Abstract
The high-mobility group box (HMGB) family includes four members: HMGB1, 2, 3 and 4. HMGB proteins have two functions. In the nucleus, HMGB proteins bind to DNA in a DNA structure-dependent but nucleotide sequence-independent manner to function in chromatin remodeling. Extracellularly, HMGB proteins function as alarmins, which are endogenous molecules released upon tissue damage to activate the immune system. HMGB1 acts as a late mediator of inflammation and contributes to prolonged and sustained systemic inflammation in subjects with rheumatoid arthritis. By contrast, Hmgb2−/− mice represent a relevant model of aging-related osteoarthritis (OA), which is associated with the suppression of HMGB2 expression in cartilage. Hmgb2 mutant mice not only develop early-onset OA but also exhibit a specific phenotype in the superficial zone (SZ) of articular cartilage. Given the similar expression and activation patterns of HMGB2 and β-catenin in articular cartilage, the loss of these pathways in the SZ of articular cartilage may lead to altered gene expression, cell death and OA-like pathogenesis. Moreover, HMGB2 regulates chondrocyte hypertrophy by mediating Runt-related transcription factor 2 expression and Wnt signaling. Therefore, one possible mechanism explaining the modulation of lymphoid enhancer binding factor 1 (LEF1)-dependent transactivation by HMGB2 is that a differential interaction between HMGB2 and nuclear factors affects the transcription of genes containing LEF1-responsive elements. The multiple functions of HMGB proteins reveal the complex roles of these proteins as innate and endogenous regulators of inflammation in joints and their cooperative roles in cartilage hypertrophy as well as in the maintenance of joint tissue homeostasis.
Keywords: HMGB protein, rheumatoid arthritis, osteoarthritis, inflammatory mediator, Wnt signaling
HMGB protein family
The high-mobility group box (HMGB) proteins are smaller than 30 kDa in size and bind to DNA in a DNA structure-dependent but nucleotide sequence-independent manner1. The HMGB family includes four members: HMGB1, 2, 3 and 4. HMGB1, 2, and 3 are more than 80% identical at the amino acid sequence level and have similar biochemical properties. These proteins comprise two DNA-binding HMG domains and an acidic tail2. HMGB4 is a mammal-specific protein that contains two HMGB-boxes but lacks the acidic tail, and its nucleotide sequence is less conserved than the sequences of the other Hmgb genes3.
Although HMGB1, 2 and 3 share high levels of amino acid sequence similarity, their expression patterns are diverse. For instance, during mouse embryonic development, both Hmgb1 and Hmgb2 are ubiquitously and expressed at high levels4, 5. By contrast, Hmgb3 expression is more localized during embryogenesis6, and Hmgb4 is detected in the developing brain and pancreas at E12.5 and E14, respectively7, 8. In adult tissues, Hmgb1 has been shown to exhibit ubiquitous and high levels of expression in whole tissue extract assays9, 10; however, other Hmgb genes show restricted expression. Hmgb2 is highly expressed in the thymus and testis5. Hmgb3 is highly expressed in hematopoietic stem cells in the bone marrow11, and Hmgb4 expression is limited to testis and neuronal cells3, 12. The results of these expression analyses suggest that Hmgb1 may have broad roles in embryonic development and in adult tissues, and other Hmgb genes may have redundant functions depending on the developmental stage and tissue in embryos and adults.
Biochemical studies have primarily been performed with HMGB1 and HMGB2, and have shown that HMGB1 and HMGB2 bind to DNA without sequence specificity13, 14. HMGB1 and HMGB2 play a role in the formation of nucleoprotein complexes by altering chromatin structures, which promotes the binding of other factors15, 16 and facilitates diverse DNA modifications1, 17. These proteins are also known to regulate various activities, such as transcription, replication, and DNA repair13. HMGB1 and HMBG2 both bind to HOX proteins, steroid hormone receptors18, and Rag1 recombinase19 and enhance the transcription and recombination activities of their partner proteins when transiently transfected in mammalian cells.
Despite their similarities in amino acid sequence, structure and biochemical characteristics, the functions of HMGB1 and HMGB2 are not completely identical5, 20. Functional studies of Hmgb genes have been performed by generating targeted mutations in mice. Hmgb1−/− mice are born without significant morphological defects but die within a day due to hypoglycemia20, which is caused by insufficient activation of a glucocorticoid receptor. Male Hmgb2−/− mice exhibit reduced fertility, which is caused by the degeneration of Sertoli cells and germ cells in seminiferous tubules and by immotile spermatozoa5. Hmgb3−/− mice exhibit erythrocythemia21. These studies show that Hmgb1, 2, and 3 are not required for embryonic development individually, with the exception that Hmgb1−/− limb long bones show delays in endochondral ossification22. The normal embryonic development of mutant mice suggests the functional redundancy of these proteins, particularly for Hmgb1 and Hmgb2, due to their high expression levels in embryos and shared biochemical characteristics.
The role of extracellular HMGB1
Regarding an extracellular role23, HMGB1 released by damaged cells acts as a chemoattractant and a proinflammatory cytokine23. The translocation of HMGB1 occurs in response to immune cell activation or cell death, and the redox state of cysteines has recently been reported to modulate the binding of HMGB1 to its receptors and its subsequent functions24. HMGB1 contains three cysteines: C23 and C45 form a disulfide bond, and C106 is unpaired. These cysteines are modified by redox reactions and give rise to three isoforms known as fully reduced HMGB1 for the all-thiol form, disulfide HMGB1 for the partially oxidized form, and sulfonyl HMGB1 for the terminally oxidized form24. Depending on the redox states of these amino acid residues, HMGB1 induces cytokine production via Toll-like receptor 4 (TLR4) or promotes chemotaxis by binding the chemokine C-X-C motif chemokine ligand 12 (CXCL12) to form a heterocomplex, which in turn binds to C-X-C chemokine receptor type 4 (CXCR4) and induces cell migration. The cysteines in HMGB1 are terminally oxidized to sulfonates; sulfonyl-HMGB1 neither is a chemoattractant nor displays cytokine-inducing activity25. HMGB1 also interacts with the receptor for advanced glycation end products (RAGE), a multifunctional transmembrane protein of the immunoglobulin superfamily26, to induce the secretion of inflammatory mediators.
During the course of inflammatory disease, HMGB1 plays a dynamic role, depending on its redox state, and functions as an alarmin, which is an endogenous molecule that is released upon tissue damage to activate the immune system27. Cell death is an important mechanism that generates alarmins, and each major type of death (necrosis, apoptosis, pyroptosis and NETosis) leads to the release of different HMGB1 isoforms28. Therefore, posttranslational modifications of HMGB1 are important for determining its role during the response to tissue injury and the course of inflammatory diseases.
HMGB1 has been reported to play a role in the pathogenesis of autoimmune and inflammatory arthritis disorders, such as rheumatoid arthritis (RA), juvenile idiopathic arthritis, osteoarthritis (OA), crystal-induced arthritis, psoriatic arthritis, and spondyloarthritis29–36 (Table 1). HMGB2 is also secreted by human THP-1 monocytic leukemia cells; extracellular HMGB2 has been detected in the blood and in other biological fluids, and it promotes the proliferation and migration of endothelial cells37. HMGB2 exerts these functions by engaging RAGE, as its blockade completely abrogates the cell responses described above.
Table 1.
The role and function of HMGB proteins in arthritis.
| HMGB protein | Disease | Location | Function | Mechanism | References |
|---|---|---|---|---|---|
| HMGB1 | rheumatoid arthritis | extracellular | synovial inflammation | binds to RAGE, TLR2, and TLR4 to activate the NF-κB | 34, 40, 41 |
| osteoarthritis | cartilage and bone destruction | pathway and signal transduction through JNK and p38 | 29, 30 | ||
| juvenile idiopathic arthritis | induction of proinflammatory cytokine and chemokine expression | 31, 32, 33 | |||
| crystal-induced arthritis | 34 | ||||
| psoriatic arthritis | 35 | ||||
| spondyloarthritis | 36 | ||||
| HMGB2 | osteoarthritis | nuclear | chondrocyte survival and homeostasis | binds to LEF1 to enhance Wnt signaling | 76 |
| maintains SZ homeostasis via EFEMP1 | 95 | ||||
| suppression of cartilage hypertrophy | binds to LEF1 and RUNX2 to suppress RUNX2 activity | 85 |
HMGB1 and RA
In patients with RA, tumor necrosis factor-alpha (TNF-α) plays a clinical role in a cytokine cascade that results in joint inflammation and destruction. TNF-α acts on macrophages to enhance phagocytosis and the production of other proinflammatory cytokines and prostaglandin E238. It also serves as a chemoattractant for neutrophils and induces chemokine expression on the endothelial cell lining to facilitate the transendothelial migration of neutrophils. TNF-α acts on fibroblast-like synoviocytes to induce their proliferation and pannus formation and upregulates matrix metalloproteinases (MMPs) such as MMP1 and MMP13, which participate in cartilage damage. In addition, TNF-α activates osteoclasts, which promote bone demineralization38. The use of anti-TNF-α therapy indicates its importance in this disease. However, many patients with RA fail to respond to this therapy or other biologics, and some patients may suffer from unexpected aggravation of joint inflammation or other autoimmune manifestations39. Collectively, these reports suggest that more complex interactions occur among TNF, interleukin-1 (IL-1), and other inflammatory mediators.
According to an in vivo analysis, HMGB1 concentrations were significantly higher in the synovial fluid (SF) from patients with RA than in patients with OA. Similarly, TNF-α levels in the SF were also significantly higher in the same patients with RA than in the same patients with OA, although HMGB1 concentrations in serum were very low, with no significant difference between patients with RA and OA40. In RA synovial tissue, HMGB1 was detected in the cytoplasm of macrophages infiltrating the sublining layer41. In vitro studies have shown that HMGB1 stimulation of synovial fluid macrophages (SFMs) obtained from patients with RA causes the release of proinflammatory cytokines, such as TNF-α, interleukin-1β (IL-1β), and IL-6. Upon stimulation with HMGB1, the levels of TNF-α released by SFMs into the supernatant peak prior to the levels of other cytokines (IL-1β, IL-6). Since TNF-α is an upstream cytokine, it might also stimulate IL-1β and IL-6 secretion42, 43. Thus, HMGB1 may induce SFMs to release high levels of not only TNF-α but also IL-1 and IL-6. By contrast, TNF-α also induces HMGB1 release from SFMs, and the HMGB1-induced release of IL-1 and IL-6 from SFMs occurred much later than TNF-α release.
Based on these results, HMGB1 acts as a late mediator of inflammation and contributes to prolonged and sustained systemic inflammation23, 44–46; in addition, a proinflammatory loop may exist between HMGB1 and TNF-α via SFMs in patients with RA.
HMGB2 and OA
Osteoarthritis (OA) is the most prevalent joint disease with a non-immune mediated pathogenesis and primarily involves cells of mesenchymal lineage, such as chondrocytes and osteoblasts. Aging is the major risk factor for this form of arthritis, which begins with disruption of the superficial zone (SZ) of cartilage, leading to progressive cartilage erosion and bone remodeling, causing disability and reduced quality of life. A link between aging and HMGB proteins has been reported in earlier studies. Phosphorylation and ADP-ribosylation of HMGB2 in the livers of old rats decreased drastically compared with young rats after spermine or sodium butyrate treatment47, 48, and dexamethasone stimulated HMGB2 methylation 12-fold in the livers of young rats, whereas this change was not observed in old rats49. Regarding HMGB2 expression during aging, Ly et al. measured mRNA levels in actively dividing dermal fibroblasts isolated from young, middle-aged, and elderly humans as well as humans with progeria50 and found that Hmgb2 was one of only 9 genes that were down-regulated in both the old age and progeria groups among the 6,000 genes examined. Recently, Aird et al. sought to investigate factors regulating chromatin reorganization during senescence and identified a decrease in HMGB2 expression as one of the most significant changes associated with senescence51.
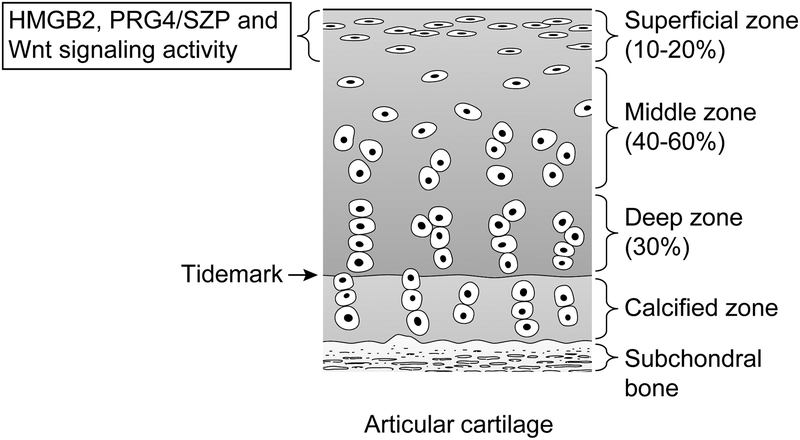
The SZ of articular cartilage is important in many respects because it forms a fluid-tissue interface of articular cartilage in the synovial cavity. The SZ possesses 3.5-fold more cells than the radial zone52, and a concomitant decrease in cell number along with surface fibrillation is the earliest indicator of OA development53. The SZ is unique because it produces lubricin, also known as proteoglycan-4/superficial zone protein (PRG4/SZP), an important joint lubricant54–56. Several recent studies have suggested that adult articular cartilage contains cells with functional properties and phenotypic markers of mesenchymal stem cells (MSCs), and these cells appear to be primarily located in the SZ57–59. HMGB2 expression is uniquely restricted to cells in the SZ in normal mature human articular cartilage60 (Figure 1), and importantly, joint aging in humans and mice leads to the loss of HMGB2 expression. Based on results from gene expression arrays, Hmgb1 expression increased, and Hmgb2 expression decreased in human OA-affected cartilage compared with normal cartilage61. PRG4/SZP has a distribution similar to that of HMGB2 and is involved in cartilage and joint homeostasis62, 63. Young Hmgb2−/− mice did not exhibit an apparent reduction in PRG4/SZP expression. With advancing age, however, Hmgb2−/− mice exhibited a substantial loss, and in some cartilage regions a complete loss, of PRG4/SZP expression in the nonmeniscus-covered weight-bearing regions of the knee joint. Joint aging in humans and mice is associated with a reduction in HMGB2 expression, and this correlates with a reduction in expression of PRG4/SZP. Mice lacking Prg4 appear normal at birth but subsequently display synovial hyperplasia, subintimal fibrosis, and an abnormal articular cartilage surface63. The correlation between the reduction of HMGB2 and loss of PRG4/SZP expression in the SZ of articular cartilage but not in the synovium suggests that HMGB2 plays a unique role in cartilage SZ cells60.
Figure 1.
Expression of HMGB2, PRG4/SZP and Wnt signaling activity in articular cartilage.
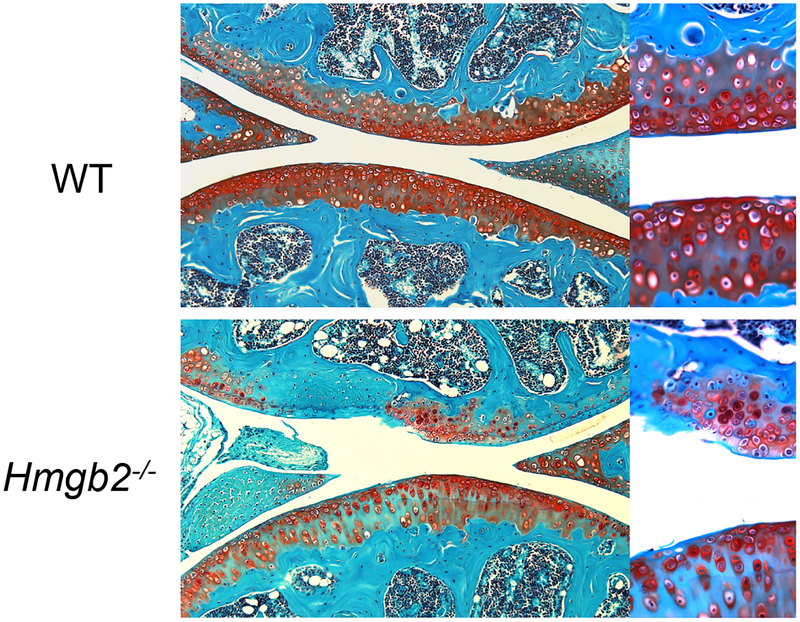
The aging-related loss of HMGB2 expression in the cartilage surface is correlated with a reduction in cellularity, and the aging-related reduction in cellularity was more remarkable in the knee joints of Hmgb2−/− mice than in wild-type mice (Figure 2). These changes were linked to increased apoptosis in Hmgb2−/− mice, and chondrocytes from Hmgb2−/− mice studied in vitro exhibited increased susceptibility to anti-Fas receptor/CD95 antibody-induced apoptosis. These findings suggest that a major function of chromatin-associated factor HMGB2 is to promote SZ chondrocyte survival. A role for HMGB proteins in protecting against cell death has been suggested in neuronal cells in subjects with polyglutamine diseases64, normal Sertoli cells5, and certain cancer cells65.
Figure 2.
Safranin O staining of the articular cartilage of knee joints from 9-month-old wild-type mice and Hmgb2−/− mice. Hmgb2−/− mice show structural cartilage defects.
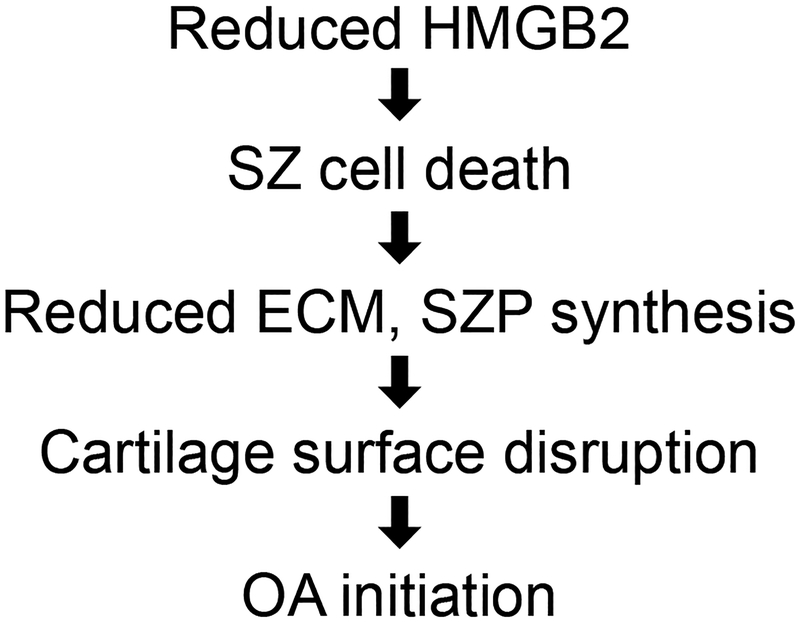
Based on the results from these studies, Hmgb2−/− mice represent a relevant model of aging-related OA. Not only do Hmgb2 mutant mice develop early-onset OA, similar to many of the previously reported mutant strains of mice66, but also, more importantly, they feature a specific phenotype in the SZ of articular cartilage. This phenotype closely resembles human OA, which first manifests in the SZ67 (Figure 3).
Figure 3.
The proposed mechanism of osteoarthritis associated with aging-related loss of HMGB2.
HMGB2 and β-catenin
Wnt proteins are secreted factors that regulate cell proliferation and differentiation during early stages of chondrogenesis68, 69. Wnt binds to Frizzled/lipoprotein receptor-related protein (LRP) receptor complexes, and signal transduction leads to the inhibition of constitutive cytoplasmic β-catenin degradation in the canonical pathway. The accumulated β-catenin translocates into the nucleus, where it acts as a co-activator for various transcription factors, such as lymphoid enhancer binding factor 1 (LEF1)70. Wnt/β-catenin signaling plays diverse and important roles in embryonic skeletal development71, 72. Recent studies have shown that increased Wnt signaling due to loss of secreted Frizzled-related protein (sFRP) function leads to the development of OA73. Similarly, overexpression of β-catenin in chondrocytes in postnatal mice stimulates the expression of matrix-degrading enzymes74. By contrast, Wnt signaling also contributes to the maintenance of normal articular cartilage function. Inhibition of β-catenin signaling by transgenic overexpression of its intracellular antagonist ICAT in articular chondrocytes causes increased cell apoptosis and articular cartilage destruction75, suggesting that the precise temporal and spatial activation of Wnt signaling in articular cartilage determines its homeostatic versus pathogenic effects.
In the embryonic stage, Wnt/β-catenin signaling is active during joint formation72. Postnatally, remarkable similarities between the localization of HMGB2 and β-catenin have been observed. HMGB2 expression and β-catenin activation have been found in all zones of articular cartilage in newborn mice; however, both become more restricted to the SZ with joint maturation, and both show aging-related loss in the SZ76. Although HMGB2 eventually becomes completely absent in the SZ of aged mouse joints, β-catenin signaling is activated in the middle and deep zones. HMGB2 enhances the expression of Wnt/β-catenin target genes in vitro60, 76.
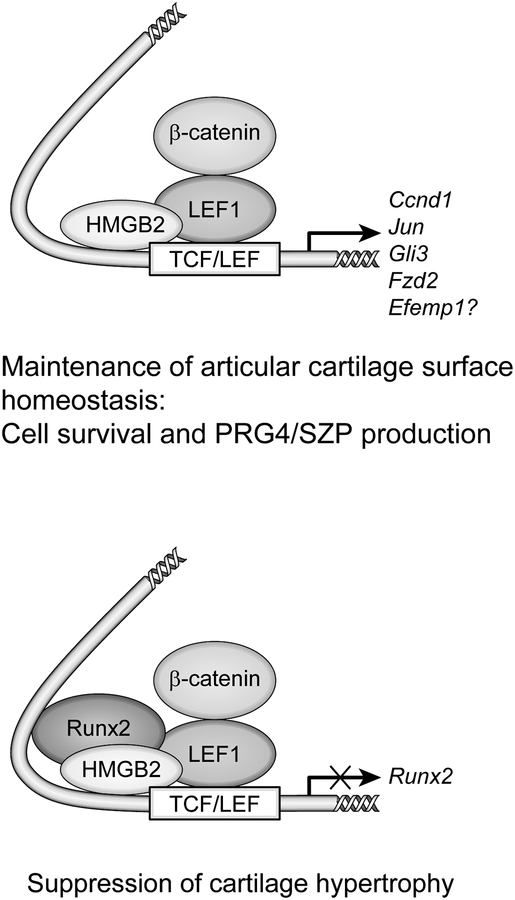
Although transfection of HMGB2 alone did not activate LEF1/T-cell factor (TCF)/β-catenin-responsive promoters, cotransfection of HMGB2 and β-catenin resulted in synergistic activation of a reporter, and this synergy was seen in chondrogenic cell types76. HMGB2 does not directly bind to regulatory DNA elements, but it augments the DNA binding of LEF1, a transcription factor that associates with β-catenin and regulates Wnt/β-catenin downstream genes, and the HMG domain of HMGB2 is responsible for interaction with LEF176. HMGB2 and β-catenin do not interact directly. HMGB2 does not alter the electrophoretic mobility of LEF1 that has been complexed with oligonucleotides in vitro, suggesting that HMGB2 dissociates from the complex after performing its architectural function17. The complex including HMGB2 β-catenin, LEF1 and probably other components leads to enhanced expression of genes containing LEF1 binding sites76.
Cell survival and the expression of cyclin D1 (Ccnd1), GLI family zinc finger 3 (Gli3) and Frizzled class receptor 2 (Fzd2), which are representative LEF1 target genes in cartilage74, 77, 78, were analyzed to determine the functional consequences of the HMGB2/LEF1 interaction in a cellular context, and the expression levels of these genes were reduced by siRNA-mediated HMGB2 knockdown in chondrocytes. Moreover, the conditional deletion of β-catenin by infecting β-catenin floxed chondrocytes with Cre-expressing adenovirus increased basal and induced apoptosis in vitro. Given the similar expression and activation patterns of HMGB2 and β-catenin in articular cartilage, the loss of these pathways in the SZ of articular cartilage may lead to altered gene expression, cell death and OA-like pathogenesis (Figure 4).
Figure 4.
Functional and physical interactions between HMGB2, RUNX2, and LEF1 on a promoter containing a TCF/ LEF motif.
HMGB2 and Runt-related transcription factor 2 (RUNX2)
Recent studies show that endochondral ossification is an essential process not only for physiological skeletal growth but also for the development of osteoarthritis79. OA articular chondrocytes express markers of hypertrophic growth plate chondrocytes such as collagen type X alpha 1 chain (COL10A1), MMP13, and RUNX2, and RUNX2 contributes to the pathogenesis of OA through chondrocyte hypertrophy and matrix breakdown after the induction of joint instability in an experimental mouse model80. In Runx2−/− mice, chondrocyte maturation is disturbed81, 82, and the overexpression of a dominant-negative form of RUNX2 in chondrocytes severely delays endochondral ossification and suppresses chondrocyte maturation83, 84. During mouse limb development, Hmgb2 is expressed in proliferating and prehypertrophic zones but not in hypertrophic cartilage, where Col10a1 is expressed robustly85. According to the results of an in vitro study, the levels of Runx2 transcripts increase in Hmgb2−/− MSCs compared with wild-type MSCs throughout chondrogenesis. Moreover, ectopic HMGB2 expression in MSCs suppresses the expression of the chondrogenesis markers Col2a1 and mitochondrial aspartate-glutamate carrier 1 (Agc1) as well as Runx285.
It has been reported that chondrocyte maturation is induced by β-catenin-mediated Wnt signaling86–88. The TCF/LEF consensus sequence is located downstream of an A/G-rich region of the Runx2 promoter in a highly conserved area that contains binding sequences for NK homeobox 2 (NKX3–2), hypoxia-inducible factor 2 (HIF-2), and vitamin D-responsive elements in addition to RUNX289–93. Runx2 is autoregulated in part by negative feedback on its own promoter; overexpression of RUNX2 protein abrogates Runx2 promoter activity, and a single RUNX2 site is sufficient for transcriptional autosuppression89. Based on the results from an in vitro physical interaction assay, HMGB2 binds to RUNX2 and LEF176, and RUNX2 also interacts with LEF194. Moreover, HMGB2 repressed the augmentation of Runx2 promoter activity mediated by co-transfection of β-catenin and LEF1 in chicken upper sternal chondrocytes, which differentiate into hypertrophic chondrocytes85. These findings support the hypothesis that the loss of HMGB2 may contribute to the pathogenesis of OA by stimulating chondrocyte hypertrophy through Runx2 transactivation. As Wnt signaling mediates chondrocyte hypertrophy through activation of Runx286, one possible mechanism to explain the observed modulation of LEF1-dependent transactivation by HMGB2 in vivo is that the differential interactions between HMGB2 and nuclear factors affect the transcription of genes containing LEF1-responsive elements (Figure 4).
EFEMP1/fibulin 3 as a target for HMGB2
According to an analysis of gene expression patterns in MSCs from wild-type and Hmgb2−/− mice, EFEMP1 (EGF-containing fibulin-like extracellular matrix protein 1)/fibulin 3 levels were significantly reduced in Hmgb2-deficient mice95. The fibulin family includes 6 ECM proteins that are localized in basal membranes, stroma, and ECM fibers, mediating cell-to-cell and cell-to-matrix communication96. Fibulins are thought to organize and stabilize ECM structures during organogenesis and vasculogenesis96. Efemp1−/− mice display early aging phenotypes97. EFEMP1 is expressed in cartilage and bone structures in the mouse embryo98, and EFEMP1 peptides are potential diagnostic biomarkers for OA99.
EFEMP1 expression is uniquely restricted to cells in the SZ in normal mature human and mouse articular cartilage. The process of joint aging in humans and mice is associated with reduced EFEMP1 expression in the articular cartilage, where it is normally expressed only in the SZ95. However, in OA-affected cartilage, cells in the chondrocyte clusters are strongly positive for EFEMP1. PRG4/SZP displays a distribution similar to EFEMP1 in the articular cartilage and is involved in cartilage and joint homeostasis62, 63. Interestingly, the number of PRG4/SZP-positive cells in Efemp1−/− mice was significantly lower than that in wild-type mice. Moreover, a correlation was observed between the reduction and loss of EFEMP1 and reduction and loss of PRG4/SZP expression in the SZ of articular cartilage. The aging-related loss of EFEMP1 expression in the cartilage surface is associated with a reduction in SZ cellularity and increased apoptosis in Efemp1−/− mice. The results of in vitro studies showed that Efemp1 knockdown in chondrocytes resulted in reduced cell viability95. In vitro studies of chondrogenesis revealed that levels of the EFEMP1 protein decreased as MSCs differentiated. The siRNA-mediated suppression of EFEMP1 expression in articular chondrocytes enhanced the expression of chondrogenic markers such as COL2A1, aggrecan (ACAN), and SRY-box 9 (SOX9), whereas SOX9 expression was decreased following EFEMP1 overexpression, suggesting that EFEMP1 plays a role in maintaining the immature status of cells in the SZ, similar to HMGB295.
Thus, EFEMP1 is specifically expressed in the SZ of mature human and mouse articular cartilage, and its expression is profoundly reduced with increasing age. EFEMP1 has a role in maintaining the immature status of cells in the cartilage SZ. The loss of EFEMP1 is linked to the loss of SZ chondrocytes and compromises the integrity of the cartilage surface. Thus, EFEMP1 may be an HMGB2 target protein, and strategies preventing the loss of EFEMP1 expression in SZ chondrocytes or restoring the normal level of expression might have therapeutic potential in the management of patients with OA.
Conclusions
HMGB1 and 2 are involved in many aspects of joint diseases, such as RA and OA, as well as normal skeletal development. Given the roles of both HMGB2 and HMGB1 in controlling inflammatory responses, studies dissecting their respective contributions to inflammation and investigating whether their combined targeting has an additive or synergistic effect would be interesting. The multiple functions of HMGB proteins in tissue injury in other systems reveal the complex roles of these proteins as innate and endogenous regulators of inflammation in cartilage and joints as well as their collaborative roles in cartilage hypertrophy and the maintenance of joint homeostasis.
Acknowledgments
We are grateful for the support from Hiroshi Asahara (Tokyo Medical and Dental University, Japan), Marco E Bianchi (San Raffaele University, Italy), Diana Brinson, Lilo Creighton and Jean Valbracht (The Scripps Research Institute, USA). This work was funded by grants from MEXT KAKENHI (grant number 15K10484 to NT) and the NIH (grant numbers AR064195 to YK and AG007996 to ML).
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
Contributor Information
Noboru Taniguchi, Department of Orthopaedic Surgery, University of Miyazaki, Miyazaki, Japan, Department of Medical Science, Tokyo Medical University, Tokyo, Japan, 5200 Kihara, Kiyotake, Miyazaki 889-1692, Japan.
Yasuhiko Kawakami, Department of Genetics, Cell Biology and Development, and Stem Cell Institute, University of Minnesota, Minneapolis, USA, 321 Church St. SE, 6-160 Jackson Hall, Minneapolis, MN 55455.
Ikuro Maruyama, Department of Systems Biology in Thromboregulation, Kagoshima University Graduate School of Medical and Dental Science, Kagoshima 8908544, Japan.
Martin Lotz, Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, CA USA, 10550 N. Torrey Pines Road, MEM 161, La Jolla, CA 92037.
References
- [1].Stros M HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010; 1799:101–13. [DOI] [PubMed] [Google Scholar]
- [2].Bustin M Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999; 19:5237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Catena R, Escoffier E, Caron C, Khochbin S, Martianov I, Davidson I. HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids. Biol Reprod. 2009; 80:358–66. [DOI] [PubMed] [Google Scholar]
- [4].Pauken CM, Nagle DL, Bucan M, Lo CW. Molecular cloning, expression analysis, and chromosomal localization of mouse Hmg1-containing sequences. Mamm Genome. 1994; 5:91–9. [DOI] [PubMed] [Google Scholar]
- [5].Ronfani L, Ferraguti M, Croci L, et al. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development. 2001; 128:1265–73. [DOI] [PubMed] [Google Scholar]
- [6].Vaccari T, Beltrame M, Ferrari S, Bianchi ME. Hmg4, a new member of the Hmg1/2gene family. Genomics. 1998; 49:247–52. [DOI] [PubMed] [Google Scholar]
- [7].Abraham AB, Bronstein R, Chen EI, et al. Members of the high mobility group B protein family are dynamically expressed in embryonic neural stem cells. Proteome Sci. 2013; 11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007; 13:103–14. [DOI] [PubMed] [Google Scholar]
- [9].Mosevitsky MI, Novitskaya VA, Iogannsen MG, Zabezhinsky MA. Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem. 1989; 185:303–10. [DOI] [PubMed] [Google Scholar]
- [10].Muller S, Ronfani L, Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med. 2004; 255:332–43. [DOI] [PubMed] [Google Scholar]
- [11].Nemeth MJ, Curtis DJ, Kirby MR, et al. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003; 102:1298–306. [DOI] [PubMed] [Google Scholar]
- [12].Rouhiainen A, Zhao X, Vanttola P, et al. HMGB4 is expressed by neuronal cells and affects the expression of genes involved in neural differentiation. Sci Rep. 2016; 6:32960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005; 15:496–506. [DOI] [PubMed] [Google Scholar]
- [14].Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007; 64:2590–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pil PM, Lippard SJ. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992; 256:234–7. [DOI] [PubMed] [Google Scholar]
- [16].Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993; 7:1521–34. [DOI] [PubMed] [Google Scholar]
- [17].Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003; 13:170–8. [DOI] [PubMed] [Google Scholar]
- [18].Boonyaratanakornkit V, Melvin V, Prendergast P, et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998; 18:4471–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi ME, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol Cell Biol. 1999; 19:6532–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Calogero S, Grassi F, Aguzzi A, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999; 22:276–80. [DOI] [PubMed] [Google Scholar]
- [21].Nemeth MJ, Cline AP, Anderson SM, Garrett-Beal LJ, Bodine DM. Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors. Blood. 2005; 105:627–34. [DOI] [PubMed] [Google Scholar]
- [22].Taniguchi N, Yoshida K, Ito T, et al. Stage-specific secretion of HMGB1 in cartilage regulates endochondral ossification. Mol Cell Biol. 2007; 27:5650–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang H, Bloom O, Zhang M, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999; 285:248–51. [DOI] [PubMed] [Google Scholar]
- [24].Antoine DJ, Harris HE, Andersson U, Tracey KJ, Bianchi ME. A systematic nomenclature for the redox states of high mobility group box (HMGB) proteins. Mol Med. 2014; 20:135–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Venereau E, Ceriotti C, Bianchi ME. DAMPs from Cell Death to New Life. Front Immunol. 2015; 6:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Parkkinen J, Raulo E, Merenmies J, et al. Amphoterin, the 30-kDa protein in a family of HMG1-type polypeptides. Enhanced expression in transformed cells, leading edge localization, and interactions with plasminogen activation. J Biol Chem. 1993; 268:19726–38. [PubMed] [Google Scholar]
- [27].Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007; 81:1–5. [DOI] [PubMed] [Google Scholar]
- [28].Magna M, Pisetsky DS. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Mol Med. 2014; 20:138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Heinola T, de Grauw JC, Virkki L, et al. Bovine chronic osteoarthritis causes minimal change in synovial fluid. J Comp Pathol. 2013; 148:335–44. [DOI] [PubMed] [Google Scholar]
- [30].Li ZC, Cheng GQ, Hu KZ, et al. Correlation of synovial fluid HMGB-1 levels with radiographic severity of knee osteoarthritis. Clin Invest Med. 2011; 34:E298. [DOI] [PubMed] [Google Scholar]
- [31].Schierbeck H, Pullerits R, Pruunsild C, et al. HMGB1 levels are increased in patients with juvenile idiopathic arthritis, correlate with early onset of disease, and are independent of disease duration. J Rheumatol. 2013; 40:1604–13. [DOI] [PubMed] [Google Scholar]
- [32].Rosenberg AM, Cordeiro DM. Relationship between sex and antibodies to high mobility group proteins 1 and 2 in juvenile idiopathic arthritis. J Rheumatol. 2000; 27:2489–93. [PubMed] [Google Scholar]
- [33].Lundback P, Stridh P, Klevenvall L, et al. Characterization of the Inflammatory Properties of Actively Released HMGB1 in Juvenile Idiopathic Arthritis. Antioxid Redox Signal. 2016; 24:605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hamada T, Torikai M, Kuwazuru A, et al. Extracellular high mobility group box chromosomal protein 1 is a coupling factor for hypoxia and inflammation in arthritis. Arthritis Rheum. 2008; 58:2675–85. [DOI] [PubMed] [Google Scholar]
- [35].Oestreicher JL, Walters IB, Kikuchi T, et al. Molecular classification of psoriasis disease-associated genes through pharmacogenomic expression profiling. Pharmacogenomics J. 2001; 1:272–87. [DOI] [PubMed] [Google Scholar]
- [36].Oktayoglu P, Em S, Tahtasiz M, et al. Elevated serum levels of high mobility group box protein 1 (HMGB1) in patients with ankylosing spondylitis and its association with disease activity and quality of life. Rheumatol Int. 2013; 33:1327–31. [DOI] [PubMed] [Google Scholar]
- [37].Pusterla T, de Marchis F, Palumbo R, Bianchi ME. High mobility group B2 is secreted by myeloid cells and has mitogenic and chemoattractant activities similar to high mobility group B1. Autoimmunity. 2009; 42:308–10. [DOI] [PubMed] [Google Scholar]
- [38].Feldmann M, Maini RN. Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned? Annu Rev Immunol. 2001; 19:163–96. [DOI] [PubMed] [Google Scholar]
- [39].Kim EY, Moudgil KD. Immunomodulation of autoimmune arthritis by pro-inflammatory cytokines. Cytokine. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Taniguchi N, Kawahara K, Yone K, et al. High mobility group box chromosomal protein 1 plays a role in the pathogenesis of rheumatoid arthritis as a novel cytokine. Arthritis Rheum. 2003; 48:971–81. [DOI] [PubMed] [Google Scholar]
- [41].Kokkola R, Sundberg E, Ulfgren AK, et al. High mobility group box chromosomal protein 1: a novel proinflammatory mediator in synovitis. Arthritis Rheum. 2002; 46:2598–603. [DOI] [PubMed] [Google Scholar]
- [42].Dinarello CA, Cannon JG, Wolff SM, et al. Tumor necrosis factor (cachectin) is an endogenous pyrogen and induces production of interleukin 1. J Exp Med. 1986; 163:1433–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Brennan FM, Chantry D, Jackson A, Maini R, Feldmann M. Inhibitory effect of TNF alpha antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet. 1989; 2:244–7. [DOI] [PubMed] [Google Scholar]
- [44].Wang H, Yang H, Czura CJ, Sama AE, Tracey KJ. HMGB1 as a late mediator of lethal systemic inflammation. Am J Respir Crit Care Med. 2001; 164:1768–73. [DOI] [PubMed] [Google Scholar]
- [45].Yang H, Wang H, Tracey KJ. HMG-1 rediscovered as a cytokine. Shock. 2001; 15:247–53. [DOI] [PubMed] [Google Scholar]
- [46].Muller S, Scaffidi P, Degryse B, et al. New EMBO members’ review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 2001; 20:4337–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Prasad S, Thakur MK. Effects of spermine and sodium butyrate on the in vitro phosphorylation of HMG non-histone proteins of the liver of young and old rats. Arch Gerontol Geriatr. 1990; 10:231–8. [DOI] [PubMed] [Google Scholar]
- [48].Thakur MK, Prasad S. ADP-ribosylation of HMG proteins and its modulation by different effectors in the liver of aging rats. Mech Ageing Dev. 1990; 53:91–100. [DOI] [PubMed] [Google Scholar]
- [49].Prasad S, Thakur MK. Differential methylation of HMG proteins by dexamethasone in the liver of aging rats. Aging (Milano). 1991; 3:333–5. [DOI] [PubMed] [Google Scholar]
- [50].Ly DH, Lockhart DJ, Lerner RA, Schultz PG. Mitotic misregulation and human aging. Science. 2000; 287:2486–92. [DOI] [PubMed] [Google Scholar]
- [51].Aird KM, Iwasaki O, Kossenkov AV, et al. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J Cell Biol. 2016; 215:325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hunziker EB, Quinn TM, Hauselmann HJ. Quantitative structural organization of normal adult human articular cartilage. Osteoarthritis Cartilage. 2002; 10:564–72. [DOI] [PubMed] [Google Scholar]
- [53].Meachim G, Collins DH. Cell counts of normal and osteoarthritic articular cartilage in relation to the uptake of sulphate (35SO4) in vitro. Ann Rheum Dis. 1962; 21:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Goldring MB. The role of the chondrocyte in osteoarthritis. Arthritis Rheum. 2000; 43:1916–26. [DOI] [PubMed] [Google Scholar]
- [55].Kuhn K, D’Lima DD, Hashimoto S, Lotz M. Cell death in cartilage. Osteoarthritis Cartilage. 2004; 12:1–16. [DOI] [PubMed] [Google Scholar]
- [56].Abramson SB. Inflammation in osteoarthritis. J Rheumatol Suppl. 2004; 70:70–6. [PubMed] [Google Scholar]
- [57].Hiraoka K, Grogan S, Olee T, Lotz M. Mesenchymal progenitor cells in adult human articular cartilage. Biorheology. 2006; 43:447–54. [PubMed] [Google Scholar]
- [58].Alsalameh S, Amin R, Gemba T, Lotz M. Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum. 2004; 50:1522–32. [DOI] [PubMed] [Google Scholar]
- [59].Dowthwaite GP, Bishop JC, Redman SN, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004; 117:889–97. [DOI] [PubMed] [Google Scholar]
- [60].Taniguchi N, Carames B, Ronfani L, et al. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc Natl Acad Sci U S A. 2009; 106:1181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Amin AR, Islam AB. Genomic analysis and differential expression of HMG and S100A family in human arthritis: upregulated expression of chemokines, IL-8 and nitric oxide by HMGB1. DNA Cell Biol. 2014; 33:550–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Flannery CR, Hughes CE, Schumacher BL, et al. Articular cartilage superficial zone protein (SZP) is homologous to megakaryocyte stimulating factor precursor and Is a multifunctional proteoglycan with potential growth-promoting, cytoprotective, and lubricating properties in cartilage metabolism. Biochem Biophys Res Commun. 1999; 254:535–41. [DOI] [PubMed] [Google Scholar]
- [63].Rhee DK, Marcelino J, Baker M, et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest. 2005; 115:622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Qi ML, Tagawa K, Enokido Y, et al. Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol. 2007; 9:402–14. [DOI] [PubMed] [Google Scholar]
- [65].Brezniceanu ML, Volp K, Bosser S, et al. HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. Faseb J. 2003; 17:1295–7. [DOI] [PubMed] [Google Scholar]
- [66].Glasson SS. In vivo osteoarthritis target validation utilizing genetically-modified mice. Curr Drug Targets. 2007; 8:367–76. [DOI] [PubMed] [Google Scholar]
- [67].Poole AR, Guilak F, Abramson S. Etiopathogenesis of osteoarthritis In: Moskowitz R, ed. Osteoarthritis. Philadelphia: Wolters Kluwer, 2007. [Google Scholar]
- [68].Johnson ML, Kamel MA. The Wnt signaling pathway and bone metabolism. Curr Opin Rheumatol. 2007; 19:376–82. [DOI] [PubMed] [Google Scholar]
- [69].Goldring SR, Goldring MB. Eating bone or adding it: the Wnt pathway decides. Nat Med. 2007; 13:133–4. [DOI] [PubMed] [Google Scholar]
- [70].Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017; 169:985–99. [DOI] [PubMed] [Google Scholar]
- [71].Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005; 8:739–50. [DOI] [PubMed] [Google Scholar]
- [72].Koyama E, Shibukawa Y, Nagayama M, et al. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008; 316:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Loughlin J, Dowling B, Chapman K, et al. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc Natl Acad Sci U S A. 2004; 101:9757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tamamura Y, Otani T, Kanatani N, et al. Developmental regulation of Wnt/beta-catenin signals is required for growth plate assembly, cartilage integrity, and endochondral ossification. J Biol Chem. 2005; 280:19185–95. [DOI] [PubMed] [Google Scholar]
- [75].Zhu M, Chen M, Zuscik M, et al. Inhibition of beta-catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheum. 2008; 58:2053–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Taniguchi N, Carames B, Kawakami Y, Amendt BA, Komiya S, Lotz M. Chromatin protein HMGB2 regulates articular cartilage surface maintenance via beta-catenin pathway. Proc Natl Acad Sci U S A. 2009; 106:16817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Robinson JA, Chatterjee-Kishore M, Yaworsky PJ, et al. Wnt/beta-catenin signaling is a normal physiological response to mechanical loading in bone. J Biol Chem. 2006; 281:31720–8. [DOI] [PubMed] [Google Scholar]
- [78].Akiyama H, Lyons JP, Mori-Akiyama Y, et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004; 18:1072–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Husa M, Liu-Bryan R, Terkeltaub R. Shifting HIFs in osteoarthritis. Nat Med. 2010; 16:641–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kamekura S, Kawasaki Y, Hoshi K, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006; 54:2462–70. [DOI] [PubMed] [Google Scholar]
- [81].Inada M, Yasui T, Nomura S, et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999; 214:279–90. [DOI] [PubMed] [Google Scholar]
- [82].Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999; 80:159–70. [DOI] [PubMed] [Google Scholar]
- [83].Ueta C, Iwamoto M, Kanatani N, et al. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J Cell Biol. 2001; 153:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Stricker S, Fundele R, Vortkamp A, Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev Biol. 2002; 245:95–108. [DOI] [PubMed] [Google Scholar]
- [85].Taniguchi N, Carames B, Hsu E, Cherqui S, Kawakami Y, Lotz M. Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation. J Biol Chem. 2011; 286:41489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dong YF, Soung do Y, Schwarz EM, O’Keefe RJ, Drissi H. Wnt induction of chondrocyte hypertrophy through the Runx2 transcription factor. J Cell Physiol. 2006; 208:77–86. [DOI] [PubMed] [Google Scholar]
- [87].Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000; 127:3141–59. [DOI] [PubMed] [Google Scholar]
- [88].Daumer KM, Tufan AC, Tuan RS. Long-term in vitro analysis of limb cartilage development: involvement of Wnt signaling. J Cell Biochem. 2004; 93:526–41. [DOI] [PubMed] [Google Scholar]
- [89].Drissi H, Luc Q, Shakoori R, et al. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000; 184:341–50. [DOI] [PubMed] [Google Scholar]
- [90].Drissi H, Pouliot A, Stein JL, van Wijnen AJ, Stein GS, Lian JB. Identification of novel protein/DNA interactions within the promoter of the bone-related transcription factor Runx2/Cbfa1. J Cell Biochem. 2002; 86:403–12. [DOI] [PubMed] [Google Scholar]
- [91].Lengner CJ, Hassan MQ, Serra RW, et al. Nkx3.2-mediated repression of Runx2 promotes chondrogenic differentiation. J Biol Chem. 2005; 280:15872–9. [DOI] [PubMed] [Google Scholar]
- [92].Tamiya H, Ikeda T, Jeong JH, et al. Analysis of the Runx2 promoter in osseous and non-osseous cells and identification of HIF2A as a potent transcription activator. Gene. 2008; 416:53–60. [DOI] [PubMed] [Google Scholar]
- [93].Drissi H, Pouliot A, Koolloos C, et al. 1,25-(OH)2-vitamin D3 suppresses the bone-related Runx2/Cbfa1 gene promoter. Exp Cell Res. 2002; 274:323–33. [DOI] [PubMed] [Google Scholar]
- [94].Kahler RA, Westendorf JJ. Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J Biol Chem. 2003; 278:11937–44. [DOI] [PubMed] [Google Scholar]
- [95].Hasegawa A, Yonezawa T, Taniguchi N, et al. Role of Fibulin 3 in Aging-Related Joint Changes and Osteoarthritis Pathogenesis in Human and Mouse Knee Cartilage. Arthritis Rheumatol. 2017; 69:576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Zhang Y, Marmorstein LY. Focus on molecules: fibulin-3 (EFEMP1). Exp Eye Res. 2010; 90:374–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].McLaughlin PJ, Bakall B, Choi J, et al. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum Mol Genet. 2007; 16:3059–70. [DOI] [PubMed] [Google Scholar]
- [98].Ehlermann J, Weber S, Pfisterer P, Schorle H. Cloning, expression and characterization of the murine Efemp1, a gene mutated in Doyne-Honeycomb retinal dystrophy. Gene Expr Patterns. 2003; 3:441–7. [DOI] [PubMed] [Google Scholar]
- [99].Henrotin Y, Gharbi M, Mazzucchelli G, Dubuc JE, De Pauw E, Deberg M. Fibulin 3 peptides Fib3–1 and Fib3–2 are potential biomarkers of osteoarthritis. Arthritis Rheum. 2012; 64:2260–7. [DOI] [PubMed] [Google Scholar]