Abstract
Recent research suggests that the frequency of polyploidy may have been underestimated in gymnosperms. One notable example is in the conifer genus Juniperus, where there are already a few reports of polyploids although data are still missing for most species. In this study, we evaluated the extent of polyploidy in Juniperus by conducting the first comprehensive screen across nearly all of the genus. Genome size data from fresh material, together with chromosome counts, were used to demonstrate that genome sizes estimated from dried material could be used as reliable proxies to uncover the extent of ploidy diversity across the genus. Our analysis revealed that 16 Juniperus taxa were polyploid, with tetraploids and one hexaploid being reported. Furthermore, by analyzing the genome size and chromosome data within a phylogenetic framework we provide the first evidence of possible lineage-specific polyploidizations within the genus. Genome downsizing following polyploidization is moderate, suggesting limited genome restructuring. This study highlights the importance of polyploidy in Juniperus, making it the first conifer genus and only the second genus in gymnosperms where polyploidy is frequent. In this sense, Juniperus represents an interesting model for investigating the genomic and ecological consequences of polyploidy in conifers.
Keywords: Juniperus, gymnosperms, conifers, polyploidy, genome size, flow cytometry
Introduction
Polyploidy or whole genome duplication (WGD) is the heritable condition of possessing more than two complete sets of chromosomes (Comai, 2005). Typically, polyploidy arises either as a result of genome duplication within a species (i.e., autopolyploidy), or from hybridization between two different species followed by chromosome doubling (allopolyploidy) (Stebbins, 1947; Comai, 2005). Most of our understanding of the consequences of polyploidy in plants has come from the study of angiosperms, where it has been shown that polyploidization generally causes a dramatic change in genomic structure, dynamics and expression, and cell organization (Tayalé and Parisod, 2013; Van de Peer et al., 2017; Wendel et al., 2018). Indeed, polyploidy is considered to have played a major role in angiosperm evolution (Blanc and Wolfe, 2004; Chen, 2007; Otto, 2007; Soltis and Soltis, 2009).
While polyploidy has been reported to occur across all major taxonomic land plant groups (Barker et al., 2016), it has been estimated to be very frequent in angiosperms with 50–80% of species being polyploid (Masterson, 1994; Otto and Whitton, 2000) and possibly all angiosperms contain at least one WGD in their ancestry (Van de Peer et al., 2017). In contrast, only 5% of all gymnosperms are reported to be polyploid based on chromosome counts (Khoshoo, 1959; Ahuja, 2005; Husband et al., 2013; Rice et al., 2015). Nevertheless, recent analyses of transcriptomic and genomic data (e.g., Li et al., 2015; Guan et al., 2016; Roodt et al., 2017) have suggested that the evolution of gymnosperms was accompanied by several ancient WGD events, including two within conifers, one at the base of Pinaceae (c. 200–342 million years ago) and one at the base of the cupressophytes (including Cupressaceae but excluding Araucaceae) (c. 210–275 million years ago). This highlights the importance of polyploidy in the very early evolution of conifers in contrast to the extreme rarity of this phenomenon among extant species [estimated to be 1.5% based on chromosome counts (Khoshoo, 1959; Husband et al., 2013; Rice et al., 2015)]. The one notable exception to the low frequency of polyploidy in extant gymnosperms is in Ephedra, which belongs to the non-coniferous lineage Gnetales. Here, polyploidy has been reported in over 65% of extant Ephedra species (Ickert-Bond et al., 2015). In this genus no evidence for any ancient WGDs has been detected in its ancestry (Li et al., 2015).
Conifers comprise the largest group of extant gymnosperms (Christenhusz et al., 2011), and from a phylogenetic perspective, they are divided into two major clades—(i) the Pinaceae and (ii) cupressophytes as they include Cupressaceae which is the most species-rich family (Lu et al., 2014; Ran et al., 2018). Within extant conifers, chromosome counts of all studied wild stands of all genera of Pinaceae are reported to be diploid (2n = 2x = 24) (Hizume, 1988; Murray, 2013) despite an exceptional genome size variation in some genera, such as Pinus L. (34.5–72.0 pg/2C) (Bogunic et al., 2003; Murray et al., 2012).
Similarly, in Cupressaceae, among ca. 91 species studied for their chromosome number to date (Hair, 1968; Murray, 2013), nearly all are diploid (2n = 2x = 22), with just three natural polyploids reported: Sequoia sempervirens is hexaploid with 2n = 6x = 66 (Ahuja and Neale, 2002; Scott et al., 2016), while Fitzroya cupressoides (Molina) I. M. Johnst. (alerce) and Juniperus thurifera L. are tetraploid with 2n = 4x = 44 (Hair, 1968; Romo et al., 2013; Vallès et al., 2015). It is also notable that within Juniperus, the study of just three species revealed each had polyploid cytotypes in some populations (Sax and Sax, 1933; Nagano et al., 2007). These findings raise the question of whether polyploidy may be common in this genus and hence whether it has played a more significant role in the evolution of Cupressaceae than previously recognized in gymnosperms as a whole, and in conifers in particular.
In this study, we focused on exploring the prevalence of polyploidy in wild populations of Juniperus. With 115 taxa (75 species with 40 varieties; Adams (2014), also see Table 1 for species and varieties), Juniperus is the most diverse genus in Cupressaceae and the second most diverse in all conifers after Pinus (Farjon, 2010; Romo et al., 2013). Juniperus has been shown to be a well-supported monophyletic genus (Mao et al., 2010; Adams and Schwarzbach, 2013; Adams, 2014), that can be divided into three monophyletic sections: (i) section Caryocedrus, with one species in the Mediterranean; (ii) sect. Juniperus, with 14 species, 12 in East Asia and the Mediterranean, and one with a circumboreal distribution (Juniperus communis L.) and one [J. jackii (Rehder) R. P. Adams] endemic to North America; and (iii) sect. Sabina, with ~60 species distributed in southwestern North America, Asia and the Mediterranean region, with outlier species in Africa and the Canary Islands. The few polyploids in wild populations noted above have all been reported to occur in species belonging to sect. Sabina. Both diploid and tetraploid cytotypes have been found in some populations of J. chinensis L. (Sax and Sax, 1933; Hall et al., 1973; Zonneveld, 2012) and in some populations of J. sabina L. (Siljak-Yakovlev et al., 2010; Farhat et al., 2019). Few sporadic triploid and tetraploid cytotypes have also been found in some ornamental cultivars. Juniperus thurifera is the only species reported to be exclusively tetraploid (2n = 4x = 44 and 40 pg/2C) (Romo et al., 2013; Vallès et al., 2015). More recently, Bou Dagher-Kharrat et al. (2013) showed that J. foetidissima Willd. had a very large genome (59.74 pg/2C), c. 3-fold larger than confirmed diploid Juniperus species which range from 19.02 to 26.40 pg/2C (Bennett and Leitch, 2012). The exceptional genome size of J. foetidissima, suggests this species may be hexaploid (Bou Dagher-Kharrat et al., 2013) but cytogenetic studies are needed to confirm this since genome size alone may be misleading as it can be highly variable between species of the same genus that have the same ploidy level (Ledig, 1998; Morse et al., 2009; Abdel Samad et al., 2014).
Table 1.
List of the Juniperus taxa studied with indication of data collection, type of material, genome size data, and chromosome numbers.
| Section | Species | var. | Coll # | Location source | Dried/Fresh | Date coll. | 2C (pg) | CV plant (%) | CV standard (%) | 2n | 1Cx (pg) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Caryocedrus | J. drupacea Labill. | Adams 14693 | Turkey | Dried | 2015 | 23.48 | 7.19 | 2.44 | 22(i) | 11.74 | |
| Juniperus | J. brevifolia (Seub.) Ant. | Adams 8152 | Azore Islands | Dried | 1997 | 22.28 | 4.42 | 2.34 | 22(i)(ii) | 11.14 | |
| J. cedrus Webb and Berthol. | Adams 11510 | La Palma | Dried | 2008 | 24.70 | 7.54 | 2.67 | 22(i)(ii) | 12.35 | ||
| J. communis L. | Adams 9035 | France | Dried | 2000 | 24.48 | 4.98 | 3.56 | 22(i) | 12.24 | ||
| J. communis L. | RBGK 1977-1318 | NA | Fresh | 2017 | 22.28 | 2.9 | 2.42 | 22(i) | 11.14 | ||
| J. communis | charlottensis R. P. Adams | Adams 10304 | Canada | Dried | 2004 | 22.32 | 4.71 | 2.28 | 22(i) | 11.16 | |
| J. communis | depressa Pursh | Adams 10940 | New Mexico | Dried | 2005 | 22.13 | 3.93 | 2.38 | 22(i)(ii) | 11.07 | |
| J. communis | hemispherica (J. and C. Presl) Parl. | Adams 9045 | Italy | Dried | 2000 | 22.66 | 3.67 | 2.09 | 22(i)(ii) | 11.33 | |
| J. communis | kamchatkensis R. P. Adams | Adams 9182-9164 | Denmark | Dried | 2000 | – | – | – | – | – | |
| J. communis | kelleyi R. P. Adams | Adams 10890 | USA | Dried | 2005 | 22.30 | 2.86 | 2.29 | 22(i) | 11.15 | |
| J. communis | megistocarpa Fernald and H. St. John | Adams 8576 | Quebec | Dried | 1998 | 22.50 | 4.2 | 2.44 | 22(i) | 11.25 | |
| J. communis | nipponica (Maxim.) E. H. Wilson | Adams 8579 | Japan | Dried | 1998 | 21.92 | 3.97 | 2.22 | 22(i) | 10.96 | |
| J. communis | oblonga hort. ex Loudon (= var. communis) | Adams 8765 | Armenia | Dried | 1999 | 22.29 | 3.72 | 2.67 | 22(i)(ii) | 11.15 | |
| J. communis | saxatilis Pall. | Adams 8686 | Japan | Dried | 1998 | 22.87 | 4.93 | 2.47 | 22(i)(ii) | 11.44 | |
| J. communis | saxatilis Pall. | Adams 10378 | Spain | Dried | 2004 | 22.30 | 4.2 | 3.1 | 22(i) | 11.15 | |
| J. communis | saxatilis Pall. | Adams 11206 | Norway | Dried | 2006 | 21.82 | 4.06 | 2.44 | 22(i) | 10.91 | |
| J. communis | saxatilis (sibirica) Pall. | Adams 7589 | Mongolia | Dried | 1995 | 23.92 | 4.22 | 2.42 | 22(i) | 11.96 | |
| J. deltoides R. P. Adams | Adams 14466 | Azerbaijan | Dried | 2014 | 22.87 | 3.87 | 2.88 | 22(i) | 11.44 | ||
| J. deltoides R. P. Adams | spilinanus (Yalt., Elicin and Terz.) Terz. | Adams 12064-12065 | Turkey | Dried | 2010 | 22.93 | 4.34 | 3.26 | 22(i) | 11.47 | |
| J. formosana Hayata | Adams 9071 | Taiwan | Dried | 2000 | 22.31 | 4.06 | 2.33 | 22(i)(ii) | 11.16 | ||
| J. formosana Hayata | RBGK 1995-2911 | NA | Fresh | 2017 | 23.03 | 3.01 | 2.44 | 22(i)(ii) | 11.52 | ||
| J. jackii (Rehder) R. P. Adams | Adams 10287 | USA | Dried | 2004 | 22.57 | 3.87 | 2.44 | 22(i) | 11.29 | ||
| J. macrocarpa Sibth. and Sm. | Adams 14047 | Turkey | Dried | 2013 | 25.74 | 4.33 | 3.2 | 22(i)(ii) | 12.87 | ||
| J. maderensis (Menezes) R. P. Adams | Adams 11497 | Madeira Island | Dried | 2008 | 22.64 | 5 | 2.15 | 22(i) | 11.32 | ||
| J. mairei Lemee and H. Leveille | Adams 6772 | China | Dried | 1991 | 23.16 | 3.9 | 2.69 | 22(i) | 11.58 | ||
| J. navicularis Gand. | Adams 8240 | Portugal | Dried | 1997 | 22.66 | 4.93 | 2.5 | 22(i) | 11.33 | ||
| J. oxycedrus L. | Adams 9039 | France | Dried | 2000 | 23.08 | 4.98 | 2.69 | 22(i)(ii) | 11.54 | ||
| J. oxycedrus | badia H. Gay | Adams 7795 | Spain | Dried | 1996 | 22.32 | 3.87 | 2.93 | 22(i)(ii) | 11.16 | |
| J. rigida Siebold and Zucc. | Adams 8544 | Japan | Dried | 1998 | 22.31 | 4.25 | 2.56 | 22(i)(ii) | 11.16 | ||
| J. rigida | conferta Parl. | Adams 8585 | Japan | Dried | 1998 | 21.81 | 3.43 | 2.13 | 22(i) | 10.91 | |
| J. taxifolia Hook. and Arn. | Adams 8448 | Japan | Dried | 1998 | 22.44 | 4.79 | 4.66 | 22(i)(ii) | 11.22 | ||
| J. taxifolia | lutchuensis (Koidz.) Satake | Adams 8541 | Japan | Dried | 1998 | 22.04 | 3.42 | 2.5 | 22(i)(ii) | 11.02 | |
| J. angosturana R. P. Adams | Adams 6881 | Mexico | Dried | 1991 | 26.79 | 3.85 | 2.51 | 22(i) | 13.4 | ||
| J. arizonica R. P. Adams | Adams 14908 | USA | Dried | 2015 | 27.64 | 3.98 | 2.42 | 22(i) | 13.82 | ||
| J. ashei Buchholz | Adams 12260 | USA | Dried | 2004 | 25.30 | 3.24 | 2.24 | 22(i)(ii) | 12.65 | ||
| J. barbadensis L. | Adams 5368 | St. Lucia | Dried | 1996 | 24.20 | 2.73 | 2.35 | 22(i) | 12.1 | ||
| J. barbadensis | lucayana Britton | Adams 11408 | Bahamas | Dried | 2008 | 24.21 | 3.16 | 2.67 | 22(i) | 12.11 | |
| J. bermudiana L. | Adams 2554 | Bermuda Island | Dried | 1995 | 25.81 | 3.2 | 2.7 | 22(i)(ii) | 12.91 | ||
| Sabina | J. bermudiana L. | RBGK 2011-1530 | NA | Fresh | 2017 | 25.51 | 3.19 | 2.71 | 22(i)(ii) | 12.76 | |
| J. blancoi Martinez | Adams 10258-10259 | Mexico | Dried | 2004 | 24.82 | 3.97 | 2.35 | 22(i) | 12.41 | ||
| J. blancoi | huehuentensis R. P. Adams, S. Gonzalez, and M. G. Elizondo | Adams 10247 | Mexico | Dried | 2004 | 24.83 | 3.95 | 2.95 | 22(i) | 12.42 | |
| J. blancoi | mucronata (R. P. Adams) Farjon | Adams 8701 | Mexico | Dried | 1998 | 25.28 | 3.15 | 2.37 | 22(i) | 12.64 | |
| J. californica Carriere | Adams 8698 | Aizona, USA | Dried | 1998 | 28.16 | 3.53 | 2.15 | 22(i) | 14.08 | ||
| J. californica Carriere | Adams 10154 | California, USA | Dried | 2004 | 28.45 | 3.16 | 2.28 | 22(i) | 14.23 | ||
| J. carinata Y. F. YU and V L. K. FU | Adams 8504 | China | Dried | 1998 | 24.30 | 3.31 | 2.21 | 22(i) | 12.15 | ||
| J. chinensis L. | Adams 8535 | Japan | Dried | 1998 | 47.51 | 3.38 | 2.78 | 44(i)/22and 44(ii) | 11.88 | ||
| J. chinensis | procumbens Sieb. ex Endl. | Adams 8683 | Japan | Dried | 1998 | 46.77 | 3.11 | 3.07 | 44(i)(ii) | 11.7 | |
| J. chinensis | sargentii A. Henry | Adams 8688 | Japan | Dried | 1998 | 49.67 | 3.77 | 2.52 | 44(i)/22(ii) | 12.42 | |
| J. coahuilensis (Martinez) Gaussen | Adams 14814 | Texas, USA | Dried | 2016 | 26.56 | 5.79 | 2.88 | 22(i) | 13.28 | ||
| J. comitana Martinez | Adams 6859 | Mexico | Dried | 1991 | 27.57 | 5.06 | 2.8 | 22(i)(ii) | 13.79 | ||
| J. convallium Rehder and Wilson | Adams 6781 | China | Dried | 1991 | 26.29 | 4.14 | 2.96 | 22(i) | 13.15 | ||
| J. coxii A. B. Jacks | Adams 8137 | Chimili Valley, Burma | Dried | 1997 | 50.70 | 4.51 | 2.11 | 44(i) | 12.68 | ||
| J. davurica Pallas | Adams 7253 | Mongolia | Dried | 1994 | 23.99 | 3.49 | 2.6 | 22(i)(ii) | 12 | ||
| J. davurica | arenaria (E. H. Wilson) R. P. Adams | Adams 10347 | China | Dried | 2004 | 24.30 | 3.38 | 2.43 | 22(i) | 12.15 | |
| J. davurica | mongolensis R. P. Adams | Adams 7254 | Mongolia | Dried | 1994 | 23.80 | 2.96 | 2.35 | 22(i) | 11.9 | |
| J. deppeana Steudel | Adams 10539 | Mexico | Dried | 2005 | 26.39 | 3 | 2.32 | 22(i) | 13.2 | ||
| J. deppeana Steudel | Adams 10927 | Arizona, USA | Dried | 2005 | 25.93 | 4.05 | 2.55 | 22(i)(ii) | 12.97 | ||
| J. deppeana | gamboana (Mart.) R. P. Adams | Adams 6869 | Mexico | Dried | 1991 | 26.36 | 4.1 | 2.7 | 22(i) | 13.18 | |
| J. deppeana | patoniana (Martinez) Zanoni | Adams 6837-11904 | Mexico | Dried | 1991 | – | – | – | – | – | |
| J. deppeana | robusta Martinez | Adams 10255 | Mexico | Dried | 2004 | 25.83 | 3.01 | 2.49 | 22(i) | 12.92 | |
| J. deppeana | robusta Martinez | Adams 10256 | Mexico | Dried | 2004 | 25.76 | 3.62 | 2.45 | 22(i) | 12.88 | |
| J. deppeana | sperryi (Correll) R. P. Adams | Adams 11494 | USA | Dried | 2008 | 25.75 | 3.91 | 2.67 | 22(i) | 12.88 | |
| J. deppeana | zacatacensis (Mart.) R. P. Adams | Adams 10517-10518 | Mexico | Dried | 2009 | 25.80 | 2.9 | 2.33 | 22(i) | 12.9 | |
| J. durangensis Martinez | Adams 10253-11929 | Mexico | Dried | 2009 | 25.54 | 3.73 | 2.58 | 22(i) | 12.77 | ||
| J. durangensis | topiensis R. P. Adams and S. Gonzalez | Adams 11923 | Mexico | Dried | 2009 | 25.64 | 4.33 | 2.12 | 22(i) | 12.82 | |
| J. erectopatens (Cheng and L. K. Fu) R. P. Adams | Adams 8532-8533-8534 | China | Dried | 1998 | – | – | – | – | – | ||
| J. excelsa M.-Bieb. | Adams 14742 | Greece | Dried | 2015 | 27.41 | 4.47 | 2.27 | 22(i)(ii)(iii) | 13.71 | ||
| J. fargesii (Rehder and Wils.) Kom. | Adams 6769 | China | Dried | 1991 | 25.33 | 3.65 | 2.27 | 22(i) | 12.67 | ||
| J. flaccida Schlecht. | Adams 6892 | Mexico | Dried | 1991 | 26.05 | 3.56 | 2.37 | 22(i) | 13.03 | ||
| J. foetidissima Willd. | Adams 14511 | Greece | Dried | 2015 | 71.32 | 3.56 | 3.15 | 66(i)(iii) | 11.89 | ||
| J. foetidissima Willd. | Adams (waiting for assignment) | Lebanon | Fresh | 2017 | 69.71 | 3.84 | 3.91 | 66(i)(iii) | 11.62 | ||
| J. foetidissima Willd. | Adams (waiting for assignment) | Turkey | Fresh | 2018 | 70.7 | 3.4 | 3.2 | 66(i)(iii) | 11.78 | ||
| J. gracilior Pilger | Adams 7664 | Dom. Rep. | Dried | 1996 | 24.97 | 3.15 | 2.29 | 22(i) | 12.49 | ||
| J. gracilior | ekmanii (Florin) R. P. Adams | Adams 7653 | Haiti | Dried | 1996 | 25.59 | 4.44 | 2.6 | 22(i) | 12.8 | |
| J. gracilior | urbaniana (Pilger and Ekman) R. P. Adams | Adams 12314 | Dom. Rep. | Dried | 2009 | 28.05 | 4.28 | 2.42 | 22(i) | 14.03 | |
| J. gracilior | saxicola (Britton and P. Wilson) R. P. Adams | Adams 5284 | Cuba | Dried | 1985 | 25.55 | 3.29 | 2.24 | 22(i) | 12.78 | |
| J. grandis R. P. Adams | Adams 11963 | California, USA | Dried | 2009 | 25.81 | 3.04 | 2.47 | 22(i) | 12.91 | ||
| J. horizontalis Moench | Adams 14381 | Canada | Dried | 2014 | 24.64 | 5.25 | 3.23 | 22(i)(ii) | 12.32 | ||
| J. indica Bertol. | Adams 8775 | Nepal | Dried | 1999 | 48.81 | 3.95 | 2.55 | 44(i)(ii) | 12.2 | ||
| J. indica Bertol. | Adams 12943 | Nepal | Dried | 2011 | 48.07 | 3.7 | 2.68 | 44(i) | 12.02 | ||
| J. indica Bertol. | RBGK 2010-2167 | NA | Fresh | 2017 | 48.85 | 2.13 | 2.03 | 44(i) | 12.21 | ||
| J. indica | caespitosa Farjon | Adams 7625-7626 | Nepal | Dried | 1995 | – | – | – | – | – | |
| J. jaliscana Martinez | Adams 15491-15492 | Mexico | Dried | 1991 | 29.50 | 3.41 | 3.02 | 22(i) | 14.75 | ||
| J. komarovii Florin | Adams 8518 | China | Dried | 1998 | 24.76 | 3.55 | 2.21 | 22(i) | 12.38 | ||
| J. maritima R. P. Adams | Adams 11056 | Vancouver Island, Canada | Dried | 2006 | 25.17 | 3.75 | 2.35 | 22(i) | 12.59 | ||
| J. martinezii Perez de la Rosa | Adams 14901 | Mexico | Dried | 2016 | 27.31 | 3.87 | 3.07 | 22(i) | 13.66 | ||
| J. microsperma (Cheng and L. K. Fu) R. P. Adams | Adams 8522 | China | Dried | 1998 | 23.66 | 3.89 | 1.97 | 22(i) | 11.83 | ||
| J. monosperma (Engelm.) Sarg. | Adams 10932 | New Mexico | Dried | 2005 | 26.96 | 4.02 | 2.12 | 22(i)(ii) | 13.48 | ||
| J. monticola Martinez | Adams 6876 | Mexico | Dried | 1991 | 24.86 | 4.82 | 2.15 | 22(i) | 12.43 | ||
| J. morrisonicola Hayata | Adams 8681 | Taiwan | Dried | 1998 | 46.61 | 2.8 | 2.62 | 44(i) | 11.65 | ||
| J. occidentalis Hook. | Adams 13546 | Oregon, USA | Dried | 2012 | 26.39 | 3.85 | 2.03 | 22(i)(ii) | 13.2 | ||
| J. osteosperma (Torr.) Little | Adams 14310 | Utah, USA | Dried | 2014 | 26.87 | 5.41 | 3.18 | 22(i) | 13.44 | ||
| J. ovata R. P. Adams | Adams 12279 | Texas, USA | Dried | 2010 | 25.48 | 4.95 | 2.9 | 22(i) | 12.74 | ||
| J. phoenicea L. | Adams 13813 | Spain | Dried | 2013 | 24.76 | 4.43 | 2.98 | 22(i)(ii) | 12.38 | ||
| J. phoenicea L. | RBGK 1996-114 | NA | Fresh | 2017 | 24.86 | 2.96 | 2.47 | 22(i)(ii) | 12.43 | ||
| J. pinchotii Sudworth | Adams 10463 | Texas, USA | Dried | 2004 | 26.24 | 3.3 | 2.04 | 22(i) | 13.12 | ||
| J. pingii Cheng and Ferre | Adams 8506 | China | Dried | 1998 | 25.49 | 3.23 | 2.16 | 22(i) | 12.75 | ||
| J. pingii | miehei Farjon | Adams 13598 | Tibet | Dried | 2000 | 29.11 | 5.55 | 1.9 | 22(i) | 14.56 | |
| J. poblana (Mart.) R. P. Adams | Adams 15208-15209 | Mexico | Dried | 2016 | 24.39 | 3.98 | 2.75 | 22(i) | 12.2 | ||
| J. poblana (Mart.) R. P. Adams | Adams 14898 | Nayarit, MX | Dried | 2016 | 26.95 | 4.42 | 2.29 | 22(i) | 13.48 | ||
| J. poblana (Mart.) R. P. Adams | decurrens R. P. Adams | Adams 11926 | Durango, Mexico | Dried | 2009 | – | – | – | – | – | |
| J. polycarpos K. Koch | Adams 14171 | Azerbaijan | Dried | 2013 | 24.92 | 4.68 | 3.09 | 22(i) | 12.46 | ||
| J. polycarpos | turcomanica (B. Fedtsch.) R. P. Adams | Adams 8757 | Turkmenistan | Dried | 1999 | 24.89 | 2.76 | 2.4 | 22(i) | 12.45 | |
| J. procera Hochst. ex. Endl. | Adams 15222-15223 | Ethiopia | Dried | 2016 | 24.44 | 4.2 | 2.46 | 22(i)(ii) | 12.22 | ||
| J. procera Hochst. ex. Endl. | RBGK 2013-277 | NA | Fresh | 2017 | 24.01 | 3.42 | 2.34 | 22(i)(ii) | 12.01 | ||
| J. przewalskii Kom. | Adams 6775 | China | Dried | 1991 | 48.90 | 3.27 | 2.38 | 44(i) | 12.23 | ||
| J. pseudosabina Fisch., Mey. and Ave-Lall. | Adams 7808 | Kazakstan | Dried | 1996 | 24.73 | 3.22 | 2.32 | 22(i)(ii) | 12.37 | ||
| J. recurva Buch.-Ham. ex D. Don. | Adams 7215 | Nepal | Dried | 1993 | 47.50 | 2.87 | 3.78 | 44(i)/22(ii) | 11.88 | ||
| J. recurva Buch.-Ham. ex D. Don. | RBGK 1976-826 | NA | Fresh | 2017 | 49.05 | 2.55 | 2.62 | 44(i)/22(ii) | 12.26 | ||
| J. rushforthiana R. P. Adams | Adams 8140 | Bhutan | Dried | 1997 | 49.94 | 4.52 | 2.2 | 44(i) | 12.49 | ||
| J. sabina L. | Adams 14316 | Azerbaijan | Dried | 2014 | 24.65 | 4.49 | 2.76 | 22(i)(ii)/44(ii) | 12.33 | ||
| J. sabina L. | balkanensis R. P. Adams and A. N. Tashev | Adams 14722 | Bulgaria | Dried | 2015 | 46.36 | – | – | 44(iiii) | – | |
| J. saltillensis M. T. Hall | Adams 6886 | Mexico | Dried | 1991 | 26.32 | 3.02 | 2.06 | 22(i) | 13.16 | ||
| J. saltuaria Rehder and Wils. | Adams 6789 | China | Dried | 1991 | 26.04 | 4.61 | 2.34 | 22(i) | 13.02 | ||
| J. scopulorum Sarg. | Adams 10895 | Utah, USA | Dried | 2005 | 25.10 | 3.34 | 2.37 | 22(i)(ii) | 12.55 | ||
| J. scopulorum Sarg. | RBGK 2004-1660 | NA | Fresh | 2017 | 25.89 | 2.78 | 2.21 | 22(i)(ii) | 12.95 | ||
| J. semiglobosa Regel | Adams 8210 | Kyrgystan | Dried | 1997 | 26.41 | 4.4 | 2.03 | 22(i) | 13.21 | ||
| J. semiglobosa Regel | jarkendensis (Kom.) R. P. Adams | Adams 7820 | China | Dried | 1996 | 24.96 | 4.06 | 2.3 | 22(i) | 12.48 | |
| J. semiglobosa Regel | talassica (Lipsky) Silba | Adams 8220-8221-8222 | Kyrgystan | Dried | 1997 | 27.24 | 4.8 | 2.1 | 22(i) | 13.62 | |
| J. seravschanica Kom. | Adams 8224 | Kazakhstan | Dried | 1997 | 48.58 | 2.89 | 2.99 | 44(i)/22(ii) | 12.15 | ||
| J. squamata Buch.-Ham. ex. D. Don in Lambert | Adams 6796 | China | Dried | 1991 | 48.55 | 4.86 | 2.74 | 44(i) | 12.14 | ||
| J. squamata | meyeri Rehder (cv.) | Adams 13547 | China | Dried | 2012 | 46.29 | 3.88 | 3.38 | 44(i) | 11.57 | |
| J. squamata | wilsonii (Rehder) R. P. Adams | Adams 12912 | China | Dried | 2012 | 25.60 | 9.33 | 3.14 | 22(i) | 12.8 | |
| J. standleyi Steyermark | Adams 15396 | Mexico | Dried | 1991 | 30.30 | 4.26 | 2.56 | 22(i) | 15.15 | ||
| J. thurifera L. | Adams 9452 | Spain | Dried | 2001 | 48.81 | 3.54 | 2.56 | 44(i)(ii) | 12.2 | ||
| J. thurifera L. | RBGK 2015-61 | NA | Fresh | 2017 | 47.14 | 2.59 | 2.31 | 44(i)(ii) | 11.79 | ||
| J. thurifera | africana Maire | Adams 9420 | Morocco | Dried | 2001 | 48.23 | 3.53 | 2.16 | 44(i)(ii) | 12.06 | |
| J. tibetica Kom. | Adams 8516 | China | Dried | 1998 | 48.27 | 2.9 | 2.26 | 44(i) | 12.07 | ||
| J. tibetica Kom. | RBGK 2013-276 | NA | Fresh | 2017 | 49.43 | 2.96 | 2.63 | 44(i) | 12.36 | ||
| J. tsukusiensis Masam. | Adams 8806 | Japan | Dried | 1999 | 23.75 | 2.94 | 2.5 | 22(i) | 11.88 | ||
| J. tsukusiensis | taiwanensis (R. P. Adams and C-F. Hsieh) | Adams 9061 | Taiwan | Dried | 2000 | 23.95 | 3.84 | 2.67 | 22(i) | 11.98 | |
| J. turbinata Guss. | Adams 7202 | Spain | Dried | 1993 | 25.28 | 4.65 | 2.8 | 22(i)(ii) | 12.64 | ||
| J. turbinata Guss. | Adams 12397 | Turkey | Dried | 2010 | 26.38 | 4.14 | 1.96 | 22(i) | 13.19 | ||
| J. uncinata R. P. Adams | Adams 7212 | Nepal | Dried | 1993 | 24.51 | 3.56 | 2.75 | 22(i) | 12.26 | ||
| J. virginiana L. | Adams 10231 | Tennessee USA | Dried | 2004 | 24.91 | 2.49 | 1.79 | 22(i)(ii) | 12.46 | ||
| J. virginiana | silicicola (Small) E. Murray | Adams 11113-11114 | Florida, USA | Dried | 2006 | 24.81 | 3.53 | 2.2 | 22(i) | 12.41 | |
| J. virginiana | silicicola (Small) E. Murray | RBGK 1984-8179 | NA | Fresh | 2017 | 24.66 | 4.6 | 3.94 | 22(i) | 12.33 | |
| J. zanonii R. P. Adams | Adams 6900 | Mexico | Dried | 1991 | 25.19 | 3.3 | 3 | 22(i) | 12.6 |
“coll #” correspond to the herbarium voucher specimens deposited at Baylor University Herbarium (BAYLU) “Adams #” or to accessions from the living collections of the Royal Botanic Gardens Kew “RBGK #,” Chromosome numbers
deduced from genome size data,
retrieved from CCDB,
directly observed in this study,
from Farhat et al. (2019), CV: coefficient of variation of the 2C values.
Altogether, these observations suggest that Juniperus may have undergone an unusual evolutionary trajectory, involving polyploidization more frequently than encountered in other conifers. This paper takes a first step toward addressing these gaps in our data to fully understand the role that polyploidization has played in the evolutionary history of Juniperus. The objective was to assess variation in genome size across the whole genus and use these data as a proxy to estimate ploidy levels. Using classical cytogenetics approaches, we also determined the ploidy level of J. foetidissima, which has the biggest genome in this genus. Finally, we used phylogenetically-informed trait evolution modeling approaches to reconstruct ancestral genome sizes for the three main clades of Juniperus and identify the occurrence of polyploidization events during the evolution of Juniperus.
Materials and Methods
Plant Material
The origins of the studied accessions are presented in Table 1. We used Robert P. Adams's worldwide collection of Juniperus leaf material, dried in silica gel and kept frozen at −20°C. This material has been stored for years (the oldest sample was collected in 1985). To address its suitability for genome size analysis and ploidy screening, we carried out measurements on both dry and fresh material for a sub-sample of 12 species which were selected to cover as much of the genus diversity at the taxonomic (representatives of sections Juniperus and Sabina), morphological (needles-like and scale leaves) and cytogenetic (species with different ploidy levels) levels. Fresh leave material was obtained from plants growing in the living collections of the Royal Botanic Gardens, Kew, UK.
Genome Size Assessments by Flow Cytometry
Nuclear DNA contents of about 3,000 stained nuclei were estimated for each sample with a CyFlowSL Partec flow cytometer (Partec GmbH) following the one-step protocol of Doležel et al. (2007) with minor modifications as described in Clark et al. (2016). We selected Allium cepa L., 2C = 34.89 pg (Doležel et al., 1998; Clark et al., 2016) and the “CyStain PI Absolute P kit” buffer (Sysmex UK) as the most appropriate internal calibration standard and nuclei isolation buffer for ploidy screening in Juniperus.
Chromosome Counts
We compiled published Juniperus chromosome numbers from the Chromosome Counts Database (CCDB; Rice et al., 2015). New chromosome counts were made for J. foetidissima and J. excelsa using 3 years old plants cultivated from seed of natural origin (from Turkey), and following Vallès et al. (2015) for protoplast preparation and Chromomycin A3 (CMA, Serva) staining.
Analyses of Genome Size and Chromosome Number Evolution
Trait evolution was modeled on the phylogenic tree of Adams (2014), pruned to the set of species and varieties with genome size data and made ultrametric with R v.3.2.2 (Team, 2016). However, five taxa with genome size estimates were not represented in the phylogeny and so they were discarded from these analyses [Juniperus communis var. kelleyi R. P. Adams, J. deltoides var. spilinanus (Yalt., Elicin and Terz.) Terz, J. durangensis var. topiensis R. P. Adams and S. Gonzalez, J. poblana var. decurrens R. P. Adams, J. semiglobosa var. talassica (Lipsky) Silba)]. The inference of ancestral genome size values was based on monoploid GS (1Cx-values) sensu Greilhuber et al. (2005). Ancestral 1Cx-values were reconstructed under ML using the “fastAnc” command and mapped onto the phylogeny with the “contMap” command of the Phytools package of R (Revell, 2012).
We used ChromEvol v.2 (Glick and Mayrose, 2014) to infer ancestral haploid (n) chromosome numbers in Juniperus. This program implements a series of likelihood models to infer duplication events, chromosome gains/losses and demi-duplications at ancestral nodes. The model that best fitted the data set was chosen under the Akaike information criterion (AIC) using default parameters.
Results
Genome Size Diversity
Genome sizes were assessed for 111 Juniperus species and varieties (Table 1), representing 96.5% of taxonomic diversity. Low differences were found between values obtained with dried and fresh material for the 12 species analyzed using both types of leaf material. Differences varied around zero with six positive (minimum = 0.6%, maximum = 9.8% and mean difference = 3.1%) and six negative percentages (minimum = −0.42%, maximum = −3.16% and mean difference = −2.15%). Overall, the genome size estimates for Juniperus ranged 3.2-fold (from 21.81 to 70.58 pg/2C) but they were seen to be distributed into three non-overlapping classes (Figure 1), class A: 21.81–30.3 pg/2C, B: 46.29–50.7 pg/2C, and C: 70.58 pg/2C.
Figure 1.
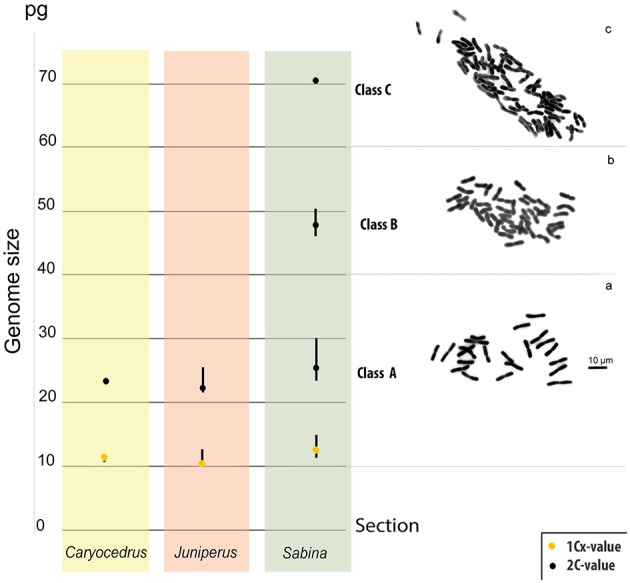
Genome size (2C-values, represented by black dots) classes in Juniperus and their unequivocal relationship with the chromosome number. Class A represents the range of genome sizes for all diploid species confirmed by published chromosome numbers. Class B represents the range of genome sizes for all tetraploid species confirmed by published chromosome number. Class C represents the genome size of the only hexaploid species so far reported (i.e., J. foetidissima). (A) Chromosomes of the diploid J. excelsa (our data); (B) chromosomes of the tetraploid J. thurifera (reproduced from Vallès et al., 2015), and (C) Chromosomes of J. foetidissima confirming its hexaploid status (our data). Monoploid genome size (1Cx-values, represented by yellow dots) of the three sections were also illustrated.
Ploidy Levels Inferred From Genome Size Data
We gathered chromosome number data from the CCDB for 41 Juniperus species and varieties (Table 1). In addition, we made the first chromosome counts for J. excels—a diploid with 2n = 22, and J. foetidissima—a hexaploid with 2n = 66 (Figures 1A,C, respectively). Ploidy levels based on chromosome numbers agreed with those inferred from genome size for all but two taxa, suggesting a strong correlation between genome size, ploidy level and chromosome number. Genome size values of class A corresponded to diploids with 2n = 2x = 22, class B to tetraploids with 2n = 4x = 44 and class C to hexaploids with 2n = 6x = 66 (Table 1; Figure 1). The two exceptions were J. seravschanica Kom. and J. chinensis var. sargentii A. Henry, which were both reported to be diploid in the CCDB but had genome size estimates indicating the samples analyzed here were tetraploid. We thus considered these taxa to have two cytotypes, as previously established for J. chinensis and J. sabina (Table 1).
Evolution of Chromosome Numbers
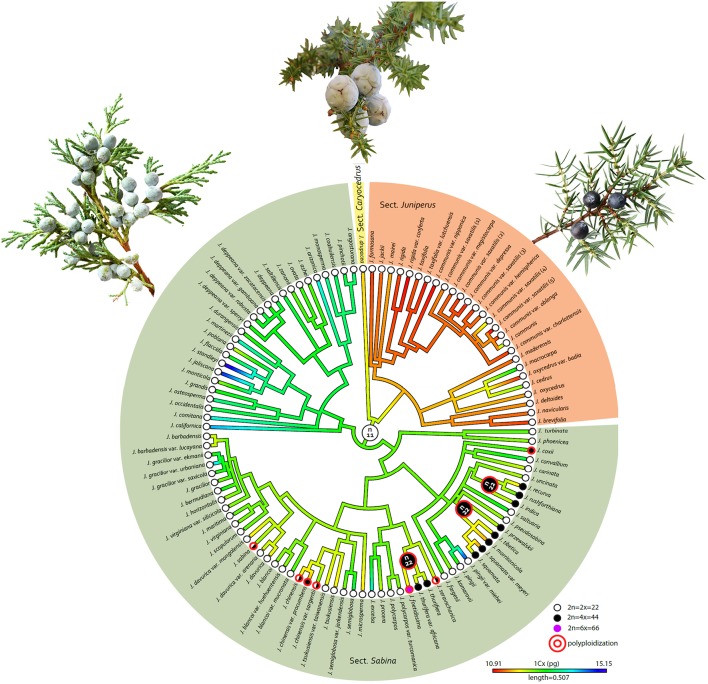
The best-fitting model in ChromEvol to explain the evolution of chromosome numbers in Juniperus was the CONST_RATE model (Supplementary Table S1), suggesting that polyploidy is the predominant mode of chromosome evolution in Juniperus. The ancestor of the whole genus was inferred to be diploid, with n = 11. It is noted that the polyploids were exclusively restricted to sect. Sabina (Figure 2). Three lineage-specific polyploidization events leading to tetraploidy were detected in the ancestors of the clades giving rise to (i) J. recurva, J. rushforthiana, J. indica, (ii) J. preswalskii, J. tibetica, J. morrisonicola, J. squamata, and (iii) J. thurifera, J. foetidissima (Figure 2). A further gain of 22 chromosomes was inferred in the lineage giving rise to the hexaploid J. foetidissima. Six species-specific or within-species polyploidization events (i.e., cytotypes) were found in J. coxii, J. sevaschanica, J. chinensis, J. chinensis var. procumbens, J. chinensis var. sargentii and J. sabina, all of which contained both diploid and tetraploid individuals (Figure 2).
Figure 2.
Ancestral state reconstruction of genome size (1Cx/pg) and chromosome number (n) on a phylogenetic tree of Juniperus reconstructed using Bayesian approaches (Adams, 2014). An illustration of the leaf shape for each section is represent by: (a) J. drupacea (sect. Caryocedrus); (b) J. communis (sect. Juniperus); (c) J. excelsa (sect. Sabina).
Evolution of Genome Size
Beside the genome size variation explained by chromosome number difference, a small variation at the 1Cx-level was detected between ploidy levels. In addition, the distribution of 1Cx-values across Juniperus presented in Figures 1, 2 showed an ancestral genome size of 12.37 pg for the whole genus and overall larger values in species belonging to sect. Sabina (mean 1Cx 12.7 pg, ancestral 1Cx 12.64 pg) compared with those of sect. Caryocedrus (mean 1Cx 11.74 pg, ancestral 1Cx 12.15 pg) and sect. Juniperus (mean 1Cx 11.38 pg, ancestral 1Cx 11.59 pg). Nevertheless, decreases in 1Cx-values were observed in several taxa from sect. Sabina, including some –but not all– polyploids. Polyploid taxa showed limited 1Cx variation relative to the value inferred for their most recent ancestors, with a maximum 1Cx downsizing of 5.70% for J. squamata var. meyeri, and a maximum 1Cx upsizing of 1.71% in J. rushfortiana (Supplementary Table S2).
Discussion
Reliability of Genome Size Estimates From Desiccated Leaf Material of Juniperus
Over the years considerable attention has focused on exploring the suitability of dried plant material for genome size and ploidy level analysis, especially given the challenges of collecting and analyzing fresh material from plants growing in remote locations. Dried material has certainly shown to be suitable for ploidy level analysis in many vascular plants (Suda and Trávníček, 2006; Schönswetter et al., 2007; Suda et al., 2007; Popp et al., 2008; Krejčíková et al., 2013; Wang and Yang, 2016). Nevertheless, the quality of data generated by flow cytometry using dried material has been shown to differ between species, buffers (Bainard et al., 2011) and type of desiccation used (Šmarda et al., 2005; Šmarda and Stančík, 2006; Suda and Trávníček, 2006) and it is now generally accepted that while desiccated material is suitable for ploidy level analysis, it is usually not reliable enough for accurate genome size estimations.
In contrast to these previous studies, our analyses of Juniperus showed that leaves dried in silica gel and stored continuously at −20°C are suitable for genome size estimations using flow cytometry, giving reasonable data quality (i.e., mean %CV = 3.9, S.D. = 0.96). This was supported by comparisons of 2C-values estimated for the same species from dried and fresh material where low differences between the two variances were found in the 12 species analyzed. We are thus confident that the genome size data generated from the desiccated material analyzed here are reliable and hence suitable for exploring genome size [but there might be a slight shift in “absolute” genome sizes (9.8% at maximum)] and ploidy diversity and evolution across Juniperus. Our results broadly agree with Bainard et al. (2011) who found that leaves desiccated immediately in the field using silica gel, was one of the most promising conservation methods, yielding reasonable quality flow cytometry peaks for some species.
Variability in Genome Size and Polyploidy in Juniperus
This study showed that junipers are characterized by possessing large genomes (mean genome size for diploid taxa = 25 pg/2C) with extensive variation between species (ranging 3.2-fold from 21.81 to 71.32 pg/2C). This large variation perfectly correspond to known ploidy levels (2x – 6x), while the variation in 1Cx is only 1.38-fold. The data considerably extend our knowledge of genome sizes in Juniperus which was previously based on data for just 19 species (Bennett and Leitch, 2012). They also show Juniperus now has the largest range in genome size so far reported for any conifer genus.
There are three main mechanisms which can lead to variation in genome size; (i) rapid loss or expansion of transposable and/or other repetitive elements, (ii) loss or gain of chromosomes (aneuploidy and dysploidy), and (iii) polyploidization, possibly followed by genome downsizing (Ramsey and Schemske, 1998; Leitch and Bennett, 2004; Greilhuber et al., 2005; Morse et al., 2009). While in Pinus the high variability in genome size (34.50–72.00 pg/2C; Murray et al., 2012) has been shown to be mainly driven by variation in copy numbers of repeats, such as retrotransposable elements (Morse et al., 2009; Kovach et al., 2010; Nystedt et al., 2013), in Juniperus, our data indicate that most of the variation in genome size is due to variation in ploidy levels. This does not exclude the occurrence of limited genome size variation within each ploidy level, but based on the data presented, it is relatively small, ranging just 1.4-fold in diploids (95 taxa) and 1.1-fold in tetraploids (15 taxa). The source of this variation is still unclear but likely to represent variation in repeat content since, to date, there have been no reports of aneuploidy in the genus (Murray, 2013).
Among the 111 taxa analyzed, just two (J. chinensis var. sargentii and J. seravschanica) showed a discrepancy between the chromosome number reported in the CCDB and the ploidy level estimated from the genome size data obtained here. This could be due to a technical error, such as misidentification of the species used for counting chromosomes and such an explanation is possible for J. seravschanica, where the synonym taxa J. macropoda Boiss. has been used to determine the ploidy level (Rice et al., 2015). Nevertheless, these exceptions could also be explained by the existence of intra-specific variability in ploidy levels (= cytotype diversity), a well-documented phenomenon encountered in many land plant lineages, especially in angiosperms and ferns (Husband et al., 2013). In contrast, cytotype diversity is rarely reported in gymnosperms, with Ephedra being the only genus where it occurs extensively (>50% of species have >1 cytotype—Ickert-Bond et al., 2015). Prior to this study, natural intraspecific variation in ploidy level in Juniperus had only been reported in a few species including in J. chinensis (2x, 4x) (Sax and Sax, 1933; Hall et al., 1973) and J. sabina (2x, 4x) (Siljak-Yakovlev et al., 2010; Farhat et al., 2019).
In view of these previous studies, the results presented here are striking—revealing a much higher frequency of polyploidy in Juniperus than hitherto detected, with 15% of taxa being tetraploid, and the discovery of an hexaploid (J. foetidissima), which is only the second hexaploid to be found in conifers. In addition, the use of ChromEvol to infer the evolution of chromosome numbers across the phylogeny of Juniperus suggests that there have been an unexpectedly high number of polyploidization events throughout its evolutionary history compared with other gymnosperm lineages (except Ephedra). Such a result suggests that mechanisms that promote polyploidization and/or the evolutionary success of polyploid species have occurred at a much higher frequency in Juniperus than in other conifers, and even in gymnosperms in general, apart from Ephedra. It is also worth noting that only one individual was analyzed for most taxa in this study. It is therefore possible that our data underestimate the importance of polyploidization in Juniperus as additional intraspecific ploidy diversity may well be uncovered when more individuals are analyzed, as already seen in J. sabina and J. chinensis.
Genome Size Evolution and Ploidy Levels of Juniper Ancestors
Studies exploring the evolution of genome size diversity across different land plant groups, have uncovered contrasting dynamics in genome size fluctuations throughout their evolution (Bainard and Villarreal, 2013; Clark et al., 2016; Soltis et al., 2018). Now that genome size data are available for almost every recognized taxa of Juniperus and that ploidy levels can be inferred given the robust relationship with genome size (Figure 1), the reconstruction of the ancestral genome size within this genus and inferred ancestral ploidy level is highly instructive. Indeed, apart from Pinus (Grotkopp et al., 2004), our study is the first to reconstruct ancestral genome size within a species-rich genus for any gymnosperm. Our analysis revealed that the ancestral ploidy level for Juniperus was diploid with an estimated genome size of 12.37 pg/1C, which fits within the range of 9–12.38 pg/1C inferred by Burleigh et al. (2012), based on a sampling including only two Juniperus species amongst 165 gymnosperm species.
Within the genus, we found evidence suggesting that fluctuations in genome size, both upsizing and downsizing, independent of polyploidy, have taken place during evolution, as also found in Pinus (Grotkopp et al., 2004) and across other gymnosperm lineages as well (Burleigh et al., 2012). However, while, in most other gymnosperm genera the shifts in genome size are likely to be driven by changes in the abundance of repetitive DNA (Nystedt et al., 2013; De La Torre et al., 2014), in Juniperus the large shifts in genome size are associated with polyploidization events, with a minimum of 10 such events predicted from our analyses (Figure 2). Whether the occurrence and frequency of polyploidy, which was seen to be restricted to sect. Sabina, contributes to the higher number of species in this section (c. 60 species) compared with the other two sections of Juniperus (sect. Juniperus = c. 13 species, sect. Caryocedrus = one species) is unclear, although previous studies pointing to higher diversification rates in some angiosperm lineages following polyploidy suggest this is possible (Wood et al., 2009; Landis et al., 2018).
Concerning the origin of the hexaploid, J. foetidissima, there are several possible pathways. It could have arisen from a triploid ancestor following one step. If so, then there are two possible routes; (i) fertilization between two unreduced triploid gametes of a triploid ancestor, or (ii) somatic doubling of a triploid, giving rise directly to the hexaploid. Alternatively, it could have arisen following two WGD events (two steps) as envisaged for the hexaploid Sequoia sempervirens (Scott et al., 2016). The first step being a WGD event either via autopolyploidy or allopolyploidy leading to the formation of a tetraploid with n = 2x, followed by hybridization with a diploid (n = x) leading to a triploid. The second step involves a WGD giving rise to a hexaploid. The reports of sporadic triploid Juniperus individuals indicate that triploids can indeed form (Hall et al., 1973). However, yet another possibility is that the origin of J. foetidissima does not involve a triploid, but instead arose from hybridization between an unreduced gamete from a tetraploid (4x) with either (a) a reduced gamete from another tetraploid (2x) or (b) an unreduced gamete from a diploid (2x). Currently, there is no information about the genomic makeup of J. foetidissima to know whether it is an auto- or allo-polyploid, or its mode of origin.
Why Is Polyploidy More Common in Juniperus Than Other Conifers?
The success of hexaploid Sequoia sempervirens and polyploid Ephedra species (4x – 8x), has been partially attributed to their capacity for vegetative propagation (Scott et al., 2016; Wu et al., 2016) and this may also contribute to the survival of polyploid Juniperus species as there is evidence that they too have the capacity for vegetative propagation [e.g., in J. sabina and J. communis (Houle and Babeux, 1994; Ronnenberg, 2005; Wesche et al., 2005; Tylkowski, 2010)]. Furthermore, the extreme longevity has been suggested to be another factor contributing to the success of polyploidy in S. sempervirens (Scott et al., 2016), and since Juniperus has been classified as long-lived (Ward, 1982; Gauquelin et al., 2012) this may also help the survival of polyploids, enabling them to become established.
Here we propose a novel hypothesis that may also contribute to higher frequency of polyploidy revealed in Juniperus—this is the high frequency of sympatry between juniper species. In contrast to most of the conifers, the geographical ranges of Juniperus species overlap considerably which opens up lots of opportunities for natural hybridization between species. For example, in Spain, hybrids between J. thurifera × J. sabina and J. thurifera × J. phoenicea and J. sabina × J. phoenicea in sympatry have been described (Rojo and Díaz, 2006, 2009; Rojo and Uribe-Echebarría, 2008). More recently, Adams et al. (2016) suggested that an ancient hybridization between J. thurifera and J. sabina gave rise to J. sabina var. balkanensis. Juniper hybrids are also common in North America between closely related species in areas of sympatry [e.g., between J. virginiana L. and J. horizontalis Moench, J. osteosperma Hook and J. occidentalis Torr. Little, J. virginiana var. silicicola, and J. bermudiana (Vasek, 1966; Palma-Otal et al., 1983; Adams and Kistler, 1991; Adams and Wingate, 2008; Adams, 2014)].
Even though the sympatry is a sine qua non condition for natural hybridization, there are few cases of conifers occurring in sympatry that do hybridize without giving rise to polyploids: e.g., Pinus taeda and P. echinata (Edwards-Burke et al., 1997). Furthermore, induced hybridization like for Cedrus species (Fady et al., 2003) produced only homoploids. Cases of unreduced gamete production were documented in Cupressaceae (Pichot and El Maâtaoui, 2000) and Ephedraceae (Wu et al., 2016). This ability to produce unreduced gametes may be the explanation for polyploidisation in Juniperus.
On the other hand, the genomic shock arising from hybridization can often be ameliorated by WGD and subsequent diploidization as it was shown in angiosperms (Hegarty et al., 2006). Given the high frequency of hybrid formation in Juniperus, and assuming that similar levels of genomic shock following hybridization also occur here, as in angiosperms, then it is possible to envisage that polyploidy may offer one potential solution to these genomic challenges, tipping the balance toward their survival in the wild. Clearly, studies are now needed at the molecular level to provide insights into whether our understanding of the genomic consequences of hybridization and polyploidization in angiosperms is also applicable to the growing list of gymnosperm polyploids.
Conclusion
Polyploidy or whole genome duplication is rare in conifers. The lack of studies on polyploidy within Juniperus prompted the present study, in which the ploidy level of 96.5% of the genus was screened in order to explore the extent of polyploidy across the genus. Silica gel-dried leaves of Juniperus were found to be highly suitable for genome size measurements using flow cytometry. This study uncovered a relatively high number of polyploidization events (at least 10) in Juniperus, compared to other conifers, and revealed that at least 15% of Juniperus taxa are tetraploids. In addition, we used both chromosome and genome size data to validate the presence of the only hexaploid in Juniperus (J. foetidissima) so far reported, and only the second hexaploid found in conifers (after Sequoia sempervirens). An analysis of the phylogenetic distribution of polyploids across Juniperus showed they were restricted to sect. Sabina and that three clades are exclusively made of polyploids (one including the hexaploid J. foetidissima), providing the first evidence of possible lineage-specific polyploidizations in the genus.
Overall, it seems clear that Juniperus is exceptional within conifers, and represents a second genus within gymnosperms where polyploidy is common. We propose that Juniperus should be considered to be a highly relevant model for studying polyploidization mechanisms and pathways in conifers, and comparisons with Ephedra will provide a comprehensive understanding of the evolutionary dynamics and consequences of polyploidy in gymnosperms.
Author Contributions
MB designed the study. RA provided the Juniperus material. PF and OH carried out the flow cytometry measurements and analyzed the data. PF and SS-Y determined the chromosome numbers. PF wrote a first draft of the manuscript that was further critically reviewed by MB, RA, OH, SS-Y, IL, TR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Royal Botanic Gardens Kew, London, UK for providing access to the flow cytometry facilities and living collections.
Footnotes
Funding. The authors thank the National Council for Scientific Research grant number CNRS-FS90—Lebanon, the Saint Joseph University Research Council (CR-USJ) FS-111 for supporting financially this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00676/full#supplementary-material
References
- Abdel Samad F., Baumel A., Juin M., Pavon D., Siljak-Yakovlev S., Médail F., et al. (2014). Phylogenetic diversity and genome sizes of Astragalus (Fabaceae) in the Lebanon biogeographical crossroad. Plant Syst. Evol. 300, 819–830. 10.1007/s00606-013-0921-8 [DOI] [Google Scholar]
- Adams R. (2014). Junipers of the World: The Genus Juniperus. Bloomington, IN: Trafford Publishing. [Google Scholar]
- Adams R., Kistler J. (1991). Hybridization between Juniperus erythrocarpa Cory and Juniperus pinchotii Sudworth in the Chisos mountains, Texas. Southwest. Nat. 36, 295–301. 10.2307/3671679 [DOI] [Google Scholar]
- Adams R., Schwarzbach A., Tashev A. (2016). Chloroplast capture by a new variety, Juniperus sabina var. balkanensis RP Adams and AN Tashev, from the Balkan peninsula: a putative stabilized relictual hybrid between J. sabina and ancestral J. thurifera. Phytologia 98, 100–111. [Google Scholar]
- Adams R., Wingate D. (2008). Hybridization between Juniperus bermudiana and J. virginiana in Bermuda. Phytologia 90, 123–213. [Google Scholar]
- Adams R. P., Schwarzbach A. E. (2013). Phylogeny of Juniperus using nrDNA and four cpDNA regions. Phytologia 95, 179–187. [Google Scholar]
- Ahuja M. R. (2005). Polyploidy in gymnosperms: revisited. Silvae Genet. 54, 59–69. 10.1515/sg-2005-0010 [DOI] [Google Scholar]
- Ahuja M. R., Neale D. B. (2002). Origins of polyploidy in coast redwood (Sequoia sempervirens (D. don) Endl. and relationship of coast redwood to other genera of Taxodiaceae. Silvae Genet. 51, 93–99. [Google Scholar]
- Bainard J. D., Husband B. C, Baldwin S., Fazekas A., Gregory T., Newmaster S., et al. (2011). The effects of rapid desiccation on estimates of plant genome size. Chromosome Res. 19, 825–842. 10.1007/s10577-011-9232-5 [DOI] [PubMed] [Google Scholar]
- Bainard J. D., Villarreal J. (2013). Genome size increases in recently diverged hornwort clades. Genome 56, 431–435. 10.1139/gen-2013-0041 [DOI] [PubMed] [Google Scholar]
- Barker M. S., Arrigo N., Baniaga A. E., Li Z., Levin D. A. (2016). On the relative abundance of autopolyploids and allopolyploids. New Phytol. 210, 391–398. 10.1111/nph.13698 [DOI] [PubMed] [Google Scholar]
- Bennett M., Leitch I. (2012). Plant DNA C-Values Database (release 6.0, Dec. 2012). Available online at: http://www.kew.org/cvalues/
- Blanc G., Wolfe K. H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. 10.1105/tpc.021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunic F., Muratovic E., Brown S., Siljak-Yakovlev S. (2003). Genome size and base composition of five Pinus species from the Balkan region. Plant Cell Rep. 22, 59–63. 10.1007/s00299-003-0653-2 [DOI] [PubMed] [Google Scholar]
- Bou Dagher-Kharrat M., Abdel-Samad N., Douaihy B., Bourge M., Fridlender A., Siljak-Yakovlev S., et al. (2013). Nuclear DNA C-values for biodiversity screening: case of the Lebanese flora. Plant Biosyst. 147, 1228–1237. 10.1080/11263504.2013.861530 [DOI] [Google Scholar]
- Burleigh J. G., Barbazuk W. B., Davis J. M., Morse A. M., Soltis P. S. (2012). Exploring diversification and genome size evolution in extant gymnosperms through phylogenetic synthesis. J. Bot. 2012:292857 10.1155/2012/292857 [DOI] [Google Scholar]
- Chen Z. J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58, 377–406. 10.1146/annurev.arplant.58.032806.103835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz M. J., Reveal J. L., Farjon A., Gardner M. F., Mill R. R., Chase M. W. (2011). A new classification and linear sequence of extant gymnosperms. Phytotaxa 19, 55–70. 10.11646/phytotaxa.19.1.3 [DOI] [Google Scholar]
- Clark J., Hidalgo O., Pellicer J., Liu H., Marquardt J., Robert Y., et al. (2016). Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 210, 1072–1082. 10.1111/nph.13833 [DOI] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. 10.1038/nrg1711 [DOI] [PubMed] [Google Scholar]
- De La Torre A., Birol I., Bousquet J., Ingvarsson P., Jansson S., Jones S. J., et al. (2014). Insights into conifer giga-genomes. Plant Physiol. 114, 1724–1732. 10.1104/pp.114.248708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J., Greilhuber J., Lucretti S., Meister A., Lysák M., Nardi L., et al. (1998). Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann. Bot. 82(Suppl_1), 17–26. 10.1093/oxfordjournals.aob.a010312 [DOI] [Google Scholar]
- Doležel J., Greilhuber J., Suda J. (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2:2233. 10.1038/nprot.2007.310 [DOI] [PubMed] [Google Scholar]
- Edwards-Burke M. A., Hamrick J. L., Price R. A. (1997). Frequency and direction of hybridization in sympatric populations of Pinus taeda and P. echinata (Pinaceae). 84, 879–886. 10.2307/2446277 [DOI] [PubMed] [Google Scholar]
- Fady B., Lefèvre F., Reynaud M., Vendramin G. G., Bou Dagher-Kharrat M., Anzidei M., et al. (2003). Gene flow among different taxonomic units: evidence from nuclear and cytoplasmic markers in Cedrus plantation forests. Theor. Appl. Genet. 107, 1132–1138. 10.1007/s00122-003-1323-z [DOI] [PubMed] [Google Scholar]
- Farhat P., Siljak-Yakovlev S., Robert A., Magda B., Robert T. (2019). Genome size variation and polyploidy in the geographical range of Juniperus sabina L. (Cupressaceae). Bot. Lett. 68, 92–96. 10.1080/00087114.2015.1024546 [DOI] [Google Scholar]
- Farjon A. (2010). A Handbook of the World's Conifers (2 Vols.). Leiden: Brill Academic Publishers. [Google Scholar]
- Gauquelin T., Chondroyannis P., Boukhdoud N., Bouyssou M., Brunel C., Danneyrolles V., et al. (2012). Le Genévrier thurifère, espèce partagée au Nord et au Sud de la Méditerranée. Forêt Méditerranéenne 33, 227–240. [Google Scholar]
- Glick L., Mayrose I. (2014). ChromEvol: assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Mol. Biol. Evol. 31, 1914–1922. 10.1093/molbev/msu122 [DOI] [PubMed] [Google Scholar]
- Greilhuber J., DoleŽel J., Lysák M. A., Bennett M. D. (2005). The origin, evolution and proposed stabilization of the terms ‘genome size'and ‘C-value'to describe nuclear DNA contents. Ann. Bot. 95, 255–260. 10.1093/aob/mci019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotkopp E., Rejmánek M., Sanderson M. J., Rost T. L. (2004). Evolution of genome size in pines (Pinus) and its life-history correlates: supertree analyses. Evolution 58, 1705–1729. 10.1111/j.0014-3820.2004.tb00456.x [DOI] [PubMed] [Google Scholar]
- Guan R., Zhao Y., Zhang H., Fan G., Liu X., Zhou W., et al. (2016). Draft genome of the living fossil Ginkgo biloba. Gigascience 5, 1–13. 10.1186/s13742-016-0154-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hair J. (1968). The chromosomes of the Cupressaceae: 1. Tetraclineae and Actinostrobeae (Callitroideae). N. Z. J. Bot. 6, 277–284. 10.1080/0028825X.1968.10428813 [DOI] [Google Scholar]
- Hall M. T., Mukherjee A., Crowley W. R. (1973). Chromosome counts in cultivated junipers. J. Arnold Arboretum 54, 369–376. [Google Scholar]
- Hegarty M. J., Barker G. L., Wilson I. D., Abbott R. J., Edwards K. J., Hiscock S. J. (2006). Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 16, 1652–1659. 10.1016/j.cub.2006.06.071 [DOI] [PubMed] [Google Scholar]
- Hizume M. (1988). Karyomorphological studies in family Pinaceae. Nat. Sci. 8, 1–108. [Google Scholar]
- Houle G., Babeux P. (1994). Variations in rooting ability of cuttings and in seed characteristics of five populations of Juniperus communis var. depressa from subarctic Quebec. Can. J. Bot. 72, 493–498. 10.1139/b94-066 [DOI] [Google Scholar]
- Husband B. C., Baldwin S. J., Suda J. (2013). The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes, in Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes, eds. Greilhuber J., Dolezel J., Wendel J. F. (Vienna: Springer Vienna; ), 255–276. 10.1007/978-3-7091-1160-4_16 [DOI] [Google Scholar]
- Ickert-Bond S., Pellicer J., Souza A., Metzgar J., Leitch I. J. (2015). Ephedra-the gymnosperm genus with the largest and most diverse genome sizes driven by a high frequency of recently derived polyploidy taxa and a lack of genome downsizing, in Annual Meeting of the Botanical Society of America, Botany 2015, Abstract ID 862 (Edmonton: ). [Google Scholar]
- Khoshoo T. (1959). Polyploidy in gymnosperms. Evolution 13, 24–39. 10.1111/j.1558-5646.1959.tb02991.x [DOI] [Google Scholar]
- Kovach A., Wegrzyn J. L., Parra G., Holt C., Bruening G. E., Loopstra C. A., et al. (2010). The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genomics 11, 1–14. 10.1186/1471-2164-11-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejčíková J., Sudová R., Lučanová M., Trávníček P., Urfus T., Vít P., et al. (2013). High ploidy diversity and distinct patterns of cytotype distribution in a widespread species of Oxalis in the Greater Cape Floristic Region. Ann. Bot. 111, 641–649. 10.1093/aob/mct030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis J. B., Soltis D., Li Z., Marx H., Barker M., Tank D., et al. (2018). Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 105, 348–363. 10.1002/ajb2.1060 [DOI] [PubMed] [Google Scholar]
- Ledig F. T. (1998). Genetic variation in Pinus, in Ecology and Biogeography of Pinus, ed. Richardson D. M. (Cambridge: Cambridge University Press; ). [Google Scholar]
- Leitch I. J., Bennett M. D. (2004). Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 82, 651–663. 10.1111/j.1095-8312.2004.00349.x [DOI] [Google Scholar]
- Li Z., Baniaga A., Sessa E., Scascitelli M., Graham S., Rieseberg L., et al. (2015). Early genome duplications in conifers and other seed plants. Sci. Adv. 1:e1501084. 10.1126/sciadv.1501084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Ran J.-H., Guo D.-M., Yang Z.-Y., Wang X.-Q. (2014). Phylogeny and divergence times of gymnosperms inferred from single-copy nuclear genes. PLoS ONE 9:e107679. 10.1371/journal.pone.0107679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Hao G., Liu J., Adams R., Milne R. (2010). Diversification and biogeography of Juniperus (Cupressaceae): variable diversification rates and multiple intercontinental dispersals. New Phytol. 188, 254–272. 10.1111/j.1469-8137.2010.03351.x [DOI] [PubMed] [Google Scholar]
- Masterson J. (1994). Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264, 421–424. 10.1126/science.264.5157.421 [DOI] [PubMed] [Google Scholar]
- Morse A. M., Peterson D. G., Islam-Faridi M. N., Smith K. E., Magbanua Z., Garcia S. A., et al. (2009). Evolution of genome size and complexity in Pinus. PLoS ONE 4:e4332. 10.1371/journal.pone.0004332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B., Leitch I. J., Bennett M. D. (2012). Gymnosperm DNA C-Values Database (release 5.0, Dec. 2012). Available online at: http://www.kew.org/cvalues/
- Murray B. G. (2013). Karyotype variation and evolution in gymnosperms, in Plant Genome Diversity, Vol. 2, eds Greilhuber I. J., Doležel J., Wendel J. (Vienna: Springer-Verlag; ), 231–243. 10.1007/978-3-7091-1160-4_14 [DOI] [Google Scholar]
- Nagano K., Matoba H., Yonemura K., Matsuda Y., Murata T., Hoshi Y. (2007). Karyotype analysis of three Juniperus species using fluorescence in situ hybridization (FISH) with two ribosomal RNA genes. Cytologia 72, 37–42. 10.1508/cytologia.72.37 [DOI] [Google Scholar]
- Nystedt B., Street N. R., Wetterbom A., Zuccolo A., Lin Y.-C., Scofield D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497:579. 10.1038/nature12211 [DOI] [PubMed] [Google Scholar]
- Otto S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. 10.1016/j.cell.2007.10.022 [DOI] [PubMed] [Google Scholar]
- Otto S. P., Whitton J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. 10.1146/annurev.genet.34.1.401 [DOI] [PubMed] [Google Scholar]
- Palma-Otal M., Moore W., Adams R., Joswiak G. (1983). Morphological, chemical, and biogeographical analyses of a hybrid zone involving Juniperus virginiana and J. horizontalis in Wisconsin. Can. J. Bot. 61, 2733–2746. 10.1139/b83-301 [DOI] [Google Scholar]
- Pichot C., El Maâtaoui M. (2000). Unreduced diploid nuclei in Cupressus dupreziana A. Camus pollen. Theor. Appl. Genet. 101, 574–579. 10.1007/s001220051518 [DOI] [Google Scholar]
- Popp M., Gizaw A., Nemomissa S., Suda J., Brochmann C. (2008). Colonization and diversification in the African ‘sky islands' by Eurasian Lychnis L. (Caryophyllaceae). J. Biogeogr. 35, 1016–1029. 10.1111/j.1365-2699.2008.01902.x [DOI] [Google Scholar]
- Ramsey J., Schemske D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. 10.1146/annurev.ecolsys.29.1.467 [DOI] [Google Scholar]
- Ran J.-H., Shen T.-T., Wang M.-M., Wang X.-Q. (2018). Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. R. Soc. 285:20181012. 10.1098/rspb.2018.1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell L. (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Rice A., Glick L., Abadi S., Einhorn M., Kopelman N. M., Salman-Minkov A., et al. (2015). The Chromosome Counts Database (CCDB)–a community resource of plant chromosome numbers. New Phytol. 206, 19–26. 10.1111/nph.13191 [DOI] [PubMed] [Google Scholar]
- Rojo J., Díaz P.-E. (2006). Juniperus × palancianus, nuevo híbrido de la provincia de castellón. Toll Negre 8, 5–8. [Google Scholar]
- Rojo J., Díaz P.-E. (2009). Juniperus × cerropastorensis, nuevo híbrido entre Juniperus sabina L. Y Juniperus thurifera L. Toll Negre 11, 6–13. [Google Scholar]
- Rojo J., Uribe-Echebarría P. (2008). Juniperus × herragudensis, otro nuevo híbrido de la provincia de Castellón. Mainhardt 60, 83–85. [Google Scholar]
- Romo A., Hidalgo O., Boratynski A., Sobierajska K., Jasinska A. K., Vallès J., et al. (2013). Genome size and ploidy levels in highly fragmented habitats: the case of western Mediterranean Juniperus (Cupressaceae) with special emphasis on J. thurifera L. Tree Genet. Genomes 9, 587–599. 10.1007/s11295-012-0581-9 [DOI] [Google Scholar]
- Ronnenberg K. (2005). Reproductive ecology of two common woody species, Juniperus sabina and Artemisia santolinifolia, in mountain steppes of southern Mongolia. Erforsch. Biol. Ress. Mongolei (Halle/Saale) 9, 207–223. [Google Scholar]
- Roodt D., Lohaus R., Sterck L., Swanepoel R., Van de Peer Y., Mizrachi E. (2017). Evidence for an ancient whole genome duplication in the cycad lineage. PLoS ONE 12:e0184454. 10.1371/journal.pone.0184454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax K., Sax H. J. (1933). Chromosome number and morphology in the conifers. J. Arnold Arboretum 14, 356–375. 10.5962/bhl.part.9959 [DOI] [Google Scholar]
- Schönswetter P., Suda J., Popp M., Weiss-Schneeweiss H., Brochmann C. (2007). Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Mol. Phylogenet. Evol. 42, 92–103. 10.1016/j.ympev.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Scott A. D., Stenz N. W., Ingvarsson P. K., Baum D. A. (2016). Whole genome duplication in coast redwood (Sequoia sempervirens) and its implications for explaining the rarity of polyploidy in conifers. New Phytol. 211, 186–193. 10.1111/nph.13930 [DOI] [PubMed] [Google Scholar]
- Siljak-Yakovlev S., Pustahija F., Šolić E., Bogunić F., Muratović E., Bašić N., et al. (2010). Towards a genome size and chromosome number database of Balkan flora: C-values in 343 taxa with novel values for 242. Adv. Sci. Lett. 3, 190–213. 10.1166/asl.2010.1115 [DOI] [Google Scholar]
- Šmarda P., Müller J., Vrána J., Kočí K. (2005). Ploidy level variability of some Central European fescues (Festuca subg. Festuca, Poaceae). Biologia (Bratislava) 60, 25–36. [Google Scholar]
- Šmarda P., Stančík D. (2006). Ploidy level variability in South American fescues (Festuca L., Poaceae): use of flow cytometry in up to 5 1/2-year-old caryopses and herbarium specimens. Plant Biol. 8, 73–80. 10.1055/s-2005-872821 [DOI] [PubMed] [Google Scholar]
- Soltis D., Soltis P., Endress P., Chase M. W., Manchester S., Judd W., et al. (2018). Phylogeny and Evolution of the Angiosperms: Revised and Updated Edition. Chicago, IL: University of Chicago Press. [Google Scholar]
- Soltis P. S., Soltis D. E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60, 561–588. 10.1146/annurev.arplant.043008.092039 [DOI] [PubMed] [Google Scholar]
- Stebbins G. L. (1947). Types of polyploids: their classification and significance. Adv. Genet. 1, 403–429. 10.1016/S0065-2660(08)60490-3 [DOI] [PubMed] [Google Scholar]
- Suda J., Trávníček P. (2006). Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry—new prospects for plant research. Cytometry Part A 69, 273–280. 10.1002/cyto.a.20253 [DOI] [PubMed] [Google Scholar]
- Suda J., Weiss-Schneeweiss H., Tribsch A., Schneeweiss G. M., Trávníček P., Schönswetter P. (2007). Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plant Senecio carniolicus (Asteraceae). Am. J. Bot. 94, 1391–1401. 10.3732/ajb.94.8.1391 [DOI] [PubMed] [Google Scholar]
- Tayalé A., Parisod C. (2013). Natural pathways to polyploidy in plants and consequences for genome reorganization. Cytogenet. Genome Res. 140, 79–96. 10.1159/000351318 [DOI] [PubMed] [Google Scholar]
- Team R. C. (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/
- Tylkowski T. (2010). Dormancy breaking in Savin juniper (Juniperus sabina L.) seeds. Acta Soc. Bot. Pol. 79, 27–29. 10.5586/asbp.2010.004 [DOI] [Google Scholar]
- Vallès J., Garnatje T., Robin O., Siljak-Yakovlev S. (2015). Molecular cytogenetic studies in western Mediterranean Juniperus (Cupressaceae): a constant model of GC-rich chromosomal regions and rDNA loci with evidences for paleopolyploidy. Tree Genet. Genomes 11, 1–8. 10.1007/s11295-015-0860-3 [DOI] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 1–14. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Vasek F. (1966). The distribution and taxonomy of three western junipers. Brittonia 18, 350–372. 10.2307/2805152 [DOI] [Google Scholar]
- Wang G., Yang Y. (2016). The effects of fresh and rapid desiccated tissue on estimates of Ophiopogoneae genome size. Plant Divers. 38, 190–193. 10.1016/j.pld.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L. K. (1982). The conservation of juniper: longevity and old age. J. Appl. Ecol. 19, 917–928. 10.2307/2403293 [DOI] [Google Scholar]
- Wendel J. F., Lisch D., Hu G., Mason A. (2018). The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49, 1–7. 10.1016/j.gde.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Wesche K., Ronnenberg K., Hensen I. (2005). Lack of sexual reproduction within mountain steppe populations of the clonal shrub Juniperus sabina L. in semi-arid southern Mongolia. J. Arid Environ. 63, 390–405. 10.1016/j.jaridenv.2005.03.014 [DOI] [Google Scholar]
- Wood T. E., Takebayashi N., Barker M. S., Mayrose I., Greenspoon P. B., Rieseberg L. H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. U.S.A. 106, 13875–13879. 10.1073/pnas.0811575106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Ma Z., Wang M. M., Qin A.-L., Ran J. H., Wang X. Q. (2016). A high frequency of allopolyploid speciation in the gymnospermous genus Ephedra and its possible association with some biological and ecological features. Mol. Ecol. 25, 1192–1210. 10.1111/mec.13538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonneveld B. (2012). Conifer genome sizes of 172 species, covering 64 of 67 genera, range from 8 to 72 picogram. Nord. J. Bot. 30, 490–502. 10.1111/j.1756-1051.2012.01516.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.