Abstract
Although classically appreciated for their role as the powerhouse of the cell, the metabolic functions of mitochondria reach far beyond bioenergetics. Mitochondria catabolize nutrients for energy, generate biosynthetic precursors for macromolecules, compartmentalize metabolites for the maintenance of redox homeostasis, and function as hubs for metabolic waste management. We discuss the importance of these roles in both normal physiology and in disease.
Introduction
The transition to a highly oxidizing atmosphere in early earth development created a selective pressure that favored organisms with respiratory capacity1,2 including heterotrophic anaerobes, which consumed aerobic prokaryotic microbes (protomitochondrion).3 Following endosymbiosis, mitochondrial signals have been synchronized with the eukaryotic cell4. This integral relationship is demonstrated by the compartmentalized nature of cellular metabolism, in which mitochondrial reactions are required components of metabolic pathways.
Mitochondria coordinate cellular adaptation to stressors such as nutrient deprivation, oxidative stress, DNA damage and ER stress.5 Although long known to be critical for bioenergetics, emerging research shows that mitochondrial metabolism is multifaceted, mirroring their diverse functions. In addition to ATP, mitochondria produce metabolic precursors for macromolecules such as lipids, proteins, DNA and RNA. Mitochondria also generate metabolic by-products, such as reactive oxygen species (ROS) and ammonia, and possess mechanisms to clear or utilize waste products.
In this review, we discuss the metabolic functions of mitochondria as bioenergetics powerhouses, biosynthetic centres, balancers of reducing equivalents, and waste management hubs. Metabolic compartmentalization is instrumental for mitochondria to perform these functions. We highlight how mitochondrial metabolism supports their diverse functions in cell biology and how metabolism is compartmentalized in normal physiology and disease. A deeper understanding of mitochondrial contributions to metabolism will further elucidate their roles in disease and may reveal co-dependent pathways to target in therapies.
Mitochondria are the powerhouses of the cell
Cells consume fuels such as sugars, amino acids and fatty acids to generate energy in the form of ATP and GTP.6 Nutrients are metabolized and shuttled into the tricarboxylic acid (TCA) cycle, and through iterative oxidations, electrons are stored in the reducing equivalents NADH and FADH2.6 These carriers deposit electrons into the electron transport chain (ETC) in the Inner Mitochondrial Membrane (IMM), and use electron flow to pump protons into the intermembrane space.7, Protons flow down their electrochemical gradient through F1F0-ATP synthase to generate ATP.8 Whereas oxidative phosphorylation is the largest source of cellular ATP, the potential energy generated by the ETC is also harnessed for biosynthetic purposes. Many diseases arise when the ETC is perturbed.9,10 We discuss how mitochondria integrate fuel metabolism to generate energy for the cell, encompassing both classical and unconventional fuel sources (Figure 1).
Figure 1: Mitochondria are the powerhouse of the cell.

Mitochondria integrate fuel metabolism to generate energy in the form of ATP. Mitochondria oxidize pyruvate (derived from glucose or lactate), fatty acids, and amino acids to harness electrons onto the carriers NADH and FADH2. NADH and FADH2 transport these electrons to the electron transport chain, in which an electrochemical gradient is formed to facilitate ATP production through oxidative phosphorylation. Enzymes have the following abbreviations: LDH: lactate dehydrogenase, VDAC: Voltage-dependent anion channel, MPC: mitochondrial pyruvate carrier, PDC: pyruvate dehydrogenase complex, PC: pyruvate carboxylase, CS: citrate synthase, IDH2: isocitrate dehydrogenase 2, OGDH: α-ketoglutarate dehydrogenase, SDH: succinate dehydrogenase, MDH2: malate dehydrogenase 2, GLS: glutaminase, GDH: glutamate dehydrogenase, BCAT2: branched chain amino transferase 2, BCKDH: branched chain ketoacid dehydrogenase, PHD3: prolyl hydroxylase 3, AMPK: adenosine monophosphate kinase, ACC: Acetyl CoA Carboxylase, , ACS: acyl CoA synthetase, CPT1/2: carnitine palmitoyltransferase 1/2. Electrons and reducing equivalents are shown in yellow.
Pyruvate
Pyruvate is generated by a number of sources, depending on nutrient availability and tissue, including glucose catabolism (thought to be a major source), and lactate.11–13 Pyruvate utilization in the cytosol versus mitochondria is one of the clearest examples of how compartmentalization is a major determinant of cellular bioenergetics. In healthy tissue, the fate of pyruvate is dependent on oxygen availability and mitochondrial respiratory capacity.14 In normoxia, pyruvate is generated via glycolysis and transported across the IMM through the Mitochondrial Pyruvate Carrier (MPC).15,16 Pyruvate is further catabolized inside mitochondria through the TCA cycle. During hypoxia, mitochondrial respiration is repressed, causing cells to adaptively sink electrons onto pyruvate through lactate dehydrogenase (LDH), generating lactate in the cytosol.17 This pathway is engaged in muscle during exercise, the intestines, and the renal medulla of the kidneys.18–20 Otto Warburg observed that cancer cells rewire glucose metabolism for lactate synthesis even in normoxia, known as the Warburg Effect.14,21 Additional studies must be performed to determine the net catalytic activity of LDH in tumors, given that metabolic tracing studies in lung cancer patients have demonstrated that lactate is a major source of TCA cycle intermediates.13 The extent of LDH-mediated pyruvate production may depend on in vitro versus in vivo models of tumor metabolism, emphasizing the need to test metabolic flux in vivo.
The critical role of pyruvate compartmentalization in bioenergetics and metabolism is highlighted by recent elegant studies of the MPC.15,16 Pharmacological inhibition of MPC represses mitochondrial pyruvate uptake, shifting reliance to glycolysis for ATP production. This shift is evident in cancer cells, which repress MPC1 to promote the Warburg Effect, and in myocytes of diabetic mice, which elevate glucose consumption in response to MPC inhibition22,23 Suppression of MPC accelerates proliferation in intestinal stem cells,24 suggesting that the role of MPC is context-dependent and sensitive to mitochondrial respiratory capacity and/or nutrient availability.
Within mitochondria, pyruvate may enter the TCA cycle via the activity of two distinct enzymes: Pyruvate Dehydrogenase Complex (PDC), which generates acetyl CoA, and Pyruvate Carboxylase (PC), which generates oxaloacetate.25 Although PDC and PC both catalyze the flux of pyruvate into the TCA cycle, their enzymatic activities can be distinguished by stable isotope tracing26,27, and their metabolic roles do not appear to be interchangeable. PDC deficiency is sufficient to rewire energy metabolism towards aerobic glycolysis despite the potential adaptive node for TCA cycle anaplerosis (a process to replenish TCA cycle intermediates), mediated by PC.28 Many cancers favor PC-mediated anaplerosis, although the factors that dictate the choice for pyruvate flux between PC and PDC are little studied.27,29,30 Therefore, these enzymes may have important functions beyond TCA cycle flux for bioenergetics.
Glutamine and Branched Chain Amino Acids (BCAAs)
The catabolism of glutamine, the most abundant amino acid, often starts in the mitochondria and its carbon and nitrogen atoms are distributed into macromolecules throughout the cell, including TCA cycle intermediates (important in bioenergetics), amino acids, nucleotides, glutathione, and lipids.31
In mitochondria, glutaminase (GLS) converts glutamine into glutamate and ammonia. Either transaminase or glutamate dehydrogenase (GDH) converts glutamate into α-ketoglutarate.32,33 Glutamine anaplerosis sustains TCA cycle intermediates in conditions of limiting glucose and MPC inhibition, demonstrating the potential flexibility of these metabolic nodes.34,35 Glutamine anaplerosis is critical for meeting the energetic requirements of proliferative cells, such as T cells during the transition from quiescent naïve T cells to effector cells, and in cancers, particularly with MYC elevation.32,36,37 GLS inhibition suppresses proliferation, and GLS inhibitors are being evaluated in clinical studies for a number of cancers.31,38,39 However, sensitivity to GLS inhibition in vitro is not always consistent in vivo, and is dependent on extracellular cystine levels.40 This emphasizes the need for investigators to study the effect of the microenvironment on metabolic dependencies and to validate experiments in vivo.
Although glutamine transporters at the plasma membrane have been identified,41 the mitochondrial glutamine transporter has not been fully characterized.42,43 This critical area of research is challenging to address because there are likely multiple mechanisms for glutamine import.
The BCAAs leucine, isoleucine, and valine are a major source of cellular energy via acetyl CoA and succinyl CoA generation.44 The tissue of origin dictates dependency on BCAA catabolism in normal physiology and in cancer.45 In normal physiology, myocytes and adipocytes activate mitochondrial BCAA catabolic enzymes to support ATP production during exercise or fasting and differentiation, respectively.46,47 BCAA catabolism is repressed in maple syrup urine disease, which is caused by mutations to branched-chain keto acid dehydrogenase (BCKDH) and causes dysfunction of immune cells, skeletal muscle and the central nervous system.48 Although mitochondrial BCAA catabolism is critical in these pathologies, it is unknown how BCAAs are imported into the mitochondria. Identifying their transport mechanisms will be critical to our understanding of mitochondrial BCAA catabolism in cellular homeostasis.
Fatty Acid Oxidation
Palmitate, a 16-carbon fatty acid (FA), stores 39KJ/g of energy compared to 16KJ/g stored in glucose.49 Therefore, FAs are a major source of cellular energy, particularly under conditions of nutrient stress. Mitochondrial FA import is a rate-determining step for fatty acid oxidation (FAO) and demonstrates how metabolic compartmentalization adapts to cellular state. As long chain FAs are unable to cross mitochondrial membranes, mitochondria have evolved an intricate set of reactions and transporter activities to allow fat to access mitochondrial β-oxidation machinery. The outer mitochondrial membrane (OMM) enzyme carnitine palmitoyl transferase 1 (CPT1) forms acylcarnitines from fatty acyl CoAs.50 Acylcarnitines are shuttled into mitochondria through the carnitine–acylcarnitine translocase (SLC25A20) in the IMM. CPT2 liberates FA from carnitine, initiating FAO.51 Acetyl CoA from FAO is used for the TCA cycle as well as for aspartate and nucleotide synthesis.52
CPT1 activity is tightly controlled by a network of metabolites, linking it to cellular nutrient status. Malonyl CoA, generated by the enzyme acetyl CoA carboxylase (ACC), represses CPT1 to inhibit acylcarnitine import.53 Malonyl CoA is the initiating metabolite for FAS, and its levels dictate the balance of fat synthesis or oxidation within a cell. In low energy conditions, AMP-activated protein kinase (AMPK) phosphorylates and inhibits ACC, decreasing malonyl CoA and increasing CPT1 activity.54 ACC2 is also hydroxylated by the dioxygenase prolyl hydroxylase 3 (PHD3).55 Hydroxylation promotes ACC2 activity in nutrient abundance. These enzymes are altered in some cancers and human diseases as the mechanism that dictates fat utilization. PHD3 is suppressed in cancers that rely on FAO, such as AML and prostate cancer, and elevated in cancers that rely on FAS such as breast and non-small-lung-cell cancer.55–57 Reciprocally, AMPK is linked to fat utilization in diseases and cancers.58,59
The dynamic regulation of FAO is key to cellular physiology. FAO is fundamental for the survival and function of memory CD8+ T cells, unlike effector cells that rely on glycolysis and glutaminolysis for energy.60,61 Likewise, FAO is activated in insulin resistance, in which free fatty acids provide a compensatory fuel for repressed glucose uptake.62,63
Mitochondria are biosynthetic hubs
Mitochondria participate in the biosynthesis of nucleotides, FAs, cholesterol, amino acids, glucose, and heme (Figure 2).64 These biosynthetic pathways are engaged in stress responses, and are often mis-regulated in disease.5 Rather than being dysfunctional, highly proliferative cells such as cancer cells and activated T cells rely on mitochondrial metabolites to form biomass.5,65 Below we review the mitochondrial compartmentalization of anabolic pathways and its role in cell stress responses and disease.
Figure 2. Mitochondria are biosynthetic hubs.

The mitochondria are a critical source of building blocks for biosynthetic pathways including nucleotide synthesis, fatty acid and cholesterol synthesis, amino acid synthesis, and glucose and heme synthesis. Compartmentalization is a key feature of biosynthetic pathways. While many of the enzymes listed are bi-directional, arrows are drawn to highlight the biosynthetic functions. Enzymes are circled in grey and brown with the following abbreviations: Nucleotide Synthesis: MTHFD1/2: methylenetetrahydrofolate dehydrogenase, SHMT1/2: serine hydroxymethyltransferase, DHODH: dihydroorotate dehydrogenase, FTDH: formate dehydrogenase. Fatty Acid and Cholesterol Synthesis: GLS: glutaminase, GDH: glutamate dehydrogenase, TA: transaminase, ACLY: ATP citrate lyase, ACC2: acetyl CoA carboxylase, PHD3: prolyl hydroxylase 3, MPC: mitochondrial pyruvate carrier. Amino Acid Synthesis: GDH: glutamate dehydrogenase, GS: glutamine synthetase, P5CS: Pyrroline-5-carboxylate synthase, PYCR1: Pyrroline-5-carboxylate reductase 1, OAT: ornithine aminotransferase, GOT2: glutamate oxaloacetate transaminase 2, GPT2: glutamate pyruvate transaminase 2, GC: glutamate carrier, AGC: aspartate-glutamate carrier, ORNT1: ornithine translocator. Glucose and Heme Synthesis: PCK1/2: phosphoenolpyruvate carboxykinase, MDH1/2: malate dehydrogenase, PC: pyruvate carboxylase, ALAS: aminolevulinate synthase, FECH: ferrochetolase, ABCB6: ATP binding cassette subfamily B member 6, FLVCR: feline leukemia virus subgroup C receptor 1.
Nucleotides
The 1C metabolic pathway involves a set of reactions that generate and transfer activated one carbon (1C) units for de novo nucleotide synthesis, compartmentalize amino acids, and contribute to redox homeostasis. The co-factor tetrahydrofolate (THF) is the carrier that mediates 1C transfer reactions for de novo nucleotide synthesis.66,67 Activated THF molecules are generated through an oxidative/reductive cycle that catabolizes serine (to generate glycine) in the mitochondria and synthesizes serine in the cytosol.
The carrier SLC25A32 imports THF into the mitochondria, where it is converted by serine hydroxymethyltransferase (SHMT2) into 5,10 methylene-THF and glycine. Like many enzymes in 1C metabolism, SHMT2 is bi-directional. SHMT2 favors production of glycine and 5,10 methylene-THF, and cells deficient in mitochondrial 1C metabolism are glycine auxotrophs.68 In the absence of SHMT2, cytosolic SHMT1 reverses flux to compensate69 demonstrating how metabolic flexibility among subcellular compartments is critical to stress adaptation.
Mitochondrial methylenetetrahydrofolate dehydrogenase (MTHFD2) converts 5,10 methylene-THF to 10-formyl-THF. MTHFD2 expression is regulated by mTORC1, and is critical for growth and proliferation.70 MTHFD2 is overexpressed in many human cancers71 and mitochondrial biogenesis and SHMT2/MTHFD2 expression are promoted during T-cell activation to support proliferation.72 10-formyl-THF has multiple fates: conversion into THF by 10-formyl-THF dehydrogenase, production of formyl-methionine for mitochondrial translation, or hydrolyzation to formate by MTHFD1L. Mitochondrial contributions to this pathway are critical, as mitochondrial formate is the main carbon source for cytosolic 1C metabolism 66
The IMM enzyme dihydroorotate dehydrogenase (DHODH), which oxidizes dihydroorotate to orotate, is required for de novo pyrimidine synthesis.73 Consistent with their reliance on 1C metabolism, T cells require DHODH for clonal expansion and differentiation into effector cells.74 DHODH is targeted in autoimmune disorders and inhibition suppresses myeloid differentiation of AML cells.75 DHODH activity is also elevated in response to DNA damage and upon genotoxic chemotherapy treatment to increase nucleotide synthesis for DNA repair.76,77
Citrate
In addition to generating electron carriers for the ETC, TCA cycle intermediates such as citrate regulate anabolic reactions. Mitochondrial citrate controls anabolic reactions by directly acting as the carbon source for FAs, cholesterol and ketone bodies through ATP citrate lyase (ACLY),78 and by allosteric modulation. Citrate is generated by citrate synthase (CS) or through the reduction of α-ketoglutarate by isocitrate dehydrogenase (IDH).79–81 Mitochondrial citrate is exported by the malate-citrate antiporter SLC25A1.82 In the cytosol, citrate is converted to acetyl CoA via ACLY, which can access many pathways, including conversion to malonyl CoA by the activity of ACC (as described above). Cytosolic citrate is a potent allosteric regulator of ACC by increasing its polymerization and activity.83
Regulation of citrate export may provide a physiological node for the cell to communicate lipid homeostasis to the mitochondria. SLC25A1 is sensitive to membrane rigidity, and high levels of cholesterol or acidic phospholipids in the IMM repress mitochondrial citrate export.84 Moreover, fasting causes a 40% reduction in mitochondrial citrate export.85 Although these studies indicate that citrate export is affected by lipid abundance, it is unknown if repression of SLC25A1-mediated citrate export affects ACC2 polymerization and FAS initiation.
Acetyl CoA is required for epigenetic modifications such as histone acetylation.86–88 Thus, fat metabolism may be intimately linked with the epigenetic state, although it is unknown whether the connection is direct. The emerging role of mitochondrial metabolism in epigenetic reprogramming may extend beyond acetyl CoA to include other mitochondrial metabolites such as succinate, fumarate, and ROS, which directly affect the activity of Fe (II)/α-KG-dependent dioxygenases, including hydroxylases, DNA demethylases and histone demethylases.89
Amino Acids
The mitochondria is a hub for amino acid synthesis, including glutamine, glutamate, alanine, proline, and aspartate. Glutamine synthetase (GS) condenses glutamate and ammonia to make glutamine.90 GS has been reported to have activity in cytosol and mitochondria, and its biological role may differ depending on its subcellular localization. GS has a “weak” mitochondrial localization sequence and is imported into the mitochondria in the liver, whereas GS is cytoplasmic in astrocytes.91 In glioblastoma, GS generates a source of glutamine for de novo purine synthesis.92 However, in breast cancer cells, GS-derived glutamine is not used for de novo nucleotide synthesis.93 One possible explanation for this difference is the subcellular localization of GS in these systems.
Glutamate is generated by and utilized as a nitrogen source for numerous reactions.94 Glutamate metabolism stratifies in proliferating and quiescent cells; proliferating cells elevate the expression of glutamate-dependent transaminases, whereas quiescent cells suppress them.95 Many of the glutamate-dependent transaminases, such as glutamate oxaloacetate transaminase (GOT) and glutamate pyruvate transaminase (GPT) have two (cytosolic and mitochondrial) isoforms.95 It will be key for future studies to elucidate the role of subcellular compartmentalization of glutamate metabolism in proliferation.
Proline and ornithine metabolism are centrally mitochondrial. The mitochondrial enzyme Pyrroline-5-carboxylate synthase (P5CS) generates pyrroline-5-carboxylate (P5C), which can be used for proline and ornithine production.96 Ornithine is made by ornithine amino transferase (OAT) and proline is produced through reduction of P5C by Pyrroline-5-carboxylate reductase (PYCR). The mechanisms underlying compartmentalization of proteinogenic amino acids, such as proline and glutamate are little studied.97
Gluconeogenesis
Gluconeogenesis is predominantly a cytosolic process, although the initiating step by PC occurs inside the mitochondria.98 PC-derived oxaloacetate is converted to malate and exported from the mitochondria for the remaining steps of gluconeogenesis.99 This export can occur through SLC25A1 (citrate-malate antiporter), SLC25A11 (α-ketoglutarate-malate antiporter) or SLC25A10 (dicarboxylate-phosphate antiporter).100 The dominant mechanism for malate export in gluconeogenesis is unknown. Furthermore, it is unclear if metabolic stressors such as nutrient deprivation or hypoxia dictate this mechanism. In the cytosol, phosphoenolpyruvate carboxykinase (PCK) converts oxaloacetate into phosphoenol pyruvate (PEP) for gluconeogenesis.101 The mitochondrial isoform of this enzyme, PCK2, has no known connections to gluconeogenesis.101
Heme
Heme metabolism illustrates an extraordinary example of metabolic compartmentalization. The committed step of the pathway is catalyzed by mitochondrial aminolevulinate synthase (ALAS), which generates ALA from glycine and succinyl CoA.102 ALA is exported via SLC25A38 and, through four cytosolic reactions, is converted into coproporphyrinogen III (CPGIII). Next, CPGIII enters the intermembrane mitochondrial space through the ATP-dependent transporter ABCB6 for further catalysis by coproporphyrinogen oxidase (CPOX).103 The intermembrane space is a region in which few metabolic reactions occur. The terminal step of heme synthesis is in the mitochondrial matrix, in which ferrochelatase (FECH) catalyzes the insertion of ferrous iron into the macrocycle.104 As heme biosynthesis generates H2O2 in the intermembrane region, we speculate that there may be direct links between heme metabolism and ROS-sensitive signaling pathways.
Mitochondria balance redox equivalents
The mitochondria and cytosol have distinct requirements for NAD+, and proper compartmentalization of redox equivalents is crucial for maintenance of cellular homeostasis and survival in response to environmental stressors.105–107 The cytosol is a more oxidizing environment in which the NAD+/NADH ratio ranges between 60-700.108 Conversely, mitochondria employ more reductive metabolic reactions, and the NAD+/NADH ratio is approximately 7-8.108 To sustain the imbalanced distribution of NAD, mammalian cells engage indirect pathways (Figure 3) because there is no known mammalian transporter for NAD+ or NADH, contrary to yeast which facilitate NAD transport through NDT1.109
Figure 3. Mitochondria balance redox equivalents.
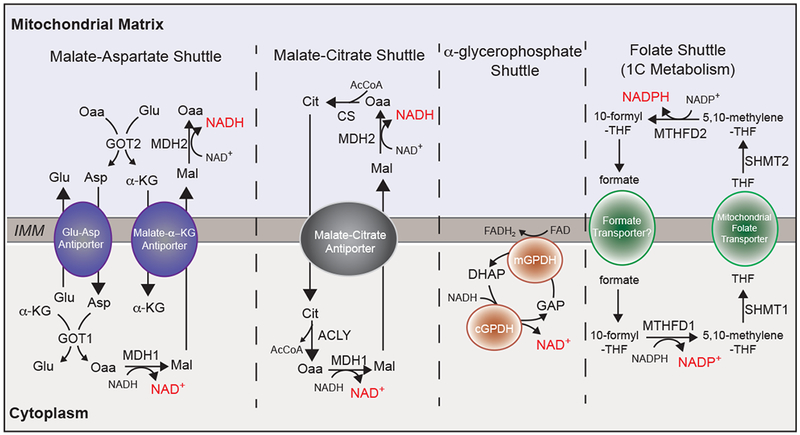
In the absence of a direct mode for NAD transport, cells rely on compartmentalized flux of metabolites to support balance of reducing equivalents NAD/NADH, and NADP/NADPH. Generally, redox shuttles favor cytosolic NAD+ synthesis and mitochondrial NADH synthesis. Enzymes and transporters have with the following abbreviations: Malate-aspartate shuttle. GOT1/2: Glutamate oxaloacetate transaminase, MDH1/2: malate dehydrogenase, ME1: malic enzyme 1, Glu-Asp antiporter: glutamate-aspartate antiporter, Malate-α-KG Antiporter: malate-α-ketoglutarate antiporter. Malate-citrate shuttle. ACLY: ATP citrate lyase, MDH1/2: malate dehydrogenase, CS: citrate synthase. α-glycerophosphate shuttle. (m/c)GPDH: mitochondrial/cytosolic glycerol-3-phosphate dehydrogenase. Folate Shuttle (1C Metabolism): MTHFD1/2: methylenetetrahydrofolate dehydrogenase, SHMT1/2: serine hydroxymethyltransferase.
Malate-Aspartate Shuttle
The malate-aspartate shuttle is ubiquitously engaged to generate cytosolic NAD+ and mitochondrial NADH.110 This cycle involves an oxidation or reduction catalyzed by malate dehydrogenase (MDH1: cytosolic, MDH2: mitochondrial), a transamination catalyzed by glutamate-oxaloacetate transaminase (GOT1:cytosolic, GOT2: mitochondrial), and two antiporters localized to the IMM (aspartate-glutamate antiporter AGC and malate α-ketoglutarate antiporter MαA).111 Compartmentalization of reducing equivalents through the malate-aspartate shuttle is key for survival in stress conditions such as exercise, in which cytosolic NAD+ is required to promote glucose catabolism and mitochondrial NADH for ATP production.112 Moreover, in PDAC cancers with oncogenic KRAS, glutamine is fluxed through the malate-aspartate shuttle to raise the NADPH/NADP+ ratio for glutathione synthesis.113 When oxidative phosphorylation is repressed, cells utilize the reverse flux of GOT1 to generate aspartate114,115. In addition to its regulation of redox balance, the malate-aspartate shuttle may also contribute to cellular amino acid compartmentalization.
Citrate-Malate Shuttle
In contrast to the malate-aspartate shuttle, the citrate-malate shuttle functions equally (with respect to reducing equivalents), but is less studied in the context of disease. Similar to malate-aspartate shuttle, the citrate-malate shuttle utilizes both isoforms of MDH. However, MDH activity is paired with CS, ACLY, and the malate-citrate antiporter (CIC).116 Rather than elevating cytosolic aspartate, the citrate-malate shuttle increases cytosolic citrate levels. Therefore, flux through the citrate-malate shuttle promotes FAS through citrate compartmentalization.117 Thus, although both the malate-aspartate and citrate-malate shuttles balance reducing equivalents through MDH activity, these shuttles are not interchangeable. The implications of cytosolic citrate accumulation in the malate-citrate shuttle are yet to be defined beyond FAS. For example, flux through the citrate-malate shuttle may also affect epigenetics through ACLY activity and acetyl CoA production.88
α-glycerophosphate Shuttle
The α-glycerophosphate shuttle is a unique redox balancing pathway, which intersects the mitochondria but does not directly affect mitochondrial NAD/NADH.118 The α-glycerophosphate shuttle is composed of cytosolic and mitochondrial α-glycerophosphate dehydrogenase (cGPDH and mGPDH). In this cycle, cGPDH utilizes NADH to reduce dihydroxyacetone phosphate (DHAP) to glycerophosphate (GAP) and generate cytosolic NAD+. GAP is subsequently oxidized to DHAP by the flavin-dependent mGPDH, which directly deposits electrons into the ETC. The α-glycerophosphate shuttle is tightly linked to glycolysis and is highly active in brown adipose tissue (BAT) to regenerate cytosolic NAD+ while simultaneously sinking electrons into the ETC for thermogenesis.118 As this pathway is engaged in highly glycolytic cells, it would be interesting for future studies to investigate the potential role of this redox shuttle in cancer.
One Carbon Metabolism
MTHFD is among the largest contributors to cellular NADPH, in addition to the pentose phosphate pathway and malic enzyme (ME).119 MTHFD isozymes are bi-directional, however, stable isotope tracing of NADPH revealed that the mitochondrial MTHFD favors NADPH production, and the cytosolic isoform favors NADP+ production.120 The 1C metabolic pathway is an adaptive mechanism to survive oxidative stress. Upon ETC inhibition, flux through the mitochondrial arm of 1C metabolism is activated for NADPH/NADP+ balance.121 NADPH is required for reduction of glutathione for clearance of ROS. In cancer cells, flux through the mitochondrial 1C pathway generates cytosolic NADPH for FAS.122
Mitochondria orchestrate waste management
The by-products of metabolic reactions are often depicted as waste. However, emerging studies have revealed a functional role for metabolic by-products such as lactate, ammonia, ROS and hydrogen sulfide (H2S).12,13,93,123,124 The study of metabolic by-products is a growing area of research, especially in cancer, in which metabolic by-products accumulate in the tumor microenvironment (TME)125 (Figure 4A). Mitochondria are indispensable in cellular waste management (Figure 4B–D). Below, we review the pathways that mitochondria utilize to re-purpose cellular waste.
Figure 4. Mitochondria orchestrate waste management.
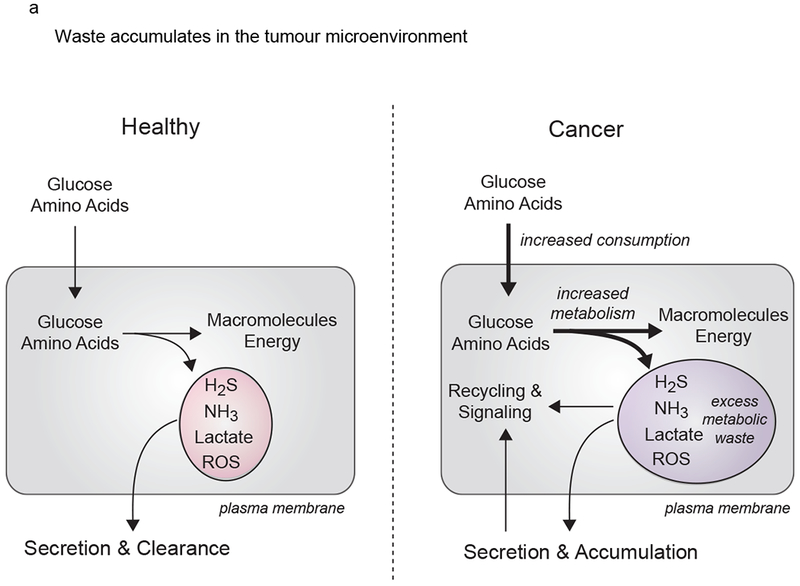

(A). Tumor cells increase nutrient consumption and metabolic fitness relative to healthy tissue, leading to accumulation of waste products in the tumor microenvironment. To manage metabolic waste, cancer cells engage recycling pathways for these metabolic by-products. (B). Ammonia. Production of and metabolic clearance of ammonia (NH3) in cell metabolism. NH3 is generated by amino acid and nucleotide catabolism. NH3 is assimilated in the mitochondria through GS (glutamine synthetase), GDH (glutamate dehydrogenase), and CPS1 (carbamoyl phosphate synthase 1). CPS1 initiates the urea cycle for production of the metabolic waste product urea. Urea can be re-catabolized by urease positive bacteria in the microbiome to regenerate NH3. AGC: aspartate-glutamate carrier, ORNT1: ornithine translocator (C). Hydrogen Sulfide. Production of and metabolic clearance of hydrogen sulfide (H2S) in cell metabolism. H2S is generated by the mammalian enzymes CBS (cystathionine β synthase), CSE (cystathionine γ lyase), 3-MST (3-mercaptopyruvate sulfurtransferase) and from the metabolic reactions in the microbiome. H2S is cleared by iterative oxidation catalyzed by sulfide quinone reductase (SQR), thiosulfate reductase (TR), and sulfite oxidase (SO). TR utilizes oxidized glutathione (GS−) as a sink for electrons. Oxidations catalyzed by SQR and SO are linked to mitochondrial ETC and oxidative phosphorylation. (D). Reactive Oxygen Species. Reactions that generate and sequester ROS (reactive oxygen species). ROS are generated in the mitochondria through the ETC and NOX4 (NADPH oxidase). SOD2 (superoxide dismutase 2) converts superoxide into a the less reactive molecule hydrogen peroxide (H2O2). In the mitochondria, H2O2 is turned over by combined functions of periodxins (Prx) and thioredoxins (Trx). H2O2 also reacts with Fe+2 (the Fenton Reaction) to generated OH. in the mitochondria. ROS inflict oxidative damage to proteins in the mitochondria and cytosol, and also function as potent mitogen signaling agents.
Ammonia
Ammonia is generated in mammalian cells by amino acid lyases and nucleotide deaminases, however, the largest contributor to ammonia in mammals is the microbiome.126 Ammonia is a neurotoxin that is sustained below 50 μM in plasma of healthy adults, and can induce seizure when plasma levels become elevated.122 Moreover, high ammonia may induce autophagy in some cultured cells.127,128 To evade toxicity, mammalian cells possess three ammonia-assimilating enzymes: carbamoyl phosphate synthetase 1 (CPS1), GS, and GDH.
The urea cycle is a sink for ammonia, ultimately generating urea, which cannot be metabolized by mammalian enzymes. CPS1 is the rate-limiting step of the urea cycle, generating carbamoyl phosphate (CP).129 N-acetyl glutamate (NAG) is an essential activator of CPS1, and congenital NAGS mutations cause hyperammonemia.130 CP is condensed with ornithine by ornithine carbamoyltransferase (OTC) to generate citrulline, which is exported through ORNT1, the citrulline-ornithine antiporter for the remaining steps of the cycle. Interestingly, in KRAS/LKB1 mutant cancer, CP from CPS1 is diverted into de novo pyrimidine synthesis.131 The mechanism of CP export from the mitochondria is unknown and may be a potential therapeutic target.
Although urea is a metabolic waste product for mammalian cells, urease-positive bacteria in the microbiome re-catabolize 15-30% of urea to regenerate ammonia.132 Consequently, similar to congenital mutations in urea cycle enzymes, the microbiome can contribute to hyperammonemia.126,133 Beyond ammonia metabolism, many microbial metabolites intersect host biology and their roles remain an active area of research.134
GDH and GS assimilate ammonia, generating glutamate and glutamine. Glutamate contributes to the urea cycle through conversion to aspartate by GOT2 and mitochondrial export via AGC1/2. GDH is a bidirectional enzyme, and high ammonia levels reverse the direction of GDH, favoring the reductive activity.135 This bi-directionality is particularly relevant in breast cancers, as ammonia accumulates in the TME, driving GDH towards glutamate synthesis.93 Beyond the TME, physiological niches with high ammonia levels (the microbiome, liver, and kidneys) may promote the reductive activity of GDH. Additionally, GDH-mediated ammonia assimilation requires NAD(P)H and therefore may contribute to redox balance.
ROS
Mitochondria generate, sequester and interconvert ROS in response to stressors such as hypoxia, nutrient availability, cytokine stimulation and changes in mitochondrial membrane potential.136 ROS are generated from the reduction of oxygen (O2) to superoxide (O2.), hydrogen peroxide (H2O2) and hydroxyl radical (OH.). Mitochondrial ROS are generated in reactions such as NADPH oxidase (NOX4) and the Fenton Reaction, and through electron leak from ETC complexes123, although NOX4 is not strictly localized to mitochondria.137 ROS are highly reactive and inflict oxidative damage to macromolecules.138
Mitochondria rely on ROS clearance to protect the concentrated iron-sulfur clusters in the ETC and iron-dependent enzymes such as aconitase. Superoxide dismutase (SOD2) converts superoxide into a less reactive molecule, H2O2.123 Cellular H2O2 can be degraded to water by catalase, glutathione peroxidase (GPx), and peroxiredoxin (Prx), however mitochondria do not have catalase and only a single splice variant of GPx4 has been demonstrated to be localized to mitochondria.139,140 Mitochondria rely on the combined activities of peroxiredoxins (Prx3 and Prx5), thioredoxins (Trx2), and thioredoxin reductase 2 (TRXR2) to decompose the locally generated H2O2.141
Beyond toxicity, ROS are potent mitogen signaling agents that foster proliferation, differentiation, and migration.123,142 Specifically, ROS oxidize cysteine residues, linking mitochondria to signaling cascades. ROS inactivates the catalytic cysteine of phosphatase 1B (PTP1B), enabling receptor tyrosine phosphorylation required for growth-factor signaling.143 ROS inactivate PTEN, which represses the PI-3 Kinase/AKT signaling cascade and PHDs to repress HIF hydroxylation.144,145 In breast cancer, low levels of the mitochondrial sirtuin 3 promote HIF stabilization through ROS, stimulating the Warburg Effect.146 In macrophages, mROS promote the antibacterial innate immune response, and mice harboring mROS-deficient macrophages are susceptible to infection.147 Similarly, mitochondria provide ROS for B-cell and T-cell activation.148,149 ROS are thus critical to proliferating systems.
Hydrogen Sulfide
H2S is produced in the microbiome by sulfur-reducing bacteria and by mammalian cells through cystathionine β synthase (CBS), cystathionine γ lyase (CSE), and 3-mercaptopyruvate sulfurtransferase (3MST).150 H2S-producing enzymes are localized to the cytosol and mitochondria, depending on the tissue type.124
High levels of H2S are toxic and repress respiration through complex IV inhibition.137 To dampen H2S toxicity, mitochondria sequentially oxidize H2S generating thiosulfate, sulfite, and ultimately sulfate.151 The first and last reactions catalyzed by flavin-dependent sulfide quinone reductase (SQR) and sulfite oxidase (SO) directly deposit electrons onto Coenzyme Q (CoQ) of the ETC.136 In CoQ deficiency, H2S oxidation is significantly repressed.152 The intermediate oxidation step of H2S is catalyzed by thiosulfate reductase (TR) and requires oxidized glutathione as an electron sink. Because the enzymes for glutathione synthesis are cytosolic, mitochondria must import glutathione for this process. Glutathione can utilize the dicarboxylate carrier SLC2510 and the α-kg carrier SLC25A11 for import, although a selective mechanism of transport remains unknown and may be pivotal for H2S clearance.153
H2S metabolism is directly linked with oxidative phosphorylation.154 Hypoxia represses H2S detoxification through respiratory chain inhibition.124 Interestingly, the microbiome, which has the highest H2S levels, is hypoxic in some regions.155 The mechanism for H2S tolerance in the microbiome remains unknown. H2S production and clearance may be critical in diseases such as cancer and diabetes, which are associated with altered respiration.
Future directions of mitochondrial metabolism in cellular homeostasis and disease
Here we discuss the multifaceted contributions of mitochondria to cell metabolism as bioenergetic powerhouses, biosynthetic centres, balancers of reducing equivalents and waste management hubs. Although mitochondrial pathways are well defined, the mechanisms by which metabolites are compartmentalized remain elusive. Identifying the transporters that coordinate metabolic flux for key pathways such as amino acid and glutathione import will be important directions for future research156. Given that mitochondrial metabolism is critical to many diseases, transporters that enable metabolic compartmentalization may be promising therapeutic targets5,65,157–159. It will also be key to consider mitochondrial metabolite concentrations, which differ from whole cell concentrations160, to better inform the kinetics of mitochondrial enzymes under different cellular stress conditions and in disease. Mitochondrial concentrations are critical when studying bi-directional enzymes such as transaminases and enzymes in 1C metabolism.
It will be important for future studies to probe the physiological contributions of mitochondria to cell biology. Metabolism is not always comparable when studying in vitro and in vivo models. These differences may dictate the efficacy of therapies, such as the glutaminase inhibitor in cancer.40 The extent to which a physiological niche alters mitochondrial contributions to metabolism and cell/tissue function has not been well explored. For example, metabolic by-products accumulate in the TME, increasing the necessity for cancer cells to engage waste management pathways13,93,125. Disparities between model systems may be avoided by performing in vitro studies in media with physiological metabolite concentrations, using model systems that represent the 3-dimensional architecture of the tissue being studied, and performing experiments in vivo161–163.Future studies in this exciting and growing field will continue to reveal the roles of mitochondrial metabolism in cellular homeostasis and disease.
Acknowledgements:
We would like to thank Sarah Tucker and Liam Kelley for editing this article. J.B.S is supported by the National Science Foundation Graduate Research Fellowship DGE1144152. MCH is supported by the Ludwig Center at Harvard and NIH grant R01CA213062.
Footnotes
Competing Interests: The authors declare no competing interests.
References
- 1.Castresana J, Saraste M Evolution of Energetic Metabolism: the respiration-early hypothesis. Trends in biochemical sciences 20, 443–448 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Lyons TW, Reinhard CT & Planavsky NJ The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315, doi: 10.1038/nature13068 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Sagan L On the Origin of Mitosing Cells. J.Theoret. Biol 14, 225–274 (1966). [DOI] [PubMed] [Google Scholar]
- 4.Timmis JN, Ayliffe MA, Huang CY & Martin W Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature reviews. Genetics 5, 123–135, doi: 10.1038/nrg1271 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Vyas S, Zaganjor E & Haigis MC Mitochondria and Cancer. Cell 166, 555–566, doi: 10.1016/j.cell.2016.07.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh CT, Tu BP & Tang Y Eight Kinetically Stable but Thermodynamically Activated Molecules that Power Cell Metabolism. Chemical reviews, doi: 10.1021/acs.chemrev.7b00510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sazanov LA A giant molecular proton pump: structure and mechanism of respiratory complex I. Nature reviews. Molecular cell biology 16, 375–388, doi: 10.1038/nrm3997 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Watt IN, Montgomery MG, Runswick MJ, Leslie AG & Walker JE Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proceedings of the National Academy of Sciences of the United States of America 107, 16823–16827, doi: 10.1073/pnas.1011099107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMauro S & Schon EA Mitochondrial respiratory-chain diseases. The New England journal of medicine 348, 2656–2668, doi: 10.1056/NEJMra022567 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Dimauro S & Rustin P A critical approach to the therapy of mitochondrial respiratory chain and oxidative phosphorylation diseases. Biochimica et biophysica acta 1792, 1159–1167, doi: 10.1016/j.bbadis.2008.10.015 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Lehninger AL, Nelson DL, Cox MM Lehninger Principles of Biochemistry. (2005).
- 12.Hui S et al. Glucose feeds the TCA cycle via circulating lactate. Nature, doi: 10.1038/nature24057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faubert B et al. Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371 e359, doi: 10.1016/j.cell.2017.09.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vander Heiden MG, Cantley LC, Thompson CB Understanding the Warburg Effect. The Metabolic Requirements of Cell Proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herzig S et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science 337, 93–96, doi: 10.1126/science.1218530 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Bricker DK et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337, 96–100, doi: 10.1126/science.1218099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papandreou I, Cairns RA, Fontana L, Lim AL & Denko NC HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell metabolism 3, 187–197, doi: 10.1016/j.cmet.2006.01.012 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Connett RJ, Honig CR, Gayeski TEJ, Brooks GA Defining hypoxia: a systems view of VO2, glycolysis, energetics and intracellular PO2. J. Appl. Physiol 68, 833–842 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Sherratt HSA The metabolism of the small intestine. Oxygen uptake and L-lactate production along the length of the small intestine of rat and guinea pig. Comp. Biochem. Physiol 24, 745–761 (1968). [DOI] [PubMed] [Google Scholar]
- 20.Gerich JE Role of the kidney in normal glucose homeostasis and in the hyperglycaemia of diabetes mellitus: therapeutic implications. Diabetic medicine : a journal of the British Diabetic Association 27, 136–142, doi: 10.1111/j.1464-5491.2009.02894.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Ruiz R, Rigoulet M & Devin A The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochimica et biophysica acta 1807, 568–576, doi: 10.1016/j.bbabio.2010.08.010 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Schell JC et al. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Molecular cell 56, 400–413, doi: 10.1016/j.molcel.2014.09.026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Divakaruni AS et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proceedings of the National Academy of Sciences of the United States of America 110, 5422–5427, doi: 10.1073/pnas.1303360110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schell JC et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nature cell biology 19, 1027–1036, doi: 10.1038/ncb3593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson KA, Schell JC & Rutter J Pyruvate and Metabolic Flexibility: Illuminating a Path Toward Selective Cancer Therapies. Trends in biochemical sciences 41, 219–230, doi: 10.1016/j.tibs.2016.01.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metallo CM, Walther JL & Stephanopoulos G Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. Journal of biotechnology 144, 167–174, doi: 10.1016/j.jbiotec.2009.07.010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sellers K et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. The Journal of clinical investigation 125, 687–698, doi: 10.1172/JCI72873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel KP, O’Brien TW, Subramony SH, Shuster J & Stacpoole PW The spectrum of pyruvate dehydrogenase complex deficiency: clinical, biochemical and genetic features in 371 patients. Mol Genet Metab 105, 34–43, doi: 10.1016/j.ymgme.2011.09.032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christen S et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell reports 17, 837–848, doi: 10.1016/j.celrep.2016.09.042 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Davidson SM et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell metabolism 23, 517–528, doi: 10.1016/j.cmet.2016.01.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeBerardinis RJ & Cheng T Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324, doi: 10.1038/onc.2009.358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeBerardinis RJ et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America 104, 19345–19350, doi: 10.1073/pnas.0709747104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haigis MC et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126, 941–954, doi: 10.1016/j.cell.2006.06.057 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Yang C et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Molecular cell 56, 414–424, doi: 10.1016/j.molcel.2014.09.025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vacanti NM et al. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Molecular cell 56, 425–435, doi: 10.1016/j.molcel.2014.09.024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882, doi: 10.1016/j.immuni.2011.09.021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wise DR et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America 105, 18782–18787, doi: 10.1073/pnas.0810199105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang JB et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer cell 18, 207–219, doi: 10.1016/j.ccr.2010.08.009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin L, Alesi GN & Kang S Glutaminolysis as a target for cancer therapy. Oncogene 35, 3619–3625, doi: 10.1038/onc.2015.447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muir A et al. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. eLife 6, doi: 10.7554/eLife.27713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhutia YD & Ganapathy V Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochimica et biophysica acta 1863, 2531–2539, doi: 10.1016/j.bbamcr.2015.12.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sastrasinh A, Sastrasinh M Glutamine Transport in Submitochondrial Particles. Am J Physiol 257, F1050–1058 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Indiveri C, Abruzzo G, Stipani I, Palmieri F Identification and purification of the reconstitutively active glutamine carrier from rat kidney mitochondria. 333, 285–290 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brosnan JT, Brosnan ME Branched-Chain Amino Acids: Metabolism, Physiological Function and Application. The journal of nutrition 136, 207S–211S (2006). [DOI] [PubMed] [Google Scholar]
- 45.Mayers JR et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165, doi: 10.1126/science.aaf5171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimomura Y, Murakami T, Nakai N, Nagasaki M, Harris RA Exercise Promotes BCAA Catabolism: Effects of BCAA Supplementation on Skeletal Muscle during Exercise. J. Nutr 134, 1583S–1587S (2004). [DOI] [PubMed] [Google Scholar]
- 47.Green CR et al. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nature chemical biology 12, 15–21, doi: 10.1038/nchembio.1961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blackburn PR et al. Maple syrup urine disease: mechanisms and management. The application of clinical genetics 10, 57–66, doi: 10.2147/TACG.S125962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohrig F & Schulze A The multifaceted roles of fatty acid synthesis in cancer. Nature reviews. Cancer 16, 732–749, doi: 10.1038/nrc.2016.89 (2016). [DOI] [PubMed] [Google Scholar]
- 50.McGarry JD, Brown NF The Mitochondrial Carnitine Palmitoyltransferase System From Concept to Molecular Analysis. Eur. J. Biochem 244, 1–14 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Houten SM, Wanders RJA A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation Journal of inherited metabolic disease 33, 469–477 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoors S et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature 520, 192–197, doi: 10.1038/nature14362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruderman N & Prentki M AMP kinase and malonyl-CoA: targets for therapy of the metabolic syndrome. Nature reviews. Drug discovery 3, 340–351, doi: 10.1038/nrd1344 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Munday MR Regulation of mammalian Acetyl-CoA carboxylase. Biochemical Society Transactions 30, 1059–1063 (2002). [DOI] [PubMed] [Google Scholar]
- 55.German NJ et al. PHD3 Loss in Cancer Enables Metabolic Reliance on Fatty Acid Oxidation via Deactivation of ACC2. Molecular cell 63, 1006–1020, doi: 10.1016/j.molcel.2016.08.014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Currie E, Schulze A, Zechner R, Walther TC & Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell metabolism 18, 153–161, doi: 10.1016/j.cmet.2013.05.017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svensson RU et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med 22, 1108–1119, doi: 10.1038/nm.4181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia D & Shaw RJ AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Molecular cell 66, 789–800, doi: 10.1016/j.molcel.2017.05.032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carracedo A, Cantley LC & Pandolfi PP Cancer metabolism: fatty acid oxidation in the limelight. Nature reviews. Cancer 13, 227–232, doi: 10.1038/nrc3483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pearce EL et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107, doi: 10.1038/nature08097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Sullivan D et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88, doi: 10.1016/j.immuni.2014.06.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stern JH, Rutkowski JM & Scherer PE Adiponectin, Leptin, and Fatty Acids in the Maintenance of Metabolic Homeostasis through Adipose Tissue Crosstalk. Cell metabolism 23, 770–784, doi: 10.1016/j.cmet.2016.04.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Serra D, Mera P, Malandrino MI, Mir JF & Herrero L Mitochondrial fatty acid oxidation in obesity. Antioxidants & redox signaling 19, 269–284, doi: 10.1089/ars.2012.4875 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahn CS & Metallo CM Mitochondria as biosynthetic factories for cancer proliferation. Cancer & metabolism 3, 1, doi: 10.1186/s40170-015-0128-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker MA, Volpi S, Sims KB, Walter JE & Traggiai E Powering the immune system: mitochondria in immune function and deficiency. Journal of immunology research 2014, 164309, doi: 10.1155/2014/164309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ducker GS & Rabinowitz JD One-Carbon Metabolism in Health and Disease. Cell metabolism 25, 27–42, doi: 10.1016/j.cmet.2016.08.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tibbetts AS & Appling DR Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annual review of nutrition 30, 57–81, doi: 10.1146/annurev.nutr.012809.104810 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Chasin LA, Feldman A, Konstam M, Urlaub G Reversion of a Chinese Hamster Cell Auxotrophic Mutant. Proceedings of the National Academy of Sciences of the United States of America 71, 718–722 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ducker GS et al. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell metabolism 23, 1140–1153, doi: 10.1016/j.cmet.2016.04.016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM & Manning BD mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733, doi: 10.1126/science.aad0489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nilsson R et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nature communications 5, 3128, doi: 10.1038/ncomms4128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ron-Harel N et al. Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell metabolism 24, 104–117, doi: 10.1016/j.cmet.2016.06.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munier-Lehmann H, Vidalain PO, Tangy F & Janin YL On dihydroorotate dehydrogenases and their inhibitors and uses. Journal of medicinal chemistry 56, 3148–3167, doi: 10.1021/jm301848w (2013). [DOI] [PubMed] [Google Scholar]
- 74.Herrmann ML, Schleyerbach R & Kirschbaum BJ Leflunomide: an immunomodulatory drug for the treatment of rheumatoid arthritis and other autoimmune diseases. Immunopharmacology 47, 273–289, doi: 10.1016/s0162-3109(00)00191-0 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Sykes DB et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell 167, 171–186 e115, doi: 10.1016/j.cell.2016.08.057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeong SM et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer cell 23, 450–463, doi: 10.1016/j.ccr.2013.02.024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown KK, Spinelli JB, Asara JM & Toker A Adaptive Reprogramming of De Novo Pyrimidine Synthesis Is a Metabolic Vulnerability in Triple-Negative Breast Cancer. Cancer discovery 7, 391–399, doi: 10.1158/2159-8290.CD-16-0611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hatzivassiliou G et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer cell 8, 311–321, doi: 10.1016/j.ccr.2005.09.008 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Metallo CM et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384, doi: 10.1038/nature10602 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mullen Andrew R., W. WW, Jin Eunsook S., Chen Pei-Hsuan, Sullivan Lucas B., Cheng Tzuling, Yang Youfeng, Linehan W. Marston, Chandel Navdeep S., and DeBerardinis Ralph J.. Reductive carboxilation supports growth in tumor cells with defective mitochondria. Nature 481, 385–388, doi: 10.1038/nature10642 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang L et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258, doi: 10.1038/nature17393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun J, Aluvila S, Kotaria R, Mayor JA, Walters ED, Kaplan R Mitochondrial and Plasma Membrane Citrate Transporters: Discovery of Selective Inhibitors and Application to Structure/Function Analysis. Mol Cell Pharmacol 2, 101–110 (2010). [PMC free article] [PubMed] [Google Scholar]
- 83.Kim CW et al. Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proceedings of the National Academy of Sciences of the United States of America 107, 9626–9631, doi: 10.1073/pnas.1001292107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.GLERUM DM, CLAEYS D, MERTENS W and AZZI A The tricarboxylate carrier from rat liver mitochondria. . European Journal of Biochemistry 194, 681–684 (1990). [DOI] [PubMed] [Google Scholar]
- 85.Zara V, Gnoni GVV Effect of starvation on the activity of the mitochondrial tricarboxylate carrier. Biochimica et biophysica acta 1239, 33–38 (1995). [DOI] [PubMed] [Google Scholar]
- 86.Lee JV et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell metabolism 20, 306–319, doi: 10.1016/j.cmet.2014.06.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Londono Gentile T et al. DNMT1 is regulated by ATP-citrate lyase and maintains methylation patterns during adipocyte differentiation. Molecular and cellular biology 33, 3864–3878, doi: 10.1128/MCB.01495-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wellen KE et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080, doi: 10.1126/science.1164097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao M et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development 26, 1326–1338, doi: 10.1101/gad.191056.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang L et al. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell metabolism 24, 685–700, doi: 10.1016/j.cmet.2016.10.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matthews GD, Gur N, Koopman WJ, Pines O & Vardimon L Weak mitochondrial targeting sequence determines tissue-specific subcellular localization of glutamine synthetase in liver and brain cells. Journal of cell science 123, 351–359, doi: 10.1242/jcs.060749 (2010). [DOI] [PubMed] [Google Scholar]
- 92.Tardito S et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nature cell biology 17, 1556–1568, doi: 10.1038/ncb3272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spinelli JB et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 358, 941–946, doi: 10.1126/science.aam9305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frigerio F, Casimir M, Carobbio S & Maechler P Tissue specificity of mitochondrial glutamate pathways and the control of metabolic homeostasis. Biochimica et biophysica acta 1777, 965–972, doi: 10.1016/j.bbabio.2008.04.031 (2008). [DOI] [PubMed] [Google Scholar]
- 95.Coloff JL et al. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell metabolism 23, 867–880, doi: 10.1016/j.cmet.2016.03.016 (2016). [DOI] [PubMed] [Google Scholar]
- 96.Perez-Arellano I, Carmona-Alvarez F, Martinez AI, Rodriguez-Diaz J & Cervera J Pyrroline-5-carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease. Protein science : a publication of the Protein Society 19, 372–382, doi: 10.1002/pro.340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Palmieri F The mitochondrial transporter family SLC25: identification, properties and physiopathology. Molecular aspects of medicine 34, 465–484, doi: 10.1016/j.mam.2012.05.005 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Bahl JJ, Matsuda M, DeFronzo RA & Bressler R In vitro and in vivo suppression of gluconeogenesis by inhibition of pyruvate carboxylase. Biochemical pharmacology 53, 67–74, doi: 10.1016/s0006-2952(96)00660-0 (1997). [DOI] [PubMed] [Google Scholar]
- 99.Kumashiro N et al. Targeting pyruvate carboxylase reduces gluconeogenesis and adiposity and improves insulin resistance. Diabetes 62, 2183–2194, doi: 10.2337/db12-1311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Monne M & Palmieri F Antiporters of the mitochondrial carrier family. Current topics in membranes 73, 289–320, doi: 10.1016/B978-0-12-800223-0.00008-6 (2014). [DOI] [PubMed] [Google Scholar]
- 101.Yang J, Kalhan SC & Hanson RW What is the metabolic role of phosphoenolpyruvate carboxykinase? The Journal of biological chemistry 284, 27025–27029, doi: 10.1074/jbc.R109.040543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dailey HA & Meissner PN Erythroid heme biosynthesis and its disorders. Cold Spring Harbor perspectives in medicine 3, a011676, doi: 10.1101/cshperspect.a011676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ulrich DL et al. ATP-dependent mitochondrial porphyrin importer ABCB6 protects against phenylhydrazine toxicity. The Journal of biological chemistry 287, 12679–12690, doi: 10.1074/jbc.M111.336180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bonkowsky HL, Bloomer JR, Ebert PS & Mahoney MJ Heme synthetase deficiency in human protoporphyria. Demonstration of the defect in liver and cultured skin fibroblasts. The Journal of clinical investigation 56, 1139–1148, doi: 10.1172/JCI108189 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang H et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130, 1095–1107, doi: 10.1016/j.cell.2007.07.035 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Titov DV et al. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352, 231–235, doi: 10.1126/science.aad4017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van de Ven RAH, Santos D & Haigis MC Mitochondrial Sirtuins and Molecular Mechanisms of Aging. Trends in molecular medicine 23, 320–331, doi: 10.1016/j.molmed.2017.02.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stein LR & Imai S The dynamic regulation of NAD metabolism in mitochondria. Trends in endocrinology and metabolism: TEM 23, 420–428, doi: 10.1016/j.tem.2012.06.005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Todisco S, Agrimi G, Castegna A & Palmieri F Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. The Journal of biological chemistry 281, 1524–1531, doi: 10.1074/jbc.M510425200 (2006). [DOI] [PubMed] [Google Scholar]
- 110.Schantz PG, Sjoberg B, Svedenhag J Malate-aspartate and alpha-glycerophosphate shuttle enzyme levels in human skeletal muscle: methodological considerations and effect of endurance training. Acta Physiol Scand 128, 397–407 (1986). [DOI] [PubMed] [Google Scholar]
- 111.Wallace DC Mitochondria and cancer. Nature reviews. Cancer 12, 685–698, doi: 10.1038/nrc3365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hayes DJ et al. An unusual metabolic myopathy: a malate—aspartate shuttle defect. Journal of the Neurological Sciences 82, 27–39, doi: 10.1016/0022-510x(87)90004-9 (1987). [DOI] [PubMed] [Google Scholar]
- 113.Son J et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105, doi: 10.1038/nature12040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sullivan LB et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563, doi: 10.1016/j.cell.2015.07.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Birsoy K et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551, doi: 10.1016/j.cell.2015.07.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gnoni GV, Priore P, Geelen MJ & Siculella L The mitochondrial citrate carrier: metabolic role and regulation of its activity and expression. IUBMB life 61, 987–994, doi: 10.1002/iub.249 (2009). [DOI] [PubMed] [Google Scholar]
- 117.Palmieri F Diseases caused by defects of mitochondrial carriers: a review. Biochimica et biophysica acta 1777, 564–578, doi: 10.1016/j.bbabio.2008.03.008 (2008). [DOI] [PubMed] [Google Scholar]
- 118.Mracek T, Drahota Z & Houstek J The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochimica et biophysica acta 1827, 401–410, doi: 10.1016/j.bbabio.2012.11.014 (2013). [DOI] [PubMed] [Google Scholar]
- 119.Fan J et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302, doi: 10.1038/nature13236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lewis CA et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular cell 55, 253–263, doi: 10.1016/j.molcel.2014.05.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bao XR et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, doi: 10.7554/eLife.10575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tedeschi PM et al. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell death & disease 4, e877, doi: 10.1038/cddis.2013.393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schieber M & Chandel NS ROS function in redox signaling and oxidative stress. Current biology : CB 24, R453–462, doi: 10.1016/j.cub.2014.03.034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wallace JL & Wang R Hydrogen sulfide-based therapeutics: exploiting a unique but ubiquitous gasotransmitter. Nature reviews. Drug discovery 14, 329–345, doi: 10.1038/nrd4433 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Lyssiotis CA & Kimmelman AC Metabolic Interactions in the Tumor Microenvironment. Trends in cell biology, doi: 10.1016/j.tcb.2017.06.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rai R, Saraswat VA & Dhiman RK Gut microbiota: its role in hepatic encephalopathy. Journal of clinical and experimental hepatology 5, S29–36, doi: 10.1016/j.jceh.2014.12.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eng CH, Yu K, Lucas J, White E & Abraham RT Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Science signaling 3, ra31, doi: 10.1126/scisignal.2000911 (2010). [DOI] [PubMed] [Google Scholar]
- 128.Braissant O, McLin VA & Cudalbu C Ammonia toxicity to the brain. Journal of inherited metabolic disease 36, 595–612, doi: 10.1007/s10545-012-9546-2 (2013). [DOI] [PubMed] [Google Scholar]
- 129.Morris SM Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annual review of nutrition 22, 87–105, doi: 10.1146/annurev.nutr.22.110801.140547 (2002). [DOI] [PubMed] [Google Scholar]
- 130.McCudden CR & Powers-Lee SG Required Allosteric Effector Site for N-Acetylglutamate on Carbamoyl-Phosphate Synthetase I. Journal of Biological Chemistry 271, 18285–18294, doi: 10.1074/jbc.271.30.18285 (1996). [DOI] [PubMed] [Google Scholar]
- 131.Kim J et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 546, 168–172, doi: 10.1038/nature22359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jones EA, Smallwood RA, Craigie A, Rosenoer VM The enterohepatic circulation of urea nitrogen. Clin Sci 37, 825–836 (1969). [PubMed] [Google Scholar]
- 133.Brusilow SW Urea cycle disorders: clinical paradigm of hyperammonemic encephalopathy. Prog Liver Dis 13, 293–309 (1995). [PubMed] [Google Scholar]
- 134.Kenny DJ & Balskus EP Engineering chemical interactions in microbial communities. Chemical Society reviews, doi: 10.1039/c7cs00664k (2017). [DOI] [PubMed] [Google Scholar]
- 135.Treberg JR, Brosnan ME, Watford M & Brosnan JT On the reversibility of glutamate dehydrogenase and the source of hyperammonemia in the hyperinsulinism/hyperammonemia syndrome. Advances in enzyme regulation 50, 34–43, doi: 10.1016/j.advenzreg.2009.10.029 (2010). [DOI] [PubMed] [Google Scholar]
- 136.Sena LA & Chandel NS Physiological roles of mitochondrial reactive oxygen species. Molecular cell 48, 158–167, doi: 10.1016/j.molcel.2012.09.025 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Block K, Gorin Y & Abboud HE Subcellular localization of Nox4 and regulation in diabetes. Proceedings of the National Academy of Sciences of the United States of America 106, 14385–14390, doi: 10.1073/pnas.0906805106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Balaban RS, Nemoto S & Finkel T Mitochondria, oxidants, and aging. Cell 120, 483–495, doi: 10.1016/j.cell.2005.02.001 (2005). [DOI] [PubMed] [Google Scholar]
- 139.Chang TS et al. Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. The Journal of biological chemistry 279, 41975–41984, doi: 10.1074/jbc.M407707200 (2004). [DOI] [PubMed] [Google Scholar]
- 140.Schneider M et al. Mitochondrial glutathione peroxidase 4 disruption causes male infertility. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 23, 3233–3242, doi: 10.1096/fj.09-132795 (2009). [DOI] [PubMed] [Google Scholar]
- 141.Cox AG, Winterbourn CC & Hampton MB Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. The Biochemical journal 425, 313–325, doi: 10.1042/BJ20091541 (2009). [DOI] [PubMed] [Google Scholar]
- 142.Rhee SG Cell signaling. H2O2, a necessary evil for cell signaling. Science 312, 1882–1883, doi: 10.1126/science.1130481 (2006). [DOI] [PubMed] [Google Scholar]
- 143.Bae YS et al. Epidermal Growth Factor (EGF)-induced Generation of Hydrogen Peroxide. Journal of Biological Chemistry 272, 217–221, doi: 10.1074/jbc.272.1.217 (1997). [DOI] [PubMed] [Google Scholar]
- 144.Leslie NR et al. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. The EMBO journal 22, 5501–5510, doi: 10.1093/emboj/cdg513 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chandel NS et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. The Journal of biological chemistry 275, 25130–25138, doi: 10.1074/jbc.M001914200 (2000). [DOI] [PubMed] [Google Scholar]
- 146.Finley LW et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer cell 19, 416–428, doi: 10.1016/j.ccr.2011.02.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.West AP, Shadel GS & Ghosh S Mitochondria in innate immune responses. Nature reviews. Immunology 11, 389–402, doi: 10.1038/nri2975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sena LA et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236, doi: 10.1016/j.immuni.2012.10.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wheeler ML & Defranco AL Prolonged production of reactive oxygen species in response to B cell receptor stimulation promotes B cell activation and proliferation. Journal of immunology 189, 4405–4416, doi: 10.4049/jimmunol.1201433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Linden DR, Levitt MD, Farrugia G & Szurszewski JH Endogenous production of H2S in the gastrointestinal tract: still in search of a physiologic function. Antioxidants & redox signaling 12, 1135–1146, doi: 10.1089/ars.2009.2885 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Levitt MD, Abdel-Rehim MS & Furne J Free and acid-labile hydrogen sulfide concentrations in mouse tissues: anomalously high free hydrogen sulfide in aortic tissue. Antioxidants & redox signaling 15, 373–378, doi: 10.1089/ars.2010.3525 (2011). [DOI] [PubMed] [Google Scholar]
- 152.Luna-Sanchez M et al. CoQ deficiency causes disruption of mitochondrial sulfide oxidation, a new pathomechanism associated with this syndrome. EMBO molecular medicine 9, 78–95, doi: 10.15252/emmm.201606345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lash LH Mitochondrial glutathione transport: physiological, pathological and toxicological implications. Chemico-biological interactions 163, 54–67, doi: 10.1016/j.cbi.2006.03.001 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Szabo C et al. Tumor-derived hydrogen sulfide, produced by cystathionine-beta-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proceedings of the National Academy of Sciences of the United States of America 110, 12474–12479, doi: 10.1073/pnas.1306241110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rigottier-Gois L Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. The ISME journal 7, 1256–1261, doi: 10.1038/ismej.2013.80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Calvo SE, Clauser KR & Mootha VK MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic acids research 44, D1251–1257, doi: 10.1093/nar/gkv1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Szendroedi J, Phielix E & Roden M The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nature reviews. Endocrinology 8, 92–103, doi: 10.1038/nrendo.2011.138 (2011). [DOI] [PubMed] [Google Scholar]
- 158.Bratic A & Larsson NG The role of mitochondria in aging. The Journal of clinical investigation 123, 951–957, doi: 10.1172/JCI64125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Weinberg SE & Chandel NS Targeting mitochondria metabolism for cancer therapy. Nature chemical biology 11, 9–15, doi: 10.1038/nchembio.1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen WW, Freinkman E, Wang T, Birsoy K & Sabatini DM Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell 166, 1324–1337 e1311, doi: 10.1016/j.cell.2016.07.040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fatehullah A, Tan SH & Barker N Organoids as an in vitro model of human development and disease. Nature cell biology 18, 246–254, doi: 10.1038/ncb3312 (2016). [DOI] [PubMed] [Google Scholar]
- 162.Debnath J, Muthuswamy SK & Brugge JS Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268, doi: 10.1016/s1046-2023(03)00032-x (2003). [DOI] [PubMed] [Google Scholar]
- 163.Cantor JR et al. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 169, 258–272 e217, doi: 10.1016/j.cell.2017.03.023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


