13°C and 23°C compared with 31°C) (Moghadam et al., 2018). Additionally, mosquito genetic background may affect Wolbachia prevalence. It has been shown that Ae. aegypti populations from Las Cruces, New Mexico, Houston, Texas, and four locations of Florida are genetically distinct (Pless et al., 2017). Further studies are needed to tease out the roles of these potential drivers of Wolbachia presence and abundance in Ae. aegypti.
Another critical question raised by our study is how Ae. aegypti acquires a strain of Wolbachia associated with Ae. albopictus and how this strain impacts Ae. aegypti life history and pathogen susceptibility. Ae. aegypti can be artificially transinfected with different Wolbachia strains, and the artificial infection can be introduced into natural populations and spread in nature (Frentiu et al., 2014; Hoffmann et al., 2011; Schmidt et al., 2017). The wAlbB has been successfully introduced into Ae. aegypti to form a line with inherited infection (Xi, Khoo, & Dobson, 2005). Interestingly, the Toll and IMD pathways favor establishment and maintenance of wAlbB infection in the line; the knockdown of Toll and IMD by RNA interference reduces the wAlbB load, while the transgenic activation of Toll and IMD increases the load (Pan et al., 2018). It appears that transinfected Wolbachia can exploit host immunity for a symbiotic association. Our survey revealed the prevalence of wAlbB in Ae. aegypti natural populations in New Mexico, and an infected colony has been established from wild‐collected mosquitoes. This provides an opportunity to study the natural Ae. aegypti ‐Wolbachia association and its impact on various mosquito life traits, such as reproductive manipulation as well as interference with viral transmission.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
AUTHORS' CONTRIBUTIONS
JX conceived the study design. KJLM, DD, KH, and MD collected mosquitoes. AK, WY, JJ, CS, and AKK conducted assays and data analysis. JX, AK, MB, and AKK drafted the manuscript. KAH, DD, MD, MB, IAH, and RX critically reviewed the manuscript. All authors read and approved the manuscript.
DATA ACCESSIBILITY
DNA sequences were deposited in GenBank under accession number MH732668‐MH732670, MH734116‐MH734121.
Supporting information
ACKNOWLEDGMENTS
We thank the participants of SouthWest Aedes Research and Mapping project (SWARM); Jennifer Corrado at Anastasia Mosquito Control District in St. Augustine, FL; and Elaine Chu at Harrison County Public Health, Mosquito and Vector Control Division in Houston, TX for their contributions to the mosquito sampling. This work was supported by New Mexico Department of Health contract to Kathryn A. Hanley, Michaela Buenemann, Immo A. Hansen, and Jiannong Xu. Concepcion Sanchez was an undergraduate research scholar supported by the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103451. The funder had no roles in the study design, data collection and analysis, or decision to publish. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the Department of Health of New Mexico.
Kulkarni A, Yu W, Jiang J, et al. Wolbachia pipientis occurs in Aedes aegypti populations in New Mexico and Florida, USA. Ecol Evol. 2019;9:6148–6156. 10.1002/ece3.5198
Data Availability Statement: DNA sequences were deposited in GenBank under accession number MH732668‐MH732670, MH734116‐MH734121.
REFERENCES
- Ahantarig, A. , Trinachartvanit, W. , & Kittayapong, P. (2008). Relative Wolbachia density of field‐collected Aedes albopictus mosquitoes in Thailand. Journal of Vector Ecology, 33(1), 173–177. 10.3376/1081-1710(2008)33[173:RWDOFA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ant, T. H. , Herd, C. S. , Geoghegan, V. , Hoffmann, A. A. , & Sinkins, S. P. (2018). The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti . PLoS Path, 14(1), e1006815 10.1371/journal.ppat.1006815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo, L. , Dunning Hotopp, J. C. , Jolley, K. A. , Bordenstein, S. R. , Biber, S. A. , Choudhury, R. R. , … Werren, J. H. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis . Applied and Environment Microbiology, 72(11), 7098–7110. 10.1128/AEM.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra, S. , Riedel T. E., Saldaña, M. A. , Hegde, S. , Pederson, N. , Hughes, G. L. , & Ellington, A. D. (2018) Direct nucleic acid analysis of mosquitoes for high fidelity species identification and detection of Wolbachia using a cellphone. PLOS Neglected Tropical Diseases, 12(8), e0006671 10.1371/journal.pntd.0006671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, G. , Xu, Y. , Lu, P. , Xie, Y. , & Xi, Z. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti . PLoS Path, 6(4), e1000833 10.1371/journal.ppat.1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagrove, M. S. , Arias‐Goeta, C. , Failloux, A. B. , & Sinkins, S. P. (2012). Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus . Proceedings of the National Academy of Sciences of the United States of America, 109(1), 255–260. 10.1073/pnas.1112021108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J. J. , & Turelli, M. (2013). Wolbachia versus dengue: Evolutionary forecasts. Evolution, Medicine, and Public Health, 2013(1), 197–207. 10.1093/emph/eot018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvitti, M. , Marini, F. , Desiderio, A. , Puggioli, A. , & Moretti, R. (2015). Wolbachia density and cytoplasmic incompatibility in Aedes albopictus: Concerns with using artificial Wolbachia infection as a vector suppression tool. PLoS ONE, 10(3), e0121813 10.1371/journal.pone.0121813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek, E. , & Teixeira, L. (2015). Mutualism breakdown by amplification of Wolbachia genes. PLoS Biology, 13(2), e1002065 10.1371/journal.pbio.1002065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon, K. L. , Brown, M. R. , & Strand, M. R. (2016). Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Molecular Ecology, 25(22), 5806–5826. 10.1111/mec.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque, A. L. , Magalhaes, T. , & Ayres, C. F. (2011). High prevalence and lack of diversity of Wolbachia pipientis in Aedes albopictus populations from Northeast Brazil. Memorias do Instituto Oswaldo Cruz, 106(6), 773–776. 10.1590/S0074-02762011000600021 [DOI] [PubMed] [Google Scholar]
- Flores, H. A. , & O'Neill, S. L. (2018). Controlling vector‐borne diseases by releasing modified mosquitoes. Nature Reviews Microbiology, 10.1038/s41579-018-0025-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu, F. D. , Zakir, T. , Walker, T. , Popovici, J. , Pyke, A. T. , van den Hurk, A. , … O'Neill, S. L. (2014). Limited dengue virus replication in field‐collected Aedes aegypti mosquitoes infected with Wolbachia . PLoS Neglected Tropical Diseases, 8(2), e2688 10.1371/journal.pntd.0002688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloria‐Soria, A. , Chiodo, T. G. , & Powell, J. R. (2018). Lack of evidence for natural Wolbachia infections in Aedes aegypti (Diptera: Culicidae). Journal of Medical Entomology, 10.1093/jme/tjy084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves Dda, S. , Cassimiro, A. P. , de Oliveira, C. D. , Rodrigues, N. B. , & Moreira, L. A. (2014). Wolbachia detection in insects through LAMP: Loop mediated isothermal amplification. Parasit Vectors, 7, 228 10.1186/1756-3305-7-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. B. , Eisen, L. , McAllister, J. , Savage, H. M. , Mutebi, J. P. , & Eisen, R. J. (2017). Updated reported distribution of Aedes (Stegomyia) aegypti and Aedes (Stegomyia) albopictus (Diptera: Culicidae) in the United States, 1995–2016. Journal of Medical Entomology, 54(5), 1420–1424. 10.1093/jme/tjx088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. B. , Eisen, R. J. , Eisen, L. , Boegler, K. A. , Moore, C. G. , McAllister, J. , … Mutebi, J. P. (2016). Reported distribution of Aedes (Stegomyia) aegypti and Aedes (Stegomyia) albopictus in the United States, 1995–2016 (Diptera: Culicidae). Journal of Medical Entomology, 53(5), 1169–1175. 10.1093/jme/tjw072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde, S. , Khanipov, K. , Albayrak, L. , Golovko, G. , Pimenova, M. , Saldaña, M. A. , … Hughes, G. L. (2018). Microbiome interaction networks and community structure from laboratory‐reared and field‐collected Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquito vectors. Frontiers in Microbiology, 9(2160), 10.3389/fmicb.2018.02160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig, M. (1936). The rickettsia, Wolbachia pipientis (gen. et sp.n.) and associated inclusions of the mosquito, Culex pipiens . Parasitology, 28(4), 453–486. [Google Scholar]
- Hertig, M. , & Wolbach, S. B. (1924). Studies on rickettsia‐like micro‐organisms in insects. Journal of Medical Research, 44(3), 329–374 327. [PMC free article] [PubMed] [Google Scholar]
- Higa, Y. , Toma, T. , Tsuda, Y. , & Miyagi, I. (2010). A multiplex PCR‐based molecular identification of five morphologically related, medically important subgenus Stegomyia mosquitoes from the genus Aedes (Diptera: Culicidae) found in the Ryukyu Archipelago, Japan. Japanese Journal of Infectious Diseases, 63(5), 312–316. [PubMed] [Google Scholar]
- Hilgenboecker, K. , Hammerstein, P. , Schlattmann, P. , Telschow, A. , & Werren, J. H. (2008). How many species are infected with Wolbachia? – A statistical analysis of current data. FEMS Microbiology Letters, 281(2), 215–220. 10.1111/j.1574-6968.2008.01110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A. A. , Montgomery, B. L. , Popovici, J. , Iturbe‐Ormaetxe, I. , Johnson, P. H. , Muzzi, F. , … O'Neill, S. L. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature, 476(7361), 454–457. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A. , Ross, P. A. , & Rasic, G. (2015). Wolbachia strains for disease control: Ecological and evolutionary considerations. Evolutionary Applications, 8(8), 751–768. 10.1111/eva.12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanne, S. , Vythilingam, I. , Yugavathy, N. , Leong, C. S. , Wong, M. L. , & AbuBakar, S. (2015). Distribution and dynamics of Wolbachia infection in Malaysian Aedes albopictus . Acta Tropica, 148, 38–45. 10.1016/j.actatropica.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Kitrayapong, P. , Baimai, V. , & O'Neill, S. L. (2002). Field prevalence of Wolbachia in the mosquito vector Aedes albopictus . American Journal of Tropical Medicine and Hygiene, 66(1), 108–111. 10.4269/ajtmh.2002.66.108 [DOI] [PubMed] [Google Scholar]
- Kittayapong, P. , Baisley, K. J. , Baimai, V. , & O'Neill, S. L. (2000). Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). Journal of Medical Entomology, 37(3), 340–345. [DOI] [PubMed] [Google Scholar]
- Mavingui, P. , Valiente Moro, C. , Tran‐Van, V. , Wisniewski‐Dye, F. , Raquin, V. , Minard, G. , … Gonzalez, V. (2012). Whole‐genome sequence of Wolbachia strain wAlbB, an endosymbiont of tiger mosquito vector Aedes albopictus . Journal of Bacteriology, 194(7), 1840 10.1128/JB.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman, C. J. , Lane, R. V. , Cass, B. N. , Fong, A. W. , Sidhu, M. , Wang, Y. F. , & O'Neill, S. L. (2009). Stable introduction of a life‐shortening Wolbachia infection into the mosquito Aedes aegypti . Science, 323(5910), 141–144. 10.1126/science.1165326 [DOI] [PubMed] [Google Scholar]
- Moghadam, N. N. , Thorshauge, P. M. , Kristensen, T. N. , de Jonge, N. , Bahrndorff, S. , Kjeldal, H. , & Nielsen, J. L. (2018). Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly (Austin), 12(1), 1–12. 10.1080/19336934.2017.1394558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, L. A. , Iturbe‐Ormaetxe, I. , Jeffery, J. A. , Lu, G. , Pyke, A. T. , Hedges, L. M. , … O'Neill, S. L. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium . Cell, 139(7), 1268–1278. 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- Notomi, T. , Okayama, H. , Masubuchi, H. , Yonekawa, T. , Watanabe, K. , Amino, N. , & Hase, T. (2000). Loop‐mediated isothermal amplification of DNA. Nucleic Acids Research, 28(12), E63 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, S. L. (2018). The use of Wolbachia by the world mosquito program to interrupt transmission of Aedes aegypti transmitted viruses. Advances in Experimental Medicine and Biology, 1062, 355–360. 10.1007/978-981-10-8727-1_24 [DOI] [PubMed] [Google Scholar]
- Pan, X. , Pike, A. , Joshi, D. , Bian, G. , McFadden, M. J. , Lu, P. , … Xi, Z. (2018). The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti . ISME Journal, 12(1), 277–288. 10.1038/ismej.2017.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless, E. , Gloria‐Soria, A. , Evans, B. R. , Kramer, V. , Bolling, B. G. , Tabachnick, W. J. , & Powell, J. R. (2017). Multiple introductions of the dengue vector, Aedes aegypti, into California. PLoS Neglected Tropical Diseases, 11(8), e0005718 10.1371/journal.pntd.0005718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T. L. , Barton, N. H. , Rašić, G. , Turley, A. P. , Montgomery, B. L. , Iturbe‐Ormaetxe, I. , … Turelli, M. (2017). Local introduction and heterogeneous spatial spread of dengue‐suppressing Wolbachia through an urban population of Aedes aegypti . PLoS Biology, 15(5), e2001894 10.1371/journal.pbio.2001894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongsripong, P. , Chandler, J. A. , Green, A. B. , Kittayapong, P. , Wilcox, B. A. , Kapan, D. D. , & Bennett, S. N. (2017). Mosquito vector‐associated microbiota: Metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod‐borne diseases. Ecology and Evolution, 8(2), 1352–1368. 10.1002/ece3.3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless, R. L. , Boelio, L. M. , Herren, J. K. , & Jaenike, J. (2009). Wolbachia as populations within individual insects: Causes and consequences of density variation in natural populations. Proceedings of the Royal Society B: Biological Sciences, 276(1668), 2805–2811. 10.1098/rspb.2009.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, T. , Johnson, P. H. , Moreira, L. A. , Iturbe‐Ormaetxe, I. , Frentiu, F. D. , McMeniman, C. J. , … Hoffmann, A. A. (2011). The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature, 476(7361), 450–453. 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Gilbreath, T. M. 3rd , Kukutla, P. , Yan, G. , & Xu, J. (2011). Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE, 6(9), e24767 10.1371/journal.pone.0024767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren, J. H. , Baldo, L. , & Clark, M. E. (2008). Wolbachia: Master manipulators of invertebrate biology. Nature Reviews Microbiology, 6(10), 741–751. 10.1038/nrmicro1969 [DOI] [PubMed] [Google Scholar]
- Xi, Z. , Khoo, C. C. , & Dobson, S. L. (2005). Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science, 310(5746), 326–328. 10.1126/science.1117607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences were deposited in GenBank under accession number MH732668‐MH732670, MH734116‐MH734121.


