Abstract
Objective:
During the treatment planning of a preclinical small animal irradiation, which has time limitations for reasons of animal wellbeing and workflow efficiency, the time consuming organ at risk (OAR) delineation is performed manually. This work aimed to develop, demonstrate, and quantitatively evaluate an automated contouring method for six OARs in a preclinical irritation treatment workflow.
Methods:
Microcone beam CT images of nine healthy mice were contoured with an in-house developed multiatlas-based image segmentation (MABIS) algorithm for six OARs: kidneys, eyes, heart, and brain. The automatic contouring was compared with the manual delineation using three quantitative metrics: the Dice Similarity Coefficient (DSC), 95th percentile Hausdorff Distance, and the centre of mass displacement.
Results:
A good agreement between manual and automatic contouring was found for OARs with sharp organ boundaries. For the brain and the heart, the median DSC was larger than 0.94, the median 95th Hausdorff Distance smaller than 0.44 mm, and the median centre of mass displacement smaller than 0.20 mm. Lower DSC values were obtained for the other OARs, but the median DSC was still larger than 0.74 for the left eye, 0.69 for the right eye, 0.89 for the left kidney and 0.80 for the right kidney.
Conclusion:
The MABIS algorithm was able to delineate six OARs with a relatively high accuracy. Segmenting OARs with sharp organ boundaries performed better than low contrast OARs.
Advances in knowledge:
A MABIS algorithm is developed, evaluated, and demonstrated in a preclinical small animal irradiation research workflow.
Introduction
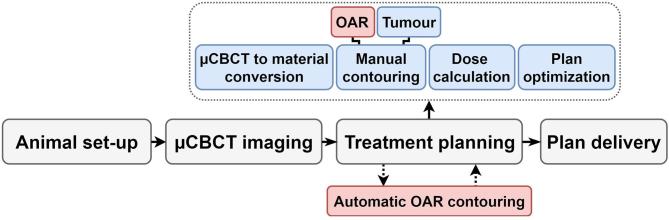
A modern pre-clinical small animal irradiation workflow, using image-guided precision irradiation cabinets, consists of several steps1 (Figure 1) that must fit into a tight time schedule, since the animal is physically constrained under deep anesthesia. In the first steps of this pre-clinical workflow, the animal is anesthetized and imaged using the onboard microcone beam CT (µCBCT) scanner, which is frequently implemented in these irradiators. After finishing the µCBCT image reconstruction, the images are loaded in a dedicated pre-clinical treatment planning system to design a quantitative treatment plan to deliver, following the pre-clinical irradiation workflow. In these treatment planning systems,2 such as SmART-ATP3 (Precision X-ray Inc., North Branford, CT), an interactive tissue segmentation is performed based on a CT number histogram, and is followed by a time consuming manual delineation of both the target (e.g. the tumor) and the organs at risk (OARs). In the final step of the planning workflow, the irradiation treatment is optimized; the dose calculation of the irradiation plan is performed and evaluated quantitatively considering the dose volume histogram parameters. Potentially also other forms of imaging, such as bioluminescence photon imaging, may be performed while the animal is under anesthesia.4 All these workflow steps should not require more than 20–60 min, for reasons of animal wellbeing and workflow efficiency.5
Figure 1.
Pre-clinical small animal image-guided precision irradiation workflow. In the treatment planning system, the time consuming manual contouring of the OARs, often performed by biologists and not delineation experts, could be replaced by automatic OAR contouring, while simultaneously other (blue) steps such as the µCBCT to material conversion, the manual tumor delineation, the dose calculation, and the plan optimization are performed. OAR, organ at risk; µCBCT, microcone beam CT.
The manual delineation of the OARs before plan delivery is one of the most time consuming tasks, especially due to the high number of axial slices resulting from the submillimetric voxel sizes used in pre-clinical µCBCT imaging. Much effort was made to automatically segment OARs for human radiation treatment planning. E.g. open source packages for automatic image segmentation were developed,6 automatic contouring competitions were organized,7 and researchers considered the future perspectives of automatic contouring in radiotherapy.8
In the literature, only a few efforts have been made to segment organs on images of rodents. Scheenstra et al9 presented an automated hybrid method to segment in vivo and ex vivo mouse brain MR images that combined affine image registration and clustering techniques. Uberti et al10 applied a semi-automatic algorithm to extract the brain from in vivo mouse MR images by user-defined constraints. To improve the pre-clinical functional imaging workflow, Lancelot et al11 evaluated a multiatlas-based rat MR image segmentation algorithm and investigated whether the algorithm was capable of automatically extracting [18F]fludeoxyglucose radioactivity from volumes of interest of positron emission tomography images in the brain.
Other researchers investigated the feasibility of automatic image segmentation using contrast-enhanced fan-beam µCT images. Yan et al12 developed a supervoxel-based machine learning algorithm that used knowledge of the different time points in a dynamic contrast-enhanced fan-beam µCT scan of mice to classify supervoxels in multiple organ categories. Baiker et al13 presented a fully automated atlas-based whole-body segmentation from low-contrast fan-beam µCT images using a four dimensional (4D) digital mouse phantom14 (MOBY) and a hierarchical atlas tree description.
Most studies in (semi-) automatic tissue segmentation of small animal organs were for MRI and contrast-enhanced fan-beam µCT. To our knowledge, there are no studies investigating the feasibility of automatic OAR segmentation in a pre-clinical irradiation workflow using µCBCT images. In this paper, we describe an in-house developed multiatlas-based image segmentation (MABIS) algorithm and investigate whether automatic OAR segmentation is feasible using a pre-clinical in vivo non-contrast-enhanced µCBCT data set of healthy mice. In MABIS algorithms, the atlas is referring to a µCBCT data set with already delineated OARs by a medical expert.
Methods and materials
Mouse data set
Nine healthy female NMRI nude mice without tumors served as the control group in a biological study were µCBCT imaged with an X-RAD 225Cx small animal radiation research system (Precision X-ray Inc., North Branford, CT) at an X-ray tube potential of 50 kVp, X-ray tube current of 5.59 mA, and a gantry rotation time of 0.5 rotations per minute.15 During the µCBCT imaging, all mice were under deep anesthesia i.e. isoflurane. The corrected projection data (flood field, dark field, and dead pixels) is reconstructed using the Feldkamp-Davis-Kress back projection algorithm implemented in the open-source Reconstruction Toolkit16 in a 512 × 512 × 1024 image matrix with a voxel dimension of 100 × 100 × 100 µm3. All nine mice were imaged at a 30 cGy imaging dose without contrast agent and in feet first-prone position including the nose cone, which is used to administer the anesthesia gas. The imaging dose level was verified using a PTW TN300012 Farmer-type ionization chamber (PTW, Freiburg, Germany) according to the AAPM TG-61 protocol.17 For each individual mouse, a medical doctor, experienced in mouse OAR delineations, contoured six OARs: the left kidney, right kidney, heart, brain, left eye, and the right eye. In this study, we assume these manually delineated contours as the ground truth.
Multiatlas-based image segmentation algorithm
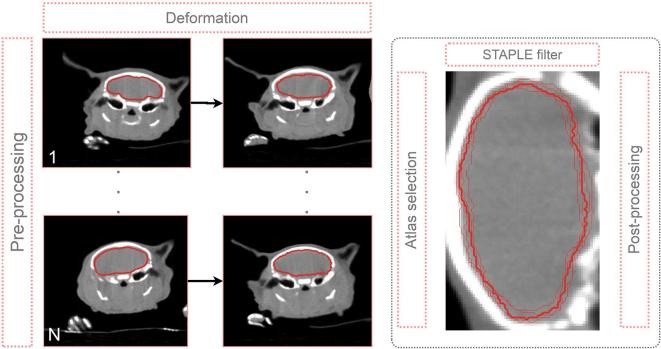
An in-house developed MABIS approach optimized at the organ level and briefly summarized in Figure 2, is adopted to automatically contour the OARs of the mouse atlas data set in a leave-one-out approach (9 mice × 8 = 72 deformations). Because the limited data set of nine mice (N = 9), the leave-one-out cross-validation approach is used to test the general performance of the MABIS algorithm.
Figure 2.
Summary of our in-house developed MABIS algorithm. A pre-processing step is completed before every atlas in the atlas database (1,2, …, N atlases) is deformed to the unsegmented mouse (in this example in the cranial body region). At the organ level, the atlases with the highest NCC coefficients are selected. The STAPLE filter algorithm calculates a consensus contour (thick contour) from the individual deformed mouse contours (thin lines) and the post-processing algorithm is executed in a last step (the right image is rotated 90 degrees for viewing purposes). MABIS, multiatlas-based image segmentation; NCC, normalized cross-correlation; STAPLE, Simultaneous Truth And Performance Level Estimation.
In the pre-processing step, the reconstructed 512 × 512 × 1024 image matrix is cropped to a 300 × 300 × 700 image matrix keeping the central part. After cropping, the µCBCT image data is denoised using the multithreaded curvature flow image filter (number of iterations = 5; time step = 0.05) implemented in the Insight Segmentation and Registration Toolkit. This edge-preserving denoising algorithm is smoothing the image to obtain a better deformation in the last stage of the multistage image deformation algorithm explained in the next section.
In the deformation step, Elastix18 and its Transformix submodule are used to apply a multistage deformable image registration (DIR). This DIR algorithm first calculates an affine transformation followed by a B-spline transformation. Here, the affine transformation eliminates the animal positioning dependency in the automatic OAR segmentation procedure. Considering the leave-one-out approach, the calculated deformation field is then used to deform the original µCBCT and segmentation data set from all mice in the remaining mouse atlas database (#8) to the unsegmented mouse that is left out. A detailed list of the selected deformation parameters of the affine transformation and B-spline transformation are listed in the Supplementary Table 1.
After deformation of the mouse atlas data set to the mouse that has been omitted, eight deformed µCBCT and eight deformed segmentation data sets could be used as input for the STAPLE filter.19 However, the MABIS algorithm performs an atlas selection for every organ, i.e. different mouse atlases could be used to contour different OARs, all depending on the performance of the deformation in the body region around the OAR. To select the best mouse atlas for every OAR, the eight deformed three-dimensional (3D) manually segmented OAR data sets are combined into a logical 4D image vector, where the fourth dimension represents the number of deformed atlases (9 minus 1 in this study). The fourth dimension is summed logically to calculate a 3D bounding box that perfectly encloses the deformed OAR in the eight deformed mouse µCBCT data sets. This 3D bounding box plus a small expansion, calculated for every OAR, is used to crop the deformed mouse µCBCT data set and the µCBCT image data of the omitted unsegmented mouse.
In the last step of the atlas selection, for every organ, the normalized cross-correlation (NCC) coefficient is calculated between eight (j = 1, …, 8) deformed cropped mouse atlases () and the cropped unsegmented mouse image data (M), where and represent the voxel intensities at voxel index . and are the mean voxel intensities in the µCBCT images A and M. The NCC coefficient is equal to one when both µCBCT data sets correlate perfectly and is equal to zero when no correlation is found.
NCCj coefficients, calculated for each deformed atlas j, are sorted in descending order for every OAR segmentation. The structure deformations, corresponding to image deformations with the highest NCCj coefficients, are used as input for the STAPLE algorithm, which calculates an OAR probability distribution. In our MABIS algorithm, the STAPLE filter implementation from Insight Segmentation and Registration Toolkit is adopted. After thresholding the OAR probability distribution, a raw automatic segmentation can be derived which is then morphologically smoothed in the post-processing step to obtain the final contour of the OAR.
The MABIS algorithm’s pre-processing step, applying the STAPLE filter for all OARs, and the post-processing step were run on a computer with two Intel® Xeon® E5-2650 v3 (2.30 GHz) processors. All Elastix and Transformix jobs were submitted to an HTCondor CPU cluster (144 cores, at least 2 GB RAM per core) connected with a one gigabit data transfer Ethernet connection.
Segmentation evaluation metrics
For evaluation of all nine automatic OAR segmentations, the Dice Similarity Coefficient (DSC), the 95th percentile Hausdorff Distance (95th HD), and the centre of mass displacement are employed.
These three quantitative evaluation metrics are calculated between the ground truth OAR segmentation by the medical doctor () and the automatic OAR segmentations from the MABIS algorithm ().
-
The DSC calculates the overlap of two contours and is equal to one if both contours perfectly overlap.
-
The 95th HD is a measure to describe the 95th percentile boundary distance between two contours, where the best outcome is 0 mm, and is quantified as follows:
The centre of mass displacement ()between the CoM of two contours is calculated in a Cartesian coordinate system . With the metric equal to zero, the mass centes of both contours perfectly overlap.
Results
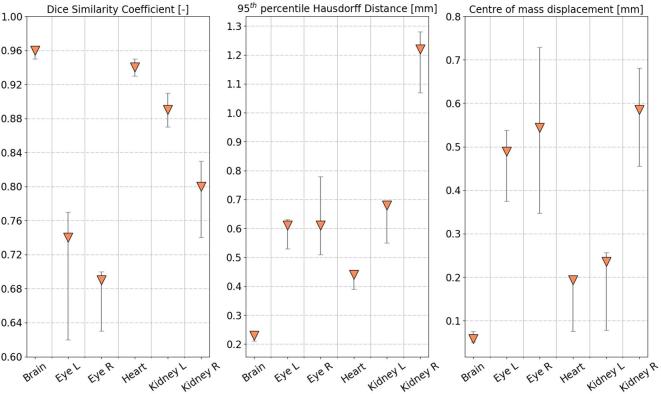
Figure 3 reports the DSC, 95th HD, and quantitative metrics for all six OARs. The automatically generated contour by the MABIS algorithm and the manual delineation, plotted on the µCBCT image, are compared in Figure 4 for each individual OAR and for all nine mice. The axial µCBCT slices shown in Figure 4 were randomly chosen around the middle of the manually delineated OAR.
Figure 3.
Quantitative evaluation of the automatic OAR segmentations in nine mice adopting the leave-one-out technique. The median evaluation metric value together with the 25th and the 75th percentiles whiskers are plotted. OAR, organ at risk.
Figure 4.
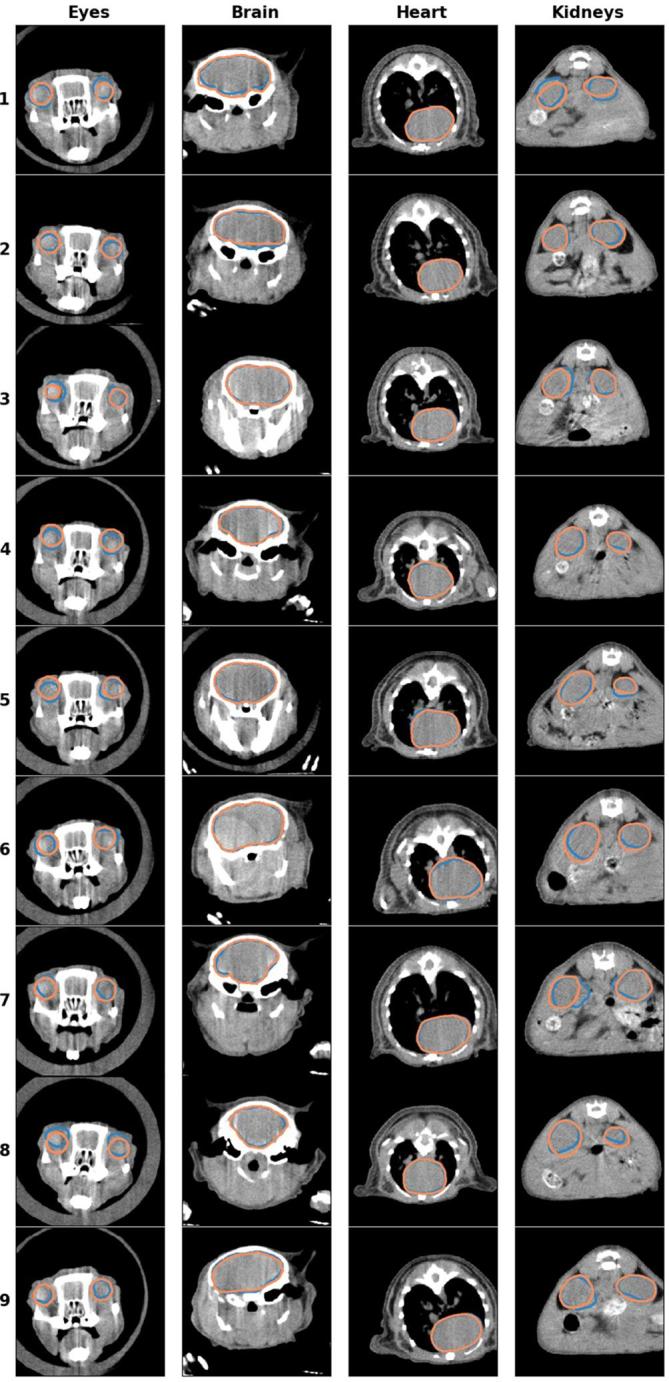
The automatic generated contour (blue) and the ground truth delineation (orange) depicted for all nine mice (rows). The axial slices were randomly selected around the middle of the manually delineated OAR. OAR, organ at risk.
The average MABIS autosegmentation time of one mouse was 12 min and 40 s. From this segmentation time, 45% of the calculation time was required to write, transfer, read, and deform the mouse atlas data set (#8), and 55% of the calculation time was needed to apply the post-processing step for six OARs. Only an average contouring time is reported in this study because the deformation calculations were submitted randomly to the HTCondor CPU cluster, which is composed of computers with different processor units.
Discussion
The in-house developed MABIS performed better for structures with sharp boundaries such as the skull (i.e. brain) or the lungs (i.e. heart). For both structures, the median DSC was larger than 0.94, the median 95th HD smaller than 0.44 mm, and the median smaller than 0.20 mm. As plotted in Figure 3, the small spread, indicated by the 25th and 75th percentile whiskers, on the nine individual DSC, 95th HD, and metrics of the brain and the heart, is noticeable.
OARs having a lower image contrast are more difficult to contour in a non-contrast enhanced µCBCT image. This difficulty applies to both the manual contouring (larger intraobserver variability) and the automatic contouring, which relies on manual contoured atlases. Organs located in low contrast body regions and subject to motion artifacts, e.g. the spleen or the liver, were not included in this automatic contouring study because of the increased intraobserver variability, which will directly give rise to non-interpretable evaluation metrics.
The absence of image contrast results in lower median DSCs than the brain or the heart, but still larger than 0.74 for the left eye, 0.69 for the right eye, 0.89 for the left kidney and 0.80 for the right kidney. The median 95th HD and are smaller for organs with sharp boundaries and larger for organs located in an abdominal region with lower tissue contrast. In particular, a larger median was obtained for the eyes and the kidneys, more specifically: 0.49 mm for the left eye, 0.54 mm for the right eye, 0.24 mm for the left kidney, and 0.59 mm for the right kidney. We expect the differences between the left kidney and the right kidney are due to the intrarater observability resulting from the less distinguishable OAR boundary.
Studies such as Baiker et al13 and Wang et al20 reported DSCs of organs that were also included in our study. Baiker et al,13 using fan-beam µCT image data, reported mean DSCs (± 1 standard deviation) to evaluate the segmentation accuracy of the kidneys (0.47 ± 0.08), the heart (0.50±0.06) [Value calculated with DataThief 3 (https://datathief.org/)], and the brain (0.73 ± 0.04). After applying the same statistical analyses in our study, using medians and percentiles, as in Baiker et al13 using means and standard deviations, our method obtained mean DSCs of 0.82 ± 0.09, 0.94 ± 0.02, and 0.96 ± 0.01 for the kidneys, heart, and brain, respectively.
The best DSC results [Value calculated with DataThief 3 (https://datathief.org/)] reported in Wang et al20 using contrast-enhanced fan-beam µCT image data, were 0.91 for the heart, 0.71 for the left kidney, and 0.81 for the right kidney. These DSCs are closer, but still lower than the results presented in our study. However, our MABIS algorithm only used eight mouse atlases to contour a mouse with lacking OAR segmentations. Considering the published results in similar type of studies, a good agreement was found between the manual and the automatically generated contours.
In addition, acquiring images in the X-RAD 225Cx small animal radiation research system can vary from 40 s up to 10 min, depending on desired resolution and number of projections. Our non-contrast-enhanced µCBCT method, imaging the mice for about 2 min, is susceptible to respiratory and cardiac motion. Animal movement is less relevant for imaging cranial body regions but is of higher relevance while performing cardiothoracic or abdominal imaging. In the µCBCT image reconstruction, animal motion causes blurred organ edges, which are not beneficial for manual contouring and our MABIS algorithm. In this study, no contrast agent was injected in the mice before imaging. Because contrast agent materials consist of elements with a higher atomic number, this will result in more X-ray attenuation and therefore, in sharper edges around organs that incorporate contrast agent. Taking into account the fact that the multistage DIR algorithm uses the mutual information metric, which assumes a relation between the probability distributions of the intensities of the atlas µCBCT and the unsegmented mouse µCBCT, our MABIS algorithm is also applicable to contrast-enhanced µCBCT image acquisitions. Applying the MABIS algorithm on contrast-enhanced image data could result in much better OAR contours resulting from the improved image contrast between organ boundaries. In small animal body regions that are susceptible to respiratory motion, the combination of contrast-enhanced µCBCT and 4D µCBCT imaging can be beneficial for the image contrast and thus the automatic image segmentation, mainly because an anesthetized animal (i.e. isoflurane) breathes with short in- and exhale time periods and much more longer rest phases, e.g. ± 0.6 and ± 2.2 s respectively.21 We expect that the µCBCT motion artifacts caused by cardiac motion will not be improved by contrast-enhanced 4D µCBCT because the periodically fast heart-beating rate of a mouse is only causing small deformations in the mouse anatomy.
To implement the MABIS segmentation method in the preclinical small animal image-guided precision irradiation workflow, the method must be accurate and the calculation time must be kept as low as possible. An accurate segmentation prevents that the operator of the treatment planning system has to correct the contour afterwards, which will increase the planning time. An average calculation time of 12 min and 40 s was required to perform all pre-processing, deformation, STAPLE filter, and post-processing for one mouse. These three steps include the edge preserving smoothing, the deformations and transformations of a 300 × 300 × 700 image matrix, and include the post-processing of six OARs. In these ~13 min, the machine operator can simultaneously perform the tissue assignment needed by the dose engine, tumor delineation, target location, and beam planning, as illustrated in Figure 1. Compared to the manual delineation time of one mouse (± 70 min), a considerable time gain was obtained using the MABIS algorithm.
In human OAR image segmentation, many anatomical degrees of freedom are involved, such as; large anatomical changes due to surgery or a medical disease, gender, patient weight, patient size, patient age, and metal artefacts (e.g. dental fillings, implants etc.). The MABIS algorithm is expected to perform better on a mouse image data set because these variations are more limited, since mice are from the same strain, usually have the same age, eat the same food etc. However, it still needs to be investigated whether the MABIS algorithm is capable to automatically contour different mouse models, although we expect the algorithm to perform well as long the anatomical variations are limited.
In this investigation, the automatic contouring of multiple OARs from different treatment sites was studied. In pre-clinical image-guided high precision small animal irradiators, the OARs are only delineated nearby the treatment site of interest because of the small millimetric irradiation beams, e.g. the brain and eyes for cranial treatments, the kidneys for abdominal treatments, the heart for thoracic treatments etc. Independent from the treatment site, the µCBCT image data can be further cropped or reconstructed in smaller image volumes which results in improved calculation times in every step of our MABIS algorithm. Furthermore, the MABIS algorithm is developed in a way that the software can also be used to automatically contour other rodents and even humans, although we expect the algorithm to perform better on rodent image data because the larger anatomical differences in humans. In future studies, the performance of our proposed MABIS algorithm should still be evaluated using a lager atlas database and in other rodent models.
Conclusion
This paper is the first study that addresses automatic OAR segmentation in mouse µCBCT images using an MABIS algorithm. This in-house developed MABIS algorithm is evaluated in a pre-clinical small animal irradiation workflow. The automatic contouring accuracy of the MABIS algorithm was quantitatively evaluated with the DSC, the 95th HD, and the for six different OARs: left kidney, right kidney, heart, brain, left eye, and right eye. Using a powerful computer cluster, the MABIS algorithm automatically contoured the six OARs in a relatively small time interval. The OARs with sharp organ boundaries are contoured very accurately, and for the OARs with less sharp organ boundaries, the MABIS algorithm produces contours that are usable but may require some manual corrections.
Contributor Information
Brent van der Heyden, Email: brent.vanderheyden@student.uhasselt.be; brent.van.der.heijden@gmail.com; brent.vanderheyden@maastro.nl.
Mark Podesta, Email: mark.a.podesta@gmail.com.
Daniëlle BP Eekers, Email: danielle.eekers@maastro.nl.
Ana Vaniqui, Email: ana.vaniqui@maastro.nl.
Isabel P Almeida, Email: isabel.dealmeida@maastro.nl.
Lotte EJR Schyns, Email: lotte.schyns@maastro.nl.
Stefan J van Hoof, Email: stefan.vanhoof@smartscientific.nl.
Frank Verhaegen, Email: frank.verhaegen@maastro.nl.
REFERENCES
- 1.Verhaegen F, Granton P, Tryggestad E. Small animal radiotherapy research platforms. Phys Med Biol 2011; 56: R55–R83. doi: 10.1088/0031-9155/56/12/R01 [DOI] [PubMed] [Google Scholar]
- 2.Verhaegen F, van Hoof S, Granton PV, Trani D. A review of treatment planning for precision image-guided photon beam pre-clinical animal radiation studies. Z Med Phys 2014; 24: 323–34. doi: 10.1016/j.zemedi.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 3.van Hoof SJ, Granton PV, Verhaegen F. Development and validation of a treatment planning system for small animal radiotherapy: SmART- Plan. Radiother Oncol 2013; 109: 361–6. doi: 10.1016/j.radonc.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 4.Yahyanejad S, Granton PV, Lieuwes NG, Gilmour L, Dubois L, Theys J, et al. Complementary use of bioluminescence imaging and contrast-enhanced micro-computed tomography in an orthotopic brain tumor model. Mol Imaging 2014; 13: 7290.2014.00038–8. doi: 10.2310/7290.2014.00038 [DOI] [PubMed] [Google Scholar]
- 5.Balvert M, van Hoof SJ, Granton PV, Trani D, den Hertog D, Hoffmann AL, et al. A framework for inverse planning of beam-on times for 3D small animal radiotherapy using interactive multi-objective optimisation. Phys Med Biol 2015; 60: 5681–98. doi: 10.1088/0031-9155/60/14/5681 [DOI] [PubMed] [Google Scholar]
- 6.Zaffino P, Raudaschl P, Fritscher K, Sharp GC, Spadea MF. Technical Note: plastimatch mabs, an open source tool for automatic image segmentation. Med Phys 2016; 43: 5155–60. doi: 10.1118/1.4961121 [DOI] [PubMed] [Google Scholar]
- 7.Raudaschl PF, Zaffino P, Sharp GC, Spadea MF, Chen A, Dawant BM, et al. Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015. Med Phys 2017; 44: 2020–36. doi: 10.1002/mp.12197 [DOI] [PubMed] [Google Scholar]
- 8.Sharp G, Fritscher KD, Pekar V, Peroni M, Shusharina N, Veeraraghavan H, et al. Vision 20/20: perspectives on automated image segmentation for radiotherapy. Med Phys 2014; 41: 050902. doi: 10.1118/1.4871620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheenstra AE, van de Ven RC, van der Weerd L, van den Maagdenberg AM, Dijkstra J, Reiber JH. Automated segmentation of in vivo and ex vivo mouse brain magnetic resonance images. Mol Imaging 2009; 8: 35–44. doi: 10.2310/7290.2009.00004 [DOI] [PubMed] [Google Scholar]
- 10.Uberti MG, Boska MD, Liu Y. A semi-automatic image segmentation method for extraction of brain volume from in vivo mouse head magnetic resonance imaging using constraint level sets. J Neurosci Methods 2009; 179: 338–44. doi: 10.1016/j.jneumeth.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancelot S, Roche R, Slimen A, Bouillot C, Levigoureux E, Langlois J-B, et al. A multi-atlas based method for automated anatomical rat brain MRI segmentation and extraction of PET activity. PLoS One 2014; 9: e109113. doi: 10.1371/journal.pone.0109113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan D, Zhang Z, Luo Q, Yang X. A novel mouse segmentation method based on dynamic contrast enhanced micro-CT images. PLoS One 2017; 12: e0169424. doi: 10.1371/journal.pone.0169424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baiker M, Milles J, Dijkstra J, Henning TD, Weber AW, Que I, et al. Atlas-based whole-body segmentation of mice from low-contrast Micro-CT data. Med Image Anal 2010; 14: 723–37. doi: 10.1016/j.media.2010.04.008 [DOI] [PubMed] [Google Scholar]
- 14.Segars W, Tsui BMW, Frey EC, Johnson GA, Berr SS. Development of a 4-D digital mouse phantom for molecular imaging research. Mol Imaging Biol 2004; 6: 149–59. doi: 10.1016/j.mibio.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 15.Verhaegen F, Dubois L, Gianolini S, Hill MA, Karger CP, Lauber K, et al. ESTRO ACROP: technology for precision small animal radiotherapy research: Optimal use and challenges. Radiother Oncol 2018; 126: 471–8. doi: 10.1016/j.radonc.2017.11.016 [DOI] [PubMed] [Google Scholar]
- 16.Rit S, Vila Oliva M, Brousmiche S, Labarbe R, Sarrut D, Sharp GC. The Reconstruction Toolkit (RTK), an open-source cone-beam CT reconstruction toolkit based on the Insight Toolkit (ITK). J Phys Conf Ser 2014; 489: 12079. doi: 10.1088/1742-6596/489/1/012079 [DOI] [Google Scholar]
- 17.Ma CM, Coffey CW, DeWerd LA, Liu C, Nath R, Seltzer SM, et al. AAPM protocol for 40-300 kV x-ray beam dosimetry in radiotherapy and radiobiology. Med Phys 2001; 28: 868–93. doi: 10.1118/1.1374247 [DOI] [PubMed] [Google Scholar]
- 18.Klein S, Staring M, Murphy K, Viergever MA, Pluim J. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010; 29: 196–205. doi: 10.1109/TMI.2009.2035616 [DOI] [PubMed] [Google Scholar]
- 19.Rohlfing T, Russakoff DB, Maurer CR. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans Med Imaging 2004; 23: 983–94. doi: 10.1109/TMI.2004.830803 [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Stout DB, Chatziioannou AF. Estimation of mouse organ locations through registration of a statistical mouse atlas with micro-CT images. IEEE Trans Med Imaging 2012; 31: 88–102. doi: 10.1109/TMI.2011.2165294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Heyden B, van Hoof SJ, Schyns LE, Verhaegen F. The influence of respiratory motion on dose delivery in a mouse lung tumour irradiation using the 4D MOBY phantom. Br J Radiol 2017; 90: 20160419. doi: 10.1259/bjr.20160419 [DOI] [PMC free article] [PubMed] [Google Scholar]