Abstract
We describe the synthesis and RNA acylation activity of a series of minimalist azidoalkanoyl imidazole reagents, with the aim of functionalizing RNA at 2’-hydroxyl groups at stoichiometric to superstoichiometric levels. We find marked effects of small structural changes on their ability to acylate and be reductively removed, and identify reagents and methods that enable efficient RNA functionalization and control.
Graphical Abstract
Table of contents text
Azidoalkanoyl imidazole agents that superstoichiometrically acylate 2’-hydroxy groups on RNA are developed for reversible RNA functionalization and control.
Development of molecular strategies for functionalization and control of RNA is an important goal to aid the study of RNA biology. Fluorescent labels are widely used in detecting, imaging, and analyzing RNAs;1 reactive groups are employed for structural mapping and footprinting of RNAs;2 and affinity labels are useful for isolation and surface immobilization.3 Removable functional groups on RNAs are also employed for caging RNA activity, using light or chemical reagents to switch on RNA function.4
A standard approach for functionalizing RNA involves incorporation of reactive groups during solid-phase RNA synthesis. Although versatile, this method is restricted to RNAs that are considerably shorter than many biological RNAs and requires expertise in monomer and oligonucleotide synthesis. Methods for functionalizing existing long or transcribed RNAs are considerably more restrictive. Notable examples involve 3’-end ligation5 and 3’-end periodate oxidation.6 These 3’-terminal modifications fail in the presence of terminal phosphate groups, and both methods are non-reversible. Prominent examples of 5’-end modification include chemical modification of 5’-phosphate with diazo compounds,7 transcriptional methods with modified guanosine or adenosine 5’-monophosphates,8 and 5’ cap modification with methyltransferases;9 all of these approaches are also irreversible. In general, end modifications are not useful for controlling or caging RNA folding.
Few practical methods exist for functionalizing the internal nucleotides of RNA. Employing modified nucleoside triphosphates during transcription is useful,10 but cannot be applied to native RNAs or those already synthesized. Recent studies have made use of methyltransferase enzymes to label RNAs, but this method requires either engineered RNA sequences and structures and/or enzymatic steps.9c,11 Another method for internal functionalization of RNA involving diazoketones can be used with long transcribed RNAs but forms unstable phosphotriesters that can lead to RNA degradation.12 None of these methods are reversible. Thus, developing simple internal functionalization methods that can be reversed and applied to existing RNAs irrespective of length remains a priority.
Here we describe the design and synthesis of simple azidoalkanoyl imidazole reagents that acylate 2’-hydroxyl groups on RNA at superstoichiometric levels. Past work on chemical RNA acylation has been aimed at sparse reaction (<<1 group per RNA strand) for the purpose of structural mapping. Two aryl isatoic anhydride reagents, NMIA and 1M7, have been employed widely in such mapping studies.2c,13 However, because they exhibit limited aqueous solubility and short half-lives (30–240 sec) in water, they are not suitable for stoichiometric RNA functionalization. More recent work established that two heterocycles activated with acylimidazole groups (FAI and NAI) can exhibit high solubility and aqueous half-lives of 30–60 min.14 Taking advantage of these properties, we recently reported that a nicotinic acylimidazole reagent, NAI-N3, can acylate RNA strands in vitro in minutes at stoichiometric (~1 acyl per strand) to superstoichiometric (>50% of hydroxyl groups reacted) levels.4c This polyacylation of 2’-OH groups (“RNA cloaking”) can be used to inhibit RNA hybridization and folding. The attached azide group enables RNA function to be restored with water-soluble phosphines that reduce the azide and de-acylate (“uncloak”) the RNA, returning it to the original state. This offers a novel post-synthetic strategy for caging and uncaging RNA activity, a field under intense study recently.4b–d,12a
To date, only one acylimidazole reagent (NAI-N3) has been shown to acylate RNA at superstoichiometric levels, and it is unknown what are the constraints on reagent structure, solubility, and reactivity that enable this reaction. In addition, little is known about how varied 2’-acyl structure affects RNA folding and hybridization. All previous RNA acylating agents have been aryl derivatives, and no simple alkyl reactive species have been identified before. Further, it is unknown how varied azide location affects the efficiency of de-acylation via Staudinger reduction.
To address these issues, we have carried out the synthesis and study of the RNA acylation of a series of minimalist azido-alkanoyl imidazole reagents. Interestingly, we find marked effects of small structural changes on their ability to acylate RNA and on their ability to be reductively removed. We also identify structures that are ideal for either reversible or permanent acylation of RNA and further modification. The results greatly expand the range and capabilities of reagents for functionalizing RNA and further our understanding of how structure modulates function of these acyl groups.
To test the simplest reversible acylation molecular scaffolds, we chose two classes of electrophiles as reactive handles for 2’ OH groups: acid imidazoles and imidazole carbamates (Fig. 1A). Acyl imidazoles have been shown to successfully react with RNAs in two previous aryl examples (NAI and FAI),14 and imidazole carbamates were employed in a photocleavable cloaking agents.4b For the new alkyl compounds, the chain length was varied (1–3) to observe possible effects on intramolecular cyclization during uncloaking, as well as length effects on hybridization of the polyacylated RNA to its complement. The beta carbon was substituted with an oxygen in 4 to determine the effects of increased solubility, flexibility, and electrophilicity on acylation/de-acylation. For the imidazole carbamate species (5–8), reactivity was tuned by altering electron withdrawing character at C2 of imidazole.4b These eight reagents were compared to previous RNA acylating reagents (NAI-N3, FAI-N3, 1M7, and NMIA). Details of synthesis and characterization of the new compounds are given in the Supporting Information (SI).
Figure 1.
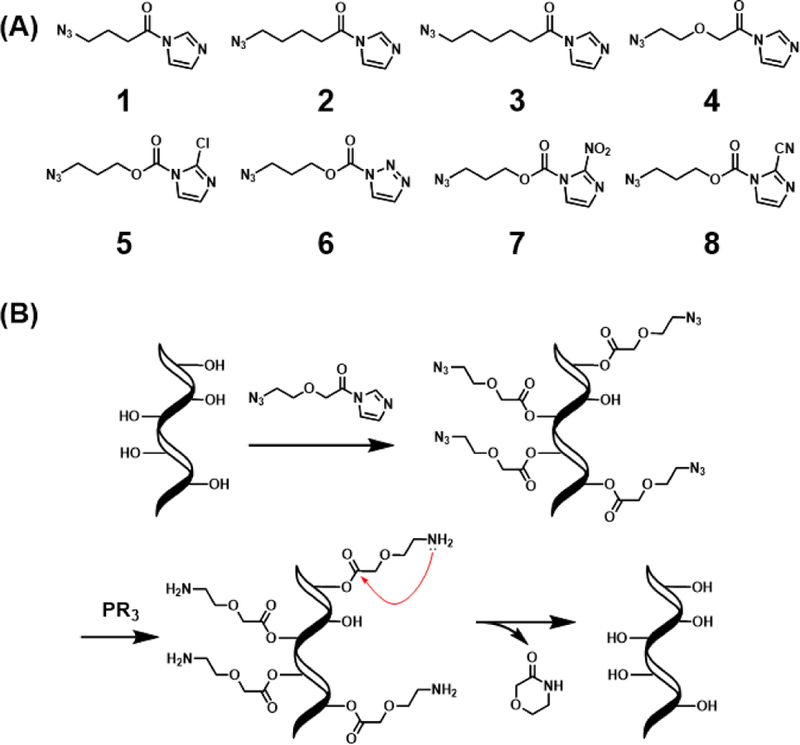
Simple azidoalkanoyl imidazoles tested for acylation and de-acylation of RNA. (A) Reagent structures studied. (B) Scheme of RNA polyacylation (cloaking) with 4 and de-acylation (uncloaking) with a phosphine (PR3).
To estimate the reactivity of the compounds, their hydrolysis half-lives in phosphate buffer were measured by 1H NMR. Half-lives of the alkanoyl imidazoles 1–3 ranged from 30 to 120 minutes (Fig. S1 and S2), falling into the general range of previous aryl RNA polyacylating reagents.4b,14 Chain length did not show a strong influence on hydrolysis rates. Interestingly, the glycolic acid derivative 4 displayed higher acyl reactivity, showing full hydrolysis in 15 minutes, likely due to the inductive effect of the beta oxygen. For the imidazole carbamates (5–8), as expected, reagents with stronger electron-withdrawing groups displayed faster hydrolysis, with half-lives ranging from 10 to 450 minutes.
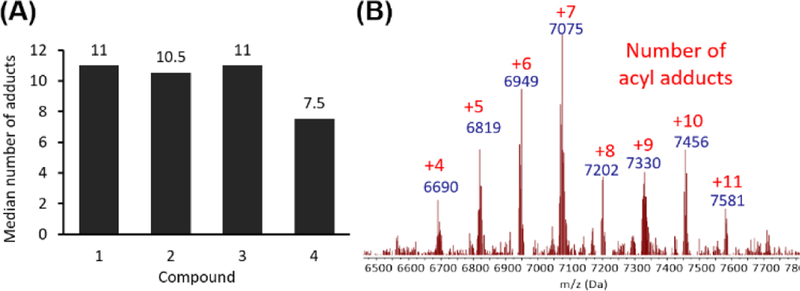
Having prepared acylating reagents with a range of chain length, oxygen substitution, and electrophilicity, we then studied their reactivity with RNA. As a model substrate, we employed a transfer RNA fragment, tRF-3005. The acylimidazole derivatives were prepared as 2 M stocks in dry DMSO, and reactions were conducted at a final concentration of 0.8 M in water at room temperature for 4 hours, after which the RNA was isolated by ethanol precipitation. The stoichiometric degree of modification was quantified by mass spectrometry. We found that all of the alkyl acid-derived acylimidazoles showed high levels of acylation, providing median modification of 11 of 18 nucleotides under these conditions, with little variation based on chain length or beta-oxygen substitution (Fig. 2 and S3). The result was surprising, as the acid imidazoles showed a broad range of reactivity. Glycolic acid derivative 4, on the other hand, had slightly lower median modification of 7.5 acylations, possibly due to its shorter lifetime in water. By performing the cloaking with 4 at 10°C, we found that median modification could be improved to 13 (Fig. S4). The 2’-OH selectivity of 4 was confirmed through comparison with the DNA version of tRF-3005, as only the RNA substrate showed considerable acylation (Fig. S5). Surprisingly, the imidazole carbamate species (5–8) all failed to acylate the test RNA significantly despite their widely varied reactivity (Fig. S3 and S4). We hypothesize that lower solubility of these carbamate compounds contributed to this behavior. Because of their poor acylating ability, the carbamate reagents were not studied further. Comparison of 1–4 with previous RNA acylating reagent NAI-N3 confirmed the previous report of high acylation activity. FAI-N3,15 not previously tested for RNA polyacylation, also showed high acylation yields, but the cloaked RNA suffered from limited solubility, which restricted its accurate quantification by MS and its practical use (Fig. S6). Other known acylating reagents, 1M7 and NMIA, gave no measurable stoichiometric acylation, likely due to their low aqueous solubility and/or short half-lives (Fig. S6).
Figure 2.
Polyacylation ability of reagents studied. (A) Median RNA acylation yields for the new acylating reagents with an 18 nt RNA. 6 μM RNA was treated with 0.8 M reagent, 40% DMSO in water for 4 h at room temp. (B) Representative MALDI-TOF mass spectrum of a polyacylated RNA (with 4). MW of Cy5-tRF-3005 = 6184 Da. Red numbers indicate the number of adducts.
With a set of four simple new alkyl acid-based reagents capable of acylating RNA at superstoichiometric levels, we then proceeded to test their de-acylation capability. In principle, all four should be reducible by phosphines, converting the azides to amines; this then potentially enables cyclization and de-acylation (Fig. 1B) forming 5-, 6-, or 7-membered lactams. Through an initial screen of phosphines, we determined that water-soluble 2-(diphenylphosphino) ethylamine (2DPPEA) was optimal for de-acylation (data not shown). Uncloaking with 20 mM 2DPPEA at 37 °C for 4 h, we found that the glycolic acid-derived acyl group resulting from 4 performed the best, giving high de-acylation yields (Fig. S7). The greater electrophilicity of the resulting 2’-O-ester likely contributed to this behavior. Surprisingly, acyl groups resulting from azidobutyryl compound 1 showed only small degree of uncloaking, while those from azidopentanoyl 2 showed considerably greater uncloaking, likely due to the lower ring strain of the δ-lactam relative to γ-lactam.16 Consistent with ring strain governing cyclization, the 7-membered chain (from 3) showed no measurable uncloaking, only reduction, making it an ideal reagent for long-term post-synthetic modification of RNA with amines. Interestingly, the acyl groups resulting from previous compound FAI-N3 were reduced but failed to show any de-acylation, apparently due to higher ring strain. Finally, the earlier reported efficient uncloaking ability of NAI-N3 was confirmed here as well (Fig. S7). However, uncloaking NAI-N3 resulted in significant unidentified side products. This issue for 4 was absent, resulting in relatively traceless uncloaking of RNA.
The effects of polyacylation of RNA by the alkanoyl acylating agents 1–4 and subsequent phosphine treatment on the RNA’s ability to form a duplex was studied with a molecular beacon (MB) assay.4c,17 The median acylation of the RNA was first normalized to approximately 12 for all acyl groups, and the cloaked RNAs were treated with phosphine to afford the uncloaked counterpart. For the cloaked RNA, all acyl imidazole probes studied showed excellent inhibition of hybridization (Fig. 3 and S8), almost completely blocking MB signal. Chain length or identity did not affect the degree of hybridization markedly. For the uncloaked RNA, consistent with the MS data, RNA acylated with 1 and 3 showed no recovery of hybridization after phosphine treatment as they displayed little or no de-acylation. For 2 there was moderate recovery (~50%) of hybridization. In contrast, uncloaked RNA treated with 4 or NAI-N3 showed nearly quantitative recovery of hybridization, thus showing robust reversible control of RNA hybridization.
Figure 3.
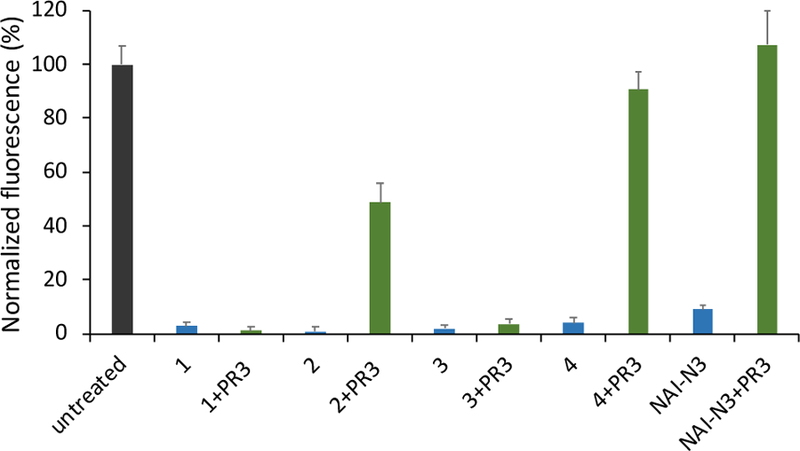
Effect of varied acyl groups on hybridization of an 18 nt RNA, using a molecular beacon fluorescence assay; positive fluorescence signals hybridization. +PR3 (green data) indicates RNA uncloaked with phosphine treatment. Fluorescence signals were obtained by subtracting the background of beacon alone and normalizing to molecular beacon signal with untreated RNA at the left. Black: untreated RNA; blue: cloaked or mock cloaked RNA; green: uncloaked or mock uncloaked RNA. Error bars represent standard deviations (n=3).
Having established that 4 displays exceptional reversibility of acylation and that it effectively modulates RNA hybridization, we investigated its ability to control RNA folding. As a model substrate, we chose Spinach, an RNA aptamer, with the hypothesis that alteration of its folding via cloaking may prevent binding to its cognate ligand DFBHI-1T and result in limited fluorescence.4c,18 We found that acylation with 100 mM 4 inhibited the fluorescence of Spinach to 10% of the original (Figs. 4, S9, and S10). After reversal of acylation with phosphine treatment, the Spinach aptamer recovered ca. 80% of its fluorescence in 60 minutes, indicating a temporal control of light-up response. The lack of full recovery upon uncloaking can be attributed to slight RNA degradation as observed by gel electrophoresis (Fig. S11). Taken together, the data show that 4 provides a useful tool for chemical control of RNA folding.
Figure 4.
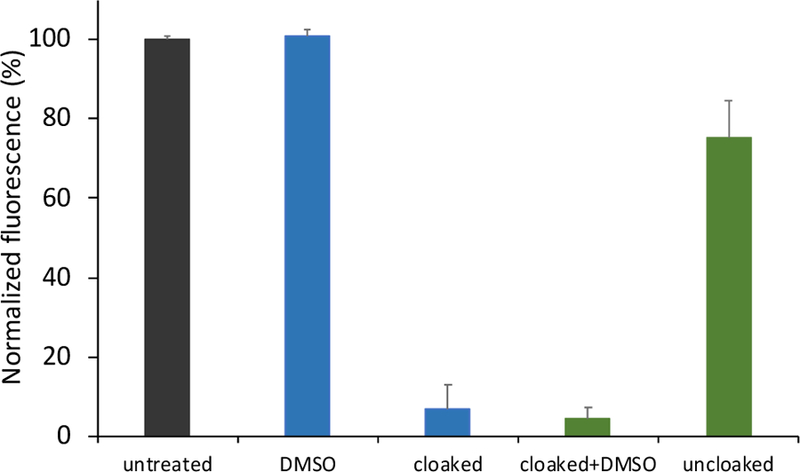
Reversible control of Spinach RNA folding through cloaking and uncloaking with 4. Spinach RNA was treated with 100 mM 4 at 10 °C for 1 h. Uncloaking was conducted with 5 mM 2DPPEA at 37 °C for 1 h. DMSO: mock DMSO treatment. Cloaked+DMSO: mock DMSO uncloaking treatment. Black: untreated RNA; blue: cloaked or mock cloaked RNA; green: uncloaked or mock uncloaked RNA. Error bars represent standard deviations (n=3).
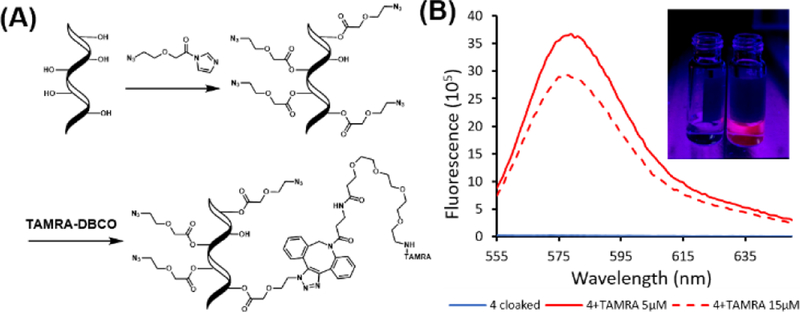
Exploring additional applications of 4, we tested the utility of azide group, once installed on RNA, as a reactive handle after RNA acylation, in a “cloak-click” strategy (Fig. 5A). First, we treated unfunctionalized tRF-3005 RNA with 0.35 M 4 to result in a median modification of 7 acyl groups per RNA strand. The azide groups on the acylated RNA were then reacted with a commercial fluorophore, TAMRA-DBCO, via strain-promoted reaction at low and high concentrations (5 or 15 µM), testing the tunability of levels of dye modification. The cloaked and clicked RNA was conveniently purified via ethanol precipitation. 5 µM treatment resulted in an average of one TAMRA per RNA, while 15 µM treatment resulted in an average of five dyes per RNA (Fig. S12). The fluorescence was characterized by fluorimeter and was visible by eye (Fig. 5B). Interestingly, the lower loading of dye yielded greater fluorescence than the higher loading, likely due to self-quenching of rhodamine at higher local concentrations, as commonly seen in antibody labeling.19 Overall, the results show that acylation followed by azide click reaction is a convenient strategy for covalent labeling of RNA with a dye, requiring two steps and only precipitation to remove excess small-molecule reagents. We envision that this method of functionalization may be widely useful for RNA labeling and bioconjugation.
Figure 5.
Fluorescence labeling of RNA acylated with 4 via a “cloak-click” strategy. (A) Scheme of the strategy. RNA is acylated in one step (“cloak”) and then reacted with an alkyne-substituted dye (“click”). Modified RNA is isolated by precipitation. (B) Fluorescence spectra of labeled RNA (100 nM) RNA with excitation at 546 nm. Inset: visual evidence of fluorescent labeling. 250 nM RNA excited on gel illuminator at 302 nm. Left: tRF-3005 RNA acylated with 4. Right: same RNA after reaction with 5 µM TAMRA-DBCO.
To further test the versatility of 4, we conducted selective 2’-hydroxy acylation analyzed by primer extension (SHAPE) experiments with SAM aptamer RNA and analyzed its structure.20 Compared to NAI-N3, an established SHAPE reagent, 4 showed identical gel electrophoresis patterns (Fig. S13). Interestingly, 4 exhibited more intense reverse transcriptase stops, allowing us to lower the concentration of the acylation reagent 2 to 4-fold relative to the previous reagent.
Taken together, the new experiments have identified a simple class of alkanoyl-based reagents that efficiently polyacylate RNA. The acylating agents in this study are prepared more readily and inexpensively than prior reagents. Furthermore, they are versatile, enabling chemical caging and uncaging or the installation of functional azide or amines groups. Our data show further applications of the compounds in control of RNA hybridization, RNA folding, in post-synthetic fluorescent labeling of RNA, and in analysis of RNA structure. Surveying the available azide-functionalized reagents, NAI-N3 is ideal for intracellular SHAPE. On the other hand, 4 shows promise for caging due to its traceless de-acylation and is capable of “cloak-click” labeling. Future studies will apply these reagents in biophysical and biological studies of RNA function.
Supplementary Material
Acknowledgments
We thank the U.S. National Institutes of Health (GM127295) for support.
Footnotes
Conflicts of interest
The authors report no conflicts of interest.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and References
- 1.(a) Bao G, Rhee WJ and Tsourkas A, Annu. Rev. Biomed. Eng, 2009, 11, 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhuang X, Acc. Chem. Res, 2005, 34, 399–414. [Google Scholar]; (c) Wachowius F and Höbartner C, ChemBioChem, 2010, 11, 469–480. [DOI] [PubMed] [Google Scholar]
- 2.(a) Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET and Chang HY, Nature, 2015, 519, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Feng C, Chan D, Joseph J, Muuronen M, Coldren WH, Dai N, Corrêa IR, Furche F, Hadad CM and Spitale RC, Nat. Chem. Biol, 2018, 14, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Merino EJ, Wilkinson KA, Coughlan JL and Weeks KM, J. Am. Chem. Soc, 2005, 127, 4223–4231. [DOI] [PubMed] [Google Scholar]; (d) Spitale RC, Flynn RA, Torre EA, Kool ET and Chang HY, Wiley Interdiscip. Rev. RNA, S2014, 5, 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Batey RT and Kieft JS, RNA, 2007, 13, 1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vincent HA, Phillips JO, Henderson CA, Roberts AJ, Stone CM, Mardle CE, Butt LE, Gowers DM, Pickford AR and Callaghan AJ, PLoS One, 2013, 8, e79142. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Balamurugan S, Obubuafo A, Soper SA and Spivak DA, Anal. Bioanal. Chem, 2008, 390, 1009–1021. [DOI] [PubMed] [Google Scholar]; (d) Martins R, Queiroz JA and Sousa F, J. Chromatogr. A, 2014, 1355, 1–14. [DOI] [PubMed] [Google Scholar]
- 4.(a) Ruble BK, Yeldell SB and Dmochowski IJ, J. Inorg. Biochem, 2015, 150, 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Velema WA, Kietrys AM and Kool ET, J. Am. Chem. Soc, 2018, 140, 3491–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kadina A, Kietrys AM and Kool ET, Angew. Chemie - Int. Ed, 2018, 57, 3059–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Deiters A, Curr. Opin. Chem. Biol, 2009, 13, 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Bruce AG and Uhlenbeck OC, Nucleic Acids Res, 1978, 5, 3665–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Paredes E, Evans M and Das SR, Methods, 2011, 54, 251–259. [DOI] [PubMed] [Google Scholar]
- 6.(a) Proudnikov D and Mirzabekov A, Nucleic Acids Res, 1996, 24, 4535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Defrancq E, Singh Y and Spinelli N, Curr. Org. Chem, 2008, 12, 263–290. [Google Scholar]
- 7.Gampe CM, Hollis-Symynkywicz M and Zécri F, Angew. Chemie - Int. Ed, 2016, 55, 10283–10286. [DOI] [PubMed] [Google Scholar]
- 8.(a) Li N, Yu C and Huang F, Nucleic Acids Res, 2005, 33, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schlatterer JC and Jäschke A, Biochem. Biophys. Res. Commun, 2006, 344, 887–892. [DOI] [PubMed] [Google Scholar]
- 9.(a) Holstein JM, Anhäuser L and Rentmeister A, Angew. Chemie - Int. Ed, 2016, 55, 10899–10903. [DOI] [PubMed] [Google Scholar]; (b) Schulz D, Holstein JM and Rentmeister A, Angew. Chemie - Int. Ed, 2013, 52, 7874–7878. [DOI] [PubMed] [Google Scholar]; (c) Holstein JM and Rentmeister A, Methods, 2016, 98, 18–25. [DOI] [PubMed] [Google Scholar]
- 10.(a) Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S and Hirao I, J. Am. Chem. Soc, 2005, 127, 17286–17295. [DOI] [PubMed] [Google Scholar]; (b) Ishizuka T, Kimoto M, Satoa A and Hirao I, Chem. Commun [DOI] [PubMed]; (c) Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P and Romesberg FE, J. Am. Chem. Soc, 2011, 133, 19878–19888. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lavergne T, Lamichhane R, Malyshev DA, Li Z, Li L, Sperling E, Williamson JR, Millar DP and Romesberg FE, ACS Chem. Biol, 2016, 11, 1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Eggert F and Kath-Schorr S, Chem. Commun, 2016, 52, 7284–7287. [DOI] [PubMed] [Google Scholar]
- 11.Tomkuviene M, Clouet-d’Orval B, Černiauskas I, Weinhold E and Klimašauskas S, Nucleic Acids Res, 2012, 40, 6765–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Ando H, Furuta T, Tsien RY and Okamoto H, Nat. Genet, 2001, 28, 317–325. [DOI] [PubMed] [Google Scholar]; (b) Blidner RA, Svoboda KR, Hammer RP and Monroe WT, Mol. Biosyst, 2008, 4, 431. [DOI] [PubMed] [Google Scholar]
- 13.Mortimer SA and Weeks KM, J. Am. Chem. Soc, 2007, 129, 4144–4145. [DOI] [PubMed] [Google Scholar]
- 14.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET and Chang HY, Nat. Chem. Biol, 2013, 9, 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan D, Beasley S, Zhen Y and Spitale RC, Bioorganic Med. Chem. Lett, 2018, 28, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bach RD and Dmitrenko O, J. Am. Chem. Soc, 2006, 128, 4598–4611. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi S and Kramer FR, Nat. Biotechnol, 1996, 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 18.(a) Warner KD, Chen MC, Song W, Strack RL, Thorn A, Jaffrey SR and Ferré-D’Amaré AR, Nat. Struct. Mol. Biol, 2014, 21, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Paige JS, Wu KY and Jaffrey SR, Science, 2011, 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabó Á, Szendi-Szatmári T, Ujlaky-Nagy L, Rádi I, Vereb G, Szöllősi J and Nagy P, Biophys. J, 2018, 114, 688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY and Batey RT, Structure, 2010, 18, 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.