Abstract
The dengue virus is considered to be a globally important human pathogen prevalent in tropical and subtropical regions of the world. According to a recent estimate, the disease burden due to DENV infections is ~390 million infections per year globally in ~100 countries including the southern US, Puerto Rico and Hawaii, resulting in nearly ~25,000 deaths mostly among children. Despite the significant morbidity and mortality that results from DENV infections, there is currently no effective chemotherapeutic treatment for DENV infections. We identified curcumin as an inhibitor of DENV2 NS2B/NS3protease in a previous high-throughput screening (HTS) campaign. We synthesized four analogues of curcumin (curcuminoids) and tested the in vitro protease activity and inhibition of replication by cell-based assays. The results revealed that curcumin is a weak inhibitor of the viral protease. However, the analogues exhibited more potent inhibition of DENV infectivity in plaque assays suggesting that the cellular pathway(s) required for viral replication and/or assembly are targeted by these compounds. Further analysis shows that inhibition of genes involved in lipid biosynthesis, and of actin polymerization by curcuminoids, are likely to be involved as their mode of action in DENV2-infected cells. Three of the curcumin derivatives possess good selectivity indices (SI) (>10) when compared to the parent curcumin.
1. Introduction:
Dengue virus (DENV) is a member of mosquito-borne flavivirus genus of Flaviviridae family which consists of > 70 human pathogens causing considerable morbidity and mortality in tropical and subtropical regions of the world (for reviews, see (Beasley, 2005; Gould and Solomon, 2008; Lindenbach et al., 2007)). There are four serotypes of dengue virus (DENV1–4) that together cause 390 million infections annually (Bhatt et al., 2013). DENV is primarily transmitted by the Aedes aegypti and Aedes albopictus mosquitos. Currently, there is no FDA-approved vaccine or antiviral drug available for control or treatment of dengue infections. Primary DENV infection by a single serotype causes self-limiting disease known as dengue fever with symptoms including fever, headache, joint pain and a skin rash, whereas a secondary infection by another serotype results in severe dengue disease through antibody dependent enhancement with symptoms including thrombocytopenia, hemoconcentration due to capillary leak leading to hemorrhage in internal organs and shock syndrome (Garcia et al., 2010; Halstead et al., 2005).
Flaviviruses have a single positive strand RNA genome flanked by highly structured 5′ and 3′ untranslated regions (UTRs) which play important roles in translation and replication of viral RNA (Lindenbach et al., 2007). RNA encodes a single polyprotein that is cleaved by viral and host proteases to produce three structural proteins (C, prM and E) and eight nonstructural proteins including the cleavage product of NS4A (NS1, NS2A, NS2B, NS3, NS4A, NS4A-2K, NS4B and NS5) (for a review, (Padmanabhan and Strongin, 2010)). Previous HTS campaign using purified DENV2 NS2B-NS3pro as a target and subsequent cherry picks for validation by secondary assays identified curcumin as a potential hit that inhibited the purified viral protease NS2B-NS3pro (unpublished results; see also (Balasubramanian et al., 2016)). Subsequently, four curcuminoids were synthesized and characterized for their antiviral activity.
Here we report the antiviral activities of curcumin and the four analogues against purified DENV2 NS2B-NS3pro as well as in stable DENV2 BHK-21/Renilla luciferase reporter replicon expressing cells and DENV2-infected BHK-21 cells. Our results indicate that curcumin and the four analogues were more potent in inhibition of viral replication and infectivity (plaque) assays than their inhibition of protease activity in vitro. The cyclopentanone analogue of curcumin (CC4) was the most active and less toxic compared to curcumin. These results suggested that curcumin and analogues might target cellular pathways required for viral replication. A previous study reported that cellular pathways, actin polymerization and fatty acid biosynthesis, are involved in endoplasmic membrane expansion and formation of viral replication factories in the infected cells (Heaton and Randall, 2011b) (Padilla et al., 2014). Curcumin inhibits hepatitis C virus replication via suppression of the transcription factor, AKt- Sterol regulatory element binding protein −1 (SREBP-1) (Kim et al., 2010). Since SREBP-1 is required for lipid biosynthesis and its downregulation affects downstream genes such as fatty acid synthase (FASN) and acetyl CoA carboxylase (ACC), in this study, we examined the role of curcuminoids on FASN and ACC gene expression by qRT-PCR in DENV2-infected cells. Our results indicate that the curcuminoids inhibit lipogenesis and thereby inhibiting DENV replication. The curcuminoids tested in this study also lead to actin depolymerization similar to curcumin (Dhar et al., 2015) in DENV2-infected cells. Actin plays important roles in entry, viral replication and assembly.
2. Materials and methods:
2.1. Curcuminoids:
Curcumin was purchased from Alfa Aesar (part of Thermo Fisher, Inc) which contained 95% total curcuminoid from turmeric, (botanical name, Rhizome) (CC1; Table 1; CAS number 458-37-7; Stock No. B21573–09). Bisdemethoxycurcumin (CC2; Table 1) was purchased from TCI America (Cat No B3347). Derivatives CC3, CC4 and CC5 (Table 1) were synthesized using the protocol described in Section 2.2.
Table 1.
Chemical structures and molecular weights of compounds used in this study.
| Label Name | Chemical Structure | Molecular Weight |
|---|---|---|
| Curcumin (CC1) |
C21H20O6 Mol. Wt. = 368.3 |
|
| Bis-demethoxy curcumin (CC2) |
C19H16O4 Mol. Wt. = 308.3 |
|
| Acyclic analogue (CC3) |
C19H18O5 Mol. Wt. = 326.35 |
|
| Cyclopentanone analogue (CC4) |
C21H20O5 Mol. Wt. = 352.39 |
|
| Cyclohexanone analogue (CC5) |
C22H22O5 Mol. Wt. = 366.41 |
2.2. General procedure for the chemical synthesis of curcuminoids CC3, CC4, and CC5
The chemical synthesis of curcuminoids was accomplished by a slight modification of the previously reported procedure (Du et al., 2006). Glacial acetic acid (50 mL) was saturated with hydrogen chloride gas for 1 h at 0 °C. A mixture of vanillin (3.04 g, 20 mmole, 2.0 equivalent) and the appropriate ketone (acetone, 580 mg, for CC3, cyclopentanone, 841 mg, for CC4, and cyclohexanone, 981 mg, for CC5, each containing 10 mmole (1.0 equivalent) was slowly added. The reaction mixture was stirred at 0 °C for 15 min, it wa s slowly brought to ambient temperature and stirred at ambient temperature for 48 h. The reaction mixture was poured into ice-cold water (200 mL). The precipitate was filtered using vacuum filtration, washed with ice-cold water (200 mL) and dried. The crude product was purified using silica gel flash column chromatography. Gradient elution (from 30% to 60% ethyl acetate in hexanes) resulted in the isolation of the desired curcuminoids as bright yellow solids in yields of 1.59 g (59%), 3.02 g (79%), 3.24 g (82%) for CC3, CC4, and CC5 (for more details on synthesis refer Supporting information). 1H-, 13C-NMR and IR spectra of CC3, CC4, and CC5 (Fig. S1–S6) were consistent with the proposed structure and previously reported literature. The spectroscopic as well as chromatographic analysis revealed that the synthetic curcuminoids were >95% pure.
2.3. In Vitro Protease Assay:
The DENV2 NS2B-NS3pro expression plasmid encoding the protease precursor used in this study contains the hydrophilic domain of NS2B cofactor (48 amino acids) and the NS3pro domain (185 residues) (Yon et al., 2005). The cloning, expression and purification of the viral NS2B-NS3pro were reported previously (Lai et al., 2014; Mueller et al., 2007; Yusof et al., 2000). In this study, we evaluated the efficacy of curcumin and four curcuminoids. Standard reaction mixture (100 μl) containing 200 mM Tris·HCl (pH 9.5), 6 mM NaCl, 30 % glycerol, 25 nM DENV2 protease, 10 μM or 25 μM inhibitors in DMSO (final DMSO concentration 1%) were incubated 15 min at 25 °C fo llowed by addition of 25 μM protease substrate (Bz)-Nle-Lys-Arg-Arg-AMC. The amount of AMC (7-Amino-4-methylcoumarin) released from the peptide substrate was quantified by measuring the fluorescence (λex.=380 nm, λem.=460 nm) in a monochromater based spectrofluorometer (SpectroMax Gemini EM Microplate reader from Molecular Devices). A potent serine protease inhibitor, aprotinin, was used as a positive control and 1% DMSO (no inhibitor; 100% activity) served as a negative control. For determination of IC50 values, standard protease assays were performed with in the presence of different inhibitor concentrations (1.56, 3.12, 6.25, 12.5, 25, 50, 75, 100, 200 μM). The percent activities after 30 min in the presence of various inhibitor concentrations were plotted using the GraphPad Prism v5.04 using the four-parameter nonlinear regression analysis (Hill slope method).
2.4. BHK/DENV2 Replicon Assay:
The replicon-based assay covers the stages of viral life cycle including viral translation, viral replication consisting of both plus- and minus-sense RNA synthesis. The assay does not include stages such as viral entry, assembly, and maturation due to the absence of genes encoding the structural proteins, C, prM, and E, which are replaced by Renilla luciferase (Rluc) reporter gene (Alvarez et al., 2005; Boonyasuppayakorn et al., 2014; Ng et al., 2007). This assay uses a Baby Hamster Kidney (BHK) cell line that stably expresses a DENV2 replicon with Rluc reporter gene (Boonyasuppayakorn et al., 2014). The replicating RNA can be quantified by measuring the luciferase activity using a kit (Promega) and a luminometer (Berthold Technologies., LB-960) (Boonyasuppayakorn et al., 2014) and the viral RNA can be quantified by quantitative reverse transcriptase-PCR (qRT-PCR).
BHK-21/DENV2 replicon-expressing cells were maintained in Minimal Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS), 100 I.U./ml penicillin, 100 μg/ml streptomycin (Penicillin + Streptomycin), and 300 μg/ml G418 (Fisher Scientific, Pittsburgh, PA). Briefly, BHK-21/DENV2 replicon expressing cells (~5 ×103 in 100 μl) were seeded onto 96-well plates and incubated at 37oC for 24 h in a humidified incubator with 5% CO2. Curcuminoids (CC1- CC5) were added at 0, 0.78, 1.56, 3.12, 6.25, 12.5, 25, 50,100 and 200 μM in 1% DMSO in triplicate wells. Cells were incubated at 37 °C for 24 h, washed, lysed, and Rluc activities were measured as described (Boonyasuppayakorn et al., 2014; Manzano et al., 2011). The results were confirmed by two independent experiments. The Rluc activities were reported as percent inhibition relative to 1% DMSO (0% inhibition) and mycophenolic acid (100% inhibition) as controls (Diamond et al., 2002; Takhampunya et al., 2006).
2.5. Cell-based Cytotoxicity Assay:
The viability of cells in the presence of varying concentration of curcuminoids was measured using the Cell Counting Kit-8 (Dojindo, Rockville, MD) and the concentration at which the cell viability was reduced by 50% (CC50) was determined. The assay is based on a highly soluble, non-toxic tetrazolium salt (WST-8), which is reduced by dehydrogenase activities in cells to give a yellow-colored formazan dye. Briefly, BHK-21 (5× 103) cells (100 μL) in wells of a 96-well plates were treated with varying concentrations (200, 100, 50, 25, 12.5, 6.25, 3.12, 1.56 and 0.78 μM) of curcuminoids (CC1-CC5). After incubation for ~24 h at 37° C under 5 % CO 2, 10 μL of 1X CCK-8 solution was directly added to all the wells. The plates were incubated further at 37° C under 5 % CO 2 for 4 h and the absorbance at 450 nm was read using a spectrophotometer (Molecular Devices). The results were confirmed by two independent experiments. CC50 values were calculated using the GraphPad Prism v5 software. Selectivity index (SI) values were calculated as the ratio of CC50/EC50.
2.6. Plaque assays
Briefly, BHK-21 cells (1 × 105) in MEM-α medium with 10 % FBS were seeded onto each well of 24-well cell culture plates. After the cells were about 80% confluent, they were infected with DENV2 in MEM-α medium supplemented with 2% FBS (multiplicity of infection, MOI of 1 for 1 h. After infection, OPTI-MEM with various drug concentrations (0.07–10 M) was added to the wells. BHK-21 cells treated with only 1% DMSO served as negative control (100% infectivity) and cells treated with 1 μM mycophenolic acid (0% infectivity) served as the positive control. After 24 h the supernatants were collected and analyzed by plaque assays as described below.
LLC-MK2 cells, plated (1× 105 cells/ml/well) in 24-well plates, were incubated at 37 °C until reaching 90% confluence. Supernatants collected as described above were serially diluted in OPTI-MEM and used for infecting duplicate wells for 2 h. Cells were overlaid with l X DMEM supplemented with 10 % FBS containing 1.5% carboxymethyl cellulose (CMC) and were incubated for 7 days at 37°C under 5 % CO2. Plaques were visually examined after fixing the cells with 4% paraformaldehyde and staining with 1% crystal violet for 30 min. The number of plaque forming units (PFU) per ml was determined (Balasubramanian et al., 2016; Boonyasuppayakorn et al., 2014). The EC50(PFU) was calculated from two independent experiments.
2.7. qRT-PCR
Human hepatocarcinoma (Huh 7.5) cells were infected with DENV2 (MOI of 1) and treated with 5 μM of a curcuminoid (CC1-CC5) for 24 h at 37° C und er 5% CO2. Uninfected Huh 7.5 cells were also treated similarly with 5 μM curcuminoids. The total RNAs were extracted using TRIzol reagent (Life technologies) according to the manufacturer’s protocol. DMSO (1%)- treated cells served as negative control. RNA concentrations were determined using BioSpec-nano (Shimadzu Corporation, Japan). Reverse transcription reaction was carried out in 20 μL containing 1μg RNA. Each qRT-PCR carried out in duplicate contained 12.5 μL of the SYBR Green Master Mix reagent (iQ supermix-Biorad), 0.5 μL of primers (20 nM) specifically designed to anneal to the in vitro transcripts (2 μL) from DENV2 NS5 (Manzano et al., 2011), human fatty acid synthase (FASN) and human acetyl-CoA carboxylase (ACC) cDNAs in 9.5 μL of diethylpyrocarbonate-treated water to a final volume of 25 μL. The PCR primers used for human ACC were: forward, CTGTTGGCTCAGATACACTC, and reverse, GCCACAGTGAAATCTCGTT. The PCR primers for human FASN were: forward, GTGGGAAGGTGTACCAGTG, and reverse, GGATGCCCTGGAAATGAG. The amplification protocol consisted of the following steps: 95°C for 2 min, followed by 50 cycles at 95°C for 10 sec, 50°C for 30 sec and 72°C for 30 sec and the melt curve analysis was carried out as described (Balasubramanian et al., 2016; Boonyasuppayakorn et al., 2014). The glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as a reference control. The primer sequences for human GAPDH were: forward, AGGGCTGCTTTTAACTCTGGT and reverse, CCCCACTTGATTTTGGAGGGA.
2.7. Oil Red O staining of lipid droplets
BHK-21 cells were maintained in MEM containing 10% FBS. The cells were infected with DENV2 at a MOI of 1 for an hour followed by incubation with MEM containing 0.1 mM oleic acid-BSA (03008, Millipore Sigma) and various concentrations of curcumin for 24 h. Cells treated with 1% DMSO were used as negative controls. The cultures were washed 3 times with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 10 min. Oil Red O (ORO) working solution was made by mixing 2.4 ml of ORO (0.3% in isopropanol) with 1.6 ml of distilled water and it was filtered (Cui et al., 2010). Cells were incubated with 0.5 mL of the ORO for 15 min, the solution was removed and rinsed three times with PBS. The images were photographed at 40× magnification. The ORO was extracted from the stained cells using 500 μl of 100% isopropanol from each well, and two aliquots of 200 μl were transferred to black-walled 96-well microtiter plates. The absorbance of the dye was measured at 405 nm using a plate reader. All tests were performed in duplicate and the means were calculated (Cui et al., 2010),
2.8. Actin Staining with Phalloidin:
BHK-21 cells were grown in MEM containing 10% FBS. Cells were seeded in 24-well plates (5 × 104/well) and incubated in a humidified CO2 incubator for 24 h. Cells were infected with DENV2 at a MOI of 1 and then treated with 5 μM of curcuminoids dissolved in DMSO (1%) for 24 h. DMSO (1%) alone treated cells were used as controls. The cultures were washed 3 times with PBS and were fixed with 4% formaldehyde for 10 min. Alexa Fluor 488 Phalloidin (Thermo Fischer Scientific) was used to stain the F-actin and the nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). The stained cells were visualized under a fluorescent microscope and images were photographed.
3. Results and Discussion:
Curcumin (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione) is derived from the spice turmeric, Curcuma longa, which is a member of the ginger family and it is a yellow- colored compound primarily found in the roots of turmeric plant (Goel et al., 2008; Jacob et al., 2007; Moghadamtousi et al., 2014). Curcumin is reported to contain various medicinal properties some of which are against cancer, diabetes, microbial infections including those caused by viruses. Curcumin has already been shown to be effective against several enveloped viruses (such as HIV, influenza A virus, herpes simplex virus, hepatitis B virus, Japanese encephalitis virus and DENV) (Chen et al., 2013) and on non-enveloped viruses, coxsackievirus B3 and enterovirus 71 (Chen et al., 2013; Qin et al., 2014). Recently curcumin has also been shown to inhibit Zika and Chikungunya viruses by inhibiting cell binding (Mounce et al., 2017). Curcumin is known to be an inhibitor of DENV2 with pre-treatment or co-treatment with the virus but failed to inhibit when added post entry (Chen et al., 2013). However, it exhibits poor stability at the plasma pH and undergoes a facile retro-aldol decomposition into ferulic acid (Equation 1) (Anand et al., 2007; Wang et al., 1997). Conversion of the β-diketone moiety in curcumin into monoketone analogues is known to minimize retro-aldol decomposition. We designed curcuminoids CC3, CC4, and CC5 in order to improve their stability at plasma pH.
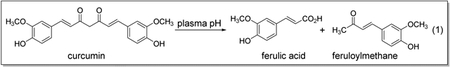 |
Equation 1 |
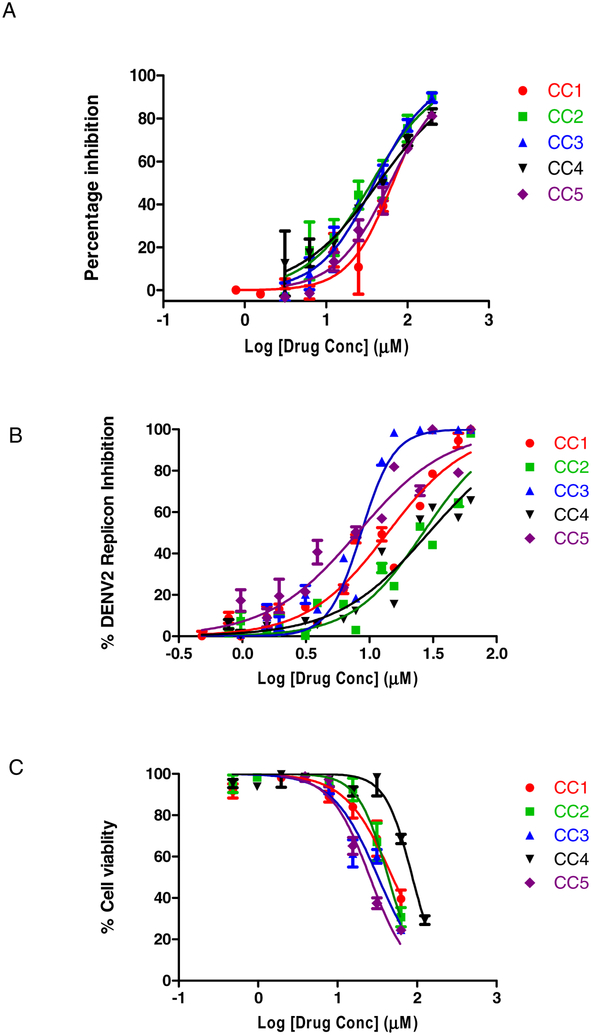
Using an in vitro protease assay, we confirmed that curcumin and its analogues show modest inhibition of protease activity. The IC50 values for the curcuminoids mean ± SD of two replicates were as follows: 66.01 ± 13.12 (CC1), 36.23 ± 9.67 (CC2), 39.17 ± 6.69 (CC3), 43.88 ± 10.14 (CC4) and 60.98 ± 8.7 (CC5) (Fig 1A), consistent with the finding of the parent compound as an HTS hit. The results show that the inhibitory activity of the parent compound, CC1, is less compared to CC2, CC3, CC4 and CC5 analogues. We analyzed the potency of the curcuminoids using the DENV2 Rluc reporter replicon-based assay as described under Materials and Methods. The EC50 values for the curcuminoids mean ± SD of three replicates each from two independent experiments were as follows: 13.91 ± 2.08 (CC1), 26.45 ± 3.67 (CC2), 8.61 ± 0.79 (CC3), 29.25 ± 4.46 (CC4) and 8.07 ± 1.52 (CC5) (Fig 1B). The CC50 values of the curcuminoids mean ± SD of three replicates each from two independent experiments were: 49.01 ± 8.24 (CC1), 43.37 ± 5.19 (CC2), 32.34 ± 4.72 (CC3), 87.40 ± 9.03 (CC4) and 25.50 ± 2.64 (CC5) (Fig 1C). Thus, in replicon assays, curcuminoids show only modest inhibition of viral replication.
Figure 1.
Inhibition of DENV2 replicon replication by curcuminoids (A) BHK-21/DENV2 replicon expressing cells (~5 ×103 in 100 μl) were seeded onto 96-well plates and incubated at 37oC for 24 h in a humidified incubator with 5% CO2. Compounds were added at 0, 0.78, 1.56, 3.12, 6.25, 12.5, 25, 50,100 and 200 μM in 1% DMSO in triplicate wells. Cells were incubated at 37 °C for 24 h, washed, lysed, and Rluc activi ties were measured. The results were confirmed by two independent experiments. (B) Cytotoxicity was measured in the same replicon expressing cells treated with the various concentrations of compounds at 24 h using the Cell Counting Kit −8 following the manufacturer’s instructions.
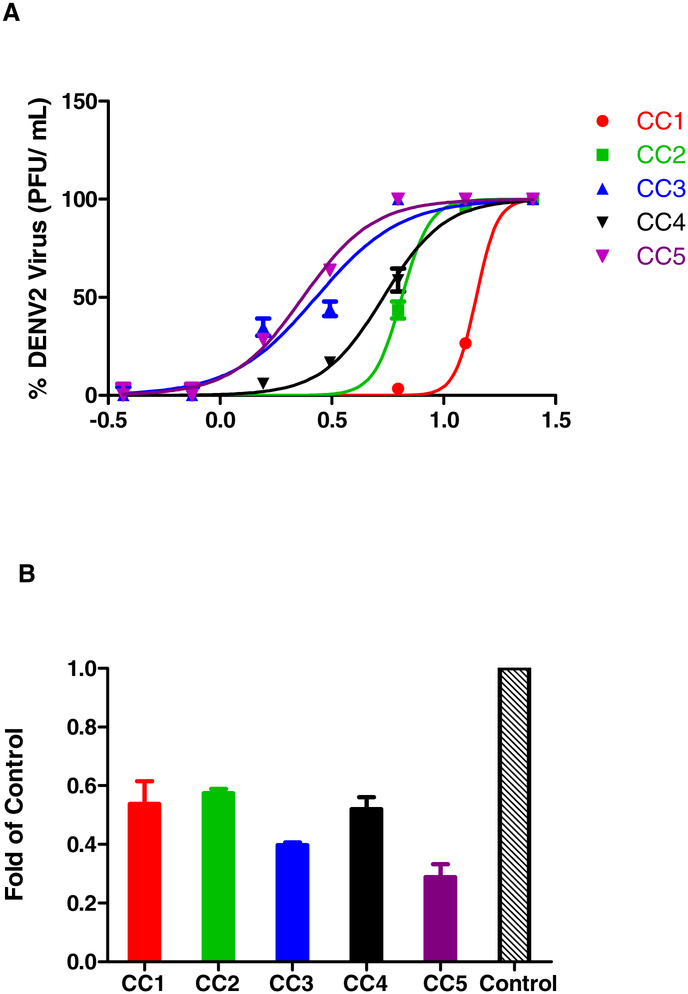
Next, we analyzed the ability of the curcuminoids to inhibit DENV2 infectivity in the formation of infectious particles measured by plaque assays as the concentrations causing 50% reduction in PFU (EC50(PFU)). The EC50(PFU) of curcuminoids were: 13.95 ± 2.57 (CC1), 6.49 ± 0.33 (CC2), 2.68 ± 0.51 (CC3), 5.37 ± 0.49 (CC4) and 2.34 ± 0.22 (CC5). All analogues showed greater inhibition in formation of infectious particles as determined by plaque assays when compared to native curcumin. But only one analogue, CC4, was less toxic than native curcumin. The selectivity indices (SI) were greater than 10 for three analogues (Fig 2A, Table 2). Next, we analysed the RNA copy number by qRT-PCR using RNAs isolated from control DENV2 infected cells in the presence of 1% DMSO alone, and DENV2-infected and curcuminoid (5 μM)-treated cells. The results indicated that in the drug-treated and DENV2-infected cells, the viral RNA copy numbers were reduced when compared to DENV2-infected control in the presence of 1% DMSO alone (Fig 2B).
Figure 2.
Inhibition of DENV2 replication analyzed by plaque assays and qRT-PCR. (A) EC50(PFU) values for inhibition of DENV2 infectivity by curcuminoids. BHK-21 cells were infected with DENV2 (MOI, of 1) and treated with varying concentrations of compounds. Incubation was done at 37 °C and the supernatants were collected at 48 h. The virus titers were determined by plaque assays using LLC- MK2 cells. EC50 values were calculated using GraphPad Prism v5 software. (B) qRT-PCR was performed as described in the Materials and Methods. The fold reduction of viral RNA copy number in DENV2-infected and drug-treated cells as compared to control DENV2-infected cells was determined.
Table 2.
Inhibition of DENV2 infectivity and DENV2 subgenomic replicon by curcuminoids
| Compound | EC50 (PFU) | CC50 | SI (PFU) | EC50 (replicon) | SI (replicon) |
|---|---|---|---|---|---|
| CC1 | 13.95±4.62 | 49.01 ± 8.24 | 3.51 | 13.91 ± 2.08 | 3.52 |
| CC2 | 6.49±0.22 | 43.37 ± 5.19 | 6.68 | 26.45 ± 3.67 | 1.63 |
| CC3 | 2.68±0.64 | 32.34 ± 4.72 | 12.06 | 8.61 ± 0.79 | 3.75 |
| CC4 | 5.37±0.62 | 87.40 ± 9.03 | 16.27 | 29.25 ± 4.46 | 2.98 |
| CC5 | 2.34±0.21 | 25.50 ± 2.64 | 10.89 | 8.07 ± 1.52 | 2.34 |
The EC50 (PFU), CC50 and EC50 (replicon) values were determined as described under Materials and Methods.
The SI (PFU) values represent the ratios of CC50/EC50 (PFU) and the SI (replicon) values represent the ratios of CC50/EC 50 (replicon).
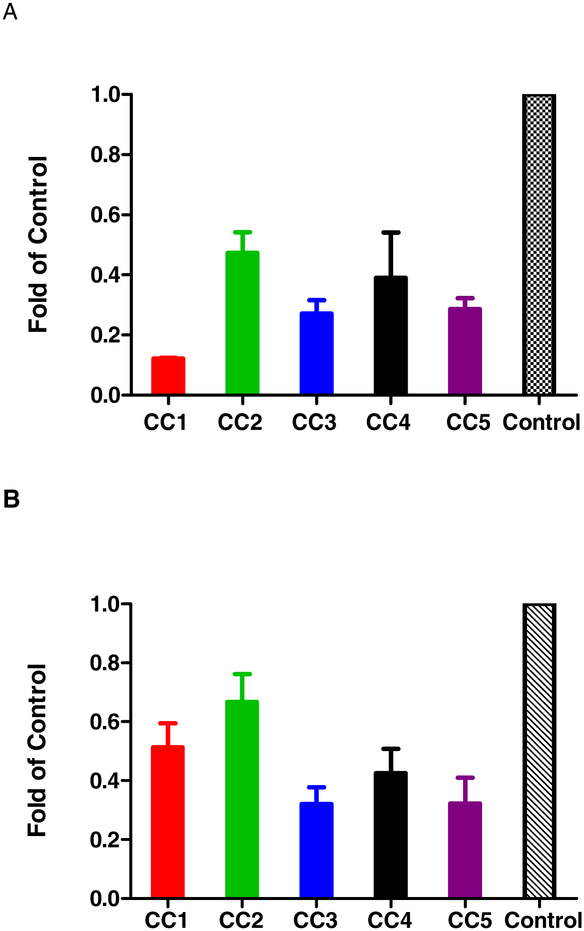
These results, taken together, suggested that curcuminoids inhibit DENV by targeting a host pathway essential for viral life cycle. Previous studies indicated that curcumin has broad spectrum antiviral activities against both RNA and DNA viruses targeting diverse pathways. For example, in one study, curcumin was shown to inhibit the hepatitis C virus entry of all genotypes (Barbosa et al., 2015) by affecting the membrane fluidity, virus binding and fusion. However, it had no effect on RNA replication. In this study, we sought to examine whether curcumin and its analogues exerted their antiviral activity through key step(s) in lipid biosynthesis and metabolism. Several studies previously indicated that HCV and DENV replication and assembly are intimately linked to lipid metabolism (Heaton and Randall, 2010, 2011b). We analyzed the gene expression of acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN) in DENV2-infected and curcuminoids-treated or untreated BHK-21 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a reference gene (Fig 3). The inhibition of gene expression of ACC and FASN was shown as fold reduction of the control in the absence of curcuminoid treatment. The reduction of DENV2 titers corresponded to the reduction in FASN and ACC. Uninfected control cells showed similar reduction in ACC and FASN (Fig S7 & S8).
Figure 3.
qRT-PCR to evaluate the effect of curcuminoids on lipogenesis: (A) qRT-PCR was performed as described in the Materials and Methods. The fold reduction of acetyl-CoA carboxylase (ACC) mRNA copy number in DENV2-infected and drug-treated cells versus the control DENV2-infected cells was determined. (B). The fold reduction of fatty acid synthase (FASN) mRNA copy number in DENV2-infected and drug-treated cells versus the control DENV2 infected cells was determined.
Flavivirus replication and assembly into immature virus particles occur in the membrane compartments of ER which undergo proliferation in infected cells to meet the demand for viral replication and assembly. Lipid droplets (LD), ER-derived lipid-rich cellular organelle, play an important role in storage of excess fatty acids and hydrolysis of neutral lipids for membrane formation and maintenance.
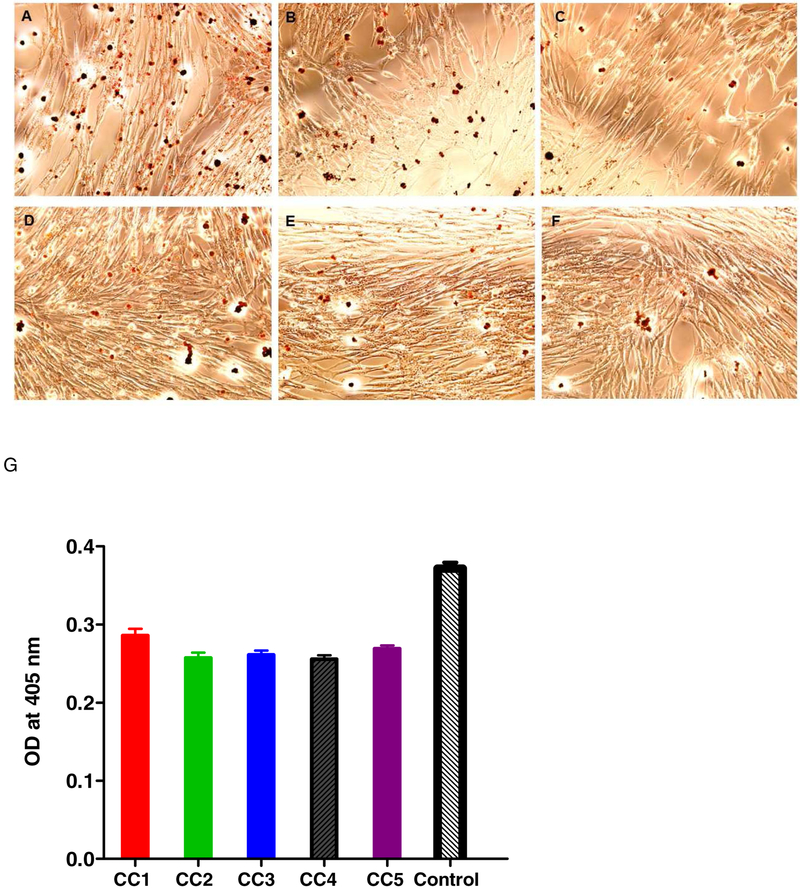
LD’s have been best described in adipocytes, they are present in all cell types and their formation can be induced by oleic acid treatment (Pol et al., 2004). Oleic acid has been shown to enhance the effects of DENV2 virus infection (Ramphan et al., 2017). Here we performed LD staining of DENV2 infected cells treated with oleic acid and with curcuminoids and showed that the curcuminoids were able to suppress the oleic acid-induced LD (Fig 4), the results from two independent experiments were plotted and were found to be statistically significant (pValue < 0.05, Turkey’s test). The LD staining was quantified as described under Materials and Methods and results indicated that curcumin affected LD levels in DENV- infected cells (Fig 4). Curcumin has also been previously shown to suppress FASN expression in adipocytes (Zhao et al., 2011) and reduce the oleic acid-induced lipid accumulation in hepatocytes (Kang et al., 2013).
Figure 4.
LD staining using Oil Red O (ORO). ORO staining protocol described under Materials and Methods was followed. The paraformaldehyde (4%)-fixed cells were stained with ORO, followed by DAPI, and were visualized by fluorescent microscopy. (A) Control oleic acid-treated cells; (B-F): 5 μM curcumioids (CC1- CC5)-treated cells; (G) Quantification of LD after extraction by absorbance at 405 nm.
Flavivirus infection influences the host cell lipids profoundly. Results from lipidomic analysis of membrane fractions associated with DENV replication revealed that 85% of lipids were significantly modified as compared to uninfected control (Perera et al., 2012). DENV infection is highly sensitive to block in the fatty acid synthesis through inhibition of FASN (Heaton et al., 2010). The inhibition of key enzymes in cholesterol synthesis pathway mevalonate diphospho decarboxylase, HMG-CoA synthase and squalene synthetase interfered with DENV replication (Rothwell et al., 2009) indicating the importance of cholesterol in DENV infection. DENV capsid has been previously shown to interact with lipid droplets (Carvalho et al., 2012). DENV virus infection triggers autophagy which release free fatty acids required to maintain high intracellular ATP favoring viral replication (Heaton and Randall, 2010). Host cell lipids have been found to regulate viral entry into cell and replication (Heaton and Randall, 2011a, b; Mazzon and Mercer, 2014). Curcumin has been demonstrated to reduce lipogenesis in high-fat diet mouse model through regulating SREBP pathway (Ding et al., 2016). Here we show that curcuminoids similarly downregulate the lipogenesis which could affect DENV infection.
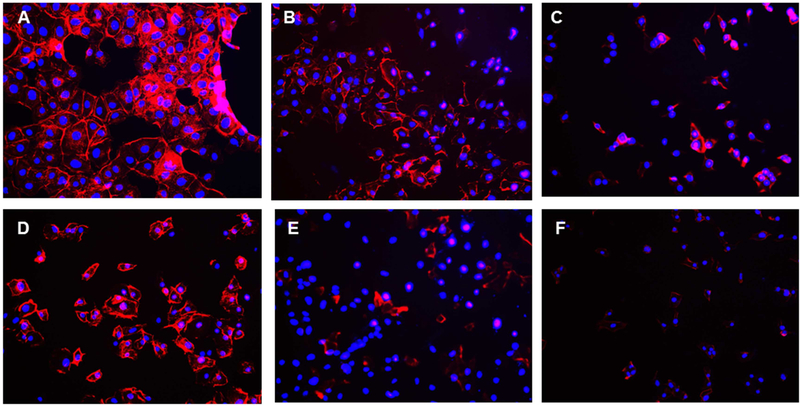
Previous studies reported that curcumin alters the actin cytoskeleton and causes disorganization of actin microfilaments concomitant with apoptosis of treated cells (Padilla et al., 2014). DENV have been shown to have close interaction with cytoskeleton elements in the replication cycle (Foo and Chee, 2015; Jitoboam et al., 2016; Kanlaya et al., 2009). Actin filament has been shown to contribute in the DENV2 entry via interaction with DENV E protein (Kanlaya et al., 2009; Yang et al., 2013). A reduction in viral replication of DENV and WNV was also observed by blocking the actin function (Ang et al., 2010; Colpitts et al., 2011). Native curcumin has been shown to inhibit actin polymerization with an IC50 of 16 μM (Dhar et al., 2015) in DENV2-infected cells. To test if the curcuminoids caused actin disorganization, phalloidin staining method was used to image F-actin (Fig 5) and the results indicate that the curcuminoids caused actin disorganization similar to the parent curcumin in DENV2 infected cells. No significant actin disorganization was observed in mock infected and curcuminoid treated cells (Fig. S9)
Figure 5.
Actin staining with Alexa Fluor 488 Phalloidin. Actin staining using Alexa Fluor 488 Phalloidin and DAPI staining were performed as described under Materials and Methods and visualized under fluorescent microscope. (A): Control DMSO treated cells; (B-F): 5 μM Curcuminoid (CC1- CC5)-treated cells.
Thus, we conclude that curcuminoids exhibit their anti-viral activities through a variety of ways on the host cells, but with modest effect in DENV protease. Modulation of host actin and lipids may be involved in regulating the viral binding and entry. Recent studies on DENV mouse models indicated that curcumin treatment reduced the DENV viremia effectively (Ichsyani et al., 2017) indicating that these analogues might be promising in antiviral therapy against DENV.
Supplementary Material
Highlights.
Curcumin, a natural product, has many medicinal properties including antiviral activities against DNA and RNA viruses.
Curcumin with a β-diketone moiety undergoes retro-aldol decomposition in plasma pH.
To improve its stability, monoketone analogues were synthesized and their anti-dengue viral activities were tested.
Three new curcuminoids had better antiviral activities than native curcumin.
Curcuminoids decreased lipogenic acetyl CoA carboxylase and fatty acid synthase gene expression.
Acknowledgments
This work was supported in whole or in part by National Institutes of Health grants R21AI109185, (to R.P.), and Raman fellowship from University Grants Commission, Govt of India, to R. Pilankatta. This project has been funded in part with Federal funds (UL1TR000101, previously UL1RR031975) from the National Center for Advancing Translational Sciences, National Institutes of Health, through the Georgetown-Howard Universities Clinical and Translational Science Awards Program (CTSA), a trademark of DHHS, part of the Roadmap Initiative, “Re-Engineering the Clinical Research Enterprise” Center for AIDS Research (D, District of Columbia C CFAR; AI117970) and Research Centers in Minority Institutions (RCMI), Howard University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV, 2005. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339, 200–212. [DOI] [PubMed] [Google Scholar]
- Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB, 2007. Bioavailability of curcumin: problems and promises. Molecular pharmaceutics 4, 807–818. [DOI] [PubMed] [Google Scholar]
- Ang F, Wong AP, Ng MM, Chu JJ, 2010. Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virology journal 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian A, Manzano M, Teramoto T, Pilankatta R, Padmanabhan R, 2016. High-throughput screening for the identification of small-molecule inhibitors of the flaviviral protease. Antiviral research 134, 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Savage DB, Siniossoglou S, 2015. Lipid droplet-organelle interactions: emerging roles in lipid metabolism. Current opinion in cell biology 35, 91–97. [DOI] [PubMed] [Google Scholar]
- Beasley DW, 2005. Recent advances in the molecular biology of west nile virus. Curr Mol Med 5, 835–850. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI, 2013. The global distribution and burden of dengue. Nature 496, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyasuppayakorn S, Reichert ED, Manzano M, Nagarajan K, Padmanabhan R, 2014. Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antiviral research 106, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Carneiro FA, Martins IC, Assuncao-Miranda I, Faustino AF, Pereira RM, Bozza PT, Castanho MA, Mohana-Borges R, Da Poian AT, Santos NC, 2012. Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. Journal of virology 86, 2096–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TY, Chen DY, Wen HW, Ou JL, Chiou SS, Chen JM, Wong ML, Hsu WL, 2013. Inhibition of enveloped viruses infectivity by curcumin. PloS one 8, e62482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts TM, Cox J, Nguyen A, Feitosa F, Krishnan MN, Fikrig E, 2011. Use of a tandem affinity purification assay to detect interactions between West Nile and dengue viral proteins and proteins of the mosquito vector. Virology 417, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Chen SL, Hu KQ, 2010. Quantification and mechanisms of oleic acid-induced steatosis in HepG2 cells. American journal of translational research 2, 95–104. [PMC free article] [PubMed] [Google Scholar]
- Dhar G, Chakravarty D, Hazra J, Dhar J, Poddar A, Pal M, Chakrabarti P, Surolia A, Bhattacharyya B, 2015. Actin-curcumin interaction: insights into the mechanism of actin polymerization inhibition. Biochemistry 54, 1132–1143. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Zachariah M, Harris E, 2002. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304, 211–221. [DOI] [PubMed] [Google Scholar]
- Ding L, Li J, Song B, Xiao X, Zhang B, Qi M, Huang W, Yang L, Wang Z, 2016. Curcumin rescues high fat diet-induced obesity and insulin sensitivity in mice through regulating SREBP pathway. Toxicology and applied pharmacology 304, 99–109. [DOI] [PubMed] [Google Scholar]
- Du ZY, Liu RR, Shao WY, Mao XP, Ma L, Gu LQ, Huang ZS, Chan AS, 2006. Alpha-glucosidase inhibition of natural curcuminoids and curcumin analogs. European journal of medicinal chemistry 41, 213–218. [DOI] [PubMed] [Google Scholar]
- Foo KY, Chee H-Y, 2015. Interaction between Flavivirus and Cytoskeleton during Virus Replication. BioMed research international 2015, 427814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G, Sierra B, Perez AB, Aguirre E, Rosado I, Gonzalez N, Izquierdo A, Pupo M, Danay Diaz DR, Sanchez L, Marcheco B, Hirayama K, Guzman MG, 2010. Asymptomatic dengue infection in a Cuban population confirms the protective role of the RR variant of the FcgammaRIIa polymorphism. The American journal of tropical medicine and hygiene 82, 1153–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Kunnumakkara AB, Aggarwal BB, 2008. Curcumin as “Curecumin”: from kitchen to clinic. Biochemical pharmacology 75, 787–809. [DOI] [PubMed] [Google Scholar]
- Gould EA, Solomon T, 2008. Pathogenic flaviviruses. Lancet 371, 500–509. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Heinz FX, Barrett AD, Roehrig JT, 2005. Dengue virus: molecular basis of cell entry and pathogenesis, 25–27 June 2003, Vienna, Austria. Vaccine 23, 849–856. [DOI] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G, 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proceedings of the National Academy of Sciences of the United States of America 107, 17345–17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G, 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell host & microbe 8, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G, 2011a. Dengue virus and autophagy. Viruses 3, 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G, 2011b. Multifaceted roles for lipids in viral infection. Trends in microbiology 19, 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichsyani M, Ridhanya A, Risanti M, Desti H, Ceria R, Putri DH, Sudiro TM, Dewi BE, 2017. Antiviral effects of Curcuma longa L. against dengue virus in vitro and in vivo. IOP Conference Series: Earth and Environmental Science 101, 012005. [Google Scholar]
- Jacob A, Wu R, Zhou M, Wang P, 2007. Mechanism of the Anti-inflammatory Effect of Curcumin: PPAR-gamma Activation. PPAR research 2007, 89369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitoboam K, Phaonakrop N, Libsittikul S, Thepparit C, Roytrakul S, Smith DR, 2016. Actin Interacts with Dengue Virus 2 and 4 Envelope Proteins. PloS one 11, e0151951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang OH, Kim SB, Seo YS, Joung DK, Mun SH, Choi JG, Lee YM, Kang DG, Lee HS, Kwon DY, 2013. Curcumin decreases oleic acid-induced lipid accumulation via AMPK phosphorylation in hepatocarcinoma cells. European review for medical and pharmacological sciences 17, 2578–2586. [PubMed] [Google Scholar]
- Kanlaya R, Pattanakitsakul SN, Sinchaikul S, Chen ST, Thongboonkerd V, 2009. Alterations in actin cytoskeletal assembly and junctional protein complexes in human endothelial cells induced by dengue virus infection and mimicry of leukocyte transendothelial migration. Journal of proteome research 8, 2551–2562. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim KH, Kim HY, Cho HK, Sakamoto N, Cheong J, 2010. Curcumin inhibits hepatitis C virus replication via suppressing the Akt-SREBP-1 pathway. FEBS letters 584, 707–712. [DOI] [PubMed] [Google Scholar]
- Lai H, Teramoto T, Padmanabhan R, 2014. Construction of dengue virus protease expression plasmid and in vitro protease assay for screening antiviral inhibitors. Methods Mol Biol 1138, 345–360. [DOI] [PubMed] [Google Scholar]
- Lindenbach D, Thiel HJ, Rice C, 2007. Flaviviridae: The viruses and their replication, in: Knipe DM, Howley PM (Eds.), Field’s Virology, 5 ed. Lippincott-Raven Publishers, Philadelphia, pp. 1101–1152. [Google Scholar]
- Manzano M, Reichert ED, Polo S, Falgout B, Kasprzak W, Shapiro BA, Padmanabhan R, 2011. Identification of cis-acting elements in the 3’-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. The Journal of biological chemistry 286, 22521–22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M, Mercer J, 2014. Lipid interactions during virus entry and infection. Cellular microbiology 16, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadamtousi SZ, Kadir HA, Hassandarvish P, Tajik H, Abubakar S, Zandi K, 2014. A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed research international 2014, 186864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounce BC, Cesaro T, Carrau L, Vallet T, Vignuzzi M, 2017. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antiviral research 142, 148–157. [DOI] [PubMed] [Google Scholar]
- Mueller NH, Yon C, Ganesh VK, Padmanabhan R, 2007. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int J Biochem Cell Biol 39, 606–614. [DOI] [PubMed] [Google Scholar]
- Ng CY, Gu F, Phong WY, Chen YL, Lim SP, Davidson A, Vasudevan SG, 2007. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antiviral research 76, 222–231. [DOI] [PubMed] [Google Scholar]
- Padilla SL, Rodriguez A, Gonzales MM, Gallego GJ, Castano OJ, 2014. Inhibitory effects of curcumin on dengue virus type 2-infected cells in vitro. Archives of virology 159, 573–579. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R, Strongin AY, 2010. Translation and processing of the dengue virus polyprotein, in: Hanley KA, Weaver SC (Eds.), Frontiers in Dengue Virus Research. Caister Academic Press, Norfolk, U.K., pp. 14–33. [Google Scholar]
- Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ, 2012. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS pathogens 8, e1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Martin S, Fernandez MA, Ferguson C, Carozzi A, Luetterforst R, Enrich C, Parton RG, 2004. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Molecular biology of the cell 15, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Lin L, Chen Y, Wu S, Si X, Wu H, Zhai X, Wang Y, Tong L, Pan B, Zhong X, Wang T, Zhao W, Zhong Z, 2014. Curcumin inhibits the replication of enterovirus 71 in vitro. Acta pharmaceutica Sinica. B 4, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramphan S, Suksathan S, Wikan N, Ounjai P, Boonthaworn K, Rimthong P, Kanjanapruthipong T, Worawichawong S, Jongkaewwattana A, Wongsiriroj N, Smith DR, 2017. Oleic acid Enhances Dengue Virus But Not Dengue Virus-Like Particle Production from Mammalian Cells. Mol Biotechnol 59, 385–393. [DOI] [PubMed] [Google Scholar]
- Rothwell C, Lebreton A, Young Ng C, Lim JY, Liu W, Vasudevan S, Labow M, Gu F, Gaither LA, 2009. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389, 8–19. [DOI] [PubMed] [Google Scholar]
- Takhampunya R, Ubol S, Houng HS, Cameron CE, Padmanabhan R, 2006. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. The Journal of general virology 87, 1947–1952. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Pan MH, Cheng AL, Lin LI, Ho YS, Hsieh CY, Lin JK, 1997. Stability of curcumin in buffer solutions and characterization of its degradation products. Journal of pharmaceutical and biomedical analysis 15, 1867–1876. [DOI] [PubMed] [Google Scholar]
- Yang J, Zou L, Hu Z, Chen W, Zhang J, Zhu J, Fang X, Yuan W, Hu X, Hu F, Rao X, 2013. Identification and characterization of a 43 kDa actin protein involved in the DENV-2 binding and infection of ECV304 cells. Microbes and infection / Institut Pasteur 15, 310–318. [DOI] [PubMed] [Google Scholar]
- Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KH, Padmanabhan R, 2005. Modulation of the nucleoside triphosphatase/RNA helicase and 5’-RNA triphosphatase activities of dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. The Journal of biological chemistry 280, 27412–27419. [DOI] [PubMed] [Google Scholar]
- Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R, 2000. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. The Journal of biological chemistry 275, 9963–9969. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun XB, Ye F, Tian WX, 2011. Suppression of fatty acid synthase, differentiation and lipid accumulation in adipocytes by curcumin. Molecular and cellular biochemistry 351, 19–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.