Abstract
Objectives
To evaluate the differences in the composition and quantities of the urine peptides in regular cannabis users and non-users by LC-MS/MS analysis.
Materials and Methods
Urine specimens from healthy control subjects and cannabis users were utilized to identify the differences in number and quantity of urine proteins LC-MS/MS analysis. Significantly altered proteins were determined by a permutation testing statistical method. Heat map, dendrogram, pathways and network analyses were performed to assess the degree of expression and potential relationships between proteins in both groups.
Results
1337 proteins were detected in both groups with 19 proteins being significantly altered in cannabis users. Innate immunity and carbohydrate metabolic pathways were highly linked with up-regulated proteins in cannabis group. Additionally, 91 proteins were present and 46 proteins were absent only in cannabis users in comparison to the control cohort. Our results suggest that regular use of cannabis is associated with significant alterations in a number of urinary peptides, with a large number of proteins present or absent only in cannabis users. Pathway analyses demonstrated an increased immune responses in cannabis users compared to controls.
Conclusions
Our observations potentially indicate activation (or inhibition) of specific signaling pathways in the lower urinary tract (LUT) during chronic exposure to exogenous cannabinoids. Our study provides initial proteomic knowledge for future investigations on potential role of exocannabinoids in development of intravesical therapies to treat LUT disorders.
Keywords: Urine proteomics, Cannabis, cannabis users, lower urinary tract, bladder function
INTRODUCTION
Cannabis consumption has significantly increased in the US since legalization in several states1. CB1 receptors are abundantly expressed in central (CNS), peripheral nervous system, and in non-neural tissues, while CB2 receptors are mostly found on immune and glial cells. Both types of cannabinoid receptors were detected in the human detrusor and urothelium, with the expression levels approximately two-fold higher in the urothelium than in the detrusor2. The relative risks or benefits of regular cannabis use on the function of the urinary bladder or expression of cannabinoid receptors in the lower urinary tract (LUT) are unknown. Prior clinical studies in multiple sclerosis (MS) patients established potential beneficial effects of systemic cannabinoids on neurogenic bladder dysfunction3, 4 In a 1994 anonymous questionnaire survey of 112 men and women with MS in the UK and USA who smoked cannabis to relieve their symptoms, 54–64% of subjects reported improvements in urinary symptoms5. In MS studies with bladder symptom-specific endpoints, cannabis extracts were found to reduce frequency, nocturia, incontinence episodes, and voids with urgency3, 4 Cannabis extract and THC displayed a significant benefit for reducing urge incontinence episodes compared to placebo in a subset trial to the multicenter, randomized, placebo-controlled cannabinoids in MS (CAMS) study3.
Reports on beneficial effects of cannabis or cannabis-based extracts on MS-related lower urinary tract symptoms (LUTS), and progress in understanding cannabinoid-related functions in LUT tissues implicate the endocannabinoid system as a putative drug target for LUTS.3, 4 However, current knowledge of cannabinoid-mediated effects on micturition does not clearly discern its sites of action, nor downstream signaling pathways activated by cannabinoids. Recent animal studies provided evidence that local activation of cannabinoid CB1 receptors in the urinary bladder reduced inflammation-induced sensitization of bladder afferent nerves6, and also inhibited increased bladder activity induced by nerve growth factor7. These preclinical studies along with the clinical results on neurogenic LUTS suggest that intravesical application of cannabinoids could be one of the promising approaches to treat LUT dysfunction locally. Therefore, quantifying unique cannabinoids in urine, establishing the impact of specific cannabinoids on LUT function, and assessment of the potential biomarkers of LUT function in cannabis users can help investigators identify specific molecular targets in the bladder for a variety of LUT diseases such as interstitial cystitis/bladder pain syndrome (IC/BPS), overactive bladder (OAB), detrusor hyperreflexia, and urinary incontinence (UI).
In this pilot study, we aimed to determine the profile of urine proteins in cannabis users in comparison to healthy controls by developing a comprehensive workflow integrating mass spectrometry-based analytical approach and advanced bioinformatics tools with the future goal of identifying potential signaling pathways in order to control LUTS via intravesical instillation of exogenous cannabinoids.
MATERIALS AND METHODS
Enrolled cohorts and sample collection
Regular cannabis users were recruited from print and media advertisements in the Denver metropolitan area. Healthy controls were selected to match cannabis users in terms of age, sex, and race/ethnicity. Both groups did not endorse the use of cigarettes. The protocol to enroll the subjects and collect urinary specimens was approved by the Colorado Multiple Institutional Review Board (COMIRB, #14–1957) and all subjects provided written informed consent. Inclusion and exclusion criteria can be found in supplementary methods.
Urine sample processing and LC-MS/MS analysis
Urine samples were processed for LC-MS/MS. The detailed methods of sample processing, data acquisition, protein identification and analysis can be found in supplementary methods.
Cannabinoid assay
Urine specimens were analyzed by a validated liquid chromatography tandem mass spectrometry (LC-MS/MS) method. Briefly, 200 μL of urine was transferred into a 1.5 mL low binding polypropylene vial (Sarstedt, Nümbrecht, Germany). Eight hundred μL of 0.2 M ZnSO4 30% water/ 70% methanol (v/v) containing the internal standards (5 μg/L) was added. Samples were vortexed for 10 min and then centrifuged (at 26,000 g, 4°C, 10 min, Sorvall Legend 23R or Thermo Scientific MR 23i). The supernatant was transferred into an HPLC vial awaiting analysis. Cannabinoids were quantified by LC-MS/MS on an ABSciex API5000 tandem mass spectrometer (ABSciex, Concord, Ontario, Canada) via a turbo V ion source operated in positive atmospheric pressure chemical ionization (APCI) mode. Limits of quantifications were 0.39 μg/L for Δ9- tetrahydrocannabinol (THC), 11-nor-9-carboxy- THC (THCCOOH) and cannabidiol (CBD); 0.78 μg/L for cannabigerol (CBG), tetrahydrocannabivarin (THCV), 11-nor-9-carboxy-THCV (THCVCOOH), cannabichromene (CBC) and cannabidivarin (CBDV); 1.56 μg/L for 11-hydroxy-THC (11-OH-THC) and cannabinol (CBN); 3.91 μg/L for THCCOOH-glucuronide. Upper limit of quantification was 400 μg/L for all analytes (except 2000 μg/L THCCOOH-glucuronide). Inter-assay accuracy and imprecision were 88.3–103 % and 2.2–11.7 %CV, respectively. Analyst software version 1.4.2 was employed for data acquisition and MultiQuant version 2.1.1 for data analysis (AB Sciex, Foster City, CA, USA).
Statistics analysis
The permutation method 8 was applied to find out the proteins with a significant difference among cannabis users and non-users. Since the two-sample t-test assumes a normal distribution, we adopted a permutation approach which uses the observed data to obtain true null distribution of the test statistic. We used t-test statistics as our test statistic and used 10000 random permutations to obtain its null distribution. The p-value was calculated as the proportion of permutations for which the absolute value of the t-statistic is greater than the absolute value of the observed t-statistic. P values ≤0.05 were considered statistically significant between the groups.
Bioinformatics analysis
Protein networks were created by using the UniProt or Entrez IDs of each protein in “STRING V10” (http://string-db.org/), LENS (http://severus.dbmi.pitt.edu/LENS/index.php/), CPDB (http://cpdb.molgen.mpg.de/) or PANTHER classification system (http://www.pantherdb.org/). The data files from PANTHER were imported into Excel sheets. Then pie charts or bar diagrams were created using either Excel sheets or GraphPad PRISM software.
RESULTS
Urinary proteomic analysis in the cannabis users
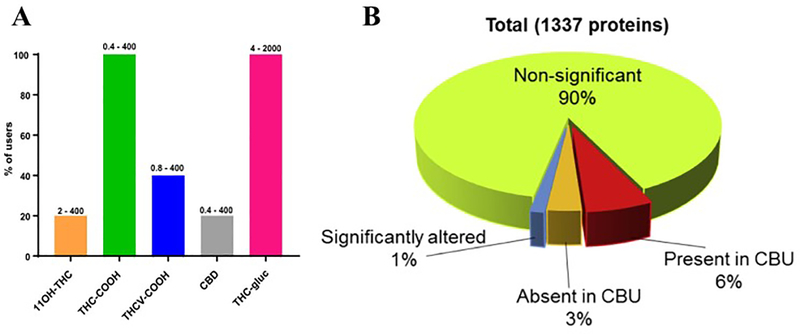
Healthy controls were selected to match cannabis users in terms of age, sex, and race/ethnicity. Both groups did not endorse the use of cigarettes and they did not have any prior medical history of any major diseases. Cannabis users smoked cannabis for 5 days/week/past 12 months for 23±6 years and they were not positive for any other illicit drug. The levels of serum creatinine and blood urea nitrogen (BUN) were similar between the groups, and confirmed the absence of detectable changes in kidney function in cannabis users (Table S1). Analysis of 11 different exocannabinoids revealed that only 5 of them were detectable in the urine of cannabis users. Of that group, 20% of specimens contained 11OH-THC, 20% had CBD, 40% contained THCV-COOH, and all specimens (100%) had a signiifcant amount of both THC-COOH and THC-gluc (Fig. 1A). Liquid chromatography tandem mass spetrometric (LC-MS/MS) quantification was performed to characterize the proteins present in the cannabis users in comparison to non-users (Fig. S1). We detected a total number of 1337 proteins using Scaffold Proteome Software, and identified 19 proteins which were significantly altered in the cannabis users when compared to non-users (p<0.05, Fig. 1B, Table S2). Interestingly, we discovered that 91 proteins were present and 46 proteins were absent only in urine specimens from the cannabis users.
Figure 1. Urinary proteomic analysis in Cannabis users.
(A) Types of cannabinoids present in the urine of cannabis users with the relative percentage of urine specimens containing each type. Different kinds of cannabinoids were measured from each cannabis user urine sample by LC-MS/MS method. Numbers on top of each bars indicate the detectable range of cannabinoid. (B) The chart indicates the number of significantly different proteins between cannabis users and controls (in blue), as well as either proteins present (in red) or absent (in orange) exclusively in cannabis users.
Network and pathway analysis for significantly altered proteins
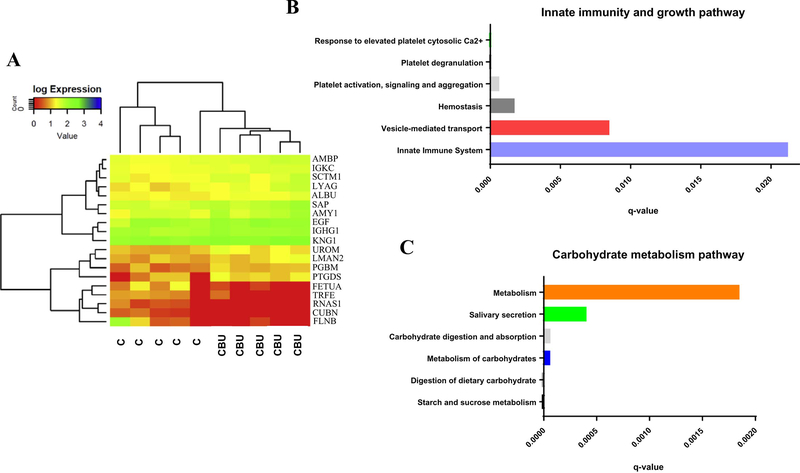
Heatmap revealed expressions of the proteins detected by the permutation approach8. The vertical axis on the heatmap demonstrates the proteins and horizontal axis indicates the samples, while the color indicates the expression level. Both samples and proteins were clustered using hierarchical clustering dendrograms. The cannabis and non-cannabis users were perfectly clustered for these 19 proteins (Fig. 2A). Fourteen proteins were significantly up-regulated (p<0.05 to non-users) and 5 proteins were significantly down-regulated (p<0.05 to non-users) in the urine of cannabis users when compared to healthy controls. The biological role of all 19 proteins is listed in Table S2. Based on the published literature, most of the up-regulated proteins are involved in the regulation of lipid metabolism, immune responses, inflammatory activity, cellular processes, antigen binding and receptor signaling proteins. These results suggest that some of these processes could be up-regulated in the cannabis users (shaded in green). On the contrary, the proteins significantly down-regulated in the cannabis users cohort mainly modulate intestinal and renal absorption, RNA and iron metabolism, neural differentiation, and tumor-related processes (shaded in red).
Figure 2. Analysis of significantly up-regulated proteins in Cannabis users.
(A) Heatmap and dendrogram analysis showing the degree of expression and relationship between the proteins and two subject groups. Significantly up-regulated proteins were searched against CPDB analytic tool for (B) Innate immunity and growth, and (C) Carbohydrate metabolism pathway.
The interactions between two or more proteins is crucial to control various biological functions. We have attempted to determine the possible and known interaction networks among the significantly altered proteins. The proteins were searched against “STRING V10”. Thirteen out of 19 proteins were identified to be involved in established interaction networks, either from the curated database or experimentally determined. Conversely, 4 out of 19 proteins did not have any known binding partner (Fig. S2A). Proteins were classified using PANTHER analytical tool, and the results indicated that majority of these proteins were hydrolases, while fewer belonged to the families of the enzyme modulators, transfer/carrier proteins, receptors and extracellular matrix proteins (Fig. S2B).
To find out the pathways linked to significantly up-regulated proteins, we performed pathway analysis using CPDB online tool (Fig. 2, B and C). We identified that innate immunity, vesicle-mediated, and carbohydrate metabolic pathways were highly linked with proteins that are specifically and significantly up-regulated in cannabis users.
Interaction network of urinary proteins present only in urine of the cannabis users
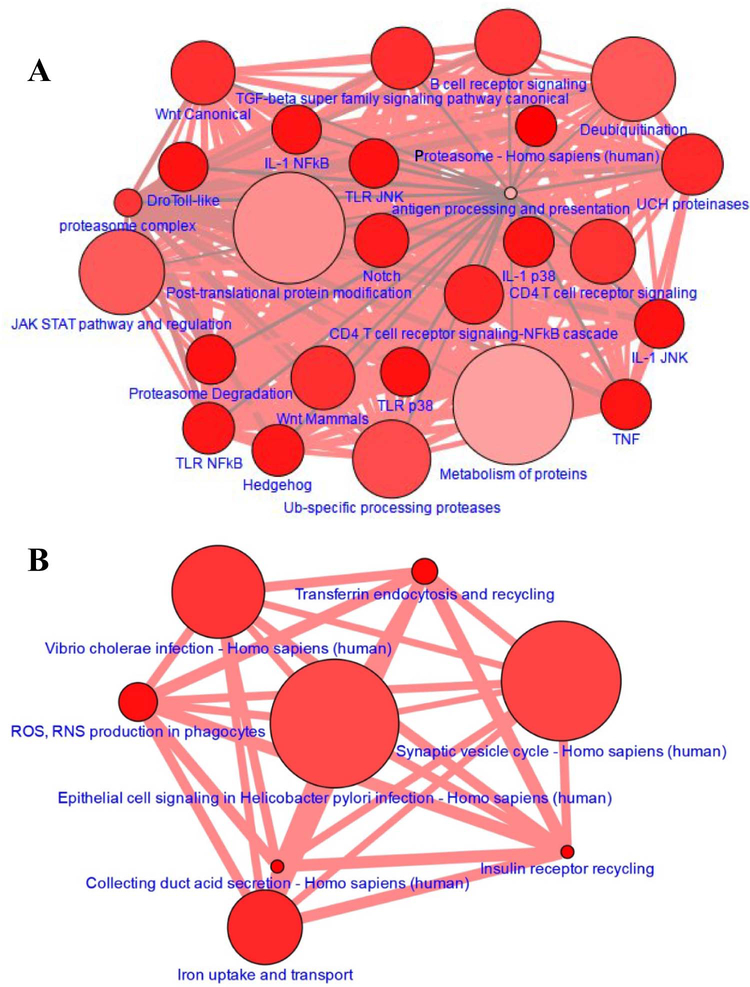
We identified 91 proteins which were present only in urine of the cannabis users but not measurable in the healthy controls (Table S3), and classified these proteins based on their biological processes using PANTHER online tool. The majority of these proteins are involved in the metabolic and cellular processes (Fig. S3, A and B). Sixty out of 91 proteins indicated established and predicted interactions with other proteins, whereas the rest of the proteins stood alone without any known or unknown interacting partner (Fig. S3, C). Pathway analysis according to CPDB demonstrates that candidate proteins were involved in number of immunological and metabolic pathways (Fig 3A). Together, these results suggest that consistent cannabis consumption affects a significant number of immunological and metabolic pathways and stimulates the release of additional proteins in urine which could be used as identifiers of the certain systemic processes in the human body.
Figure 3. Pathway analysis for the proteins that are specific to Cannabis users group.
(A) Proteins present in at least one cannabis user but not in any control were selected and pathways were analyzed using CPDB online tool. (B) Pathway analysis was performed using CPDB tool for the proteins that were absent in all cannabis users. The node is based on the number of entities in the pathway and the edge thickness represents the number of genes shared between each pathway (darker red = more genes).
Network of proteins absent in the urine of the cannabis users
We also identified 46 proteins which were absent in urine of the cannabis users but present in the control group (Table S4). We used the PANTHER classification system to identify the signaling pathways for these proteins. More than two proteins belonged to the integrin signaling group, inflammation pathways mediated by chemokine and cytokine signaling, PI3 kinase pathway, nicotinic receptor signaling, and Huntington disease pathway (Fig. S4, A and B). Using STRING database, we investigated interaction networks among the proteins. Seventeen out of 46 proteins fell into some known and predicted interactions with other targets, while the rest of 31 proteins stood alone without any known partner (Fig. S4, C). Pathway analysis using CPDB demonstrates that candidate proteins were involved in synaptic vesicle cycle, collecting duct acid secretion and insulin receptor recycling pathways (Fig. 3B). These results suggest that the cannabis consumption causes a depletion in a significant number of urinary proteins in comparison to healthy non-users.
Comparative analysis between cannabis users and controls
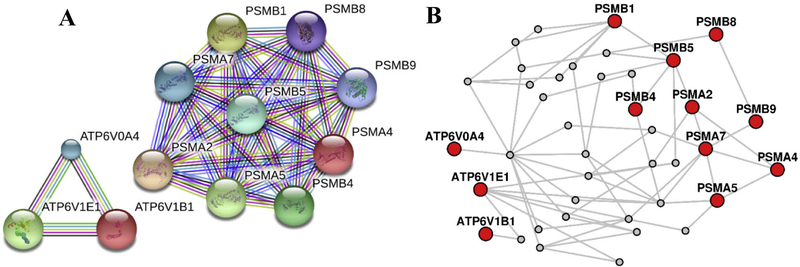
We compared the identified pathways between cannabis users and controls, and perceived that the number of immunological pathways were completely absent in controls in comparison to users (Fig. 3). Then, we chose the candidate proteins involved in both groups, and searched the protein-protein interaction networks using STRING and LENS online tools. We observed no overlapping between the candidate proteins specific and non-specific to the users (Fig. 4). Overall, these results demonstrate that there is an increased immune responses in cannabis users compared to controls.
Figure 4. Protein-protein interaction network established through STRING and LENS online tool for the candidate protein.
Proteins that are specific (PSMs) and not-specific (ATP6Vs) to cannabis users were selected and network analysis was performed using STRING (A) and LENS (B) online tools.
DISCUSSION
Exogenous cannabinoids elicit their well-known diverse effects by activating numerous signaling pathways in tissues and organs. Chronic systemic exposure to cannabis plant compounds can modulate the release of a variety of proteins in urine. Urinary proteins originate from different sources, and are often used as clinical biomarkers of ongoing physiological or pathological processes, and to monitor clinical outcomes in response to therapeutic treatments. Soluble urinary proteins come mainly from plasma by glomerular filtration, or they could be proteolytically cleaved membrane bound proteins, secreted by the thick ascending limb of Henle loop (e.g. uromodulin). Solid phase elements consist of slough epithelial cells and casts, small membrane fragments from shedding of microvilli or by apoptosis, and urinary exosomes 9 Due to extensive psychotropic effects of systemic phytocannabinoids, the latest cannabinoid-based therapeutic strategies mainly target the organs and tissues locally. In case of the urinary bladder, local delivery of drugs can be achieved by the intravesical route. Therefore, the detailed analysis of urinary proteomics in the cannabis users may prove helpful to identify bladder signaling pathways affected by exogenous cannabinoids.
Our study found 1337 urinary proteins using LC-MS/MS, and analyzed the degree of expression and networks of 19 significantly altered proteins. This data is in concord with previous results which identified more than 1500 proteins in urine of healthy individuals 10 Out of 19 significantly different proteins, 5 proteins (cubilin, pancreatic ribonuclease, serotransferrin, alpha-2-HS-glycoprotein and filamin B) were significantly down-regulated in urine of cannabis users. Cubilin is an endocytic receptor participating in intestinal absorption of B12 complex, and reabsorption of proteins in proximal renal tubules11 including albumin12. Alpha-2-HS-glycoprotein (AHSG) was reported as a potential biomarker for colorectal, hypopharyngeal squamous cell and breast cancers13, 14 Due to secretion of AHSG mainly by the liver, it also could serve as a marker of liver dysfunction, cardiovascular disease and disorders associated with metabolic syndrome15. Additional studies suggested that transferrin-like factor acts as an autocrine factor for bladder cancer cells16, and filamin A has a tumor-promoting effect by interacting with signaling molecules17. Down-regulation of these proteins in cannabis users may be indicative of alleviating effects of exocannabinoids in some types of cancer.
A significantly larger number of the urinary proteins (fifteen) were up-regulated in urine of the cannabis users. Kininogen-1 is a protein with antimicrobial activity, participating also in blood coagulation and is usually down-regulated in acute kidney injury18, and renal cell carcinoma19. Uromodulin also modulates cell adhesion and signal transduction, inhibits the aggregation of calcium oxalate crystals, and provides a defense against urinary tract infections due to antimicrobial activity20. Therefore, an up-regulation of urinary uromodulin in the cannabis users may point towards a protective role of exo-cannabinoids in inflammation and infections of the LUT. Pro-epidermal growth factor regulates cell growth and proliferation, and was shown to be down-regulated in macro albuminuria and renal carcinomas21,22 Prostaglandin-H2D-isomerase (PTGDS) is a glutathione-independent prostaglandin D synthase that is involved in various physiological functions, such as vasodilation, thermoregulation, smooth muscle contraction and relaxation, sleep induction and sedation, regulation of immune response, mediation of cellular homeostasis and modulation of nociception. Its overexpression may be particularly relevant as a pathogenic mechanism for occurrence of hyperalgesia and pain23. The identified role of mannose-binding lectin-associated serine protease 2, up-regulated in urine of cannabis users, is consistent with the two-edged sword theory on complement activation in infectious diseases24. A less readily activated complement system leads to increased susceptibility to infection but reduced severity of inflammation. We also detected an up-regulation of enzymes and extracellular matrix proteins, such as Ig kappa, and Ig gamma-1 in urine of the cannabis users, which are involved in immune and inflammatory responses25. We also detected an increased concentration of perlecan in urine of the cannabis users. This proteoglycan is involved in regulation of cell adhesion, endocytosis, bone and cartilage formation, lipid metabolism, inflammation and would healing, thrombosis, cancer angiogenesis, cardiovascular development, and autophagy.19, 26
Cannabis usage is known to affect cortico-striatal networks that are essential for generating movements27. Cannabis compounds easily cross the placenta, and are found in breast milk which may cause problems in neurological development28 and changes in the grey matter associated with cannabis usage29. Consistent with the above reports, we observed that cannabis users specifically excreted proteins involved in Parkinson’s disease and proteins related to locomotion and reproduction. It was previously reported that cannabis-derivative substances might have anti-inflammatory and anti-fibrotic effects30.
We acknowledge several limitations of our study. First, since we used biobanked urine specimens, we could not evaluate LUTS cannabis cohort to link urinary peptide levels with the presence or absence of LUTS. Second, the concentration of proteins in urine may be affected by diet and /or exercise, which were not recorded in this study. The sample size was also relatively limited in this pilot investigation. However, in an effort to truly minimize additional confounders in the data, all cannabis using subjects and controls were otherwise healthy, individuals without chronic medical conditions, confirmed by history, physical exams and laboratory testing.
CONCLUSIONS
In summary, our findings identified differentially up- and down-regulated proteins in urine of cannabis users and non-users, as well as many urinary proteins that are present or absent only in the cannabis group in comparison to control cohort. The evaluation of exclusive proteins of LUT function in cannabis users may help identify specific molecular targets in the bladder for a variety of LUT diseases in the future studies, and provide knowledge for developing intravesical therapeutics to treat LUTS locally.
Supplementary Material
Figure S1. Workflow of the project
Figure S2. Network and pathway analysis of significantly altered proteins
Figure S3. Network and pathway analysis of CBU specific proteins
Figure S4. Network and pathway analysis of proteins absent in CBU
ACKNOWLEDGEMENTS
We thank the research staff of the Colorado Pulmonary-Alcohol Research Collaborative (CoPARC) for timely procurement, processing and delivery of urinary specimens. We also thank Dr. Monika Dzieciatkowska from Mass Spectrometry Facility in our campus. We highly appreciate the help of Cristina Sempio and Jost Klawitter from the iC42 Clinical Research and Development laboratory at our Campus for identifying exogenous cannabinoids.
GRANT SUPPORT
This work was supported by DK095817 grant (A.P.M.) and R24 AA019661 (E.L.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Choi NG, DiNitto DM, Marti CN. Older marijuana users: Life stressors and perceived social support. Drug Alcohol Depend. 2016;169:56–63. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi V, Philips BJ, Su R, et al. Differential expression of functional cannabinoid receptors in human bladder detrusor and urothelium. J Urol. 2009;181:1932–1938. [DOI] [PubMed] [Google Scholar]
- 3.Freeman RM, Adekanmi O, Waterfield MR, Waterfield AE, Wright D, Zajicek J. The effect of cannabis on urge incontinence in patients with multiple sclerosis: a multicentre, randomised placebo-controlled trial (CAMS-LUTS). Int Urogynecol J Pelvic Floor Dysfunct. 2006;17:636–641. [DOI] [PubMed] [Google Scholar]
- 4.Brady CM, DasGupta R, Dalton C, Wiseman OJ, Berkley KJ, Fowler CJ. An open-label pilot study of cannabis-based extracts for bladder dysfunction in advanced multiple sclerosis. Mult Scler. 2004;10:425–433. [DOI] [PubMed] [Google Scholar]
- 5.Consroe P, Musty R, Rein J, Tillery W, Pertwee R. The perceived effects of smoked cannabis on patients with multiple sclerosis. Eur Neurol. 1997;38:44–48. [DOI] [PubMed] [Google Scholar]
- 6.Walczak JS, Cervero F. Local activation of cannabinoid CB(1) receptors in the urinary bladder reduces the inflammation-induced sensitization of bladder afferents. Mol Pain. 2011;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ZY, Wang P, Bjorling DE. Activation of cannabinoid receptor 1 inhibits increased bladder activity induced by nerve growth factor. Neurosci Lett. 2015;589:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgington ESaO P Randomization tests. Fourth Edition ed: Chapman & Hall/CRC, Boca Raton, FL; 2007. [Google Scholar]
- 9.Pejcic M, Stojnev S, Stefanovic V. Urinary proteomics--a tool for biomarker discovery. Renal failure. 2010;32:259–268. [DOI] [PubMed] [Google Scholar]
- 10.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome biology. 2006;7:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilmer MJ, Christensen EI, van den Heuvel LP, Monnens LA, Levtchenko EN. Urinary protein excretion pattern and renal expression of megalin and cubilin in nephropathic cystinosis. Am J Kidney Dis. 2008;51:893–903. [DOI] [PubMed] [Google Scholar]
- 12.Christensen EI, Nielsen R, Birn H. From bowel to kidneys: the role of cubilin in physiology and disease. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2013;28:274–281. [DOI] [PubMed] [Google Scholar]
- 13.Fan NJ, Kang R, Ge XY, et al. Identification alpha-2-HS-glycoprotein precursor and tubulin beta chain as serology diagnosis biomarker of colorectal cancer. Diagn Pathol. 2014;9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi JK, Chang JW, Han W, et al. Autoantibody to tumor antigen, alpha 2-HS glycoprotein: a novel biomarker of breast cancer screening and diagnosis. Cancer Epidemiol Biomarkers Prev. 2009;18:1357–1364. [DOI] [PubMed] [Google Scholar]
- 15.Dabrowska AM, Tarach JS, Wojtysiak-Duma B, Duma D. Fetuin-A (AHSG) and its usefulness in clinical practice. Review of the literature. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia. 2015;159:352–359. [DOI] [PubMed] [Google Scholar]
- 16.Tanoguchi H, Tachibana M, Murai M. Autocrine growth induced by transferrin-like substance in bladder carcinoma cells. Br J Cancer. 1997;76:1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao QQ, Zhang TP, Zhao WJ, et al. Filamin A: Insights into its Exact Role in Cancers. Pathology oncology research: POR. 2016;22:245–252. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Calero L, Martin-Lorenzo M, Ramos-Barron A, et al. Urinary Kininogen-1 and Retinol binding protein-4 respond to Acute Kidney Injury: predictors of patient prognosis? Scientific reports. 2016;6:19667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandim V, Pereira Dde A, Kalume DE, et al. Proteomic analysis reveals differentially secreted proteins in the urine from patients with clear cell renal cell carcinoma. Urologic oncology. 2016;34:5.e11–25. [DOI] [PubMed] [Google Scholar]
- 20.Devuyst O, Dahan K, Pirson Y. Tamm-Horsfall protein or uromodulin: new ideas about an old molecule. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2005;20:1290–1294. [DOI] [PubMed] [Google Scholar]
- 21.Petrides PE, Bock S, Bovens J, Hofmann R, Jakse G. Modulation of pro-epidermal growth factor, pro-transforming growth factor alpha and epidermal growth factor receptor gene expression in human renal carcinomas. Cancer research. 1990;50:3934–3939. [PubMed] [Google Scholar]
- 22.Guo Z, Liu X, Li M, et al. Differential urinary glycoproteome analysis of type 2 diabetic nephropathy using 2D-LC-MS/MS and iTRAQ quantification. Journal of translational medicine. 2015;13:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellei E, Vilella A, Monari E, et al. Serum protein changes in a rat model of chronic pain show a correlation between animal and humans. Scientific reports. 2017;7:41723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasanmoentalib ES, Valls Seron M, Ferwerda B, et al. Mannose-binding lectin-associated serine protease 2 (MASP-2) contributes to poor disease outcome in humans and mice with pneumococcal meningitis. Journal of neuroinflammation. 2017;14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Souza GA, Godoy LM, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome biology. 2006;7:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarosiek I, Schicho R, Blandon P, Bashashati M. Urinary metabolites as noninvasive biomarkers of gastrointestinal diseases: A clinical review. World journal of gastrointestinal oncology. 2016;8:459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prashad S, Filbey FM. Cognitive motor deficits in cannabis users. Curr Opin Behav Sci. 2017;13:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metz TD, Stickrath EH. Marijuana use in pregnancy and lactation: a review of the evidence. Am J Obstet Gynecol. 2015;213:761–778. [DOI] [PubMed] [Google Scholar]
- 29.Koenders L, Cousijn J, Vingerhoets WA, et al. Grey Matter Changes Associated with Heavy Cannabis Use: A Longitudinal sMRI Study. PLoS One. 2016;11:e0152482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zurier RB, Burstein SH. Cannabinoids, inflammation, and fibrosis. Faseb j. 2016;30:3682–3689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Workflow of the project
Figure S2. Network and pathway analysis of significantly altered proteins
Figure S3. Network and pathway analysis of CBU specific proteins
Figure S4. Network and pathway analysis of proteins absent in CBU