ctDNA monitoring during EGFR-TKI treatment is useful for detecting T790M mutation. However, the efficacy of osimertinib treatment based on T790M status in plasma ctDNA remains to be established.
Keywords: liquid biopsy, EGFR-TKI, ctDNA, T790M, osimertinib
Abstract
Background
Osimertinib, a third generation epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor (TKI), is active against EGFR-mutant non-small cell lung cancer (NSCLC) resistant to first-/second-generation EGFR-TKIs with the T790M mutation. T790M monitoring in plasma circulating tumor DNA (ctDNA) in patients receiving EGFR-TKIs is less invasive than re-biopsy and could provide valuable clinical information.
Methods
Patients with advanced or postoperative recurrent NSCLC with sensitizing EGFR mutations who were planned to receive or were receiving first-/second-generation EGFR-TKI treatment without disease progression were eligible for enrollment. Plasma samples at baseline and every 1–2 months thereafter were analyzed for EGFR mutation status using the cobas®EGFR Mutation Test v2.
Results
Between September 2016 and March 2017, 122 patients at 15 Japanese institutions were enrolled. In August 2018, 1291 plasma samples from 121 patients were analyzed for EGFR mutation status. At baseline, a sensitizing EGFR mutation was detected in 29 (23.9%) of 121 patients and T790M mutation was detected in three (2.5%). At follow-up, 66 (54.5%) patients experienced disease progression and 64 (52.9%) discontinued first-line EGFR-TKI treatment. Twenty-two (18.2%) patients showed T790M in plasma ctDNA, of which 15(68.2%) received osimertinib. Although 31 patients received re-biopsy to examine EGFR status at disease progression, T790M was detected in only nine (22.0%) patients, of which 7 (77.8%) received osimertinib.
Conclusions
ctDNA monitoring during EGFR-TKI treatment is useful for detecting T790M mutation. The efficacy of osimertinib treatment based on T790M status in plasma ctDNA remains to be established, warranting further research.
Introduction
Treatment with first- or second-generation epidermal growth factor (EGFR)-tyrosine kinase inhibitors (TKIs) is effective for non-small cell lung cancer (NSCLC) patients harboring a sensitizing EGFR mutation. However, acquired resistance is inevitable after 9–14 months (1–6).
The most common mechanism of resistance to first- and second-generation EGFR-TKIs in the first-line setting is the EGFR T790M mutation, which accounts for approximately 60% of cases (7,8). Osimertinib, a third-generation EGFR-TKI targeting EGFR T790M mutation, is reported to be highly active against T790M-positive NSCLC (9). To detect the T790M mutation in patients progressing during EGFR-TKI treatment, tumor re-biopsy is necessary. However, re-biopsies with invasive procedures (bronchoscopy or needle biopsy) are often infeasible in standard care of NSCLC patients (10,11).
Circulating tumor DNA (ctDNA) detected in plasma is recognized as a noninvasive biomarker for the molecular analysis of NSCLC (12). The cobas® EGFR Mutation Test (Roche Diagnostics K.K., Switzerland.) is a companion diagnostic test for the detection of EGFR mutations in plasma specimens and has been approved to identify such patients with NSCLC (13–15). T790M monitoring in plasma ctDNA of patients receiving EGFR-TKIs could yield valuable clinical information. We conducted an observational study to estimate the usefulness of plasma ctDNA monitoring in NSCLC patients with EGFR mutations receiving EGFR-TKIs.
Patients and methods
Study population
Patients with histologically and/or cytologically confirmed advanced or postoperative recurrent NSCLC harboring sensitizing EGFR mutations were eligible if they were at least 20 years old and were receiving or planned to receive first-line EGFR-TKIs (gefitinib, erlotinib, or afatinib). Sensitizing EGFR mutations were defined as follows: (1) Exon 19 deletion; (2) Exon 21 L858R; and (3) other minor mutation (i.e., Exon 18 G719X). The co-existence of T790M was not excluded. Patients were required to have an Eastern Cooperative Oncology Group performance status of 0 to 2. Patients were excluded if they had undergone prior EGFR-TKI treatment with disease progression or had hepatitis B virus (HBsAg), hepatitis C virus (HCV-RNA), or HIV.
Plasma ctDNA analysis
Plasma to assess the EGFR genotype of circulating ctDNA was collected at baseline and every 1–2 months. The following events were particularly noted: (1) radiological disease progression; (2) clinical disease progression; (3) re-biopsy at disease progression; and (4) treatment change. At each institution, 10-mL samples of blood were centrifuged within 4 hours of plasma collection. Plasma ctDNA was analyzed at SRL Laboratory (Tokyo, Japan) using the cobas® EGFR Mutation Test version 2 (v2) to detect sensitizing EGFR mutations and the T790M resistance mutation.
Re-biopsy and EGFR mutation analysis
When disease had progressed, re-biopsy was recommended. The EGFR genotypes of re-biopsied materials were analyzed at each hospital using the peptide nucleic acid-locked nucleic acid clamp method (16) or the cobas® EGFR Mutation Test v2.
Clinical data collection
Case report forms (CRFs) were collected at 6 and 12 months after registration. The CRF included clinical information about radiological disease progression (PD; date, site of disease progression), clinical PD (date, pattern of PD), survival (date last verified), death (date), cause of death, and adverse events. Radiological PD was assessed according to the Response Evaluation Criteria in Solid Tumors v1.1 at each institution. Clinical PD was defined as follows: clinical symptoms with disease progression; worsening of performance status due to disease progression; main organ dysfunction (lymphangitis carcinomatosa, bone marrow metastasis, meningitis carcinomatosa, or liver metastasis with hepatic dysfunction); and other clinically meaningful multiple metastasis. Adverse events were assessed according to the Common Terminology Criteria for Adverse Events v4.0.
Statistical analysis
This study is an observational study to estimate the usefulness of ctDNA monitoring in NSCLC patients with EGFR mutations who received first-line EGFR-TKIs. The primary analysis was designed to estimate the plasma ctDNA T790M-positivity rates using the cobas® EGFR Mutation Test in patients with T790M-positive tumors and at each clinical point. In the prior CSPOR-LC02 study (observational study of treatment of EGFR mutation-positive advanced or recurrent NSCLC: UMIN 000010538), radiological PD was documented in approximately 80% of the patients (17). Among the patients (80%) who acquired resistance to EGFR-TKIs, approximately 60% were presumed to have T790M. This study used descriptive statistics and was set to 120 cases in consideration of feasibility of research. Median time to progression was estimated based on the Kaplan–Meier method.
Ethical considerations
This study protocol was approved by the institutional review board at each participating institution. Declaration of Helsinki ethical standards and local and national regulations were followed. All patients provided written informed consent before participation.
Results
Patients
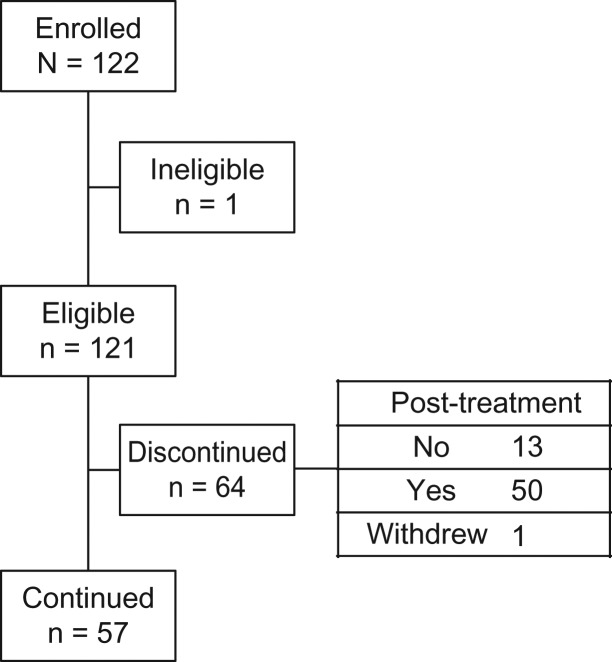
A total of 122 Japanese patients were enrolled between September 2016 and March 2017 at 15 sites in Japan. One patient was ineligible because of first-line EFGR-TKI treatment failure. A total of 121 patients were registered. Patient characteristics are shown in Table 1. At the data cut-off date of this study (30 August 2018), CRFs were collected from 121 and 108 patients at 6 and 12 months after registration, respectively. Median(range) follow-up time was 369 (9–438) days. During the follow-up period, 66 (54.5%) patients experienced disease progression and 64 (52.9%) discontinued first-line EGFR-TKI treatment (Fig. 1). Median (95% CI) time to progression, which was defined as radiological or clinical PD since first-line EGFR-TKI treatment, was 663 (512–916) days.
Table 1.
Patient characteristics
| N = 121 | |||
|---|---|---|---|
| Age | Median (range) | 72 | (40–92) |
| Sex | Male | 42 | (34.7) |
| Female | 79 | (65.0) | |
| PS | 0 | 64 | (52.9) |
| 1 | 54 | (44.6) | |
| 2 | 3 | (2.5) | |
| Smoking status | Never | 80 | (66.1) |
| Current/former | 39 | (32.3) | |
| Unknown | 2 | (1.7) | |
| Histology | Adenocarcinoma | 118 | (97.5) |
| Others | 3 | (2.5) | |
| EGFR genotype | Ex 19 del | 61 | (50.4) |
| Ex 21 L858R | 55 | (45.5) | |
| Others | 4 | (3.36) | |
| Ex 21 L858R + other | 1 | (0.8) | |
| Clinical stage of NSCLC | IIIA | 1 | (0.8) |
| IIIB | 3 | (2.5) | |
| IV | 78 | (64.5) | |
| recurrence | 39 | (32.2) | |
| EGFR-TKI | Gefitinib | 50 | (41.3) |
| Erlotinib | 40 | (33.1) | |
| Afatinib | 31 | (25.6) | |
Data are n (%), unless otherwise stated.
EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer; PS, performance status; TKI, tyrosine kinase inhibitor.
Figure 1.
CONSORT flow diagram of this study.
Frequency of T790M detection in re-biopsied samples
Forty-one samples obtained from 33 patients (at disease progression during first-line EGFR-TKI treatment, n = 31, and at disease progression during post-discontinuation treatment, n = 2) were collected to analyze the EGFR genotype. DNA was extracted from various materials including lung tissue (n = 19), lymph nodes (n = 4), pleural effusions (n = 8), and other tissues (n = 10). A sensitizing EGFR mutation and T790M were detected in 24 (72.7%) and nine (27.3%) patients, respectively. EGFR mutations were not detected in four (12.1%) patients. There was insufficient DNA to analyze the EGFR genotype in the samples from four (12.1%) patients. Only two (22.2%) of nine patients with T790M detected in re-biopsied materials showed T790M in plasma. The sensitivity and specificity of the cobas® EGFR Mutation Test v2 to detect T790M mutation in re-biopsied materials by plasma ctDNA in this study were 2/9 (22.2%) and 15/22(68.2%), respectively. The concordance rate of T790M detection in re-biopsied materials and plasma was 54.8%.
Frequency of T790M detection in plasma ctDNA
Because one patient withdrew from this study after registration, a total of 1291 plasma samples were collected from 120 patients. At baseline, a sensitizing EGFR mutation (Ex 19 del n = 14, L858R n = 15) was detected in 29 (24.2%) of 120 patients and T790M was detected in three (2.5%) patients. At 12 months, a sensitizing EGFR mutation and T790M was detected in the plasma ctDNA of 57 (47.5%) and 22 (18.3%) patients, respectively. The frequency of T790M detection in plasma ctDNA at 12 months was higher in patients with Ex 19 deletions detected in plasma (15/29, 51.7%) than with the L858R mutation (6/28, 21.4%) (chi-square test, P = 0.017). Other EGFR mutations except Ex 19 deletion, L858R, and T790M were not detected in plasma ctDNA.
Timing of T790M detection in plasma ctDNA
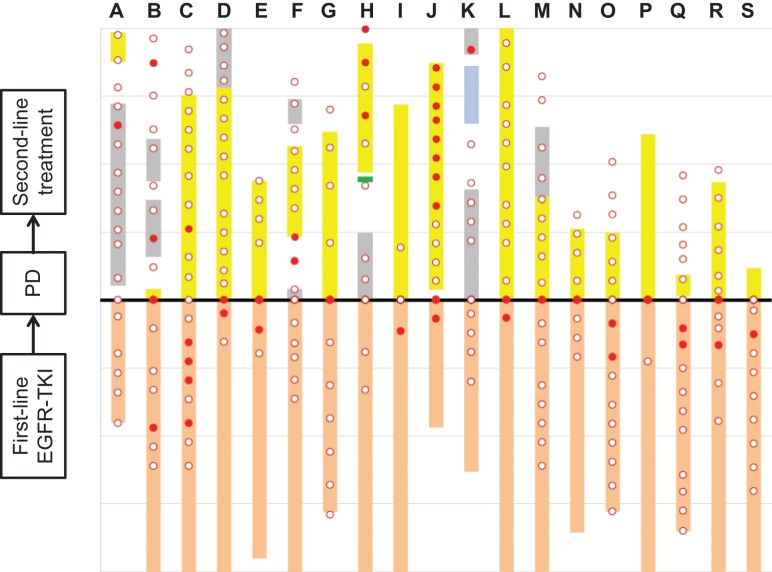
Of 22 patients with T790M detected in plasma ctDNA, 19 patients experienced disease progression and discontinued first-line EGFR-TKI treatment. Three patients with T790M detected in plasma did not experience clinical/radiological disease progression and continued first-line EGFR-TKI at 12 months. The timing of T790M detection in plasma ctDNA in 19 patients with disease progression is shown in Figure 2. T790M detection in plasma ctDNA preceded and/or appeared at disease progression in 15 (78.9%) of 19 patients; however, in four (21.1%) patients, T790M was detected for the first time after the disease had progressed.
Figure 2.
Timing of T790M detection in plasma ctDNA in patients with disease progression. Timing of T790M detection (●) and no T790M detection (○); duration of first-line EGFR-TKI treatment (orange bar), osimertinib (yellow bar), afatinib(green),cytotoxic chemotherapy (gray bar), and immune checkpoint inhibitor (blue bar). Letters A–S represent each patient.
Treatment after first-line EGFR-TKI failure and T790M detection
Among the 64 patients who discontinued first-line EGFR-TKI treatment, 50 received post-discontinuation treatment. Of these patients, 20 (40%) received osimertinib as the second-line treatment following first-line EGFR-TKI failure. Of nine patients with T790M detected in re-biopsied materials, seven (77.8%) received osimertinib. Of 22 patients with T790M detected in plasma ctDNA, 15 (68.1%) received osimertinib, three (13.6%) continued first-line EGFR-TKI therapy, and two (9.0%) switched to platinum-based chemotherapy.
Discussion
At a median follow-up of 1 year, T790M was detected in 29 patients. T790M was detected in the plasma ctDNA only, re-biopsied materials only, and both the plasma ctDNA and the re-biopsied materials of 20, 7, and 2 patients, respectively. The concordance rate of T790M detection in re-biopsied materials and plasma was 54.8%. T790M detection in plasma ctDNA preceded and/or appeared at disease progression in 15 (78.9%) of 19 patients with disease progression.
Repeated monitoring of circulating ctDNA in this study increased the frequency of T790M detection and the proportion of osimertinib treatment in the second-line setting of patients with advanced EGFR mutation-positive NSCLC. Seto et al. reported in the REMEDY study that T790M were detected in only 19.7% of plasma samples and the frequency of T790M detection and the proportion of the patients treated with osimertinib were approximately 25.8% and 23.7%, respectively, in the real-world setting (18). In this study, T790M was detected in 22 (33.3%) of 66 patients at disease progression during first-line EGFR-TKI treatment and 20 (30.3%) patients received osimertinib treatment in the second-line setting. Although we analyzed plasma ctDNA using the PCR-based cobas® EGFR Mutation Test v2, next-generation sequencing (NGS) can reach higher values of sensitivity compared with PCR-based methods (19,20). The NGS concordance rate with tumor tissue for EGFR alterations is very high. According to the International Association for the Study of Lung Cancer, an NGS multiplex panel is preferred and recommended over PCR-based methods as it is capable of detecting not only the common resistance mutation T790M but also a spectrum of alterations (19). The considerable advantage of NGS methods over PCR-based methods could potentially be useful for increasing the frequency of T790M detection and ensuring the appropriate delivery of osimertinib in patients with first- and second-generation EGFR-TKI failure.
T790M detection in plasma could precede radiological progressive disease and could be potentially used to monitor response to first- and second-generation EGFR-TKIs (21); however, T790M detection appeared at unexpected sites and at unexpected moments. T790M detection in plasma ctDNA preceded and/or appeared at disease progression in 15 (78.9%) of 19 patients with disease progression and after disease progression during discontinuation of first-line EGFR-TKI treatment in four (21.1%) patients in this study. Zheng et al. reported that 45% of patients harboring a T790M mutation could have this alteration detected before progressive disease through ddPCR assays (22). Further investigation is necessary to elucidate the clinical benefits of switching to osimertinib from first-/second-generation EGFR-TKIs when T790M is detected in patients’ plasma ctDNA without disease progression.
As first-line use of osimertinib becomes the standard of care in the first-line setting of advanced EGFR mutation-positive NSCLC with the results of the FLAURA trial (23), the frequency of T790M will decline, but understanding the mechanisms of resistance to osimertinib will likely be of clinical utility to patients in the near future. Oxnard et al. reported that the persistent existence of T790M at osimertinib treatment failure is a good prognostic factor of patients with T790M mutation treated with osimertinib (24). Del Re et al. reported that the T790M/activating EGFR mutant allele frequency ratio is a prognostic factor of osimertinib treatment (25). These data suggest that ctDNA monitoring of sensitizing EGFR mutations and T790M may be useful in monitoring the efficacy of osimertinib treatment.
This study has several limitations. First, this study was an observational study rather than one designed to define treatment strategies when T790M is detected in plasma. Second, tissue re-biopsy upon disease progression was not mandatory but recommended in this study, and hence the frequency of re-biopsy to estimate EGFR status (31/66, 47.0%) and T790M detection in re-biopsied materials (9/31, 29.0%) was low. Additionally, the sample size in this study is small for assessing the concordance rate of T790M detection in tissue and plasma. Finally, we could not assess the levels of sensitizing EGFR mutations and T790M as well as other resistance mechanisms of first- and second-generation EGFR-TKIs including MET and erb-b2 receptor tyrosine kinase 2 amplification.
In conclusion, plasma ctDNA monitoring using the cobas® EGFR Mutation Test v2 increased the frequency of T790M detection in patients with a sensitizing EGFR mutation during first- and/or second-generation EGFR-TKI treatment. Further investigation is necessary to evaluate the clinical usefulness of starting treatment with the third-generation EGFR-TKI osimertinib based on T790M detection in plasma ctDNA.
Acknowledgments
We thank all patients, their families, care-givers and staff at all institutions.
Sites participating in this study were as follows:
Fujisawa City Hospital
Gunma Prefectural Cancer Center
Iwate Prefectural Central Hospital
Japanese Red Cross Medical Center Japanese Red Cross Saitama Hospital
Kansai Electric Power Hospital
Kasukabe Medical Center
Kyorin University Hospital
Mitsui Memorial Hospital
National Hospital Organization Shibukawa Medical Center
National Center for Global Health and Medicine
NTT Medical Center Tokyo
Showa University Hospital
Showa University Fujigaoka Hospital
Toranomon Hospital
Funding
This work (Clinical Trial Number: UMIN 000023248) was supported by AstraZeneca. The study funding was provided to the Comprehensive Support Project for Oncology Research (CSPOR) with support from an investigator-sponsored study program of AstraZeneca.
Conflict of interest statement
Dr.Naka has received personal fees from AstraZeneca. Dr.Ohashi has received personal fees from Taiho. Dr.Kunitoh has received personal fees from AstraZeneca,Boehringer Ingelheim, Chugai, Taiho, Daiichi-sankyo, Johnsonand Johnson. All remaining authors have declared no conflicts of interest.
References
- 1. Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harboring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomized phase 3 trial. Lancet Oncol 2010;11:121–8. [DOI] [PubMed] [Google Scholar]
- 2. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. [DOI] [PubMed] [Google Scholar]
- 3. Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicenter, open-label, randomized phase 3 trial. Lancet Oncol 2012;13:239–46. [DOI] [PubMed] [Google Scholar]
- 4. Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicenter, open-label, randomized, phase 3 study. Lancet Oncol 2011;12:735–42. [DOI] [PubMed] [Google Scholar]
- 5. Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213–22. [DOI] [PubMed] [Google Scholar]
- 6. Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141–51. [DOI] [PubMed] [Google Scholar]
- 7. Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortot AB, Janne PA. Molecular mechanisms of resistance in epidermal growth factor receptor-mutant adenocarcinomas. Eur Respir Rev 2014;23:356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bosc C, Ferretti GR, Cadranel J, et al. Rebiopsy during disease progression in patients treated by TKI for oncogene-addicted NSCLC. Target Oncol 2015;10:247–53. [DOI] [PubMed] [Google Scholar]
- 11. Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re-biopsy in advanced non-small-cell lung cancer: a prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014;86:170–3. [DOI] [PubMed] [Google Scholar]
- 12. Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD 9291. Lung Cancer 2015;90:509–15. [DOI] [PubMed] [Google Scholar]
- 14. Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA analysis for detection of EGFR T790M mutation in patients (pts) with EGFR mutation-positive advanced non-small cell lung cancer (aNSCLC). J Thorac Oncol 2017;12:1061–70. [DOI] [PubMed] [Google Scholar]
- 15. Zhou C, Wang M, Cheng Y, et al. 1311P Detection of EGFR 790M in Asia-Pacific patients (pts) with EGFR mutation-positive advanced non-small cell lung cancer (NSCLC): circulating tumor (ct) DNA analysis across 3 platforms. Ann Oncol 2017;28:v460–96. 10.1093/annonc/mdx380. [DOI] [Google Scholar]
- 16. Sutani A, Nagai Y, Udagawa K, et al. Gefitinib for nonsmall- cell lung cancer patients with epidermal growth factor receptor gene mutations screened by peptide nucleic acidlocked nucleic acid PCR clamp. Br J Cancer 2006;95:1483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goto Y, Tanai C, Yoh K, et al. Continuing EGFR-TKI beyond radiological progression in patients with advanced or recurrent, EGFR mutation-positive non-small-cell lung cancer: an observational study. ESMO Open 2017;2:e000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seto T, Nogami N, Yamamoto N, et al. Real-world EGFR T790M testing in advanced non-small-cell lung cancer: a prospective observational study in Japan. Oncol Ther 2018;6:203–15. 10.1007/s40487-018-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rolfo C, Mack PC, Scagliotti GV, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the ISALC. J Thorac Oncol 2017;13:1248–68. [DOI] [PubMed] [Google Scholar]
- 20. Sacher AG, Komatsubara KM, Oxnard GR. Application of plasma genotyping technologies in non-small cell lung cancer: a practical review. J Thorac Oncol 2017;12:1344–56. [DOI] [PubMed] [Google Scholar]
- 21. Lee JY, Qing X, Xiumin W, et al. Longitudinal monitoring of EGFR mutation in plasma predicts outcomes of NSCLC patients treated with EGFR-TKIs: Korean Lung Cancer Consortium (KLCC 12-02). Oncotarget 2016;7:6984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 2016;6:20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 24. Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol 2018;4:1527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Del Re M, Bordi P, Rofi E, et al. The amount of activating EGFR mutations in circulating cell-free DNA is a marker to monitor osimertinib response. Br J Cancer 2018;119:1252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]