Abstract
Endobronchial stents are increasingly used to treat airway complications in multiple conditions including lung transplantation but little is known about the biofilms that form on these devices. We applied deep sequencing to profile luminal biofilms of 46 endobronchial stents removed from 20 subjects primarily with lung transplantation-associated airway compromise. Microbial communities were analyzed by bacterial 16S rRNA and fungal ITS marker gene sequencing. Corynebacterium was the most common bacterial taxa across biofilm communities. Clustering analysis revealed three bacterial biofilm types: one low diversity and dominated by Corynebacterium; another was polymicrobial and characterized by Staphylococcus; and the third was polymicrobial and associated with Pseudomonas, Streptococcus, and Prevotella. Biofilm type was significantly correlated with stent material: covered metal with the Staphylococcus-type biofilm, silicone with the Corynebacterium-dominated biofilm, and uncovered metal with the polymicrobial biofilm. Subjects with sequential stents had frequent transitions between community types. Fungal analysis found Candida was most prevalent, Aspergillus was common and highly enriched in two of three stents associated with airway anastomotic dehiscence, and fungal taxa not typically considered pathogens were highly enriched in some stents. Thus, molecular analysis revealed a complex and dynamic endobronchial stent biofilm with three bacterial types that associate with stent material, a central role for Corynebacterium, and that both expected and unexpected fungi inhabit this unique niche. The current work provides a foundation for studies to investigate the relationship between stent biofilm composition and clinical outcomes, mechanisms of biofilm establishment, and strategies for improved stent technology and use in airway compromise.
Introduction
Endobronchial stents are frequently implanted to treat airway complications, with lung transplantation being a particularly common setting for indications including dehiscence, stenosis, and malacia [1,2]. Stents are composed of various materials, including uncovered metal, covered metal, and silicone [3]. Despite their value in treating medical conditions, implanted biomaterials typically become colonized via biofilms. In other biomedical devices, biofilms have been linked to device failure, especially in immunosuppressed and critically ill hosts [4]. Bacterial biofilms may also contain potential pathogens, and organisms within biofilms are more resistant to host immunity and antimicrobials than free-floating planktonic forms [5]. Biofilms on central venous catheters, intra-cardiac devices, and orthopedic devices has been explored extensively [4,5]. Understanding the nature of biofilms that form on implanted devices is an essential first step in assessing biofilm impact on outcome and/or optimizing such devices. Remarkably little is known about biofilms on endobronchial stents, however.
Emerging culture-independent molecular tools have revolutionized our ability to understand the composition of microbial communities [6–8]. Such sequence-based approaches can comprehensively define entire communities, are highly quantitative, and do not depend on a priori knowledge of suspected constituents or the ability to culture individual organisms. These methods have recently been applied to oral [9–11] and endotracheal tube biofilms[12–16]. Here we present the first sequence-based analysis of luminal biofilms of endobronchial stents used for clinical treatment of critical airway compromise, most of which were in the setting of lung transplant-associated airway complications.
Materials and methods
Subjects and sample collection
Endobronchial stents were collected from March 2014 through November 2016 from subjects who were undergoing clinically-indicated bronchoscopic removal due to need for stent replacement or to resolution of the disease process. Stents were initially placed in the airway and subsequently retrieved through rigid bronchoscopy by a team of interventional pulmonary specialists. Following removal, stents were placed in sterile specimen cups containing sterile saline for transport to the lab (15–30 minutes) and then gently rinsed again to dissociate poorly adherent material. The lumen was then vigorously swabbed circumferentially (Copan Diagnostics), and swabs kept at -80°C. In four cases, bronchoalveolar lavage (BAL) was available from the same bronchoscopy. Clinical data was extracted from the electronic medical record. All subjects gave written informed consent under protocols approved by the University of Pennsylvania IRB (protocol #823558, #812748, #817513, #820073).
Bacterial and fungal amplification, sequencing and analysis
DNA extraction, PCR amplification, and sequencing details are included in supplemental materials. Briefly, DNA was extracted from swabs by combined chemical and mechanical (BioSpec Mini-Beadbeater-16) methods using PowerSoil DNA isolation kit (MoBio) with an additional 95°C incubation to improve DNA recovery from fungi. Extracted DNA was amplified using barcode-labelled primers (27F forward and 338R reverse) directed at the bacterial 16S rRNA gene V1-V2 and fungal ITS-1 regions (ITS1F forward and ITS2 reverse), and sequenced on the Illumina MiSeq platform with 250-bp paired-end reads [17]. ITS amplification was assessed using automated electrophoresis with Agilent 22000 TapeStation system (Agilent Technologies, Santa Clara, CA) and if no detectable amplification occurred, or if the amplification was less than that of primer dimers, these samples were not pooled for sequencing. As a control for environmental background, sterile swabs exposed to saline in collection cups were analyzed in parallel.
Bacterial 16S rRNA gene sequences were clustered into de novo operational taxonomic units (OTUs) at 97% sequence similarity using UCLUST in the QIIME 1.91 pipeline [18], and then aligned to the GreenGenes reference database (v13_8) using PyNAST. Samples with more than 1000 reads (which included all stent swab samples) and OTU’s with 2 or more hits across samples were kept for downstream analysis. Fungal ITS sequence data were processed with the PIPITS pipeline, which uses ITSx software that employs hidden Markov models to extract known ITS sequences from the amplicons. This was followed by annotation of 97% percent identity OTU representative sequences with BROCC and manual BLASTn results [19,20]. Bacterial analysis was based on relative abundances, except when comparing BAL with bronchoscopic pre-wash samples where we also examined absolute read counts so that taxa within low microbial biomass pre-wash samples are not exaggerated [6,21]. Fungal analysis was carried out using absolute read numbers rather than relative abundances given the high variability in fungal quantity between samples, also to avoid exaggeration of taxa with high proportions in specimens with very low fungal content [22]. Sequence data are available in the NCBI Short Read Archives (Project SRP154880) and our code and metadata are available at: https://github.com/johnmcginniss/stent/.
Statistical analysis
All figures and statistical tests were conducted in R (v.3.4.4; http://www.r-project.org). Alpha diversity (richness, Shannon index) was calculated using the vegan package. Beta diversity (UniFrac distances) was calculated in the QIIME pipeline (v.1.91; http://qiime.org) and used to perform principal coordinate analysis (PCoA) [23]. Clinical data were summarized using range, median, and interquartile range. Pairwise Wilcoxon rank sum test was used to compare between-group differences with Bonferroni-Holm correction for multiple testing. Fisher’s exact test was used to analyze categorical data. Permutational multivariate analysis of variance (PERMANOVA) with 999 permutations was used to calculate group differences in weighted UniFrac distances. We used the hclust function from the R stats package for hierarchical clustering and the pam function from the cluster package for partitioning around medoids analysis (PAM) of weighted UniFrac data [24], with further details in the S1 File.
Results
Subjects and stent samples
We analyzed the luminal biofilm of 46 airway stents from 20 subjects (Table 1). Nineteen were treated for airway complication following lung transplantation: 3 for anastomotic dehiscence, and 14 for stenosis or dynamic airway collapse (indication was not available for 2 subjects). All transplant subjects were maintained on an immunosuppression regimen of multiple agents (generally tacrolimus, mycophenolate, and prednisone). One subject had non-transplant-associated dynamic airway collapse. Stents were silicone (n = 20), uncovered metal (n = 18) or covered metal (n = 8).
Table 1. Subject and stent characteristics.
| Subject | Lung DiagnosisA | Trans-plant | Stent type | Stent location | Day Post-TransplantB | Stent duration (days) | Stent Top Taxa by OTU CountC | Concurrent Bronchoscopic CultureD | ITS AnalysisE |
|---|---|---|---|---|---|---|---|---|---|
| 0002 | IPF | Left | Silicone | Left | 474 | 371 | 1. Anaerococcus (g) 2. Porphyromonas (g) |
NA | + |
| 0018 | COPD | Left | Silicone | Left | 301 | 42 | 1. Streptococcus (g) 2. Veillonella (g) |
NA | + |
| Covered | Left | 310 | 9 | 1. Streptococcus (g) 2. Staphylococcus (g) |
NA | + | |||
| Silicone | Left | 353 | 53 | 1. Streptococcus (g) 2. Actinomyces (g) 10. Pseudomonas (g) |
1. Pseudonomas aeruginosa 2. Mouth flora |
+ | |||
| Covered | Left | 548 | 195 | 1. Nesseriaceae (f) 2. Pseudomonas (g) |
NA | + | |||
| 0079 | Non-IPF ILD | Left | Silicone | Left | 454 | 348 | 1. Corynebacterium (g) 2. Anaerococcous (g) |
NA | BT |
| 0720 | CF | BLT | Silicone | Left | 785 | 21 | 1. Corynbacterium (g) 2. Enterobacteriaceae (f) |
NA | + |
| Silicone | Right | 785 | 21 | 1. Corynbacterium (g) 2. Enterobacteriaceae (f) |
NA | + | |||
| 0777 | COPD | BLT | Uncovered | Right | 67 | 42 | 1. Prevotella (g) 2. Fusobacterium (g) |
NA | + |
| Uncovered | Right | 98 | 31 | 1. Fusobacterium (g) 2. Prevotella (g) 3. Pseudomonas (g) |
1. Pseudomonas aeruginosa 2. Aspergillus spp. |
+ | |||
| 0778 | COPD | BLT | Uncovered | Left | 20 | 4 | 1. Pseudomonas (g) 2. Ureaplasma (g) |
NA | BT |
| 0877 | IPF | Right | Silicone | Right | 330 | 55 | 1. Corynebacterium (g) 2. Finegoldia (g) |
1. Mouth flora | + |
| 0895 | IPF | Right | Silicone | Right | 157 | 14 | 1. Corynebacterium (g) 2. Streptococcus (g) |
1. Mouth flora 2. Mycobacterium avium intracellulare |
+ |
| Silicone | Right | 214 | 28 | 1. Corynebacterium (g) 2. Prevotella (g) |
NA | BT | |||
| 0935 | Non-CF bronchiectasis | BLT | Uncovered | Left | 79 | 23 | 1. Mycoplasma (g) 2. Streptococcus (g) |
NA | + |
| Uncovered | Right | 79 | 27 | 1. Mycoplasma (g) 2. Streptococcus (g) |
NA | + | |||
| 0937 | COPD | BLT | Silicone | Right | 163 | 72 | 1. Staphylococcus (g) 2. Streptococcus (g) |
NA | + |
| 0985 | IPF | Left | Silicone | Left | 130 | 8 | 1. Enterobacteriaceae (f) 2. Streptococcus (g) |
NA | + |
| 0988 | IPF | BLT | Uncovered | Right | 51 | 15 | 1. Pseudomonas (g) 2. Enterobacteriaceae (f) |
NA | + |
| Uncovered | Right | 72 | 9 | 1. Pseudomonas (g) 2. Enterococcus (g) |
NA | BT | |||
| Uncovered | Left | 85 | 13 | 1. Streptococcus (g) 2. Corynebacterium (g) |
NA | BT | |||
| Uncovered | Right | 85 | 6 | 1. Corynebacterium (g) 2. Streptococcus (g) |
NA | + | |||
| Uncovered | Left | 93 | 7 | 1. Streptococcus (g) 2. Corynebacterium (g) |
NA | BT | |||
| Uncovered | Left | 99 | 13 | 1. Pseudomonas (g) 2. Corynebacterium (g) |
NA | BT | |||
| 0991 | IPF | BLT | Silicone | Right | 128 | 51 | 1. Corynebacterium (g) 2. Staphylococcus (g) |
NA | BT |
| Silicone | Left | 189 | 112 | 1. Prevotella (g) 2. Veillonella (g) |
NA | BT | |||
| Covered | Left | 247 | 58 | 1. Staphylococcus (g) 2. Pseudomonas (g) |
NA | BT | |||
| Silicone | Left | 317 | 51 | 1. Stapylococcus (g) 2. Corynebacterium (g) |
NA | + | |||
| 1000 | IPF | Left | Uncovered | Left | 98 | 16 | 1. Prevotella (g) 2. Campylobacter (g) 11. S. aureus (s)* 32. Enterobacteriacea (f) |
1. Staphylococcus aureus 2. Serratia marcescenans |
BT |
| 8001 | COPD | BLT | Silicone | Left | 1932 | 1455 | 1. Fusobacterium (g) 2. Corynebacterium (g) |
NA | BT |
| Silicone | Right | 1932 | 723 | 1. Corynebacterium (g) 2. Pseudomonas (g) |
NA | + | |||
| 8002 | COPD (non-transplant) | NA | Silicone | Left | NA | 304 | 1. Corynebacterium (g) 2. Actinomyces (g) |
NA | BT |
| 8003 | COPD | BLT | Silicone | Left | 649 | 436 | 1. Corynebacterium (g) 2. Anaerococcus (g) |
NA | BT |
| 8004 | IPF | Left | Silicone | Right | 979 | 248 | 1. Corynebacterium (g) 2. Pseudomonas (g) |
NA | BT |
| 0048 | IPF | Left | Covered | Left | 287 | 87 | 1. Staphylococcus (g) 2. Peptoniphilus (g) |
NA | + |
| Covered | Left | 475 | 188 | 1. Staphylococcus (g) 2. Anaerococcus (g) |
1. Mouth flora | + | |||
| 0099 | Non-IPF ILD | BLT | Uncovered | Left | 143 | 14 | 1. Parvimonas (g) 2. Campylobacter (g) |
NA | + |
| Uncovered | Right | 143 | 14 | 1. Parvimonas (g) 2. Campylobacter (g) |
NA | BT | |||
| Covered | Left | 151 | 8 | 1. Veillonella (g) 2. Bulleidia (g) |
NA | + | |||
| Uncovered | Right | 151 | 8 | 1. Parvimonas (g) 2. Campylobacter (g) |
NA | BT | |||
| Uncovered | Right | 199 | 48 | 1. Parvimonas (g) 2. Corynebacterium (g) |
1. Mouth flora | + | |||
| Covered | Left | 253 | 33 | 1. Corynebacterium (g) 2. Anaerococcus (g) |
NA | BT | |||
| Uncovered | Right | 253 | 33 | 1. Corynebacterium (g) 2. Anaerococcus (g) |
NA | BT | |||
| Covered | Left | 262 | 9 | 1. Corynebacterium (g) 2. Anaerococcus (g) 3. Enterococcus (g) |
1. Enterococcus faecalis 2. Mouth flora |
+ | |||
| Uncovered | Right | 262 | 9 | 1. Corynebacterium (g) 2. Prevotella (g) |
1. Enterococcus faecalis 2. Mouth flora |
BT | |||
| Silicone | Right | 267 | 5 | 1. Corynebacterium (g) 2. Enterococcus (g) |
1. Mouth flora | BT |
ALung Diagnosis is the disease leading to transplantation. One subject (#8002) was a non-transplant subject.
BDay post-transplant refers to the number of days post-transplant on which the stent was removed.
CFor each sample, the OTU-based taxa were collapsed into the lowest taxonomic assignment available in our analysis. These were then ranked on absolute read count. Reported are the top two taxa per sample. If the cultured organism was not in the top two it is reported with its respective rank.
DCulture is from bronchoalveolar lavage or tissue. NA denotes no culture taken at the time of stent removal.
E+ denotes sample that underwent ITS sequencing; samples without sufficient ITS amplicon for sequencing are indicated as below threshold (BT).
* 2/3 of reads assigned to Staphylococcus in this sample were able to be identified as S.aureus using BLASTn.
Bacterial 16S rRNA gene sequence analysis of biofilm communities
Stent biofilm samples were subject to bacterial 16S rRNA gene amplification using V1-V2 primers followed by Illumina sequencing. After quality filtering we had approximately 6.88x106 reads in total, with a median read count per stent biofilm sample of 53,471 (IQR 46,252–64,614). After 97% clustering this yielded 13,769 de novo OTUs in the swab samples.
The amount of 16S amplicon, as a surrogate for bacterial biomass [22,25], was markedly greater in stent biofilms compared to several types of controls (p<0.001 comparing biofilm samples to laboratory background and bronchoscope prewash; S1 Fig). The S2 Fig shows a heatmap of taxa in stent and control samples.
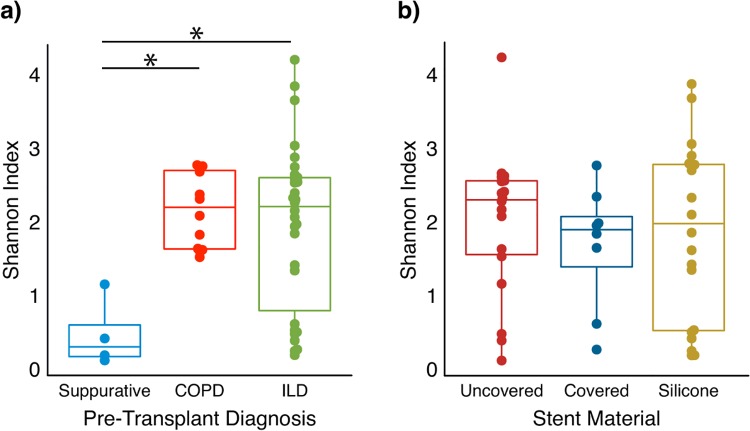
Subjects transplanted for suppurative lung disease had lower within-sample diversity (alpha-diversity as measured by the Shannon index) compared to those transplanted for ILD (p = 0.0134) or COPD (p = 0.0033) (Fig 1A). In contrast, alpha-diversity did not differ across the three stent materials (Fig 1B).
Fig 1. Diversity of stent biofilm bacterial communities by underlying disease and stent material.
Biofilm communities were grouped by underlying pulmonary diagnosis (A) and stent material (B), and within-sample bacterial diversity was calculated using the Shannon Index. Each dot represents a sample, the boxplot represents the median, 75th and 25th percentiles, and points outside the whiskers are outliers. Subjects with underlying suppurative lung disease (cystic fibrosis (CF) and non-CF bronchiectasis) had lower alpha diversity compared to COPD and ILD (p < 0.05, pairwise comparison Wilcoxon rank sum test). Stent material did not impact alpha diversity.
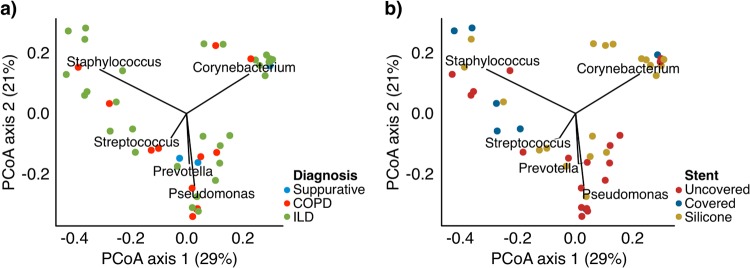
We examined between-sample diversity (beta-diversity) using a principal coordinate analysis (PCoA) of weighted UniFrac distances (Fig 2). UniFrac compares communities based on shared phylogenetic lineages of constituent taxa, and the weighted approach accounts for relative abundances of constituent taxa. The taxa present in greater >5% mean relative abundance driving the distribution on the PCoA were Corynebacterium, Staphylococcus, Pseudomonas, Prevotella, and Streptococcus. There was a statistically significant relationship between stent material and composition on the weighted UniFrac analysis (Fig 2B; R2 = 0.09, p = 0.016; PERMANOVA;). In contrast, there was only a nonsignificant trend towards clustering by diagnosis (Fig 2A; R2 = 0.07, p = 0.06).
Fig 2. Principal coordinate analysis of stent microbial communities.
Biofilm communities were analyzed by Principal Coordinate Analysis (PCoA), using weighted UniFrac. Each sample is represented by a dot. Panel (A) is colored by underlying diagnosis and panel (B) is by stent material. The vectors represent the genus-level bacteria present above 5% mean relative abundance that explain the ordination on the PCoA. The length of the vector is proportional to its explanatory power.
To examine whether alternative analytical approaches might change the nature of these results, we carried out the same analysis after raising the threshold for inclusion of an OTU to ≥10 reads across all samples (reducing the total to 3199 OTUs) and excluding OTUs that could potentially be derived from contamination sources [26] (leaving 3167 OTUs). We also applied rarefaction to 10,000 reads per sample. However, as shown in S3 Fig, this alternative analysis did not change the associations between biofilm diversity and clinical or stent features, nor impact the ordination on the PCoA plot. Thus, these findings are robust to different methodological approaches. Subsequent analyses therefore employed the more complete dataset.
When we examined taxa at the 1% mean relative abundance level (S4 Fig), Corynebacterium continued to define one dimension, suggesting a dominant effect on these communities, whereas multiple additional taxa contributed to other dimensions of the PCoA plot. We repeated this analysis without 10 specimens from one highly sampled subject, #0099, which revealed a similar pattern (S5 Fig), indicating that this relationship is not driven by subject oversampling.
Within the Corynebacterium genus there were 865 de novo OTUs with more than 10 reads assigned. However, >90% of reads were assigned to two OTUs, accounting for 71.8% and 18.9%, respectively. The top hits by BLASTn search of the NCBI 16S rRNA database for these OTUs were C. striatum (99% coverage and 100% identity) and C. xerosis (99% coverage and 99% identity), respectively; however, there is a high degree of homology in this region among Corynebacterium, and other species also had slightly lower but still >97% coverage and identity for these two OTU’s, thus precluding definitive assignment at the species level. Within the Staphylococcus genus there were 3 de novo OTUs that accounted for >95% of reads. The two most prevalent Staphylococcal OTUs could not be assigned at the genus level, while the top five BLASTn hits for the third most prevalent OTU (21% of Staphylococcal reads) were to S. aureus (100% coverage and 99% sequence identity), with other Staphylococcal species having lower coverage and/or identity.
Temporal dynamics and subject-level factors influencing biofilms
We asked whether stent biofilms characteristics were linked to the duration that stents were in place. Duration of stent was not related to biofilm total bacterial biomass (S6 Fig; r = 0.18, p = 0.24), nor PCoA axes 1 or 2 (axis 1: r = 0.06, p = 0.69; axis 2: r = -0.01, p = 0.97). This suggests that factors other than stent duration determine community composition.
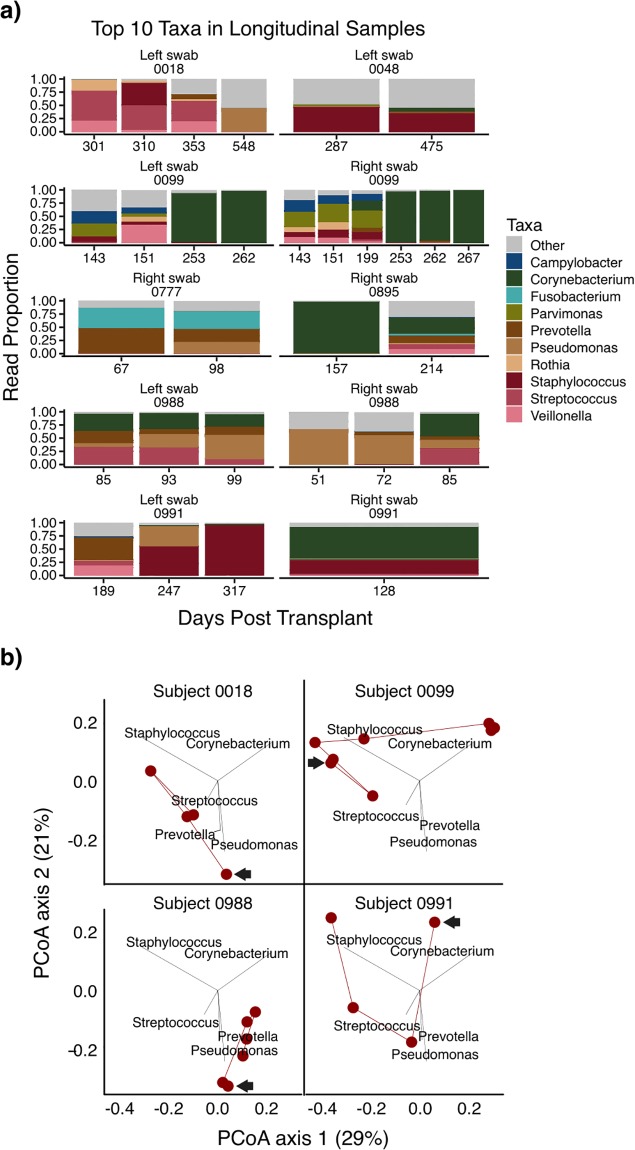
Seven subjects had ≥2 stents available, including two with contemporaneous bilateral stents (Fig 3A). Sequential stents often showed consistent biofilm composition, but sometimes this was interrupted by gradual (#0991) or abrupt (#0099, #0988) compositional changes. In contrast, when bilateral stents were present, the communities were highly concordant (e.g. #0099 days 143, 151, 253, 262; #0988 day 85). We also visualized the 4 subjects with ≥3 stents on the PCoA plot (Fig 3B). Most showed considerable change. Together these results indicate that subject-level variables such as pre-transplant lung disease, and temporal factors such as time post-transplant or stent duration, did not have a primary role in determining stent biofilm composition.
Fig 3. Change in stent biofilm composition over time in subjects with serial stents.
(A) Biofilm communities from subjects with two or more stent samples; if bilateral stents were sampled, the right and left stents are shown separately. Stacked bar graphs represent the proportion of reads assigned the top ten most abundant bacterial genera across samples. The height of a segment of each bar is proportional to its relative abundance and its color reflects the assigned bacterial genus. The number below each sample indicates the number of days since lung transplant that the stent was removed. (B) Stent biofilm communities of four subjects who were sampled at three or more time points are shown on a weighted UniFrac PCoA of the entire sample set. The arrow indicates the first sample and the lines then connect samples sequentially. Three of the four subjects with longitudinal samples had stent biofilms that moved between different clusters over time.
Identification of biofilm community types and their relationship to stent materials
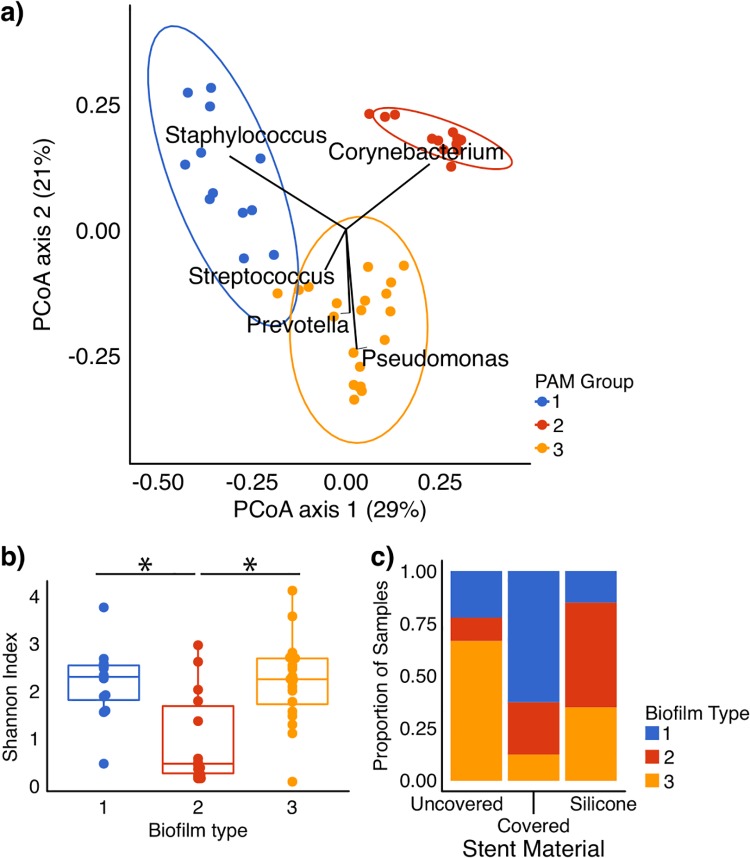
We applied two machine learning algorithms to investigate weighted UniFrac distance clustering: agglomerative hierarchical clustering (h-clust) and partitioning around medoids (PAM). Both found 3 clusters best explained the data (Fig 4). PAM biofilm type 1 was defined by Staphylococcus, type 2 by Corynebacterium, and type 3 by polymicrobial communities. Biofilm type 2 was clearly distinct, while types 1 and 3 reflected more of a continuum between community types (Fig 4A). The three PAM biofilm types had similar bacterial biomass based on 16S amplicon concentrations and were markedly greater than background (S1B Fig), with the Corynebacterium-dominated type 2 being slightly but non-significantly greater (p = 0.07, ANOVA). This result indicates that none of the biofilms reflect background or contamination.
Fig 4. Partitioning about medoids (PAM) analysis of stent biofilm bacterial communities.
(A) Weighted UniFrac PCoA, colored by biofilm type as determined by PAM analysis, analyzed at the genus level, which identified three groups with high confidence. Group 1 is driven primarily by Staphylococcus, Group 2 by Corynebacterium, and Group 3 by several bacteria including Streptococcus, Prevotella, and Pseudomonas. Ellipses indicate 95% confidence intervals for the data distribution within the categorical variable. (B) Shannon diversity of biofilm types defined by PAM. Biofilm type 2 (the Corynebacterium dominant type) had significantly lower diversity than the other types (p<0.05 for both, pairwise Wilcoxon test). (C) Stacked bar chart showing the proportion of biofilm samples of each biofilm type by stent material. Stent material was significantly associated with PAM group (p < 0.05, Fisher’ exact test and Chi Square Test). Silicone stents tended to have a higher proportion of Corynebacterium-dominated biofilm type 2, uncovered metal had more type 3, and covered metal had more type 1.
We queried the taxa present above 1% mean relative abundance within each PAM biofilm type (Table 2). Corynebacterium was widely present. Furthermore, Corynebacterium dominated biofilm type 2, accounting for 86% of all reads. In contrast, PAM types 1 and 3 had more taxa above 1%. Concordantly, biofilm type 2 had lower diversity than the other types (Fig 4B).
Table 2. Top taxa within endobronchial stent biofilm PAM types.
| Biofilm Type | > 1% Taxa | Proportion of Total Reads | |
|---|---|---|---|
| 1 | Staphylococcus (g) | 0.289 | |
| Parvimonas (g) | 0.119 | ||
| Campylobacter (g) | 0.0957 | ||
| Anaerococcus (g) | 0.0859 | ||
| Peptoniphilus (g) | 0.0684 | ||
| Streptococcus (g) | 0.0594 | ||
| Veillonella (g) | 0.0547 | ||
| Rothia (g) | 0.0331 | ||
| Pseudomonas (g) | 0.0290 | ||
| Oribacterium (g) | 0.0266 | ||
| Corynebacterium (g) | 0.0246 | ||
| Porphyromonas (g) | 0.0243 | ||
| Finegoldia (g) | 0.0189 | ||
| Bulleidia (g) | 0.0185 | ||
| Actinomyces (g) | 0.0184 | ||
| 2 | Corynebacterium (g) | 0.860 | |
| Anaerococcus (g) | 0.0469 | ||
| Actinomyces (g) | 0.0241 | ||
| Staphylococcus (g) | 0.0141 | ||
| 3 | Pseudomonas (g) | 0.162 | |
| Streptococcus (g) | 0.146 | ||
| Prevotella (g) | 0.121 | ||
| Corynebacterium (g) | 0.106 | ||
| Fusobacterium (g) | 0.0745 | ||
| Mycoplasma (g) | 0.0744 | ||
| Veillonella (g) | 0.0568 | ||
| Enterobacteriaceae (f) | 0.0391 | ||
| Actinomyces (g) | 0.0267 | ||
| Neisseriaceae (f) | 0.0260 | ||
| Rothia (g) | 0.0209 | ||
| Campylobacter (g) | 0.0182 | ||
| Neisseria (g) | 0.0142 | ||
| Parvimonas (g) | 0.0133 | ||
| Enterococcus (g) | 0.0133 | ||
| Anaerococcus (g) | 0.0130 | ||
Taxa reflecting >1% of reads across all stents within the biofilm type are listed, with their relative abundances, grouped at the genus (g) or family (f) level.
Stent material was significantly associated with biofilm type (Fig 4C; p = 0.008, Fisher’s exact test). Specifically, covered metal was associated with type 1 (Staphylococcus), silicone with type 2 (Corynebacterium), and uncovered metal with type 3 (polymicrobial). In contrast, biofilm type did not correlate with pre-transplant diagnosis.
Stent biofilm relationship to the bronchoalveolar lavage (BAL) microbiome and environmental controls
Four subjects had BAL performed during the stent removal bronchoscopy. We compared the stent biofilm, BAL, and washes of the bronchoscope working channel prior to the procedure (S7 Fig). The stent biofilm and BAL bacterial composition were similar, and distinct from the environmental controls. In biofilms with high Corynebacterium, this bacterium was abundant in the BAL but not in environmental controls, indicating it was authentically present in the stent biofilm and lower respiratory tract, and not derived from environmental contamination.
Fungal microbiome in stent biofilms
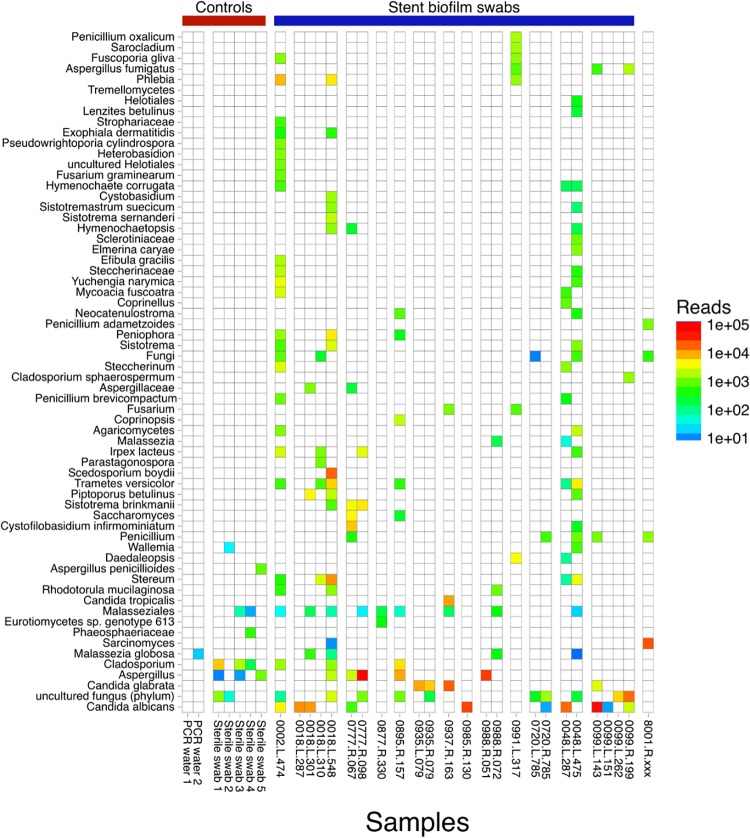
We used ITS sequencing to identify fungal taxa in the stent biofilm. Of the 46 specimens, 21 had no definitive fungal amplification using ITS primers and were not pooled for sequencing. We first visualized fungal taxa in the positive samples on a heat map (Fig 5). Because the presence of fungi is highly variable in lower respiratory tract specimens [22,27], we analyzed fungi by read count numbers rather than relative abundances, which can exaggerate the appearance of taxa in samples with few fungal reads [22].
Fig 5. Fungal taxa within stent biofilms samples as determined by ITS sequencing.
Twenty-five biofilm swab samples and seven control samples had fungal ITS amplification sufficient for sequencing. In the heat map, rows reflect fungal taxa at the lowest level assignment possible (down to the species level). Each column is a unique biofilm sample, grouped by subject. The name is in the format of: subject ID—side of stent—day post-transplant. Because the total fungal biomass is highly variable among samples, the color scale reflects absolute read counts rather than relative abundances to avoid exaggeration of taxa in low fungal content specimens. Taxa with fewer than 10 reads in any sample are excluded.
Several stents yielded high numbers of ITS reads aligning to Aspergillus and Candida, and in some samples, Scedosporium, Stereum, and Sarcinomyces (Fig 5, Table 3). Candida species included C. albicans, C. glabrata, and C. tropicalis. Among samples with Aspergillus, three were confidently assigned to A. fumigatus, while the remainder could only be assigned to Aspergillus at the genus level. Scedosporium is occasionally recognized as an opportunistic pathogen. However, Stereum and Sarcinomyces were unexpected because they are typically considered environmental contaminants, yet they were present at high read numbers within biofilms and absent from our environmental controls. Environmental control specimens had very low numbers of fungal reads, suggesting that fungi identified at high read count in stent biofilms are authentic constituents rather than contamination-derived.
Table 3. Fungal reads identified in endobronchial stents.
| Sample IDA | Taxonomic AssignmentB,C | Read CountC |
|---|---|---|
| 0002-L-474 | Candida albicans (s) | 5190 |
| Phlebia (g) | 4982 | |
| Yuchengia narymica (s) | 4018 | |
| Steccherinum (g) | 3502 | |
| Mycoacia fuscoatra (s) | 3273 | |
| Fungi (k) | 3245 | |
| Trametes gibbosa (s) | 3075 | |
| Irpex lacteus (s) | 3005 | |
| 0018-L-287 | Candida albicans (s) *2 OTU’s | 12654 |
| 0018-L-301 | Candida albicans (s) | 9824 |
| Piptoporus betulinus (s) | 5023 | |
| 0018-L-548 | Scedosporium boydii (s) | 20098 |
| Stereum (g) | 10012 | |
| Trametes versicolor (s) | 7086 | |
| Hypholoma sublateritium (s) | 5106 | |
| Trechispora stellulata (s) | 4802 | |
| Phlebia (g) | 4245 | |
| Peniophora (g) | 3864 | |
| Sisotrema (g) | 3325 | |
| Burgoa verzuoliana (s) | 3232 | |
| Phlebia radiate (s) | 3038 | |
| 0048-L-287 | Candida albicans (s) | 17057 |
| 0048-L-475 | Trametes versicolor (s) | 5510 |
| Stereum (g) | 4070 | |
| 0777-R-67 | Cystofilobasidiium infirmominiatum (s) | 7915 |
| Saccharomyces (g) | 4956 | |
| Sistotrema brinkmannii (s) | 4166 | |
| 0777-R-098 | Aspergillus (g) | 111651 |
| Sistotrema brinkmannii (s) | 5813 | |
| Irpex lacteus (s) | 3707 | |
| 8001-R-XXXX | Sarcinomyces (g) | 23361 |
| 0895-R-157 | Aspergillus (g) | 7842 |
| Cladosporium (g) | 4243 | |
| 0935-L-079 | Candida glabrata (s) | 10599 |
| 0935-R-079 | Candida glabrata (s) | 7582 |
| 0937-R-163 | Candida glabrata (s) | 21282 |
| Candida tropicalis (s) | 11226 | |
| 0985-R-130 | Candida albicans (s) | 24303 |
| 0988-R-051 | Aspergillus (g) | 29169 |
| 0099-L-143 | Candida albicans (s) | 41296 |
| Candida glabrata (s) | 3552 | |
| 0099-L-262 | Uncultured fungus (p) | 6987 |
| 0099-R-199 | Uncultured fungus (p) | 18322 |
| 0991-L-317 | Daedaleopsis (g) | 4511 |
| Sterileswab1 | Cladosporium (g) *2 OTU’s | 7817 |
Taxa with ≥3000 reads in a sample are shown, along with the number of reads.
ASample ID corresponds to those of Fig 5.
BTaxa are shown at the most precise level that could be assigned, as species (s), genus (g), family (f), phylum (p), or in one case no better than kingdom (k).
CTaxa with greater than 10,000 reads in a sample are shown in bold.
In an analysis of bacterial/fungal co-variation, Candida showed a significant inverse correlation with the most abundant bacterial genus, Corynebacterium (S8 Fig; ρ = -0.48, p = 0.017, Spearman’s rank correlation). In contrast, there was no relationship between Candida and Streptococcus or other prevalent taxa.
Relationship between biofilm composition, airway cultures, and clinical features
Ten subjects had airway cultures concurrent with stent removal; six cultures revealed respiratory pathogens (Table 1). In five out of the six positive cultures, the cultured bacteria were identified in the stent biofilm. Importantly, the respiratory pathogen was not the most abundant 16S rRNA sequence, but rather was part of a community that included anaerobic or upper respiratory-type bacteria that are not usually cultured. One subject (#0895) grew Mycobacterium avium-intracellulare (MAI), which was not identified by sequencing. This suggests that either MAI does not participate in stent biofilm communities even if present in the lower respiratory tract, or these molecular methods are less sensitive than culture for mycobacteria [27], even though our previous work suggests that this pipeline can detect mycobacteria [17]. Conversely, we detected Enterococcus by sequencing but not culture (#0099, day 267) five days after a prior positive airway culture for this organism.
Three subjects had stents placed for anastomotic dehiscence, and two of these had the highest numbers of Aspergillus reads in our study (#0777, #0988; Fig 5 and Table 3). Subject 0077 (day 98) had a contemporaneous fungal culture, which was positive for Aspergillus. Candida spp. were detected by sequencing in many stents, including several at high levels. Candida spp. are typically reported in respiratory cultures as yeast with no further identification by our clinical microbiology lab. The stents that had Scedosporium, Stereum, and Sarcinomyces did not have contemporaneous BAL cultures.
All but one subject was on at least one antibiotic at the time of stent removal and most were also exposed to anti- fungal medications (S1 Table). The ubiquity of the exposure and between-subject heterogeneity limits statistical analysis of the impact of specific antimicrobials on biofilm composition, but indicates that, overall, biofilm formation is robust to antimicrobial treatment. While the goal of this study was to understand the composition of luminal biofilms established, we also queried the relationship to clinical outcome, data for which was available in 13 subjects. We asked if biofilm type (limited to the final stent in subjects with >1 stent) was related to outcome categorized as resolution of the airway problem motivating stent placement, presence of excessive granulation tissue, and stent mucus plugging. Within these categories, there was no significant relationship between stent biofilm type and outcomes (p = 1, Fisher’s Exact Test), though the number of subjects in any individual outcome group was small in this cohort. Thus, the impact of biofilm types identified here on outcomes will require future prospective studies.
Discussion
Here we report the first molecular investigation of the airway stent biofilm. In this predominantly post-transplant cohort, we identified three bacterial community types—one low diversity and dominated by Corynebacterium, while the other two have greater diversity and are characterized by Staphylococcus or by a mixed population including Pseudomonas, Prevotella, and Streptococcus. Biofilm type correlated with stent material. When individuals had serial stents, the biofilm communities frequently shifted, suggesting that device-related, procedural and/or contemporaneous clinical factors, but not pre-transplant lung disease, influence biofilm formation. Fungal sequences were frequently detected, sometimes at high read counts.
Corynebacterium was the most prevalent bacteria. It dominated biofilm type 2, and was present at substantial albeit lower abundance in other biofilm types. Corynebacteria are common skin flora and are generally not considered pathogens in the respiratory tract [6,28], although there have been reports of C. striatum respiratory tract infection in immunocompromised hosts[29–32]. One previous study identified Corynebacterium in airway cultures from 4.6% of patients after lung transplantation, and within this population the presence of airway stents was associated with Corynebacterium persistence [33]. Our data suggests a potentially important role for Corynebacterium is its interactions and competition with other bacteria and fungi in the biofilm. Supporting this notion, Corynebacterium spp. have been shown to antagonize Staphylococcus aureus and Streptococcus pneumoniae in the anterior nares [34–36]. Furthermore, Corynebacterium spp. have recently been identified as foundational taxa in dental biofilms, serving as a nidus of nucleation for other bacteria in dental plaque [11]. Thus, in the post-transplant stent biofilm Corynebacterium may be derived from the oral cavity and thrive within the biofilm, provide a scaffold for some taxa to persist while competing with others, and play a central role in endobronchial stent biofilm structure and composition.
Despite their importance for treating airway compromise, stents may themselves increase risk of lower respiratory tract complications, including infection, granulation tissue formation and mucous plugging [37,38]. Stent colonization has been postulated to contribute to these consequences [39–41], and so understanding biofilms that form on these devices could be important to optimizing their development and use. We found stent material was significantly correlated with biofilm type, with silicone stents favoring the Corynebacterium-dominated biofilms, covered metal favoring Staphylococcus-type biofilms, and uncovered metal associated with polymicrobial biofilms. Although the association was imperfect and explained only part of the biofilm composition, this information if further substantiated could help guide stent design or selection for the purposes of minimizing infectious and inflammatory complications. Such knowledge could be important in reducing the risk of colonization by potential pathogens. Future studies could address how antimicrobial-impregnated stent materials impact biofilm development.
Fungi were common within stent biofilms, with Candida being the most frequently identified, including pathogenic species C. albicans, C. glabrata, and C. tropicalis. While Candida are often considered contaminants in respiratory cultures, they have a propensity to establish biofilms [4,42,43]. Our observation of high read numbers of Candida spp. suggest they are bona fide inhabitants of these post-transplant airway stent biofilms. Aspergillus is often suspected as a cause of anastomotic complications requiring stent insertion [1], and we found a preponderance of Aspergillus reads in dehiscence cases. The presence of Aspergillus in these biofilms is not only consistent with such an association, but raises the possibility that the biofilm might also contribute to fungal persistence. Several other fungi were found in individual samples at high abundance, including Scedosporium, Stereum, and Sarcinomyces. Scedosporium is recognized as an opportunistic pathogen. In contrast, the latter two fungi are generally not considered human pathogens. Further study is needed to determine whether these biofilm inhabitants are linked to anastomotic tissue infections, either a cause or a consequence, or might perpetuate inflammation, impede healing, or cause stent/airway complications.
The fact that stent biofilms were markedly higher in bacterial biomass and differed in composition from environmental controls (S1 and S2 Figs) indicates that these findings authentically reflect biofilm and not contamination. In addition, the major findings are robust to different analytical approaches. Nevertheless, our study has several limitations. The study was observational so there was inherent heterogeneity in exposures, although accurately reflects clinical practice in complex patients. Sampling was done at the time of stent removal, so it is difficult to make inferences into the how the stent microbiome changed in situ. We investigated a relatively large number of stents (n = 46), but the number of any single stent material, underlying lung disease, and outcome group was modest. Finally, our study was done in a predominantly post-transplant population, which has distinct exposures to antibiotics and immunosuppression that may limit generalizability to stent biofilms that form in non-transplant populations.
In summary, this is the first study to comprehensive, unbiased molecular methods to interrogate the biofilm of airway stents. We identify a central role for Corynebacterium in the stent biofilm, three broad types of biofilm communities, evidence for unexpected uncultured fungi and fungal/bacterial community co-variation, a possible link between fungi and anastomotic dehiscence, and an association between stent material and biofilm type. Understanding the nature and composition of airway stent biofilms sets the stage for future studies to determine how luminal biofilms influence outcomes, identification of low-risk or high-risk biofilms and better understanding of mechanisms involved in their establishment. This knowledge will offer opportunities for improved stent technology and decision-making around stent selection and management.
Supporting information
(DOCX)
(A) Stent swabs have a higher biomass using 16S amplicon quantification compared to bronchoscope pre-wash and lab controls (p < 0.001 for both comparisons); (B) PAM groups are not statistically different in biomass (p = 0.23 comparing groups 1–2 and 2–3, p = 0.48 comparing 1–3).
(TIF)
Each row reflects a taxon with greater than 2000 reads assigned to it across samples. Each column is a sample with an annotation above noting its sample type and the 16S amplicon quantification through PicoGreen.
(TIF)
Diversity analyses were repeated after filtering OTUs with less than 10 reads across samples, filtering OTUs flagged by the decontam package, and rarefaction to of samples to 10,000 reads. (A, B) Shannon diversity remained significantly lower in stent swabs from subjects with underlying suppurative disease compared to COPD (p = 0.0033) and ILD (p = 0.021). (C, D) Biofilm composition remains significantly related to stent material (PERMANOVA, R2 = 0.09, p = 0.019) but not diagnosis (R2 = 0.07, p = 0.09) by weighted UniFrac.
(TIF)
Weighted UniFrac PCoA analysis of stent biofilm communities showing vectors representing the genus-level bacteria present above 1% relative abundance that explain the ordination on the PCoA. Panel (A) is colored by diagnosis and panel (B) is colored by stent material.
(TIF)
Weighted UniFrac PCoA of stent biofilm samples, excluding 10 samples from subject 0099. Vectors show bacterial taxa driving the ordination that are present at greater than 5% relative abundance. After removal of the oversampled subject’s data, similar clusters form and are driven by the same bacterial genera as in the full dataset, suggesting that this subject’s samples did not disproportionately skew community clustering.
(TIF)
Biofilm samples were grouped based on the duration stent was in place prior to removal: from 0 to 30 days, 31 to 60 days, and greater than 60 days. Panel (A) shows the relationship between stent duration and bacterial biomass as assessed by 16S amplicon quantity, and panel (B) proportion of samples belonging to each biofilm PAM group.
(TIF)
Four subjects had bronchoalveolar lavage (BAL) carried out contemporaneous with stent removal, with samples shown as heatmaps, along with matched bronchoscope pre-wash samples as a background control. Each column is a sample, each row is a bacterial taxon, and each group represents a different subject. The left-hand panel shows the number of matched reads per sample; because the environmental controls are low biomass samples we used absolute read counts rather than relative abundances so taxa with low numbers of reads within a low microbial biomass sample are not exaggerated. The right-hand panel shows the relative abundances within each sample. Within each subject grouping, the first column is a BAL sample (B), second is a stent biofilm (S), and third is a bronchoscope pre-wash environmental control (P).
(TIF)
For all samples with at least 500 fungal reads (n = 22), the absolute reads on a log10 scale of Candida within the ITS dataset (x-axis) were plotted against the proportion Corynebacterium and Streptococcus (y-axis). The line is fit with a general linear regression model with 95% confidence intervals shown in gray. There was a significant inverse relationship between Candida and Corynebacterium (ρ = -0.48, p = 0.017, Spearman’s rank correlation) but no correlation with Streptococcus relative abundance. The regression line is derived from a generalized linear regression model with 95% confidence interval in gray.
(TIF)
(DOCX)
Acknowledgments
We thank subjects who consented for this study, members of the Interventional Pulmonary team who assisted with collections, and L. Glaser of the Clinical Microbiology Lab for helpful discussion. We acknowledge valuable assistance from the Penn Center for AIDS Research and the Penn-CHOP Microbiome Program.
Abbreviations
- 16S rRNA gene
16S ribosomal ribonucleic acid gene
- BAL
bronchoalveolar lavage
- CF
cystic fibrosis
- COPD
chronic obstructive pulmonary disease
- hclust
hierarchical clustering
- ILD
interstitial lung disease
- IPF
idiopathic pulmonary fibrosis
- ITS
internal transcribed spacer
- PAM
partitioning around medoids
- PCoA
principal coordinate analysis
- QIIME
quantitative insights into microbial ecology
- V1-V2
hypervariable regions 1 & 2 of the 16S rRNA gene
Data Availability
All the 16S rRNA and ITS sequences are deposited with metadata in the NIH SRA (accession number PRJNA482386) and additional code and metadata at: https://github.com/johnmcginniss/stent.
Funding Statement
Funded by RGC, FDC, JDC: NIH National Heart, Lung, and Blood Institute grant R01-HL113252; http://grantome.com/grant/NIH/R01-HL113252-04. RGC: NIH National Institute of Allergy and Infectious Diseases grant P30-AI045008; http://grantome.com/grant/NIH/P30-AI045008-20. JEM: NIH National Heart, Lung, and Blood Institute training grant T32-HL007586; http://grantome.com/grant/NIH/T32-HL007586-33. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Crespo MM, McCarthy DP, Hopkins PM, Clark SC, Budev M, Bermudez CA, et al. ISHLT Consensus Statement on adult and pediatric airway complications after lung transplantation: Definitions, grading system, and therapeutics. J Hear Lung Transplant. 2018; 10.1016/j.healun.2018.01.1309 [DOI] [PubMed] [Google Scholar]
- 2.Lee P, Kupeli E, Mehta AC. Airway Stents. Clinics in Chest Medicine. 2010. pp. 141–150. 10.1016/j.ccm.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 3.Herth FJF, Eberhardt R. Airway stent. Curr Opin Pulm Med. 2016;22: 252–256. 10.1097/MCP.0000000000000266 [DOI] [PubMed] [Google Scholar]
- 4.Veerachamy S, Yarlagadda T, Manivasagam G, Yarlagadda PK. Bacterial adherence and biofilm formation on medical implants: A review. Proc Inst Mech Eng Part H J Eng Med. 2014;228: 1083–1099. 10.1177/0954411914556137 [DOI] [PubMed] [Google Scholar]
- 5.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3: a010306 10.1101/cshperspect.a010306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184: 957–963. 10.1164/rccm.201104-0655OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial Topography of the Healthy Human Lower Respiratory Tract. MBio. 2017;8: 1–12. 10.1128/mBio.02287-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huttenhower C, Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486: 207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. Nature Publishing Group; 2014;8: 1659–1672. 10.1038/ismej.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson SN, Meissner T, Su AI, Snesrud E, Ong AC, Schork NJ, et al. Functional expression of dental plaque microbiota. Front Cell Infect Microbiol. 2014;4: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci. 2016;113: E791–E800. 10.1073/pnas.1522149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danin P-E, Girou E, Legrand P, Louis B, Fodil R, Christov C, et al. Description and Microbiology of Endotracheal Tube Biofilm in Mechanically Ventilated Subjects. Respir Care. 2015;60: 21–29. 10.4187/respcare.02722 [DOI] [PubMed] [Google Scholar]
- 13.Pan Y, Du L, Ai Q, Song S, Tang X, Zhu D, et al. Microbial investigations in throat swab and tracheal aspirate specimens are beneficial to predict the corresponding endotracheal tube biofilm flora among intubated neonates with ventilator‑associated pneumonia. Exp Ther Med. 2017; 1450–1458. 10.3892/etm.2017.4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marino PJ, Wise MP, Smith A, Marchesi JR, Riggio MP, Lewis MAO, et al. Community analysis of dental plaque and endotracheal tube biofilms from mechanically ventilated patients. J Crit Care. Elsevier Inc.; 2017;39: 149–155. 10.1016/j.jcrc.2017.02.020 [DOI] [PubMed] [Google Scholar]
- 15.Sands KM, Wilson MJ, Lewis MAO, Wise MP, Palmer N, Hayes AJ, et al. Respiratory pathogen colonization of dental plaque, the lower airways, and endotracheal tube biofilms during mechanical ventilation. J Crit Care. Elsevier Inc.; 2017;37: 30–37. 10.1016/j.jcrc.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 16.Zakharkina T, Martin-Loeches I, Matamoros S, Povoa P, Torres A, Kastelijn JB, et al. The dynamics of the pulmonary microbiome during mechanical ventilation in the intensive care unit and the association with occurrence of pneumonia. Thorax. 2017;72: 803–810. 10.1136/thoraxjnl-2016-209158 [DOI] [PubMed] [Google Scholar]
- 17.Clarke EL, Lauder AP, Hofstaedter CE, Hwang Y, Fitzgerald AS, Imai I, et al. Microbial lineages in sarcoidosis a metagenomic analysis tailored for low-microbial content samples. Am J Respir Crit Care Med. 2018;197: 225–234. 10.1164/rccm.201705-0891OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7: 335–6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gweon HS, Oliver A, Taylor J, Booth T, Gibbs M, Read DS, et al. PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol Evol. 2015;6: 973–980. 10.1111/2041-210X.12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13: R60 10.1186/gb-2012-13-7-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5: 52 10.1186/s40168-017-0267-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bittinger K, Charlson ES, Loy E, Shirley DJ, Haas AR, Laughlin A, et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 2014;15: 487 10.1186/s13059-014-0487-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71: 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynolds AP, Richards G, De La Iglesia B, Rayward-Smith VJ. Clustering rules: A comparison of partitioning and hierarchical clustering algorithms. J Math Model Algorithms. 2006;5: 475–504. 10.1007/s10852-005-9022-1 [DOI] [Google Scholar]
- 25.Knecht VR, McGinniss JE, Shankar HM, Clarke EL, Kelly BJ, Imai I, et al. Molecular analysis of bacterial contamination on stethoscopes in an intensive care unit. Infect Control Hosp Epidemiol. 2019;40: 171–177. 10.1017/ice.2018.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome; 2018; 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186: 536–545. 10.1164/rccm.201204-0693OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, et al. Assessing Bacterial Populations in the Lung by Replicate Analysis of Samples from the Upper and Lower Respiratory Tracts. PLoS One. 2012;7: 1–12. 10.1371/journal.pone.0042786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Díez-Aguilar M, Ruiz-Garbajosa P, Fernández-Olmos A, Guisado P, Del Campo R, Quereda C, et al. Non-diphtheriae Corynebacterium species: An emerging respiratory pathogen. Eur J Clin Microbiol Infect Dis. 2013;32: 769–772. 10.1007/s10096-012-1805-5 [DOI] [PubMed] [Google Scholar]
- 30.Renom F, Garau M, Rubí M, Ramis F, Galmés A, Soriano JB. Nosocomial outbreak of Corynebacterium striatum infection in patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2007;45: 2064–2067. 10.1128/JCM.00152-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee PP, Ferguson DA, Sarubbi FA. Corynebacterium striatum: An underappreciated community and nosocomial pathogen. Journal of Infection. 2005. pp. 338–343. 10.1016/j.jinf.2004.05.005 [DOI] [PubMed] [Google Scholar]
- 32.Renom F, Gomila M, Garau M, Gallegos MDC, Guerrero D, Lalucat J, et al. Respiratory infection by Corynebacterium striatum: Epidemiological and clinical determinants. New Microbes New Infect. 2014;2: 106–114. 10.1002/nmi2.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Los-Arcos I, Len O, Martín-Gómez MT, Baroja A, Berastegui C, Deu M, et al. Clinical Characteristics and Outcome of Lung Transplant Recipients with Respiratory Isolation of Corynebacterium spp. J Clin Microbiol. 2018;56: 1–8. 10.1128/JCM.00142-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus Shifts toward commensalism in response to corynebacterium species. Front Microbiol. 2016;7: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalili D, Amini M, Faramarzi MA, Fazeli MR, Khoshayand MR, Samadi N. Isolation and structural characterization of Coryxin, a novel cyclic lipopeptide from Corynebacterium xerosis NS5 having emulsifying and anti-biofilm activity. Colloids Surfaces B Biointerfaces. 2015;135: 425–432. 10.1016/j.colsurfb.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 36.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. MBio. 2016;7: e01725–15. 10.1128/mBio.01725-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agrafiotis M, Siempos II, Falagas ME. Infections related to airway stenting: a systematic review. Respiration. 2009;78: 69–74. 10.1159/000213244 [DOI] [PubMed] [Google Scholar]
- 38.Grosu HB, Eapen GA, Morice RC, Jimenez CA, Casal RF, Almeida FA, et al. Stents are associated with increased risk of respiratory infections in patients undergoing airway interventions for malignant airways disease. Chest. 2013;144: 441–449. 10.1378/chest.12-1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noppen M, Piérard D, Meysman M, Claes I, Vincken W. Bacterial colonization of central airways after stenting. Am J Respir Crit Care Med. 1999;160: 672–677. 10.1164/ajrccm.160.2.9812081 [DOI] [PubMed] [Google Scholar]
- 40.Ost DE, Shah AM, Lei X, Godoy MCB, Jimenez CA, Eapen GA, et al. Respiratory infections increase the risk of granulation tissue formation following airway stenting in patients with malignant airway obstruction. Chest. 2012;141: 1473–1481. 10.1378/chest.11-2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nouraei SAR, Petrou MA, Randhawa PS, Singh A, Howard DJ, Sandhu GS. Bacterial Colonization of Airway Stents. Arch Otolaryngol Neck Surg. 2006;132: 1086 10.1001/archotol.132.10.1086 [DOI] [PubMed] [Google Scholar]
- 42.Kim D, Sengupta A, Niepa THR, Lee BH, Weljie A, Freitas-Blanco VS, et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep. Nature Publishing Group; 2017;7: 1–14. 10.1038/srep41332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lohse MB, Gulati M, Johnson AD, Nobile CJ. Development and regulation of single-and multi-species Candida albicans biofilms. Nature Reviews Microbiology. 2018. pp. 19–31. 10.1038/nrmicro.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(A) Stent swabs have a higher biomass using 16S amplicon quantification compared to bronchoscope pre-wash and lab controls (p < 0.001 for both comparisons); (B) PAM groups are not statistically different in biomass (p = 0.23 comparing groups 1–2 and 2–3, p = 0.48 comparing 1–3).
(TIF)
Each row reflects a taxon with greater than 2000 reads assigned to it across samples. Each column is a sample with an annotation above noting its sample type and the 16S amplicon quantification through PicoGreen.
(TIF)
Diversity analyses were repeated after filtering OTUs with less than 10 reads across samples, filtering OTUs flagged by the decontam package, and rarefaction to of samples to 10,000 reads. (A, B) Shannon diversity remained significantly lower in stent swabs from subjects with underlying suppurative disease compared to COPD (p = 0.0033) and ILD (p = 0.021). (C, D) Biofilm composition remains significantly related to stent material (PERMANOVA, R2 = 0.09, p = 0.019) but not diagnosis (R2 = 0.07, p = 0.09) by weighted UniFrac.
(TIF)
Weighted UniFrac PCoA analysis of stent biofilm communities showing vectors representing the genus-level bacteria present above 1% relative abundance that explain the ordination on the PCoA. Panel (A) is colored by diagnosis and panel (B) is colored by stent material.
(TIF)
Weighted UniFrac PCoA of stent biofilm samples, excluding 10 samples from subject 0099. Vectors show bacterial taxa driving the ordination that are present at greater than 5% relative abundance. After removal of the oversampled subject’s data, similar clusters form and are driven by the same bacterial genera as in the full dataset, suggesting that this subject’s samples did not disproportionately skew community clustering.
(TIF)
Biofilm samples were grouped based on the duration stent was in place prior to removal: from 0 to 30 days, 31 to 60 days, and greater than 60 days. Panel (A) shows the relationship between stent duration and bacterial biomass as assessed by 16S amplicon quantity, and panel (B) proportion of samples belonging to each biofilm PAM group.
(TIF)
Four subjects had bronchoalveolar lavage (BAL) carried out contemporaneous with stent removal, with samples shown as heatmaps, along with matched bronchoscope pre-wash samples as a background control. Each column is a sample, each row is a bacterial taxon, and each group represents a different subject. The left-hand panel shows the number of matched reads per sample; because the environmental controls are low biomass samples we used absolute read counts rather than relative abundances so taxa with low numbers of reads within a low microbial biomass sample are not exaggerated. The right-hand panel shows the relative abundances within each sample. Within each subject grouping, the first column is a BAL sample (B), second is a stent biofilm (S), and third is a bronchoscope pre-wash environmental control (P).
(TIF)
For all samples with at least 500 fungal reads (n = 22), the absolute reads on a log10 scale of Candida within the ITS dataset (x-axis) were plotted against the proportion Corynebacterium and Streptococcus (y-axis). The line is fit with a general linear regression model with 95% confidence intervals shown in gray. There was a significant inverse relationship between Candida and Corynebacterium (ρ = -0.48, p = 0.017, Spearman’s rank correlation) but no correlation with Streptococcus relative abundance. The regression line is derived from a generalized linear regression model with 95% confidence interval in gray.
(TIF)
(DOCX)
Data Availability Statement
All the 16S rRNA and ITS sequences are deposited with metadata in the NIH SRA (accession number PRJNA482386) and additional code and metadata at: https://github.com/johnmcginniss/stent.