Unstructured abstract
Adoptive immunotherapy with engineered T cells is at the forefront of cancer treatment. T cells can be engineered to express T cell receptors (TCR) specific for tumor-associated antigens (TAAs) derived from intracellular or cell surface proteins. T cells engineered with TCRs (TCR-T) allow for targeting diverse types of TAAs, including proteins overexpressed in malignant cells, those with lineage-restricted expression, cancer-testis antigens, and neoantigens created from abnormal, malignancy-restricted proteins. Minor histocompatibility antigens can also serve as TAAs for TCR-T to treat relapsed hematologic malignancies after allogeneic hematopoietic cell transplantation. Moreover, TCR constructs can be modified to improve safety and enhance function and persistence of TCR-T. TCR-T therapies targeting three different TAAs are in early phase clinical trials for treatment of hematologic malignancies. Preclinical studies of TCR-T specific for many other TAAs are underway and offer great promise as safe and effective therapies for a wide range of cancers.
Keywords: T cell immunotherapy, engineered T cell receptor, hematologic malignancies, tumor-associated antigen
Introduction
The importance of T cells in cancer immunity is well-established. T cells recognize antigens that are short peptides derived from intracellular or cell surface proteins, presented in complex with major histocompatibility complex (MHC) molecules, also referred to as human leukocyte antigens (HLA) on human cells. Spontaneous T cell responses to a variety of cancer antigens have been observed, including in early murine models where exposure to viable or lethally irradiated tumor induced protective immunity against subsequent tumor exposure.1 Endogenous T cell responses also occur in patients, but are often incompletely effective against advanced malignancies. Multiple mechanisms mediate tumor escape and/or block the formation of efficacious anti-tumor immune responses. These mechanisms include induction of a metabolically hostile tumor microenvironment, recruitment of suppressor cells, such as macrophages, regulatory T cells and myeloid-derived suppressor cells, production of immunosuppressive cytokines, expression of T cell inhibitory ligands by tumor or associated cells, and deletion of antigen-specific T cells.2–4 Acute myeloid leukemia (AML), for example, produces an immunologically hostile environment5 through multiple mechanisms such as overexpressing indoleamine 2,3 dioxygenase 16 and secreting arginase II7 to metabolically suppress T cells, overexpressing the PD-L1 T cell-inhibitory molecule,8 and blocking transcription factor activities needed for T cell activation and proliferation.9 Immunosuppressive microenvironments have also been observed in other hematologic malignancies, including chronic myeloid leukemia (CML)10 and chronic lymphocytic leukemia (CLL).11 Consequently, T cells that could mediate anti-tumor immune responses become quantitatively or qualitatively defective. Adoptive transfer of T cells with tumor specificity is one approach to overcoming such deficiencies in endogenous anti-tumor immunity.
Adoptive T cell therapy for cancer treatment
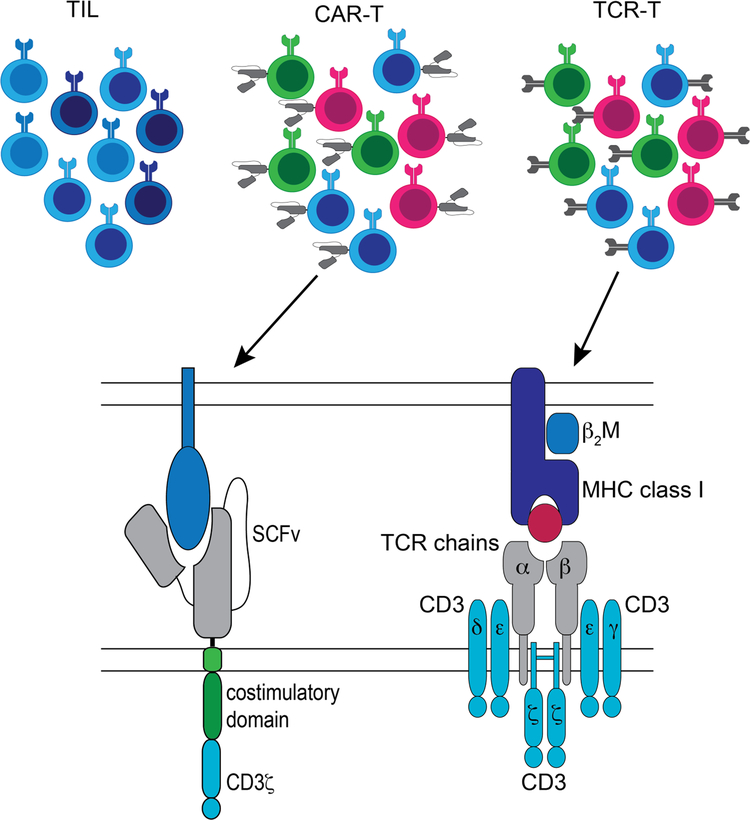
To take advantage of naturally occurring T cell responses, one can isolate tumor-infiltrating lymphocytes (TIL) from tumor, activate and expand the T cells ex vivo, and re-infuse the resulting TIL product,12–15 producing clinical responses in some patients. The technology is limited primarily to solid tumors with surgically accessible lesions, although marrow-infiltrating lymphocytes (MIL) are being tested as a therapeutic for hematologic malignancies.16,17 However, neither TIL nor MIL can be generated for all patients.18,19 Moreover, the specificities of infused TIL/MIL are generally undefined, and a minority of the T cells are tumor-reactive.20 In contrast, T cells can be modified to express cell surface receptors that confer specific recognition of a malignant cell target, allowing administration of a T cell product with a defined specificity and composition.21,22 Chimeric antigen receptors (CARs) are artificial antigen-specific receptors consisting of an extracellular ligand-binding domain linked to a CD3ζ chain along with one or more costimulatory domains23,24 that can be transferred into T cells to produce an engineered T cell (CAR-T) with defined specificity for a cell surface molecule. Alternatively, T cells can be engineered to express a transgenic T cell receptor (TCR-T) specific for a tumor-associated antigen (TAA) formed from the complex of a tumor peptide and an MHC molecule (pMHC). Natural TCRs, from which transgenic TCRs are derived, consist of heterodimers of alpha and beta chains expressed in complex with CD3 proteins (Figure 1), and associate with CD4 or CD8 co-receptors during pMHC engagement and often with costimulatory molecules, such as CD28.
Figure 1.
Types of adoptive T cell therapy
Both CAR-T and TCR-T are adoptive cellular therapies with a defined, pre-selected target; both are often employed with a defined cell composition and can be generated for most patients in which the appropriate target is expressed. Each approach has advantages and disadvantages (Table 1). The major advantage for TIL/MIL and TCR-T over CAR-T is the ability to target peptides derived from intracellular proteins or cell surface proteins. Not all malignancies will have a suitable target for CAR-T therapy; that is, a surface molecule with high expression on tumor but low or absent expression on normal tissue, unless the normal tissue is dispensable. In contrast, TCR-T can allow access to the intracellular malignant proteome. CAR-T, however, are MHC/HLA-independent and therefore can be used in patients of all HLA types. CARs are expressed at higher surface levels than TCRs, but have less efficient signaling kinetics that do not properly recapitulate physiological TCR signaling leading to lower sensitivity to antigen.25,26 Harris and colleagues engineered two TCRs with high affinity for two distinct TAAs and compared the function of the TCRs either as conventional αβ TCRs or as the ligand-binding domain of CAR constructs. Although the CAR constructs had similar binding affinity to αβ TCRs, primary T cells expressing the TCRs secreted cytokine in response to 100-fold lower peptide concentrations than T cells expressing the equivalent CAR.25 Additionally, at high antigen density, CAR-T mediate greater maximal release of some cytokines, such as interleukin (IL)-2 and IL-6,25 which can initiate a cascade leading to toxic cytokine release syndrome (CRS). TCR-Ts harness an endogenous T cell response that has been fine-tuned by the evolution of the immune system and are rarely associated with clinically significant CRS.
Table 1.
Approaches to adoptive T cell immunotherapies
| Advantages | Disadvantages | |
|---|---|---|
| TIL | Polyclonal (reduce potential for escape through antigen loss); targets from intracellular and cell surface proteins | T cell specificity generally unknown; cannot be generated for all patients; not well established for hematologic malignancies; patient specific |
| TCR-T | Targets derived from intracellular and cell surface proteins; defined specificity | HLA restriction; tumor escape through antigen loss (altered processing of peptides, HLA downregulation) |
| CAR-T | No HLA-restriction; defined specificity | Cell surface targets only; tumor escape through antigen loss (downregulation or loss of target protein) |
Antigen targets of T cell receptors for immunotherapy
Antigen processing and presentation
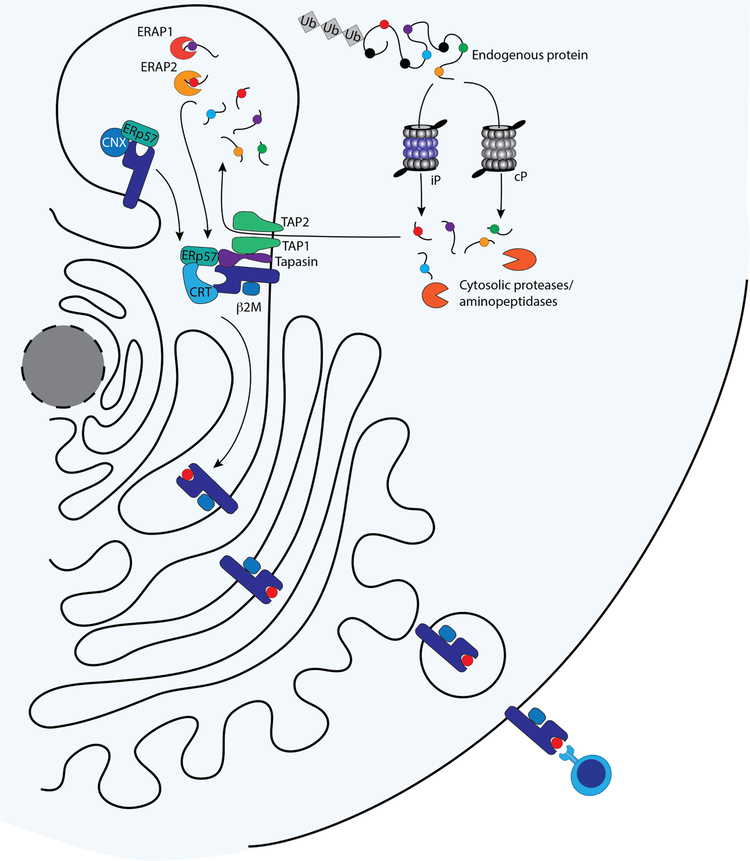
T cell antigens are generated from full-length proteins through a multi-step intracellular process (Figure 2) that has been extensively reviewed.27–29 CD8+ T cell antigens are primarily derived from endogenous proteins that are degraded in the cytoplasm by proteasomes and cytosolic aminopeptidases. The resulting peptides are transported into the endoplasmic reticulum (ER) through the transporter associated with antigen processing (TAP) complex, where they are trimmed by ER aminopeptidases (ERAPs) into lengths of 8–10 amino acids. While we have a detailed understanding of how processing occurs, the exact rules of trimming and processing are not well understood. Trimmed peptides are assembled with MHC class I heavy chain and beta-2 microglobulin via a peptide-loading complex. The pMHC class I complex then transits from the ER to the plasma membrane, where it is presented externally. CD8+ T cell antigens can also be generated from viral proteins in infected cells and from misfolded or improperly synthesized proteins. Additionally, certain antigen-presenting cells can present peptides from internalized exogenous proteins on MHC class I molecules through cross-presentation. TAA are potentially present on all malignant cells, regardless of the tissue of origin, because MHC class I molecules are ubiquitously expressed on all nucleated cells. However, downregulation and loss of class I MHC expression are established mechanisms of immune evasion by tumors.30–38 CD8+ T cells are directly cytotoxic, making MHC class I restricted TAA particularly attractive as immunotherapy targets.
Figure 2.
Antigen processing and presentation
In contrast, CD4+ T cells recognize peptides presented on MHC class II molecules. Peptides are loaded onto MHC Class II after degradation of internalized exogenous proteins or autophagy of endogenous proteins.39 Class II molecules are normally expressed on antigen-presenting and hematopoietic cells, including AML. This review will focus on class I-restricted CD8+ T cell antigens, since these are the majority of targets in preclinical and clinical development for TCR-T therapies for hematologic malignancies. However, class II restricted antigens also appear to have a role in antitumor immunity. Downregulation of MHC class II expression40 or complete loss of a mismatched MHC haplotype41 can occur in leukemic relapses after allogeneic hematopoietic stem cell transplantation (allo-HCT), suggesting that class II antigens contribute to disease control after allo-HCT. Upregulation of class II expression been described for some solid tumors,42–45 and, in melanoma, is associated with increased probability of response to immune checkpoint blockade therapy.46 Adoptive transfer of TIL enriched for CD4+ T cells specific for a mutation in erbb2-interacting protein produced striking clinical responses in one patient with metastatic cholangiocarcinoma.47 As we learn more about class II-restricted TAAs, these too may prove to be relevant targets in hematologic malignancies.
Classes of T cell antigens for TCR-based immunotherapy
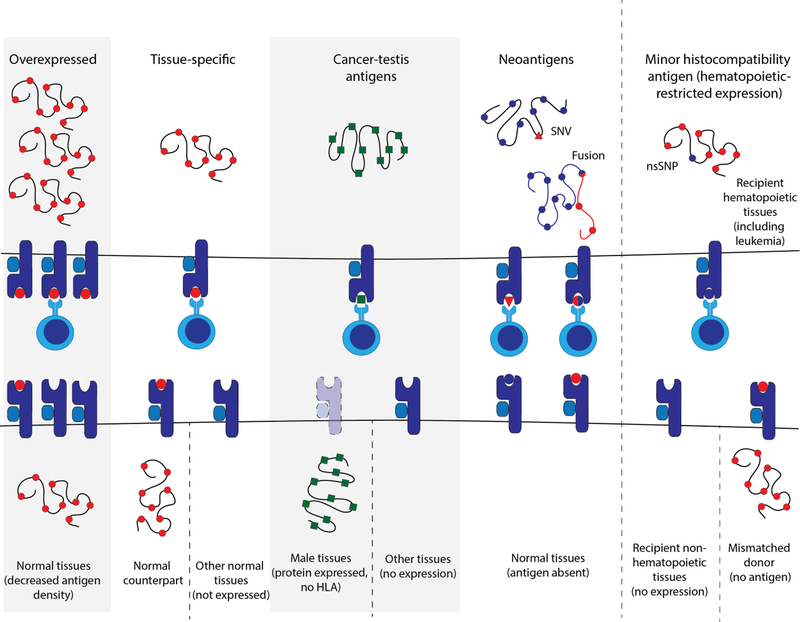
An ideal target for any cell-based immunotherapy is one that is selectively presented on malignant cells, necessary for survival of the tumor cells, and shared amongst patients. Four major TAA classes have been considered as targets for TCR-T immunotherapy in solid and liquid tumors: 1) overexpressed antigens derived from wild-type proteins with relatively high expression in malignant cells; 2) lineage-restricted antigens that are also presented on the normal counterparts of the malignant cells; 3) cancer-testis antigens (CTA), which are normally expressed in germline tissues and aberrantly expressed in malignant cells; and 4) neoantigens created from abnormal proteins (mutations, fusions, frameshifts, or novel isoforms) or abnormal peptides specific to the malignant cells. Antigen specificity for malignant versus healthy cells varies among the categories of TAAs (Table 2), and the potential risk of on-target, off-tumor toxicity inversely corresponds to that specificity. Specific examples of TAAs in each category are shown in Table 3.
Table 2.
Categories of TAAs for the development of TCR-T immunotherapy
| Overexpressed | Lineage-restricted | Cancer-testis antigens | Neoantigens | Minor histocompatibility (H) antigens | |
|---|---|---|---|---|---|
| Antigen derived from | Wild-type protein with relatively increased expression in malignant cells | Wild-type protein expressed in malignant cells and normal counterparts | Wild-type protein expressed only in malignant cells and germline cells | Abnormal protein created by cancer-specific mutation, gene fusion, frame shift or abnormal splicing, or peptide created by abnormal antigen processing | Polymorphic normal peptides created by polymorphisms that differ between donor and recipient in allogeneic hematopoietic cell transplantation (HCT) |
| Specificity for malignant cells | Lowest | Low | Moderate | Highest | Hematopoietic-resetricted minor H antigens become leukemia-specific following allo-HCT |
| Potential for on-target off tumor toxicity | High | Moderate/high (may be acceptable if normal counterpart is dispensable) | Low (Testis/germline lissues lack HLA) |
Lowest | Variable |
| Breadth of potential applicability | High | High | Moderate/high | Low | Moderate/high (only for relapses after allo-HCT) |
Table 3.
Examples of TAA in hematologic malignancies and solid tumors
| Category of TAA | Cancer types | Examples | Diseases | Other tissues | References |
|---|---|---|---|---|---|
| Overexpressed | Hematologic | WT1 | AML, MDS | Kidney (podocytes), CD34+ cells | 91 |
| Survivin | AML, ALL, MDS | None in adult tissues | 3 | ||
| hTERT | ALL, AML, CLL | Ovaries, testis | 113–115 | ||
| BOB1 | B cell leukemias and lymphomas, multiple myeloma | Normal B cells | 121 | ||
| Lineage-restricted | Hematologic | CD20 | Lymphoma | Normal B cells | 122 |
| CD22 | ALL | 123 | |||
| Solid tumor | MART-1/MelanA | Melanoma | Normal melanocytes | 141,142 | |
| Tyrosinase | Reviewed in 143 | ||||
| gp100 | |||||
| Cancer-testis antigens | Hematologic | PRAME | AML, ALL, MDS, multiple solid tumors | Adrenals, ovaries, endometrium, testis | 144,102 |
| Aurora kinase A | AML, CML, ATL | Testis | 125,126 127 | ||
| Solid tumor | MAGE family | Melanoma, lung, breast, colorectal, prostate | Testis, brain (MAGEA12) | Reviewed in 143 | |
| Neoantigens | Hematologic | NPM1 | AML | None | 130 |
| CBFB-MYH11 | 129 | ||||
| Leukemia-associated minor histocompatibility (H) antigens | Hematologic | HA-1 | Relapsed hematologic malignancies after allo-HCT | Hematopoietic | 69 |
In hematologic malignancies, minor histocompatibility (H) antigens are a fifth important class of TAAs. Minor H antigens are MHC/HLA-bound polymorphic peptides that differ between allo-HCT recipient and donor as a result of genetic polymorphisms. Once full donor normal hematopoietic chimerism is achieved after allo-HCT, hematopoietic-restricted minor H antigens are present only on residual recipient malignant hematopoietic cells, providing significant specificity when donor and recipient are mismatched for the polymorphism. Like lineage-restricted antigens, the specificity of minor H antigens for malignant cells depends on how tightly restricted expression of the parent protein is to hematopoietic cells. Therapies targeting minor H antigens could potentially treat multiple hematologic malignancies, since the antigens are not disease specific. However, varying polymorphism frequencies across populations and the requirement for donor-recipient mismatch currently limits the applicability of minor H-directed immunotherapy to a subset of individuals who relapse after allo-HCT.
General considerations in developing TCR immunotherapy
Transgenic TCR development begins by discovering and cloning a naturally occurring TCR specific for a suitable target. Generally, there are three starting pools of cells in which to identify a relevant TCR: patient TIL/MILs, patient peripheral blood T cells, and healthy donor peripheral blood T cells requiring primary in vitro stimulation.48,49 TIL/MILs can be used as a source of T cells potentially enriched for TAA-specific TCRs, but T cells derived from immunosuppressive tumor environments are often dysfunctional.50 Stimulating healthy donor T cells with known or predicted novel TAAs in vitro to isolate reactive T cells and their TCRs can circumvent T cell dysfunction.
To determine reactivity to a specific target, binding and functionality are tested by measuring pMHC multimer binding, cytotoxicity, and/or cytokine production by ELISPOT or flow cytometry. Newer methods like barcoded dextramer staining51 allow screening hundreds of pMHC complexes from one sample. Once responding T cells are found, the TCR sequence must be determined and cloned. Rapid amplification of cDNA ends before polymerase chain reaction (RACE-PCR) allows identification of the TCR α and β chains of a reactive T cell clone. Contemporary approaches like single-cell sequencing can directly identify the TCR sequences of individual clones in a bulk population. The Wu group paired single-cell sequencing with a library of cloning plasmids for each TCRα/TCRβ chain to reconstruct TCRs from a bulk population and rapidly deconvolute TCR specificity and avidity for a target antigen,52 allowing TCRs to be screened against relevant targets at an accelerated pace compared to traditional methods.
After sequencing a reactive TCR, the receptor must be transferred to a new T cell. Transduction with a viral vector encoding a polycistronic construct of both α and β chains of the transgenic TCR separated by a ribosomal skip motif, such as a 2A self-cleaving peptide, is most commonly used. Nonviral techniques for TCR gene transfer, such as transposon-based technologies53–55 and nanoparticles,56 are also in development. In any case, conventional T cells that are genetically modified with a transgenic TCR have their own endogenous TCRs, making mispairing of the transferred and endogenous TCRs a concern. In preclinical studies using an OT-I TCR murine model, mispairing of the endogenous TCR with the transferred TCR led to lethal graft-versus-host disease (GVHD),57 although GVHD due to TCR mispairing has not been observed in human trials to date. The risk of mispairing can be mitigated by introducing cysteine modifications to the transferred TCR α and β chains to favor pairing of the introduced chains to each other. Other potential modifications to the TCR construct include codon optimization and minimal murinization of the constant region58 to enhance expression of the transgenic TCR and encourage correct pairing of the introduced chains. Alternatively, using small interfering RNA (siRNA) and CRISPR/Cas9 to disrupt the endogenous TCR has been shown to limit toxicity and increase T cell activity in humanized mouse models.59 While eliminating the endogenous TCR prevents mispairing, it is unclear whether retaining an endogenous TCR might help transferred T cells persist when antigen burden is low. However, it is clear that designing transgenic TCRs for exclusive self-pairing is paramount to limiting autoimmunity and toxicity.
Many native TCRs recognize overexpressed self-antigens with inherently low affinity due to tolerance mechanisms. In vitro affinity maturation can enhance target recognition,60–62 although with increased risk of cross-reactivity and off-target toxicity. In a clinical trial of an affinity-matured MAGE-A3-specific TCR, transgenic T cells also recognized the Titin cardiac protein and led to the death of two patients.63 The native, non-matured TCR did not recognize cardiac tissue,64 highlighting the relationship between augmented affinity and potential cross-reactivity. Which complementary-determining regions (CDRs) are altered in affinity maturation may influence the risk of cross-reactivity and toxicity. CDR1 and CDR2 interact predominantly with MHC; alterations in these regions can increase TCR binding of MHC regardless of peptide.65,66 In contrast, CDR3 interacts predominantly with peptide,67 so affinity maturation targeting this region might produce higher affinity TCRs with less likelihood of cross-reactivity, although experimental data is limited. A recent approach to developing TCRs with highly diverse CDR3s used in vitro antigen-driven differentiation of progenitor T cells to generate higher affinity TCRs without changes to CDR1/2.68 Safety testing of TCRs generated in this manner may lead to better understanding of the relationship between cross-reactivity and affinity.
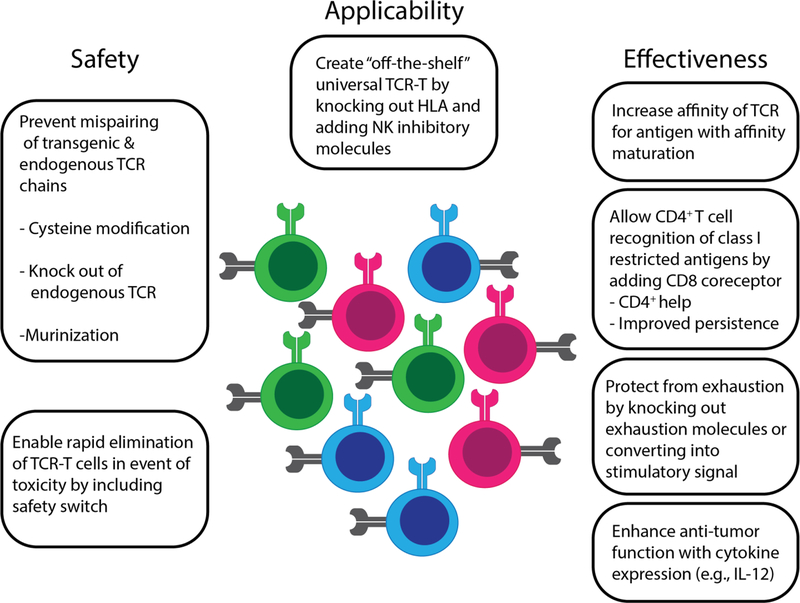
As our understanding of natural and synthetic T cell biology increases, investigators are testing numerous modifications that might improve the functionality, persistence, and safety of transferred T cells (Figure 4). Introducing a CD8 co-receptor can facilitate recognition of class I antigens by CD4+ T cells, and enhance ‘help’ for cytotoxic CD8+ T cells.69–74 Safety switches, including inducible pro-death proteins, can be used to rapidly remove transferred T cells if toxicities such as CRS or GVHD become an issue.75–80 The ability to secrete IL-12 and other cytokines can increase activity of so-called ‘armored’ CAR-T or TCR-T cells and create a proinflammatory environment that enables antigen presentation,81–83 albeit with an increased potential for toxicity if cytokine secretion is not tightly regulated.84 Function of transferred T cells can also be increased by siRNA or CRISPR-Cas9 elimination of inhibitory molecule genes, such as PD-1,85,86 or by converting negative malignant cell-derived signals into activation signals for engineered T cells, for example by fusing the inhibitory receptor CD200R to a costimulatory CD28 domain.87 Normal signaling pathways, such as thrombopoietin and c-MPL,88 can also be co-opted to deliver an activating signal to engineered T cells through the transgene construct. ‘Off-the-shelf’ versions of engineered cells that are HLA negative and express natural killer (NK) cell inhibitory molecules could be used as universal donor cells,89 potentially eliminating the need for autologous cell collection from patients, although silencing the endogenous TCR will likely be required in this situation to prevent GVHD.
Figure 4.
Construct modifications to enhance TCR-T cells
TCR immunotherapy in hematologic malignancies
A recent systematic review estimated that only 16% of previous and current TCR-T clinical trials were aimed at treating hematologic malignancies.90 However, numerous preclinical studies are presently underway. Suitable TCR targets must be highly expressed on malignant cells, including blasts, progenitor and cancer stem cells, and have little or ideally no expression on healthy non-hematopoietic tissues. However, expression of the target antigen on normal hematopoietic cells may be acceptable in select circumstances; for example, if: 1) expression is sufficiently low that high-avidity T cells will not recognize normal cells; 2) the normal cells are relatively dispensable, as in the case of normal B cells; 3) the normal cells do not express HLA molecules (e.g. testis); or, 4) TCR immunotherapy expected to cause myelolablation will be used prior to allo-HCT and the TCR-T cells are designed to be short-lived. Because the field is still evolving and the optimal antigens for TCR immunotherapy are as-yet unknown, target candidates from all TAA categories are being explored (Table 3).
Clinical development of TCR immunotherapies
WT1 TCR immunotherapy
Wilm’s tumor 1 (WT1) protein is overexpressed in acute leukemias and myelodysplastic syndromes (MDS)91 and has limited expression on normal tissues, including normal CD34+ hematopoietic stem cells. T cells specific for WT1 epitopes that are presented on HLA-A*24:0292 or -A*02:0193,94 recognize primary leukemic blasts, making WT1 an attractive therapeutic target. Donor-derived T cell responses to WT1 have been observed in vivo after allo-HCT.94 WT1-specific CD8+ T cell responses can be stimulated ex vivo, and retain their activity in vivo.95,96 The Chapuis and Greenberg group adoptively transferred ex vivo expanded, donor-derived CD8+ T cells specific for an HLA-A*02:01-restricted WT1 epitope in a cohort of eleven patients with relapsed or high-risk acute leukemia or MDS after allo-HCT (NCT00052520).97 Transferred T cells were well tolerated, indicating minimal on-target off-tumor toxicity, persisted and showed direct (disease response) or indirect (absence of relapse) anti-leukemic activity in five patients. Other clinical trials of ex vivo expanded or sensitized WT1-specific T cells for high-risk leukemias and multiple myeloma are ongoing (NCT00620633, NCT01758328).
Chapuis, Greenberg and coworkers next developed transgenic TCR-T cells directed against an HLA-A*02:01-restricted WT1 epitope for prevention or treatment of AML relapse after allo-HCT. To generate WT1-specific T cells, cytomegalovirus- or Epstein Barr virus-specific T cells from the HCT donor were transduced with a native, high-affinity, WT1-specific TCR identified from the peripheral repertoires of a healthy HLA-A*02:01+ individual. In a phase 1 clinical trial (NCT01640301), eleven patients with high-risk AML were treated prophylactically with WT1 TCR-T cells, and none had relapsed at a median follow-up of 21.3 months after allo-HCT, compared to 27% relapse at 16 months among matched controls.98,99 No survival advantage over standard of care was seen in patients treated with WT1 TCR T cells at relapse in preliminary findings. The Tawara group developed a retroviral construct encoding a high-affinity WT1/HLA-A*24:02-specific TCR identified from the peripheral repertoire of a healthy individual,92 along with siRNAs to eliminate expression of endogenous TCR chains.100 In a phase 1 clinical trial (UMIN000011519), eight patients with chemotherapy-refractory AML or high-risk MDS were treated with escalating doses (1.2–3.5× 108 cells per infusion) of autologous T cells expressing the WT1 TCR construct. No toxicity was observed, WT1 TCR T cells persisted in five patients, and transient decreases were noted in the percentage of bone marrow leukemic blasts in two patients. These data suggest that adoptively transferred WT1 TCR T cells directed against either HLA-A*02:01 or -A*24:02-restricted WT1 epitopes are safe, well-tolerated and may have anti-leukemic activity in vivo.
PRAME TCR immunotherapy
The PRAME CTA is overexpressed on solid tumors and in acute lymphoblastic leukemia (ALL), AML and MDS.101,102 A high-avidity TCR specific for an HLA-A*02:01-restricted, PRAME-derived epitope was identified from the T cell repertoire of an HLA-A*02:01-negative donor after transplantation and subsequent donor lymphocyte infusion into an HLA-A*02:01-positive recipient.103 The parental clone from which this TCR was isolated could recognize primary AML and ALL samples, as well as solid tumor cell lines, but did not recognize normal tissues except for mature dendritic cells and kidney epithelial cells. Retroviral transfer of the TCR conferred functional avidity and recognition of target cell lines similar to that measured for the parental clone. An early phase clinical trial of adoptive transfer of PRAME/A*02:01 TCR-transduced autologous T cells (BPX-701, Bellicum Pharmaceuticals) to treat AML, MDS and uveal melanoma opened in 2017 (NCT02743611). A second PRAME/A*02:01-specific TCR was isolated independently by another group;104 an early phase clinical trial testing autologous T cells expressing this TCR for treatment of high-risk AML, MDS and multiple myeloma opened in 2017 in Germany (NCT03503968).
HA-1 T cell immunotherapy
Hematopoietic-restricted minor H antigens can function as TAA in the context of recurrence after allo-HCT, assuming that donor and recipient have a suitable genotype mismatch. The minor H antigen HA-1H (hereafter referred to as HA-1) is encoded by a DNA sequence spanning a single nucleotide polymorphism (RS_1801284) within the HMHA1 gene. The resulting immunogenic peptide, VLHDDLLEA, is efficiently presented by HLA-A*02:01; the corresponding allelic variant peptide, VLRDDLLEA, has lower affinity for and unstable binding to HLA-A*02:01. The HMHA1 protein product is expressed in normal and malignant hematopoietic cells, but not in non-hematopoietic cells. In HA-1-mismatched allo-HCT, high-avidity HA-1-specific T cells from a HA-1-negative donor can be primed against HA-1 and mediate a graft-versus-tumor effect, as evidenced by decreased relapse rates in HA-1 mismatched allo-HCT (donor negative, recipient positive) and expansion of HA-1-specific T cells after donor lymphocyte infusion for post-allo-HCT relapses.105 A high-avidity, HA-1-specific TCR was identified from the repertoire of a healthy HLA-A*02:01-positive HA-1-negative individual and was sequenced and cloned into a lentiviral vector, along with a selection marker, safety switch (iCasp9) and CD8 co-receptor, to allow TCR-transduced CD4+ T cells to recognize the MHC class I-restricted epitope. CD8+ and CD4+ T cells expressing the HA-1 TCR construct were highly functional against leukemia cell lines and primary leukemia in vitro and could be rapidly eliminated using the safety switch.69 A phase 1 clinical trial of CD8+ and CD4+ T cells transduced with this HA-1 TCR construct is now open to patients with hematologic malignancies (NCT03326921). HA-1 TCR T cell immunotherapy is also being evaluated in the Netherlands, using a different HA-1-specific TCR, a different transgene (without CD8 coreceptor) and viral vector (retroviral rather than lentiviral), and a different cell product (EudraCT number 2010–024625–20).
Preclinical development of TCR immunotherapies
Overexpressed antigens
Survivin
Survivin is a member of the inhibitors of apoptosis protein family that is overexpressed in numerous cancers, including AML106–108 and various lymphomas,109,110 where its overexpression correlates with poor prognosis. High-affinity TCRs specific for an HLA-A*02:01-restricted survivin epitope were identified by stimulating HLA-A*02:01-negative CD8+ T cells with autologous dendritic cells transfected to express HLA-A*02:01. While transfer of TCRs isolated by this method enabled recognition and lysis of survivin-positive tumor cell lines, transfer of high-affinity TCRs also produced HLA-restricted fratricide of HLA-A*02:01-positive T cells.111 To overcome this obvious limitation, another group subsequently used stimulation with autologous dendritic cells to identify survivin-specific, HLA-A*02:01-restricted T cells from HLA-A*02:01-positive individuals.112 Adoptive transfer of T cells transduced to express survivin-specific TCRs thus isolated showed antileukemic activity and prolonged survival in immunodeficient murine xenograft models of AML.112
Human telomerase reverse transcriptase (hTERT)
hTERT is a catalytic subunit for telomere elongation that is expressed in numerous hematologic malignancies, but not in normal tissues.113–115 Two hTERT epitopes have been investigated as targets for transgenic TCR immunotherapy. A high-avidity TCR specific for an HLA-A*24:02-specific hTERT epitope116 was identified from the repertoire of a healthy individual by autologous stimulation, and subsequently cloned into a retroviral vector that also included siRNAs to eliminate expression of endogenous TCR chains.117 Adoptive transfer of hTERT TCR-T cells into an immunodeficient murine xenograft model of HTLV-associated T cell leukemia (ATL) inhibited tumor outgrowth for >6 months. In separate studies, a murine TCR specific for an HLA-A*02:01-restricted hTERT epitope was isolated by hTERT vaccination of HLA-A*02:01-expressing transgenic mice118 and cloned into a retroviral vector.119 Adoptive transfer of human T cells transduced with the hTERT-specific TCR showed in vivo activity against CLL119, B cell ALL, and AML120 in murine models. Although we are not aware of any TCR-T clinical trials targeting hTERT, clinical trials of hTERT vaccination are underway in the U.S. and Europe.
BOB1
BOB1 is a B cell-specific transcription factor that is highly expressed in B cell leukemias and lymphomas, as well as in multiple myeloma. A TCR specific for an HLA-B*07:02-restricted BOB1 epitope was identified from the alloreactive repertoire of a healthy individual and transferred into a retroviral vector.121 The transferred TCR enabled selective recognition and killing of BOB1-positive primary B cell leukemia, mantle cell lymphoma and multiple myeloma cells in vitro. Adoptive transfer of BOB1 TCR-transduced CD8+ T cells also controlled tumor outgrowth in an immunodeficient murine xenograft model of established myeloma.121
Lineage-restricted
Targeting lineage-restricted antigens in myeloid malignancies can cause myeloablation, necessitating allo-HCT to rescue hematopoiesis. However, in B cell malignancies, targeting of lineage-restricted antigens is more feasible because healthy B cells are dispensable. While this capacity has been exploited most thoroughly in the development of CAR-T cell therapy (CD19-, CD22-, and CD20-directed), B cell-restricted antigens can also be targeted by TCR-based approaches. Transfer of an HLA-B*07:02-restricted CD20 epitope-specific TCR enabled recognition of CD20-positive malignant cell lines and primary tumors (CLL, ALL, and mantle cell lymphoma), and suggested that TCR-based approaches may be effective in low expression CD20-positive B cell malignancies.122 T cells transduced with a different TCR specific for an HLA-B*07:02-restricted CD22 epitope killed primary B cell leukemia cells, but also killed healthy dendritic cells and macrophages,123 illustrating both the potential promise and limitation of targeting lineage-restricted antigens.
Cancer-testis antigens
Aurora kinase A (AurA) is a serine-threonine kinase involved in mitotic cell division, which among normal tissues is only detected in testis only, but is expressed at high levels in leukemias.124,125 An epitope derived from amino acids 207–215 of AurA is presented on HLA-A*02:01 and -A*24:02 on primary myeloid leukemia, and CD8+ T cells specific for this epitope lyse leukemia cells in vitro.125 A TCR specific for the HLA-A*02:01-restricted AurA epitope was identified in a healthy donor, and retroviral transfer of this TCR enabled in vitro recognition of leukemia by transgenic CD4+ and CD8+ T cells.126 The CD8+ AurA TCR-T cells also controlled leukemia progression in vivo in an immunodeficient murine xenograft model. Of note, AurA TCR-T cells may also be applicable as immunotherapy in ATL.127
Neoantigens
In contrast to solid tumors, which may carry hundreds or even thousands of mutations in an individual patient,128 most hematologic malignancies have relatively few protein-coding mutations or gene fusions, and thus fewer potential neoantigens. However, for TCR-T immunotherapy, a small number of neoantigens will suffice, as long as those neoantigens are shared among patients and ideally occur early in oncogenesis or are essential in maintaining the malignant phenotype. For example, the type A variant of the CBFB-MYH11 fusion is critical in leukemogenesis, and occurs in ~10% of individuals with AML and ~90% of AML patients with the inv(16) or t(16;16) cytogenetic abnormalities. An epitope spanning the CBFB-MYH11 fusion region is presented on HLA-B*40:01 and enables selective recognition and killing of leukemic blasts by epitope-specific T cells. Lentiviral transfer of a high-avidity TCR confers epitope specificity and antileukemic cytotoxicity in vitro, suggesting that CBFB-MYH11 is a viable target for TCR-based immunotherapy for a subset of patients.129 Targeting the fusion may enable eradication of the cell of origin, as it is known to play a key role in maintaining the leukemic phenotype. An epitope derived from a highly recurrent frameshift mutation in exon 12 of nucleophosmin1 (NPM1) is another potential neoantigen target for TCR immunotherapy. NPM1 mutations occur in 30–35% of AML cases and are early events that persist across the disease course. CD8+ T cells specific for an HLA-A*02:01-restricted NPM1 epitope can selectively recognize NPM1-mutated leukemic blasts, and adoptive transfer of NPM1/HLA-A*02:01 specific TCR-transduced T cells controlled tumor outgrowth and prolonged survival in an immunodeficient murine xenograft model.130 These early studies suggest a role for shared neoantigens in TCR-based immunotherapy of AML and other hematologic malignancies.
Future of TCR immunotherapy in hematologic malignancies
TCR-T cell immunotherapy is a very young field but is evolving quickly. Advances in the fields of basic immunology, protein science, synthetic biology, genomics, and cell and genome engineering should allow us to overcome many previously-recognized obstacles and facilitate the development of TCR-T therapies. One major bottleneck has been identifying bona fide target epitopes with sufficient cancer-specificity for safe targeting in patients. In silico algorithms can effectively predict binding of peptides to MHC/HLA molecules, but not whether the peptides are processed and presented on cell surfaces. With the wave of ‘omic’ approaches, some have attempted to determine the ‘peptidome’ of malignant cells by immunoprecipitating MHC complexes, eluting peptides from these complexes and identifying peptides by tandem liquid chromatography–mass spectrometry.131–133 This unbiased approach can identify peptides naturally processed and presented on cells of interest, but also has technical hurdles.134–136 Improving techniques to identify and characterize the peptidome of malignant cells, such as the use of monoallelic cells,134 has the potential to fast-track discovery of physiologically-relevant targets for TCR-T immunotherapy. TCR sequencing techniques are rapidly evolving, as is our understanding of how to predict which pMHC complexes functionally engage specific TCR sequences.137 Although these technologies are early in development, understanding the biophysical and structural rules that govern TCR-pMHC binding will also improve prediction of cross-reactivity to other pMHC complexes and thus uncover potential off-target toxicities, and may even allow rational design of synthetic target-specific TCRs in the future.
Currently, all TCR-T immunotherapies target single antigens, running the risk the cancer may simply escape recognition by downregulating expression of the target protein, as seen in CD19 CAR-T cell trials.138,139 TCR-T targeting of proteins that are essential to maintaining the leukemic phenotype, like leukemia-initiating fusions, should avoid this escape mechanism. For targets that are less essential to the malignant cell, transferring transgenic TCR-T cells with multiple specificities could reduce the probability of antigen escape. One approach would be to make multiple TCR-T products with different specificities separately and infuse them together into the patient. Another would be to modify T cells to express multiple TCRs, although ensuring correct pairing of introduced TCR chains would be challenging. TCR mimic monoclonal antibodies, which bind specifically to pMHC complexes, can be used as CARs140 and might be more readily multiplexed.
Conclusions
Much progress has been made in the field of TCR-T immunotherapy generally and for hematologic malignancies specifically. Despite the challenges of identifying suitable targets for TCR-T therapies, a number of promising TAAs are under preclinical investigation as targets for TCR-T immunotherapy. Encouragingly, four different TCR-T immunotherapies are now in early phase clinical trials, targeting: WT1 epitopes restricted to HLA-A*02:01 and A*24:02; an HLA-A*02:01-restricted PRAME epitope; and the HLA-A*02:01-restricted minor H antigen HA-1. Along with improvements in T cell antigen discovery and cell engineering techniques, gained in part from experience with CAR-T cell therapies, these studies set the stage for development of TCR-T immunotherapies with potent curative potential in the hematologic malignancies.
Figure 3.
Categories of tumor-associated antigens
Footnotes
Conflict of interest statement: M.B. (Bleakley) has received compensation from Miltenyi Biotec for presentations at conferences and corporate symposia pertaining to research unrelated to that presented in the current manuscript. M.B. (Bleakley) and M.A.B. have filed a provisional patent application number 62/616,261 covering applications of T cell immunotherapy for CBF AML. M.B. (Bleakley) has filed a provisional patent application number 62/399,291 covering applications of engineered T cell receptor for targeting minor histocompatibility (H) antigen HA-1 and compositions for treating leukemia and PCT/US2017/053112 covering applications of TCRs specific for minor histocompatibility (H) antigen HA-1 and uses thereof.
REFERENCES
- 1.Gross L Intradermal Immunization of C3H Mice against a Sarcoma That Originated in an Animal of the Same Line. Cancer Res 1943;3:326–33. [Google Scholar]
- 2.Lemos H, Huang L, Prendergast GC, Mellor AL. Immune control by amino acid catabolism during tumorigenesis and therapy. Nat Rev Cancer 2019;19:162–75. [DOI] [PubMed] [Google Scholar]
- 3.van der Burg SH, Arens R, Ossendorp F, van Hall T, Melief CJ. Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat Rev Cancer 2016;16:219–33. [DOI] [PubMed] [Google Scholar]
- 4.Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer 2017;17:209–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teague RM, Kline J. Immune evasion in acute myeloid leukemia: current concepts and future directions. J Immunother Cancer 2013;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuno K, Hara T, Tsurumi H, et al. Expression of indoleamine 2,3-dioxygenase in leukemic cells indicates an unfavorable prognosis in acute myeloid leukemia patients with intermediate-risk cytogenetics. Leuk Lymphoma 2015;56:1398–405. [DOI] [PubMed] [Google Scholar]
- 7.Mussai F, De Santo C, Abu-Dayyeh I, et al. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood 2013;122:749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Gajewski TF, Kline J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood 2009;114:1545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buggins AG, Milojkovic D, Arno MJ, et al. Microenvironment produced by acute myeloid leukemia cells prevents T cell activation and proliferation by inhibition of NF-kappaB, c-Myc, and pRb pathways. J Immunol 2001;167:6021–30. [DOI] [PubMed] [Google Scholar]
- 10.Bruck O, Blom S, Dufva O, et al. Immune cell contexture in the bone marrow tumor microenvironment impacts therapy response in CML. Leukemia 2018;32:1643–56. [DOI] [PubMed] [Google Scholar]
- 11.Burger JA, Gribben JG. The microenvironment in chronic lymphocytic leukemia (CLL) and other B cell malignancies: insight into disease biology and new targeted therapies. Semin Cancer Biol 2014;24:71–81. [DOI] [PubMed] [Google Scholar]
- 12.Dudley ME, Wunderlich JR, Robbins PF, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 2002;298:850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley ME, Wunderlich JR, Yang JC, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol 2005;23:2346–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg SA, Packard BS, Aebersold PM, et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N Engl J Med 1988;319:1676–80. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg SA, Yannelli JR, Yang JC, et al. Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin 2. J Natl Cancer Inst 1994;86:1159–66. [DOI] [PubMed] [Google Scholar]
- 16.Noonan K, Matsui W, Serafini P, et al. Activated marrow-infiltrating lymphocytes effectively target plasma cells and their clonogenic precursors. Cancer Res 2005;65:2026–34. [DOI] [PubMed] [Google Scholar]
- 17.Noonan KA, Huff CA, Davis J, et al. Adoptive transfer of activated marrow-infiltrating lymphocytes induces measurable antitumor immunity in the bone marrow in multiple myeloma. Sci Transl Med 2015;7:288ra78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley ME, Gross CA, Langhan MM, et al. CD8+ enriched “young” tumor infiltrating lymphocytes can mediate regression of metastatic melanoma. Clin Cancer Res 2010;16:6122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geukes Foppen MH, Donia M, Svane IM, Haanen JB. Tumor-infiltrating lymphocytes for the treatment of metastatic cancer. Mol Oncol 2015;9:1918–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheper W, Kelderman S, Fanchi LF, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med 2019;25:89–94. [DOI] [PubMed] [Google Scholar]
- 21.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science 2018;359:1361–5. [DOI] [PubMed] [Google Scholar]
- 22.Turtle CJ, Hanafi LA, Berger C, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 2016;126:2123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kershaw MH, Westwood JA, Darcy PK. Gene-engineered T cells for cancer therapy. Nat Rev Cancer 2013;13:525–41. [DOI] [PubMed] [Google Scholar]
- 24.Salter AI, Pont MJ, Riddell SR. Chimeric antigen receptor-modified T cells: CD19 and the road beyond. Blood 2018;131:2621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris DT, Hager MV, Smith SN, et al. Comparison of T Cell Activities Mediated by Human TCRs and CARs That Use the Same Recognition Domains. J Immunol 2018;200:1088–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salter AI, Ivey RG, Kennedy JJ, et al. Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci Signal 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cresswell P Antigen processing and presentation. Immunol Rev 2005;207:5–7. [DOI] [PubMed] [Google Scholar]
- 28.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol 2008;8:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol 2013;31:443–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koopman LA, Corver WE, van der Slik AR, Giphart MJ, Fleuren GJ. Multiple genetic alterations cause frequent and heterogeneous human histocompatibility leukocyte antigen class I loss in cervical cancer. J Exp Med 2000;191:961–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campoli M, Chang CC, Ferrone S. HLA class I antigen loss, tumor immune escape and immune selection. Vaccine 2002;20 Suppl 4:A40–5. [DOI] [PubMed] [Google Scholar]
- 32.Garrido F, Ruiz-Cabello F, Aptsiauri N. Rejection versus escape: the tumor MHC dilemma. Cancer Immunol Immunother 2017;66:259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla SA, Rooney MS, Rajasagi M, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol 2015;33:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benitez R, Godelaine D, Lopez-Nevot MA, et al. Mutations of the beta2-microglobulin gene result in a lack of HLA class I molecules on melanoma cells of two patients immunized with MAGE peptides. Tissue Antigens 1998;52:520–9. [DOI] [PubMed] [Google Scholar]
- 35.Sade-Feldman M, Jiao YJ, Chen JH, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 2017;8:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gettinger S, Choi J, Hastings K, et al. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov 2017;7:1420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGranahan N, Rosenthal R, Hiley CT, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell 2017;171:1259–71 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulson KG, Voillet V, McAfee MS, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun 2018;9:3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crotzer VL, Blum JS. Autophagy and its role in MHC-mediated antigen presentation. J Immunol 2009;182:3335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christopher MJ, Petti AA, Rettig MP, et al. Immune Escape of Relapsed AML Cells after Allogeneic Transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vago L, Perna SK, Zanussi M, et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med 2009;361:478–88. [DOI] [PubMed] [Google Scholar]
- 42.Pollack MS, Heagney SD, Livingston PO, Fogh J. HLA-A, B, C and DR alloantigen expression on forty-six cultured human tumor cell lines. J Natl Cancer Inst 1981;66:1003–12. [DOI] [PubMed] [Google Scholar]
- 43.Forero A, Li Y, Chen D, et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol Res 2016;4:390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCaw TR, Li M, Starenki D, et al. The expression of MHC class II molecules on murine breast tumors delays T-cell exhaustion, expands the T-cell repertoire, and slows tumor growth. Cancer Immunol Immunother 2019;68:175–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovig T, Andersen SN, Thorstensen L, et al. Strong HLA-DR expression in microsatellite stable carcinomas of the large bowel is associated with good prognosis. Br J Cancer 2002;87:756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson DB, Estrada MV, Salgado R, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun 2016;7:10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran E, Turcotte S, Gros A, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014;344:641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stronen E, Toebes M, Kelderman S, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 2016;352:1337–41. [DOI] [PubMed] [Google Scholar]
- 49.Bleakley M, Otterud BE, Richardt JL, et al. Leukemia-associated minor histocompatibility antigen discovery using T-cell clones isolated by in vitro stimulation of naive CD8+ T cells. Blood 2010;115:4923–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med 2010;207:2187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bentzen AK, Marquard AM, Lyngaa R, et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat Biotechnol 2016;34:1037–45. [DOI] [PubMed] [Google Scholar]
- 52.Hu Z, Anandappa AJ, Sun J, et al. A cloning and expression system to probe T-cell receptor specificity and assess functional avidity to neoantigens. Blood 2018;132:1911–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kebriaei P, Singh H, Huls MH, et al. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J Clin Invest 2016;126:3363–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deniger DC, Pasetto A, Tran E, et al. Stable, Nonviral Expression of Mutated Tumor Neoantigen-specific T-cell Receptors Using the Sleeping Beauty Transposon/Transposase System. Mol Ther 2016;24:1078–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bishop DC, Xu N, Tse B, et al. PiggyBac-Engineered T Cells Expressing CD19-Specific CARs that Lack IgG1 Fc Spacers Have Potent Activity against B-ALL Xenografts. Mol Ther 2018;26:1883–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith TT, Stephan SB, Moffett HF, et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat Nanotechnol 2017;12:813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bendle GM, Linnemann C, Hooijkaas AI, et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat Med 2010;16:565–70, 1p following 70. [DOI] [PubMed] [Google Scholar]
- 58.Sommermeyer D, Uckert W. Minimal amino acid exchange in human TCR constant regions fosters improved function of TCR gene-modified T cells. J Immunol 2010;184:6223–31. [DOI] [PubMed] [Google Scholar]
- 59.Ochi T, Fujiwara H, Okamoto S, et al. Novel adoptive T-cell immunotherapy using a WT1-specific TCR vector encoding silencers for endogenous TCRs shows marked antileukemia reactivity and safety. Blood 2011;118:1495–503. [DOI] [PubMed] [Google Scholar]
- 60.Manning TC, Schlueter CJ, Brodnicki TC, et al. Alanine scanning mutagenesis of an alphabeta T cell receptor: mapping the energy of antigen recognition. Immunity 1998;8:413–25. [DOI] [PubMed] [Google Scholar]
- 61.Udyavar A, Alli R, Nguyen P, Baker L, Geiger TL. Subtle affinity-enhancing mutations in a myelin oligodendrocyte glycoprotein-specific TCR alter specificity and generate new self-reactivity. J Immunol 2009;182:4439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmitt TM, Ragnarsson GB, Greenberg PD. T cell receptor gene therapy for cancer. Hum Gene Ther 2009;20:1240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linette GP, Stadtmauer EA, Maus MV, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013;122:863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cameron BJ, Gerry AB, Dukes J, et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med 2013;5:197ra03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol 2006;7:1191–9. [DOI] [PubMed] [Google Scholar]
- 66.Stadinski BD, Trenh P, Smith RL, et al. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity 2011;35:694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES, Call ME. Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb Perspect Biol 2010;2:a005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmitt TM, Aggen DH, Ishida-Tsubota K, Ochsenreither S, Kranz DM, Greenberg PD. Generation of higher affinity T cell receptors by antigen-driven differentiation of progenitor T cells in vitro. Nat Biotechnol 2017;35:1188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dossa RG, Cunningham T, Sommermeyer D, et al. Development of T-cell immunotherapy for hematopoietic stem cell transplantation recipients at risk of leukemia relapse. Blood 2018;131:108–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giuntoli RL 2nd, Lu J, Kobayashi H, Kennedy R, Celis E. Direct costimulation of tumor-reactive CTL by helper T cells potentiate their proliferation, survival, and effector function. Clin Cancer Res 2002;8:922–31. [PubMed] [Google Scholar]
- 71.Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4(+) T cells in the antitumor immune response. J Exp Med 1998;188:2357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kennedy R, Celis E. Multiple roles for CD4+ T cells in anti-tumor immune responses. Immunol Rev 2008;222:129–44. [DOI] [PubMed] [Google Scholar]
- 73.Morris EC, Tsallios A, Bendle GM, Xue SA, Stauss HJ. A critical role of T cell antigen receptor-transduced MHC class I-restricted helper T cells in tumor protection. Proc Natl Acad Sci U S A 2005;102:7934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Veken LT, Hagedoorn RS, van Loenen MM, Willemze R, Falkenburg JH, Heemskerk MH. Alphabeta T-cell receptor engineered gammadelta T cells mediate effective antileukemic reactivity. Cancer Res 2006;66:3331–7. [DOI] [PubMed] [Google Scholar]
- 75.Di Stasi A, Tey SK, Dotti G, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med 2011;365:1673–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramos CA, Asgari Z, Liu E, et al. An inducible caspase 9 suicide gene to improve the safety of mesenchymal stromal cell therapies. Stem Cells 2010;28:1107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Straathof KC, Pule MA, Yotnda P, et al. An inducible caspase 9 safety switch for T-cell therapy. Blood 2005;105:4247–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tey SK, Dotti G, Rooney CM, Heslop HE, Brenner MK. Inducible caspase 9 suicide gene to improve the safety of allodepleted T cells after haploidentical stem cell transplantation. Biol Blood Marrow Transplant 2007;13:913–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou X, Di Stasi A, Tey SK, et al. Long-term outcome after haploidentical stem cell transplant and infusion of T cells expressing the inducible caspase 9 safety transgene. Blood 2014;123:3895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou X, Naik S, Dakhova O, Dotti G, Heslop HE, Brenner MK. Serial Activation of the Inducible Caspase 9 Safety Switch After Human Stem Cell Transplantation. Mol Ther 2016;24:823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kerkar SP, Muranski P, Kaiser A, et al. Tumor-specific CD8+ T cells expressing interleukin-12 eradicate established cancers in lymphodepleted hosts. Cancer Res 2010;70:6725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pegram HJ, Lee JC, Hayman EG, et al. Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood 2012;119:4133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang L, Morgan RA, Beane JD, et al. Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin Cancer Res 2015;21:2278–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alsaieedi A, Holler A, Velica P, Bendle G, Stauss HJ. Safety and efficacy of Tet-regulated IL-12 expression in cancer-specific T cells. Oncoimmunology 2019;8:1542917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Borkner L, Kaiser A, van de Kasteele W, et al. RNA interference targeting programmed death receptor-1 improves immune functions of tumor-specific T cells. Cancer Immunol Immunother 2010;59:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Su S, Hu B, Shao J, et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci Rep 2016;6:20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oda SK, Daman AW, Garcia NM, et al. A CD200R-CD28 fusion protein appropriates an inhibitory signal to enhance T-cell function and therapy of murine leukemia. Blood 2017;130:2410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishimura CD, Brenner DA, Mukherjee M, et al. c-MPL provides tumor-targeted T-cell receptor-transgenic T cells with costimulation and cytokine signals. Blood 2017;130:2739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Torikai H, Reik A, Soldner F, et al. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood 2013;122:1341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang J, Wang L. The Emerging World of TCR-T Cell Trials Against Cancer: A Systematic Review. Technol Cancer Res Treat 2019;18:1533033819831068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inoue K, Ogawa H, Sonoda Y, et al. Aberrant overexpression of the Wilms tumor gene (WT1) in human leukemia. Blood 1997;89:1405–12. [PubMed] [Google Scholar]
- 92.Ohminami H, Yasukawa M, Fujita S. HLA class I-restricted lysis of leukemia cells by a CD8(+) cytotoxic T-lymphocyte clone specific for WT1 peptide. Blood 2000;95:286–93. [PubMed] [Google Scholar]
- 93.Gao L, Bellantuono I, Elsasser A, et al. Selective elimination of leukemic CD34(+) progenitor cells by cytotoxic T lymphocytes specific for WT1. Blood 2000;95:2198–203. [PubMed] [Google Scholar]
- 94.Rezvani K, Yong AS, Savani BN, et al. Graft-versus-leukemia effects associated with detectable Wilms tumor-1 specific T lymphocytes after allogeneic stem-cell transplantation for acute lymphoblastic leukemia. Blood 2007;110:1924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ho WY, Nguyen HN, Wolfl M, Kuball J, Greenberg PD. In vitro methods for generating CD8+ T-cell clones for immunotherapy from the naive repertoire. J Immunol Methods 2006;310:40–52. [DOI] [PubMed] [Google Scholar]
- 96.Weber G, Gerdemann U, Caruana I, et al. Generation of multi-leukemia antigen-specific T cells to enhance the graft-versus-leukemia effect after allogeneic stem cell transplant. Leukemia 2013;27:1538–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chapuis AG, Ragnarsson GB, Nguyen HN, et al. Transferred WT1-reactive CD8+ T cells can mediate antileukemic activity and persist in post-transplant patients. Sci Transl Med 2013;5:174ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chapuis AG, Egan D, Bar M, et al. EBV-Specific Donor Cells Transduced to Express a High-Affinity WT1 TCR Can Prevent Recurrence in Post-HCT Patients with High-Risk AML. Blood 2016;128:1001. [Google Scholar]
- 99.Chapuis AG, Egan D, Bar M, et al. T Cell Receptor Gene Therapy Targeting WT1 Prevents Acute Myeloid Leukemia Relapse Post-Transplant. Nat Med 2019. (accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tawara I, Kageyama S, Miyahara Y, et al. Safety and persistence of WT1-specific T-cell receptor gene-transduced lymphocytes in patients with AML and MDS. Blood 2017;130:1985–94. [DOI] [PubMed] [Google Scholar]
- 101.Qin Y, Zhu H, Jiang B, et al. Expression patterns of WT1 and PRAME in acute myeloid leukemia patients and their usefulness for monitoring minimal residual disease. Leuk Res 2009;33:384–90. [DOI] [PubMed] [Google Scholar]
- 102.van Baren N, Chambost H, Ferrant A, et al. PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br J Haematol 1998;102:1376–9. [DOI] [PubMed] [Google Scholar]
- 103.Amir AL, van der Steen DM, van Loenen MM, et al. PRAME-specific Allo-HLA-restricted T cells with potent antitumor reactivity useful for therapeutic T-cell receptor gene transfer. Clin Cancer Res 2011;17:5615–25. [DOI] [PubMed] [Google Scholar]
- 104.Weis MW C; Ellinger C; Wilde S; Schendel DJ Isolation and characterization of a PRAME-specific TCR with high avidity, potent antitumor efficacy and a favorable preclinical safety profile. American Association for Cancer Research Annual Meeting 2017;Abstract 4977. [Google Scholar]
- 105.Marijt WA, Heemskerk MH, Kloosterboer FM, et al. Hematopoiesis-restricted minor histocompatibility antigens HA-1- or HA-2-specific T cells can induce complete remissions of relapsed leukemia. Proc Natl Acad Sci U S A 2003;100:2742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adida C, Recher C, Raffoux E, et al. Expression and prognostic significance of survivin in de novo acute myeloid leukaemia. Br J Haematol 2000;111:196–203. [DOI] [PubMed] [Google Scholar]
- 107.Carter BZ, Qiu Y, Huang X, et al. Survivin is highly expressed in CD34(+)38(−) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML. Blood 2012;120:173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tamm I, Richter S, Oltersdorf D, et al. High expression levels of x-linked inhibitor of apoptosis protein and survivin correlate with poor overall survival in childhood de novo acute myeloid leukemia. Clin Cancer Res 2004;10:3737–44. [DOI] [PubMed] [Google Scholar]
- 109.Adida C, Haioun C, Gaulard P, et al. Prognostic significance of survivin expression in diffuse large B-cell lymphomas. Blood 2000;96:1921–5. [PubMed] [Google Scholar]
- 110.Schlette EJ, Medeiros LJ, Goy A, Lai R, Rassidakis GZ. Survivin expression predicts poorer prognosis in anaplastic large-cell lymphoma. J Clin Oncol 2004;22:1682–8. [DOI] [PubMed] [Google Scholar]
- 111.Leisegang M, Wilde S, Spranger S, et al. MHC-restricted fratricide of human lymphocytes expressing survivin-specific transgenic T cell receptors. J Clin Invest 2010;120:3869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arber C, Feng X, Abhyankar H, et al. Survivin-specific T cell receptor targets tumor but not T cells. J Clin Invest 2015;125:157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science 1994;266:2011–5. [DOI] [PubMed] [Google Scholar]
- 114.Counter CM, Gupta J, Harley CB, Leber B, Bacchetti S. Telomerase activity in normal leukocytes and in hematologic malignancies. Blood 1995;85:2315–20. [PubMed] [Google Scholar]
- 115.Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci U S A 1995;92:9082–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tajima K, Ito Y, Demachi A, et al. Interferon-gamma differentially regulates susceptibility of lung cancer cells to telomerase-specific cytotoxic T lymphocytes. Int J Cancer 2004;110:403–12. [DOI] [PubMed] [Google Scholar]
- 117.Miyazaki Y, Fujiwara H, Asai H, et al. Development of a novel redirected T-cell-based adoptive immunotherapy targeting human telomerase reverse transcriptase for adult T-cell leukemia. Blood 2013;121:4894–901. [DOI] [PubMed] [Google Scholar]
- 118.Ugel S, Scarselli E, Iezzi M, et al. Autoimmune B-cell lymphopenia after successful adoptive therapy with telomerase-specific T lymphocytes. Blood 2010;115:1374–84. [DOI] [PubMed] [Google Scholar]
- 119.Sandri S, Bobisse S, Moxley K, et al. Feasibility of Telomerase-Specific Adoptive T-cell Therapy for B-cell Chronic Lymphocytic Leukemia and Solid Malignancies. Cancer Res 2016;76:2540–51. [DOI] [PubMed] [Google Scholar]
- 120.Sandri S, De Sanctis F, Lamolinara A, et al. Effective control of acute myeloid leukaemia and acute lymphoblastic leukaemia progression by telomerase specific adoptive T-cell therapy. Oncotarget 2017;8:86987–7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jahn L, Hombrink P, Hagedoorn RS, et al. TCR-based therapy for multiple myeloma and other B-cell malignancies targeting intracellular transcription factor BOB1. Blood 2017;129:1284–95. [DOI] [PubMed] [Google Scholar]
- 122.Jahn L, van der Steen DM, Hagedoorn RS, et al. Generation of CD20-specific TCRs for TCR gene therapy of CD20low B-cell malignancies insusceptible to CD20-targeting antibodies. Oncotarget 2016;7:77021–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jahn L, Hagedoorn RS, van der Steen DM, et al. A CD22-reactive TCR from the T-cell allorepertoire for the treatment of acute lymphoblastic leukemia by TCR gene transfer. Oncotarget 2016;7:71536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hamada M, Yakushijin Y, Ohtsuka M, Kakimoto M, Yasukawa M, Fujita S. Aurora2/BTAK/STK15 is involved in cell cycle checkpoint and cell survival of aggressive non-Hodgkin’s lymphoma. Br J Haematol 2003;121:439–47. [DOI] [PubMed] [Google Scholar]
- 125.Ochi T, Fujiwara H, Suemori K, et al. Aurora-A kinase: a novel target of cellular immunotherapy for leukemia. Blood 2009;113:66–74. [DOI] [PubMed] [Google Scholar]
- 126.Nagai K, Ochi T, Fujiwara H, et al. Aurora kinase A-specific T-cell receptor gene transfer redirects T lymphocytes to display effective antileukemia reactivity. Blood 2012;119:368–76. [DOI] [PubMed] [Google Scholar]
- 127.Casey NP, Fujiwara H, Ochi T, Yasukawa M. Novel immunotherapy for adult T-cell leukemia/lymphoma: Targeting aurora kinase A. Oncoimmunology 2016;5:e1239006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Biernacki MA, Foster KA, Coon ME, et al. An Epitope from the Recurrent CBFB-MYH11 Fusion Protein Is a Highly Immunogenic AML Neoantigen In: Transplantation and Cellular Therapies Annual Meeting; 2019. February 20-24; Houston, TX: Abstract LBA29. [Google Scholar]
- 130.van der Lee DI, Reijmers RM, Honders MW, et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J Clin Invest 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Berlin C, Kowalewski DJ, Schuster H, et al. Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia 2015;29:647–59. [DOI] [PubMed] [Google Scholar]
- 132.Bilich T, Nelde A, Bichmann L, et al. The HLA ligandome landscape of chronic myeloid leukemia delineates novel T-cell epitopes for immunotherapy. Blood 2019;133:550–65. [DOI] [PubMed] [Google Scholar]
- 133.Nelde A, Kowalewski DJ, Backert L, et al. HLA ligandome analysis of primary chronic lymphocytic leukemia (CLL) cells under lenalidomide treatment confirms the suitability of lenalidomide for combination with T-cell-based immunotherapy. Oncoimmunology 2018;7:e1316438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abelin JG, Keskin DB, Sarkizova S, et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 2017;46:315–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hawkins OE, Vangundy RS, Eckerd AM, et al. Identification of breast cancer peptide epitopes presented by HLA-A*0201. J Proteome Res 2008;7:1445–57. [DOI] [PubMed] [Google Scholar]
- 136.Trolle T, McMurtrey CP, Sidney J, et al. The Length Distribution of Class I-Restricted T Cell Epitopes Is Determined by Both Peptide Supply and MHC Allele-Specific Binding Preference. J Immunol 2016;196:1480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dash P, Fiore-Gartland AJ, Hertz T, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 2017;547:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Grupp SA, Kalos M, Barrett D, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med 2013;368:1509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ma Q, Garber HR, Lu S, et al. A novel TCR-like CAR with specificity for PR1/HLA-A2 effectively targets myeloid leukemia in vitro when expressed in human adult peripheral blood and cord blood T cells. Cytotherapy 2016;18:985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Romero P, Dunbar PR, Valmori D, et al. Ex vivo staining of metastatic lymph nodes by class I major histocompatibility complex tetramers reveals high numbers of antigen-experienced tumor-specific cytolytic T lymphocytes. J Exp Med 1998;188:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zippelius A, Pittet MJ, Batard P, et al. Thymic selection generates a large T cell pool recognizing a self-peptide in humans. J Exp Med 2002;195:485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135–46. [DOI] [PubMed] [Google Scholar]
- 144.Ikeda H, Lethe B, Lehmann F, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 1997;6:199–208. [DOI] [PubMed] [Google Scholar]