Abstract
Current therapeutic approaches are ineffective in many patients with established Diabetic Kidney Disease (DKD) disease, an epidemic affecting one in three patients with diabetes. Early identification of patients at high risk for progression and individualizing therapies have the potential to mitigate kidney complications due to diabetes. To achieve this, a better understanding of the complex pathophysiology of DKD is needed. A system biology approach integrating large scale omic data is well suited to unravel the molecular mechanisms driving DKD and may offer new perspectives how to personalize therapy. Recent studies indeed demonstrate that integrating genome scale data sets generated from prospectively designed clinical cohort studies with model systems using innovative bioinformatics analysis revealed critical molecular pathways in DKD and led to the development of candidate prognostic molecular biomarkers. This review seeks to provide an overview of the recent progress in the application of the integrative systems biology approaches specifically in the field of molecular biomarkers for DKD. We will mainly focus the discussion on how to use integrative system biology approach to firstly identify patients at high risk of progression, and secondly to identify patients who may or may not respond to treatment. Challenges and opportunities in applying precision medicine in DKD will also be discussed.
Keywords: Type 2 diabetes, chronic kidney disease, biomarkers, systems biology
Introduction
DKD is affecting one in three patients with diabetes.[1] The progression of DKD in these patients is often undetected and the main cause of DKD resulting in end-stage renal disease (ESRD), with substantial decreased quality of life and increased mortality and morbidity.[2] Early identification of patients at highest risk will allow early intervention which can delay disease onset and reduce incidence of ESRD.[3] The prediction of DKD has mainly relied on evaluation of the classical risk factors, such as blood pressure, estimated glomerular filtration rate (eGFR) and proteinuria. However, these factors have been inadequate at identifying patients at high risk of progression and predicting if a patient will respond to a drug intervention.[4]
DKD is a complex multifactorial disease with many deregulated processes that initiate and sustain the progression of the disease including but not limited to cell respiration, oxidative stress, metabolic disorders, vascular and endothelial factors, inflammation and fibrosis, angiogenesis, and regeneration potential.[5] Research efforts that have targeted single pathways and molecules have had limited impact on our ability to effectively alter the course of the disease course in the past 25 years.[6,7]Novel alternative approaches are needed to gain a comprehensive understanding of the early disease pathophysiology at the molecular level to improve the clinical impact of basic and translational research.
Systems biology approach seeks to discover how dynamic interactions determine function, and addresses the missing links between molecules and physiology. The dominant top-down approach, which has been thoroughly reviewed by Bruggeman and Westerhoff,[8] starts from the view of the whole system by acquiring genome-wide experimental data, followed by data analysis and integration, and then by functional characterization leading to the formation of a testable hypotheses. The bottom-up systems biology approach examines the mechanisms through which functional properties arise in the interactions of known components.[9] Systems biology has been successfully applied in a wide spectrum of biomedical research and has improved our understanding of neurodegenerative,[10] cancer,[11] inflammatory,[12]and genetic diseases.[13]
A system biology approach based on integrating multiple omics data types along the genome-phenome regulatory cascades can be a well-suited approach for DKD since it provides a comprehensive view of the disease pathophysiology cutting across multiple organ system in a chronic disease process. Though systems biology approaches have just recently been deployed to DKD, studies so far have made an impact by advancing the understanding of the molecular mechanisms underlying DKD pathogenesis. This has in turn lead to the identification of critical molecular abnormalities in DKD and to the development of novel treatments and potential molecular biomarkers.[14–18]
Several reviews have outlined the specifics of the underlying techniques used in systems biology with a focus on novel therapeutics development.[6,7,19,20] Here, we will describe recent advances in the discovery and validation of non-invasive biomarkers for predicting disease progression and response to treatment in DKD. Specifically, we will focus on how systems biology, through more comprehensive understanding of the complexity of DKD, will improve clinical outcome by facilitating the discovery of novel plasma or urinary molecular biomarkers to identify patients at high risk of progression (prognostic markers); and to predict the patients’ responses to treatment (predictive markers). In addition, we will also review challenges and strategies to apply integrative systems biology approach for precision medicine in DKD.
The use of systems biology approach to identify patients at high risk of progression
Current clinical in-use markers for kidney disease progression
To identify targeted and untargeted molecular features that associate with DKD progression, the nephropathy progression phenotype is defined using changes in eGFR and albuminuria.[21] These markers correlate with risk of developing ESRD. Table 1 summarized the strengths and challenges of currently widely used definitions of progression endpoints.
Table 1:
Progression and treatment response phenotype.
| Endpoint | Challenges | Advantages |
|---|---|---|
| ESRD | ● Insensitive ● Long follow-up ● Low number of events |
● Gold standard ● Clinically relevant for patient |
| eGFR decline 30 - 57 % |
● Inter-lab assay variation ● Influenced by drugs hemodynamics ● Diet and hydration status influence assay ● Variation attributed to nephron loss only reliable in advanced DKD |
● Higher precision in advanced DKD ● Strong association with ESRD |
| eGFR slope | ● Laboratory issues as described above ● Non linearity in eGFR slopes complicates interpretation ● Accuracy relies on multiple values over time ● Cut-off value for steep or shallow slope is arbitrary |
● Can be used as continuous variable or a cut-off value can be applied to allow dichotomous comparison ● Can be assessed earlier than 40% or 57% eGFR decline ● Allow studies in patients in early stages DKD |
| UACR / Albuminuria /Proteinuria increase of 30% |
● Reliability differs across assays ● Large day-to-day variation ● Unclear if decreased UACR necessarily improve clinical outcomes for all interventions |
● Non-invasive ● High precision in early DKD |
The clinical markers perform well predicting risk of late stage chronic kidney disease (CKD) or DKD. A model using routinely obtained laboratory tests, including eGFR and albuminuria, has been shown to accurately predict progression to kidney failure in patients with CKD stages 3 to 5.[22] However, predicting progression at early CKD stage when treatment can make a difference remains challenging. Thus molecular biomarkers that are related to the pathogenesis of the disease and can predict the risk of progression better or add incremental value to the clinical markers are desired.
The identification of novel molecular markers associated with DKD progression
High throughput multi-omics approaches (transcriptomics, proteomics, metabolomics and lipidomics) have been used to profile patients’ biospecimen (kidney biopsy, plasma and urine samples). These multi-omics datasets contributed to the discovery of biomarker candidates that are associated with disease progression.[18,23–30] Table 2 includes molecular biomarkers that were discovered using systems biology approaches with demonstrated association with kidney outcomes in patient-related studies.
Table 2:
Candidate biomarkers yield from –omics studies and have shown association with DKD progression.
| Marker | Outcome measure | Pathway | Integration | Study |
|---|---|---|---|---|
| Urinary EGF | Composite endpoint of ESRD or 40% eGFR reduction, eGFR slope |
Cell growth & Regeneration |
Transcriptomics & protein & quantitative morphometry |
Ju et al.[23] Nair et al.[25] |
| Rapid decline in eGFR, onset of eGFR less than 60 |
Cell growth & Regeneration |
Proteomics from rodent model & validate in patient study |
Betz et al [18]. | |
| Urinary CKD273 classifier |
Onset of microalbuminuria, Rapid decline in eGFR |
Fibrosis & ECM degradation |
Proteomics | Lindhardt et al. [30] Pontillo[69] |
| Urinary TCA cycle metabolites |
Onset of eGFR <65 | Mitochondrial metabolism |
Transcriptomics & metabolomics rodent model & validation in patient study |
Sas et al.[26] |
| Plasma uremic solutes and essential amino acids |
ESRD | Purine metabolism and Amino acids metabolism |
Metabolomics | Niewszas et al.[28] |
| Serum 7 modified metabolites |
ESRD, eGFR decline | Metabolism Underlie acetylation or c- glycosylation |
Metabolomics | Niewczas et al/[27] |
| Serum diacyglycerols and monoacylglyxerol |
ESRD | Dysregulation of Classical pathways of lipolysis |
Lipidomics | Afshinnia et al. [29] |
Candidate molecular biomarkers identified from plasma and urine can be secreted or shed from the kidney and other non-renal tissue and cells. For many candidate molecules, kidney secretion is only a small proportion of the amount detected and this raises major concern about the relevance of these markers to kidney pathophysiology. Privileged by minimal tissue sample requirement, transcriptomic data is by far the most utilized -omics data that can help to bridge the kidney pathophysiology and molecular signatures obtained in urine or plasma. The feasibility of this type of approach is illustrated by our recent study, in which a transcriptomic analysis identified a list of biomarkers associated with CKD (including DKD) pathogenesis. The protein products of these RNA transcripts were then assessed in urine to develop non-invasive biomarkers.[23] The most promising candidate was epidermal growth factor (EGF) because of its robust correlation with eGFR in multiple cohorts, kidney-tissue specificity, and the tight correlation between urinary EGF protein levels (uEGF) and intrarenal EGF mRNA levels in matching patient biospecimen. Interestingly, using a very different approach, Betz and colleagues also identified uEGF as a prognostic biomarker for DKD progression.[18] They first performed a urinary peptidomic analysis using a rat model with hyperglycemia and hypertension, reflecting the renal pathologic changes observed in human DKD.[31] Subsequently, they filtered the list of candidate biomarkers based on the differential expression of their associated mRNA in the kidney of patients with DKD using the data mining tool in the transcriptomic database Nephroseq (https://www.nephroseq.com). In normoalbuminuric patients with preserved renal function at baseline, the authors found that lower urine EGF/Cr was independently associated with a higher risk of both eGFR outcomes (eGFR <60 ml/min/1.73 m2 or eGFR decline of >5% per year ) after adjustment for established renal risk factors. These two independent studies support the further development of uEGF as prognostic biomarker of DKD progression especially in the clinical setting.
In addition, Betz’s study demonstrated the feasibility of first using an animal model as a primary discovery step to identify candidate markers and then applying cross-species validation in patient cohort studies. This is particularly relevant when there is a lack of access to patient samples. It is also worth noting that both studies utilized kidney tissue transcriptomic data to facilitate the prioritization of the biomarker candidates for further validation, highlighting the importance of evaluating biomarkers in the context of kidney pathophysiology.
However, the lack of kidney biopsy samples from patients with early stage DKD is a big barrier to the discovery of novel biomarkers. Recent work published by our group tackled this challenge by providing transcriptomic data of protocol kidney biopsies derived from Pima Indian participants with type II diabetes.[25] These individuals did not have overt clinical evidence of DKD at the time of biopsy but the ultrastructure lesion was observed morphologically. Integrative analysis of the transcriptomic data derived from tubular tissue with the quantitative morphometric analysis of the ultrastructural lesion indicated that the genes significantly correlated with tubule-interstitial damage are enriched for pathways involving in mitochondrial dysfunction, inflammation, migratory mechanisms, and tubular metabolic functions suggesting ongoing disease processes. Importantly, the expression of these genes at time of biopsy showed significant association with the urine albumin to creatinine ratio (uACR) and measured eGFR 10 years after biopsy, indicating a link between the early molecular events and long-term disease progression.[25] These transcriptomic data, along with rich longitudinal clinical follow-up information and the plasma and urine samples collected following standard procedures, provides a valuable resource which will continuously contribute to future identification and validation of novel therapeutic targets and non-invasive biomarkers of early DKD.
Given the strengths and limitations of each -omics technology, and the various interdependent and interactive processes measured by these -omics technologies, a comprehensive integration of this type of data is more likely to reveal more relevant biomarker candidates. An elegant example of this multi-scalar integration was recent study published by Sas et al, in which they applied transcriptomics, metabolomics, and metabolic flux analysis using diabetic rodent models and observed tissue-specific differences in glucose and fatty acid metabolism in the kidney, retina, and nerve tissues.[26] To confirm their findings from the animal model, the authors investigated diabetic kidney transcriptomic data and urinary metabolites from the Pima Indian cohort patients, and found increased expression in TCA cycle and associated metabolic cofactor pathways and observed that urinary TCA cycle intermediates are consistently increased in patients with DKD in comparison with healthy controls. The authors also demonstrated that levels of most TCA cycle metabolites in baseline urine samples were significantly higher in progressors versus non-progressors, supporting their potential as prognostic biomarkers of DKD progression.
Finally, a panel of 273 urinary peptides that was identified from capillary electrophoresis-mass spectrometry (MS) analysis of urine samples of patients with CKD was observed as another prognostic biomarker capable of predicting onset of microalbuminuria in normoalbuminuric in type 2 diabetic patients.[32][30] A letter of support from the US Food and Drug Administration (FDA) was issued to CKD273 to encourage the further development its clinical utility in ‘prognostic enrichment, drug development and study design considerations’ in early stage DKD.
Together, these studies illustrate the success of systems biology approaches at identifying novel markers of DKD progression and providing mechanistic insights involved in progressive CKD and early DKD.
The use of systems biology approach to predict patients’ response to treatment
Currently DKD clinical guidelines include a one-size fits all approach by recommending the same drug treatment for all patients regardless of their molecular heterogeneity. Clinical trials in patients with DKD focus on response in the overall population but do not take into account how patients respond to the drug individually. Careful subgroup analyses of recent trials in DKD, albeit post-hoc, suggested that disease molecular pathology differed between subgroups and disease subgroups associated with response, such that a subgroup of patients benefitted whereas other subgroups did not or were even harmed.[33–35][36,37] The failure of almost all trials in the last decade indicates that this one-size fits all approach is inadequate. Further subgroup analyses are needed to understand how the molecular heterogeneity of DKD can be incorporated in the treatment algorithm for DKD patients especially if the analysis can identify molecular markers to improve patient stratification strategies.
To improve DKD patient care, an individualized molecular-based treatment algorithm is needed to ensure that treatment recommendations are based on an individual’s molecular disease process. Systems biology approaches can comprehensively evaluate the molecular mechanisms driving the diverse responses to therapies as well as identify biomarkers that are able to predict the response to a therapy before treatment is initiated (predictive marker) or select individuals more likely to benefit from subsequent clinical outcomes after a few weeks’ of therapy (dynamic marker).[38–40] An exemplary study in developing a predictive response to therapy was carried out by Lindhardt et al. in patients with type 2 diabetes and therapy-resistant hypertension.[40] They investigated the potential of a marker panel at predicting the treatment response to mineralocorticoid receptor antagonists (MRA) spironolactone, an effective treatment for slowing the progression of renal function decline and reducing blood pressure and albuminuria.[40,41] The researchers found that higher CKD273, an aforementioned a proteomic classifier, is associated with a larger reduction in UACR in the spironolactone group than the placebo group. Further investigations in large prospective study cohorts are required to validate the finding.
Systems biology approaches can facilitate better prediction of patient’s response to current approved drugs for treating DKD, through integration of 1) drug-specific molecular profile or mechanism of action (MOA); 2) patient-specific non-invasive molecular profile that reflects individual kidney progression pathogenesis and 3) bioinformatics tools to match the patient-specific non-invasive molecular profile with the drug molecular profile. Drug-specific molecular profiles for renoprotective drugs under investigation can be generated using multi-omics profiling of treatment responses in in vitro and in vivo model systems.[42–45] Relevant kidney-specific models include key kidney cell types (primary cells are preferred as they reflect in vivo condition better than established cell lines), organoids, and in vivo DKD rodent models. These molecular profiles provide insights to the MOA of tested drugs and can be associated with morphometric and phenotypic outcomes. Integration of intrarenal transcriptomic data with patient-specific non-invasive molecular profile will allow generation of signatures that reflects individual’s kidney progression pathogenesis. Pattern-matching tools, which have been helpful in understanding disease and advancing the discovery of novel drugs in oncology can be applied to detect similarities among gene expression signatures.[46–48][49] The drug-specific molecular profile with the highest correlation to the patient’s molecular profile is hypothesized to be the most appropriate candidate. This type of integrative systems biology approach is “the right drug for the right patients” approach physician will need to advance treatment in DKD. To the best of our knowledge, no report has yet utilized above described integrative approach in DKD, however, several projects from the Biomarker Enterprise to Attack Diabetic Kidney Disease (BEAt-DKD) Consortium are ongoing. The results from this project are awaited with interest.
The caveat is that animal and cell culture models sometimes fail to recapitulate many aspects of DKD found in humans and thus the limitations of model systems need to be taken into consideration.[50] Human biospecimen from adequately powered and well–designed clinical trial studies are therefore necessary. Post-hoc comparison of the molecular profiles of responders versus non-responders can identify candidate markers that are associated with drug response in patients. These candidate markers, together with the above described drug molecular signatures, will provide a powerful resource to allow for more accurate prediction of treatment.
Strategies to meet existing challenges for discovery of prognostic and predictive biomarkers
The integration of multi-scalar datasets shows promise in advancing the discovery of novel biomarkers for the prediction of DKD progression and response to treatment. Many challenges exist that could hinder the success of applying integrative systems biology approach for precision medicine in DKD, including but certainly not limited to: 1) lack of a well-powered early DKD study cohort where multi-omics data is available for individual patients; 2) lack of information about tissue/cell- specificity to reduce the nonrenal confounding processes; 3) lack of appropriate algorithms and theoretical methods to turn big data into knowledge that supports the discovery, development and clinical implementation of the molecular markers for risk prediction and patients stratification. Below we describe our view of potential strategies to address these challenges.
Expand biosample availability for research
Well-powered early DKD study cohorts with kidney biopsy, urine and plasma sample from the same patients are needed to adequately perform integrative, multi-omics study. At the present time unfortunately, renal biopsies are only indicated for patients with diabetes under the suspicion of the presence of nephropathies other than DKD.[51] As mentioned before, the lack of kidney biopsy makes it more difficult to associate non-invasive biomarkers from urine or plasma with kidney outcomes.
A global effort to develop well-designed DKD study cohorts are currently ongoing. Aforementioned BEAt-DKD and the Diabetes studY of Nephropathy And other Microvascular complications (DYNAMO) aim to deliver tools and knowledge that will facilitate the development of new personalized treatments for DKD. In the near future, more and more DKD cohorts are expected to become available under the support of these international collaborative consortia. Study cohorts of patients with DKD, with several layers of the multi-omics data from biospecimen of the same patient, may offer unique and valuable resources to study molecular mechanisms underlying early DKD progression and to identify molecular biomarkers and therapeutic targets.
Identify tissue- and cell- specific molecular markers
Kidney- specific biomarker are more likely to provide superior diagnostic specificity and less likely to be confounded by nonrenal processes.[23,52] Single-cell technology is considered a powerful tool to identify cell/tissue-specific biomarker in a disease-specific setting to diagnose and monitor the incidence and progression, as well as the responses of patients to the treatment.[53,54] Transcriptomic analysis using single-cell technology, such as single-cell RNA-seq (scRNA-seq) has been applied to mouse kidney [55–57] and human kidney biopsies [58] and have led to the discovery of genes and their associated pathways that are cell type specific and that associate with kidney disease pathogenesis. Despite the rapid advances in this field, significant challenges remain in the analysis, integration, and interpretation of single-cell omics data, including extracting signal from noise especially in data derived from kidney biopsies.[59]
Until single cell data becomes reliable and widely available, there are other resources that can help facilitate cell/tissue specificity at the bulk mRNA level such as BioGPS (http://biogps.org), a catalogue of tissue specific mRNA gene expression in a panel of 79 human and 61 mouse tissues. BioGPS allows comparison of gene expression among various tissues and identification of tissue-specific pattern of mRNA expression.[60] The Epithelial Systems Biology Laboratory (https://hpcwebapps.cit.nih.gov/ESBL/Database/Targets/TranscriptomicData.html) contains RNA-seq data from microdissected rat glomeruli and 14 structurally definable kidney tubular subunits which allows for comprehensive analysis of nephron segment–specific gene expression. [61] In silico nano-dissection (http://nano.princeton.edu), an computational approach applied to bulk transcriptomic data from tissue homogenates, can accurately predict genes specifically expressed or enriched in specific cell types. [63] This method is particularly helpful for identification of genes specifically expressed in human cell types that are difficult to isolate.
Apply innovative bioinformatics tools
A significant body of literature is dedicated to developing novel algorithms and theoretical methods for multi-omic data integration which is necessary to extract meaning from big data.[64,65] Precision medicine has made its greatest impact in the field of oncology and the methods used in oncology may be useful in the analysis of multi-omics data in DKD studies. Non-negative matrix factorization (NMF) method was applied to gene expression data from one of the largest most heterogeneous cancer cohort (The Cancer Genome Atlas (TCGA)) and it helped identify molecularly distinct subtypes that were clinically meaningful.[66,67] Matrix factorization approaches (iCluster) has also been successfully utilized to integrated multiple types of data including methylation, mRNA and microRNA data.[68] In DKD, these type of approaches can be applied to -omics data to help identify functionally relevant disease subsets and could also represent an approach to merge disparate data types in DKD study.
In a landmark paper, Iorio and colleagues leveraged multi-omic data to identify predictive biomarkers of response in cell line models.[68] The group used the machine learning algorithm LOBICO (a logic optimization for binary input to continuous input), to assess the contribution of multiple biomarker from different types of data in models predicting drug response. The multi-predictor models were better at predicting response to drugs in cell lines than single-predictor models and these observations further underscored the importance of integrative analytic approaches. Finally, the group successfully integrated public available molecular characterization data with their own experiment data to not only further advance and validate their discoveries but to increase the statistical power of their study. These approaches can be applied to collate similar data types in DKD from literature to appropriately increase the statistical power of a study.
Summary and outlook
Systems biology research in DKD is helping to transform one-size-fits-all healthcare into a precision medicine based paradigm (Figure 1). Based on their molecular profiles, patients can be classified into disease subgroups; sensitive and specific prognostic biomarkers with better accuracy can be used to identify patients at high risk of progression at early stage of the disease; specific predictive biomarkers using innovative bioinformatics tool can be used to match patients’ molecular signature with the molecular profiles of drugs to pinpoint the most effective treatment option for individual patients. For clinical trials as well as drug use in clinical practice, systems biology can help to enrich trials through identifying the “right patients” thereby leading to ethical and efficient smaller, less complex and more efficient clinical trial conduct and drug use in practice.
Figure 1:
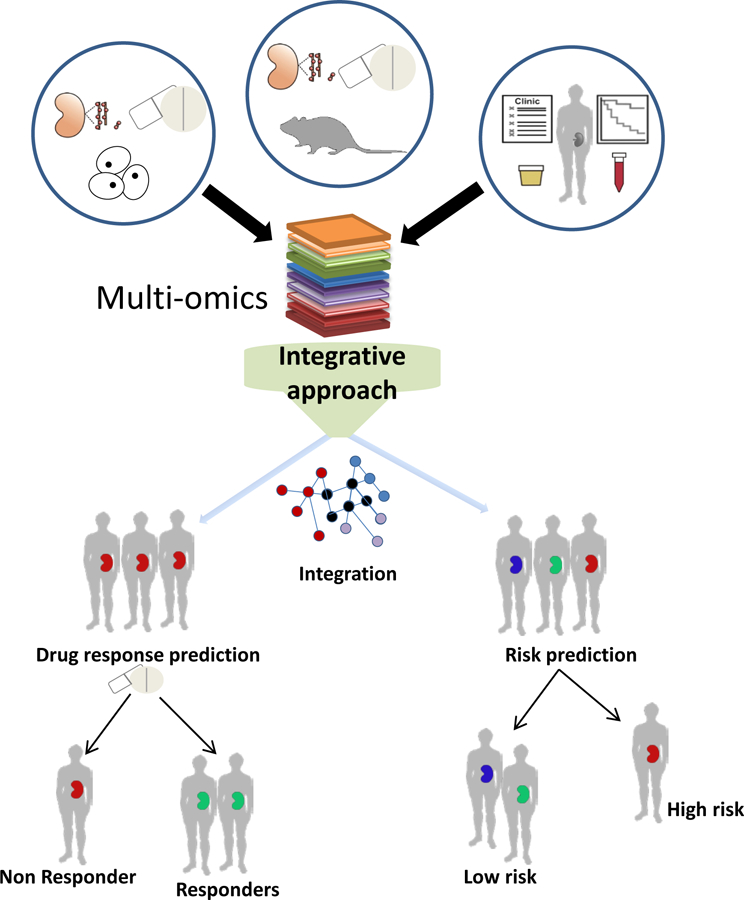
Systems biology approach facilitates the application of precision medicine in diabetic kidney disease. A systems biology approach applies bioinformatics tools on multi-omics datasets derived from in vitro and in vivo DKD associated model systems and patient studies with the aim to facilitate molecular characterization of DKD pathophysiology and stratification of patients based on the risk of progression and the response to treatment.
Acknowledgement
This work is partially supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (P30 DK081943, to M.K.) and Innovative Medicines Initiative Joint Undertaking under grant agreement [No. 115974 (BEAt-DKD)]. The authors acknowledge Dr. Jennifer Harder and Dr. Kuang He for critical review of the paper. The Precision Medicine Symposium which took place in December 2017 in Groningen was endorsed by the BEAt-DKD project. The BEAt-DKD project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 115974. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA.
References
- [1].Drey N, Roderick P, Mullee M, Rogerson M. A population-based study of the incidence and outcomes of diagnosed chronic kidney disease. Am J Kidney Dis 2003;42:677–84. 10.1016/S0272-6386(03)00916-8. [DOI] [PubMed] [Google Scholar]
- [2].Eckardt K-U, Coresh J, Devuyst O, Johnson RJ, Köttgen A, Levey AS, et al. Global Kidney Disease 1 Evolving importance of kidney disease: from subspecialty to global health burden. Ser 158 WwwThelancetCom 2013. 10.1016/S0140-6736(13)60439-0. [DOI] [PubMed] [Google Scholar]
- [3].Atthobari J, Asselbergs FW, Boersma C, de Vries R, Hillege HL, van Gilst WH, et al. Cost-effectiveness of screening for albuminuria with subsequent fosinopril treatment to prevent cardiovascular events: A pharmacoeconomic analysis linked to the prevention of renal and vascular endstage disease (PREVEND) study and the prevention of renal . Clin Ther 2006;28:432–44. 10.1016/j.clinthera.2006.03.012. [DOI] [PubMed] [Google Scholar]
- [4].Uwaezuoke S The role of novel biomarkers in predicting diabetic nephropathy: a review. Int J Nephrol Renovasc Dis 2017;Volume 10:221–31. 10.2147/IJNRD.S143186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mayer B Using systems biology to evaluate targets and mechanism of action of drugs for diabetes comorbidities vol. 59 Springer; Berlin Heidelberg; 2016. 10.1007/s00125-016-4032-2. [DOI] [PubMed] [Google Scholar]
- [6].Harder JL, Hodgin JB, Kretzler M. Integrative Biology of Diabetic Kidney Disease. Kidney Dis 2015;1:194–203. 10.1159/000439196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brosius FC, Ju W. The Promise of Systems Biology for Diabetic Kidney Disease. Adv Chronic Kidney Dis 2018;25:202–13. 10.1053/j.ackd.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kesić S Systems biology, emergence and antireductionism. Saudi J Biol Sci 2016;23:584–91. 10.1016/J.SJBS.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends Microbiol 2007;15:45–50. [DOI] [PubMed] [Google Scholar]
- [10].Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet 2015;16:441–58. 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tomita M, Kami K. Systems Biology, Metabolomics, and Cancer Metabolism. Science (80- ) 2012;336:990–1. 10.1126/science.1223066. [DOI] [PubMed] [Google Scholar]
- [12].Zak DE, Aderem A. Systems biology of innate immunity. Immunol Rev 2009;227:264–82. 10.1111/j.1600-065X.2008.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fernandes M, Husi H. Integrative Systems Biology Investigation of Fabry Disease. Diseases 2016;4:35 10.3390/diseases4040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berthier CC, Zhang H, Schin M, Henger A, Nelson RG, Yee B, et al. Enhanced Expression of Janus Kinase-Signal Transducer and Activator of Transcription Pathway Members in Human Diabetic Nephropathy. Diabetes 2009;58:469–77. 10.2337/db08-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, et al. Modular Activation of Nuclear Factor- B Transcriptional Programs in Human Diabetic Nephropathy. Diabetes 2006;55:2993–3003. 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- [16].Zhang H, Nair V, Saha J, Atkins KB, Hodgin JB, Saunders TL, et al. Podocyte-specific JAK2 overexpression worsens diabetic kidney disease in mice. Kidney Int 2017;92:909–21. 10.1016/j.kint.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gilbert RE. Proximal Tubulopathy: Prime Mover and Key Therapeutic Target in Diabetic Kidney Disease. Diabetes 2017;66:791–800. 10.2337/db16-0796. [DOI] [PubMed] [Google Scholar]
- [18].Betz BB, Jenks SJ, Cronshaw AD, Lamont DJ, Cairns C, Manning JR, et al. Urinary peptidomics in a rodent model of diabetic nephropathy highlights epidermal growth factor as a biomarker for renal deterioration in patients with type 2 diabetes. Kidney Int 2016;89:1125–35. 10.1016/j.kint.2016.01.015. [DOI] [PubMed] [Google Scholar]
- [19].Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and Diabetes: Analytical and Computational Approaches. Diabetes 2015;64:718–32. 10.2337/db14-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cijiang He J, Chuang PY, Ma’Ayan A, Iyengar R. Systems biology of kidney diseases. Kidney Int 2012;81:22–39. 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Levey AS, Coresh J, Balk E, Kausz AT, Levin A, Steffes MW, et al. National Kidney Foundation Practice Guidelines for Chronic Kidney Disease: Evaluation, Classification, and Stratification. Ann Intern Med 2003;139:137 10.7326/0003-4819-139-2-200307150-00013. [DOI] [PubMed] [Google Scholar]
- [22].Tangri N, Stevens LA, Griffith J, Tighiouart H, Djurdjev O, Naimark D, et al. A Predictive Model for Progression of Chronic Kidney Disease to Kidney Failure. JAMA 2011;305:1553 10.1001/jama.2011.451. [DOI] [PubMed] [Google Scholar]
- [23].Ju W, Nair V, Smith S, Zhu L, Shedden K, Song PXKK, et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med 2015;7: 10.1126/scitranslmed.aac7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bringans SD, Ito J, Stoll T, Winfield K, Phillips M, Peters K, et al. Comprehensive mass spectrometry based biomarker discovery and validation platform as applied to diabetic kidney disease. EuPA Open Proteomics 2017;14:1–10. 10.1016/J.EUPROT.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nair V, Komorowsky CV, Weil EJ, Yee B, Hodgin J, Harder JL, et al. A molecular morphometric approach to diabetic kidney disease can link structure to function and outcome. Kidney Int 2018;93:439–49. 10.1016/j.kint.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, et al. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight 2016;1:e86976 10.1172/jci.insight.86976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Niewczas MA, Mathew AV, Croall S, Byun J, Major M, Sabisetti VS, et al. Circulating Modified Metabolites and a Risk of ESRD in Patients With Type 1 Diabetes and Chronic Kidney Disease. Diabetes Care 2017;40:383–90. 10.2337/dc16-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int 2014;85:1214–24. 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Afshinnia F, Rajendiran TM, Karnovsky A, Soni T, Wang X, Xie D, et al. Lipidomic Signature of Progression of Chronic Kidney Disease in the Chronic Renal Insufficiency Cohort. Kidney Int Reports 2016;1:256–68. 10.1016/J.EKIR.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lindhardt M, Persson F, Zürbig P, Stalmach A, Mischak H, de Zeeuw D, et al. Urinary proteomics predict onset of microalbuminuria in normoalbuminuric type 2 diabetic patients, a sub-study of the DIRECT-Protect 2 study. Nephrol Dial Transplant 2016;32: 10.1093/ndt/gfw292. [DOI] [PubMed] [Google Scholar]
- [31].Conway BR, Rennie J, Bailey MA, Dunbar DR, Manning JR, Bellamy CO, et al. Hyperglycemia and renin-dependent hypertension synergize to model diabetic nephropathy. J Am Soc Nephrol 2012;23:405–11. 10.1681/ASN.2011060577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Good DM, Zürbig P, Argilés À, Bauer HW, Behrens G, Coon JJ, et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics 2010;9:2424–37. 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hoekman J, Lambers Heerspink HJ, Viberti G, Green D, Mann JFE, de Zeeuw D. Predictors of congestive heart failure after treatment with an endothelin receptor antagonist. Clin J Am Soc Nephrol 2014;9:490–8. 10.2215/CJN.07040713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Solomon SD, Uno H, Lewis EF, Eckardt K- U, Lin J, Burdmann EA, et al. Erythropoietic Response and Outcomes in Kidney Disease and Type 2 Diabetes. N Engl J Med 2010;363:1146–55. 10.1056/NEJMoa1005109. [DOI] [PubMed] [Google Scholar]
- [35].Heerspink HJL, Ninomiya T, Persson F, Brenner BM, Brunel P, Chaturvedi N, et al. Is a reduction in albuminuria associated with renal and cardiovascular protection? A post hoc analysis of the ALTITUDE trial. Diabetes, Obes Metab 2016;18:169–77. 10.1111/dom.12600. [DOI] [PubMed] [Google Scholar]
- [36].Lonergan M, Senn SJ, McNamee C, Daly AK, Sutton R, Hattersley A, et al. Defining drug response for stratified medicine. Drug Discov Today 2016;00:1–7. 10.1016/j.drudis.2016.10.016. [DOI] [PubMed] [Google Scholar]
- [37].Schievink B, de Zeeuw D, Parving H-H, Rossing P, Lambers Heerspink HJ. The renal protective effect of angiotensin receptor blockers depends on intra-individual response variation in multiple risk markers. Br J Clin Pharmacol 2015;80:678–86. 10.1111/bcp.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pena MJ, Mischak H, Heerspink HJL. Proteomics for prediction of disease progression and response to therapy in diabetic kidney disease. Diabetologia 2016;59:1819–31. 10.1007/s00125-016-4001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dekkers CCJ, Petrykiv S, Laverman G, Cherney DZ, Gansevoort RT, Heerspink HJL. Effects of the SGLT-2 inhibitor dapagliflozin on glomerular and tubular injury markers. Diabetes, Obes Metab 2018. 10.1111/dom.13301. [DOI] [PMC free article] [PubMed]
- [40].Lindhardt M, Persson F, Oxlund C, Jacobsen IA, Zürbig P, Mischak H, et al. Predicting albuminuria response to spironolactone treatment with urinary proteomics in patients with type 2 diabetes and hypertension. Nephrol Dial Transplant 2017;33: 10.1093/ndt/gfw406. [DOI] [PubMed] [Google Scholar]
- [41].Schanstra JP, Zurbig P, Alkhalaf A, Argiles A, Bakker SJL, Beige J, et al. Diagnosis and Prediction of CKD Progression by Assessment of Urinary Peptides. J Am Soc Nephrol 2015;26:1999–2010. 10.1681/ASN.2014050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aschauer C, Perco P, Heinzel A, Sunzenauer J, Oberbauer R. Positioning of tacrolimus for the treatment of diabetic nephropathy based on computational network analysis. PLoS One 2017;12:e0169518 10.1371/journal.pone.0169518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jin H, Zhang HN, Hou XL, Zhang B, Wu J, Zhang HB. Clinical study of double dose of valsartan combined with tacrolimus in treatment of diabetic nephropathy. Eur Rev Med Pharmacol Sci 2016;20:174–9. [PubMed] [Google Scholar]
- [44].Qi XM, Wu YG, Liang C, Zhang P, Dong J, Ren KJ, et al. FK506 ameliorates renal injury in early experimental diabetic rats induced by streptozotocin. Int Immunopharmacol 2011;11:1613–9. 10.1016/j.intimp.2011.05.023. [DOI] [PubMed] [Google Scholar]
- [45].Wu Y, Wang Y, Qi X, Zhang P, Zhang C, Zhang W. Increased Macrophage Activation Inhibited by Tacrolimus in the Kidney of Diabetic Rats. Nephron - Exp Nephrol 2014;128:46–56. 10.1159/000366446. [DOI] [PubMed] [Google Scholar]
- [46].Shi J, Jiang S, Qiu D, Le W, Wang X, Lu Y, et al. Rapid Identification of Potential Drugs for Diabetic Nephropathy Using Whole-Genome Expression Profiles of Glomeruli. Biomed Res Int 2016;2016:1–13. 10.1155/2016/1634730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang J, Cao J. Gene Expression Analysis in Tubule Interstitial Compartments Reveals Candidate Agents for IgA Nephropathy. Kidney Blood Press Res 2014;39:361–8. 10.1159/000355814. [DOI] [PubMed] [Google Scholar]
- [48].Li L, Greene I, Readhead B, Menon MC, Kidd BA, Uzilov AV., et al. Novel Therapeutics Identification for Fibrosis in Renal Allograft Using Integrative Informatics Approach. Sci Rep 2017;7:39487 10.1038/srep39487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science (80- ) 2006;313:1929–35. 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- [50].Brosius FC, Alpers CE, Bottinger EP, Breyer MD, Coffman TM, Gurley SB, et al. Mouse Models of Diabetic Nephropathy. J Am Soc Nephrol 2009;20:2503–12. 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gonzalez Suarez ML, Thomas DB, Barisoni L, Fornoni A. Diabetic nephropathy: Is it time yet for routine kidney biopsy? World J Diabetes 2013;4:245–55. 10.4239/wjd.v4.i6.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fukuda A, Wickman LT, Venkatareddy MP, Wang SQ, Chowdhury MA, Wiggins JE, et al. Urine podocin:nephrin mRNA ratio (PNR) as a podocyte stress biomarker. Nephrol Dial Transplant 2012;27:4079–87. 10.1093/ndt/gfs313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhu Z, Wang DC, Popescu LM, Wang X. Single-cell transcriptome in the identification of disease biomarkers: opportunities and challenges. J Transl Med 2014;12:212 10.1186/s12967-014-0212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu R, Wang X, Aihara K, Chen L. Early Diagnosis of Complex Diseases by Molecular Biomarkers, Network Biomarkers, and Dynamical Network Biomarkers. Med Res Rev 2014;34:455–78. 10.1002/med.21293. [DOI] [PubMed] [Google Scholar]
- [55].Lu Y, Ye Y, Bao W, Yang Q, Wang J, Liu Z, et al. Genome-wide identification of genes essential for podocyte cytoskeletons based on single-cell RNA sequencing. Kidney Int 2017;92:1119–29. 10.1016/j.kint.2017.04.022. [DOI] [PubMed] [Google Scholar]
- [56].Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 2018: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed]
- [57].Lu Y, Ye Y, Bao W, Yang Q, Wang J, Liu Z, et al. Genome-wide identification of genes essential for podocyte cytoskeletons based on single-cell RNA sequencing. Kidney Int 2017;92:1119–29. 10.1016/j.kint.2017.04.022. [DOI] [PubMed] [Google Scholar]
- [58].Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2017;2 10.1172/jci.insight.93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yuan G-C, Cai L, Elowitz M, Enver T, Fan G, Guo G, et al. Challenges and emerging directions in single-cell analysis. Genome Biol 2017;18:84 10.1186/s13059-017-1218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 2009;10:R130 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lee JW, Chou C-L, Knepper MA. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am Soc Nephrol 2015;26:2669–77. 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Stevens LA, Coresh J, Greene T, Levey AS. Assessing Kidney Function — Measured and Estimated Glomerular Filtration Rate. N Engl J Med 2006;354:2473–83. 10.1056/NEJMra054415. [DOI] [PubMed] [Google Scholar]
- [63].Ju W, Greene CS, Eichinger F, Nair V, Hodgin JB, Bitzer M, et al. Defining cell-type specificity at the transcriptional level in human disease. Genome Res 2013;23:1862–73. 10.1101/gr.155697.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Golubnitschaja O, Filep N, Yeghiazaryan K, Blom HJ, Hofmann-Apitius M, Kuhn W. Multi-omic approach decodes paradoxes of the triple-negative breast cancer: lessons for predictive, preventive and personalised medicine. Amino Acids 2017. 10.1007/s00726-017-2524-0. [DOI] [PubMed]
- [65].Meng C, Kuster B, Culhane AC, Gholami AM. A multivariate approach to the integration of multi-omics datasets. BMC Bioinformatics 2014;15:162 10.1186/1471-2105-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Winterhoff B, Hamidi H, Wang C, Kalli KR, Fridley BL, Dering J, et al. Molecular classification of high grade endometrioid and clear cell ovarian cancer using TCGA gene expression signatures. Gynecol Oncol 2016;141:95–100. 10.1016/J.YGYNO.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016;166:740–54. 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lindhardt M, Persson F, Currie G, Pontillo C, Beige J, Delles C, et al. Proteomic prediction and Renin angiotensin aldosterone system Inhibition prevention Of early diabetic nephRopathy in TYpe 2 diabetic patients with normoalbuminuria (PRIORITY): essential study design and rationale of a randomised clinical multicentre trial. BMJ Open 2016;6:e010310 10.1136/bmjopen-2015-010310. [DOI] [PMC free article] [PubMed] [Google Scholar]


