Mammals evolved testis-specific Mage-a genes to protect the male germline under starvation stress and are co-opted in cancer.
Abstract
Ensuring robust gamete production even in the face of environmental stress is of utmost importance for species survival, especially in mammals that have low reproductive rates. Here, we describe a family of genes called melanoma antigens (MAGEs) that evolved in eutherian mammals and are normally restricted to expression in the testis (http://MAGE.stjude.org) but are often aberrantly activated in cancer. Depletion of Mage-a genes disrupts spermatogonial stem cell maintenance and impairs repopulation efficiency in vivo. Exposure of Mage-a knockout mice to genotoxic stress or long-term starvation that mimics famine in nature causes defects in spermatogenesis, decreased testis weights, diminished sperm production, and reduced fertility. Last, human MAGE-As are activated in many cancers where they promote fuel switching and growth of cells. These results suggest that mammalian-specific MAGE genes have evolved to protect the male germline against environmental stress, ensure reproductive success under non-optimal conditions, and are hijacked by cancer cells.
INTRODUCTION
Mammals have a low reproductive rate with relatively small litters of offspring and long intervals between births compared to other animals (1). Therefore, it is critically important for mammals to ensure robust gametogenesis and produce high-quality gametes. In males, an intricate developmental process called spermatogenesis has evolved to maintain a constant and uninterrupted supply of gametes (sperm) throughout reproductive life (2, 3). Mammalian spermatogenesis is an energetically costly process in which diploid spermatogonia generate haploid spermatozoa through a series of highly coordinated and cyclic events. First, spermatogonial stem cells (SSCs) give rise to extensive numbers of undifferentiated spermatogonia cells. Upon retinoic acid (RA) trigger, undifferentiated spermatogonia undergo differentiation and further rounds of division to give rise to spermatocytes, which enter the meiotic process that results in generation of haploid spermatids. Last, spermatids develop from round cells into highly specialized elongated mature spermatozoa, ultimately able to fertilize the egg (2–4).
Continuous spermatogenesis is dependent on SSCs, which balance self-renewing divisions that maintain the stem cell pool with differentiating divisions that sustain continuous sperm production. A key determinant of mature sperm output is the size of the spermatogonial population and the extent of premeiotic expansion (5). In addition to maintaining spermatogenesis in normal conditions, for long-term survival of the species, gamete production under nonfavorable conditions and spermatogenesis recovery after stress must be prioritized. One of the major environmental factors affecting species survival and reproductive capacity is nutrient availability (6). In addition to famine that drives natural selection to cope with nutritional stress, other environmental stressors, such as carcinogens and heat, can affect spermatogenesis, resulting in decreased sperm output and transgenerationally inherited genetic or epigenetic alterations (7, 8). However, the specific mechanisms that have evolved to protect the male germline under stress are largely unknown, especially in mammals.
As a highly orchestrated developmental process, spermatogenesis requires tightly regulated stage- and cell-specific gene expression that is achieved by unique chromatin remodeling, transcriptional control, and the expression of testis-specific genes or isoforms. It is estimated that approximately 50% of the genome is expressed in testis some time from birth to adulthood. Furthermore, one-third of the mouse genome is differentially expressed in testis during development. Many male germ cell–specific or testis-predominant genes (approximately 2 to 4% of mouse/rat transcriptome) have evolved to coordinate the remarkably complex and robust process of spermatogenesis (9). A large number of these genes, normally restricted to expression in male germ cells, become aberrantly activated in cancer and can have oncogenic properties. Furthermore, these gene products can often induce an immune response in patients with cancer and thus are classified as cancer-testis antigens (CTAs).
Many CTAs have recently evolved and reside on the X chromosome as multicopy gene families, such as the melanoma antigen (MAGE) gene family. Most of the MAGE genes reside on the X chromosome (fig. S1A), which has previously been shown to be enriched for genes expressed in early spermatogenesis (10, 11). MAGE genes encode proteins that share a common MAGE homology domain (fig. S1B) (12). Although only a single MAGE gene exists in lower eukaryotes, the MAGE family rapidly expanded in eutherians and consists of more than 40 genes in humans. Mechanistically, MAGEs assemble with E3 RING ubiquitin ligases to form MAGE-RING ligases and act as regulators of ubiquitination by modulating ligase activity and substrate specification (13, 14). MAGEs have been studied as biomarkers of various malignancies and in the context of cancer immunotherapy (15), but little is known about their normal physiological functions. Here, we show that mammalian-specific MAGE-A genes have evolved to protect the male germline against nutrient and genotoxic stress and have been hijacked in cancer.
RESULTS
MAGEs are mammalian-specific genes that are restricted to expression in defined cell types of the testis
We aimed to comprehensively identify genes that have evolved specifically in eutherian (placental) mammals and are restricted to expression in the testis. We evaluated global gene expression data from the Genotype-Tissue Expression (GTEx) Consortium (16) comprising 43 human tissues and 18,059 transcripts and found 185 highly expressed, testis-specific transcripts (Fig. 1A and table S1). To focus our study on those genes that have recently evolved and may play a role specifically in reproduction of eutherian mammals, we performed ortholog analysis using OrthoDB (17) and found that 26 of the 185 transcripts appear specifically in eutherian mammals (Fig. 1B and table S2). Ten of these genes belonged to the MAGE gene family (Fig. 1B). Although only a single MAGE gene exists in lower eukaryotes, the MAGE family rapidly expanded in eutherians and consists of more than 40 genes in humans. Although MAGEs have been studied in the context of cancer immunotherapy (15), very little is known about their normal physiological functions. To address this question, we systematically characterized the expression patterns of all MAGE genes in a variety of tissues and cell types from mouse and human to ultimately generate a comparative anatomical expression atlas of 38 human and 38 mouse MAGE genes (Fig. 1, C and D, and figs. S1 to S3). Note that some of these genes are not necessarily 1:1 orthologs, with some evolving independently in mouse and human. These results, along with other datasets described herein (totaling approximately 50,000 data points), are presented in a web-searchable resource (http://MAGE.stjude.org). As predicted, many MAGE genes are highly enriched in the testis of both humans and mice (Fig. 1, C and D). To understand when and where MAGE proteins are expressed in the testis, we performed an in-depth analysis of the developing mouse testis in which time-dependent appearance of spermatogonia, spermatocytes, and spermatids during the first wave of spermatogenesis provides a valuable model system (18). Age-dependent patterns of MAGE family expression following initiation of postnatal spermatogenesis revealed premeiotic, meiotic, and postmeiotic germline transcripts (Fig. 1E and fig. S4). Isolation of specific cell types from mouse testes and analysis of KitlSl/KitlSl-d mice, which are defective in spermatogenesis (fig. S4), confirmed that MAGEs are expressed in germ cells during distinct steps of spermatogenesis, from undifferentiated spermatogonia to haploid spermatids (Fig. 1F).
Fig. 1. MAGEs are mammalian-specific genes that are restricted to expression in defined cell types of the testis.
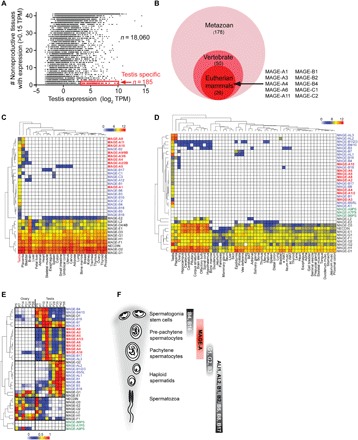
(A) Identification of testis-specific transcripts. (B) Many MAGE genes evolved in eutherian mammals. (C and D) Unsupervised hierarchical clustering of human (C) and mouse (D) MAGE genes based on reverse transcription quantitative polymerase chain reaction (RT-QPCR) expression data. Color key indicates relative log2 expression (0 to 12). (E) Unsupervised hierarchical clustering of MAGE genes during the first wave of spermatogenesis in the mouse testis (P5 to P56) as measured by RT-QPCR. Color key indicates relative expression (0 to 1). (F) Summary of differential expression of MAGE genes during spermatogenesis.
Mage-a genes promote the maintenance of SSCs
To validate the results obtained by reverse transcription quantitative polymerase chain reaction (RT-QPCR) and visualize specific cell types within intact testis tissue, we performed in situ hybridization and immunohistochemistry on sections of mouse and human testis. MAGE-A mRNA and protein are enriched in premeiotic germ cells, including spermatogonia and premeiotic spermatocytes (Fig. 2, A and B, and fig. S5A). Staging of the tubules in the hematoxylin-stained sections revealed that Mage-a expression was the highest in stages VIII to XI, suggesting that RA may induce Mage-a expression (19, 20). Addition of RA to primary mouse spermatogonial cultures induced the expression of Mage-a genes (Fig. 2C and fig. S5B). Consistently, Mage-a protein expression was highest in Stra8 and Kit-positive spermatogonia (fig. S5C). To determine whether expression of Mage-a genes is important in SSCs (2), we used primary cultures of undifferentiated spermatogonia from mice carrying an Id4-eGfp reporter transgene that exhibits high levels of expression in the SSC population (21). We found that partial knockdown of Mage-a genes (Fig. 2D) decreased the percentage of EGFPBright SSCs and increased the percentage of cells in the EGFPDim progenitor pool (Fig. 2, E and F). Small interfering RNA (siRNA) targeting Rb1 was used as a positive control given our previous findings of its importance in SSC maintenance (22). siRNA targeting endogenous ID4 was used as a positive control to increase EGFPBright cells due to compensatory up-regulation of the Id4-eGfp reporter transgene (23). To assess stem cell function in vivo, we depleted Mage-a genes from primary spermatogonia cultures carrying a Rosa26-LacZ transgene before transplantation into the testes of recipient males that have been depleted of germ cells. The efficiency of testis repopulation was analyzed by counting the number of LacZ-positive colonies, which are clonally derived from an individual SSC (24). Knockdown of Mage-a genes in SSCs trended toward reduced efficiency of testis repopulation after transplantation into mice (40% compared to control), although it did not quite reach statistical significance (Fig. 2, G and H). Thus, Mage-a genes are important for maintenance and differentiation of SSCs when manipulated ex vivo.
Fig. 2. Mage-a genes promote the maintenance of SSCs.
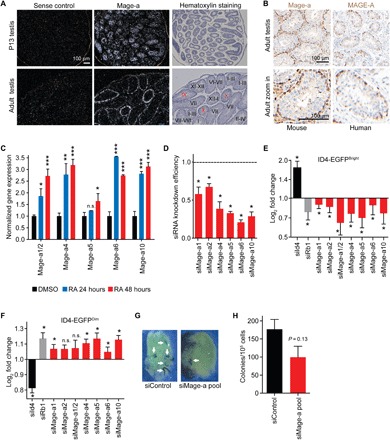
(A and B) In situ hybridization (A) and immunohistochemistry (B) show that Mage-a genes are expressed in early stages of spermatogenesis. Staining was performed using human and mouse anti-MAGE-A antibodies that recognize multiple MAGE-A proteins, including mouse Mage-a1/Mage-a2/Mage-a3/Mage-a5/Mage-a6/Mage-a8. (C) Primary spermatogonial stem cell cultures were treated with dimethyl sulfoxide (DMSO) or RA for 24 or 48 hours before expression of Mage-a genes were detected by RT-QPCR (n = 3). Data are means ± SEM. (D to F) Mage-a genes are required to maintain ID4-EGFPbright stem cells in primary SSC cultures. Knockdown efficiency after a 24-hour transfection is shown (D). Log2 fold change of ID4-EGFPbright (E) and ID4-EGFPdim (F) cells after knockdown of indicated genes (n = 3 biological replicates on a single ID4-EGFP SSC cell line). Data are means ± SD. (G and H) Mage-a genes are required for robust stem cell repopulation of testis (n = 3 technical replicates). Note that siMAGE-A depletes multiple Mage-a genes. Data shown are mean ± SEM. P values determined by Student’s t test, *P < 0.05, **P < 0.01, ***P <0.001. n.s., not significant.
Male germline of Mage-a knockout mice is hypersensitive to genotoxic stress
To test the function of these genes in vivo, we generated Mage-a knockout (KO) mice. Given the high homology and similar expression patterns of the eight Mage-a genes (fig. S6, A and B), we created an allelic series using CRISPR-Cas9 to produce animals with mutation of two Mage-a genes (Δ2), deletion of six Mage-a genes (Δ6), or the combination resulting in loss of all eight Mage-a genes (Δ8; Fig. 3A). Genotyping, immunoblotting, and immunohistochemistry confirmed loss of Mage-a proteins in the testis (Fig. 3, B to D, and fig. S6C). Mage-a Δ2, Δ6, and Δ8 mice were born at the expected Mendelian ratios and appeared phenotypically normal (fig. S6, D to H). Analysis of testes from all genotypes revealed similar weights and histology (Fig. 3, E and I, and fig. S6I). Mating tests, including pregnancy rate, litter size, and interlitter latency, revealed no defect in fertility of male Mage-a Δ2, Δ6, and Δ8 mice (Fig. 3, F and G, and fig. S6J). Thus, under normal laboratory conditions, Mage-a genes are not required for male fertility, which is consistent with their relatively late appearance in evolution. Alternatively, additional Mage genes may have functional redundancy with the eight Mage-a genes deleted. However, we saw no difference in the expression of other Mage genes in the testis of Mage-a KO mice (fig. S6K).
Fig. 3. Male germline of Mage-a KO mice is hypersensitive to genotoxic stress.
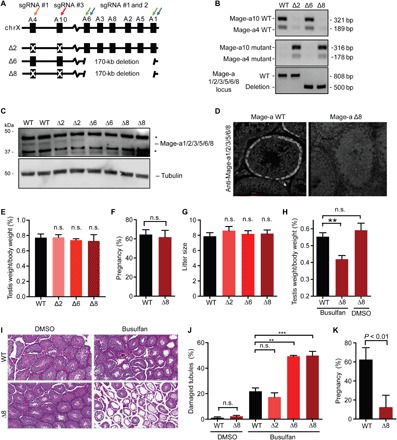
(A) Schematic of allelic series deleting 2, 6, or all 8 mouse Mage-a genes through CRISPR-Cas9 technology. (B to D) Confirmation of Mage-a KO by genotyping (B), immunoblotting of testis lysates (C), and immunostaining of testis (D). WT, wild type. (E to G) Mage-a genes are not required for fertility under basal conditions; testis weights (n ≥ 6), pregnancy rate (n = 6), and litter size (n ≥ 16) are shown. (H to K) Mage-a genes are required to recover (8 weeks) from busulfan (20 mg/kg) genotoxic stress in mice. Testis weights (n = 4) (H), histology (I), and quantification (n > 200 tubules) (J) are shown. Fertility of mice from weeks 8 to 10 after busulfan treatment was determined (n = 4) (K). Data are means ± SEM. P values determined by Student’s t test, **P < 0.01 and ***P <0.001.
To determine whether Mage-a genes are important for spermatogenesis and fertility after challenge, we treated Mage-a wild-type Δ2, Δ6, and Δ8 mice with the genotoxic agent busulfan and waited 8 weeks for mice to recover (25). Consistent with increased germ cell apoptosis after short-term N-ethyl-N-nitrosourea treatment of Mage-a Δ6 mice (26), both Mage-a Δ6 and Δ8 mice had significant defects in recovery after busulfan treatment, resulting in reduced testis weights (Fig. 3H), increased damaged seminiferous tubules (Fig. 3, I and J), and reduced fertility (Fig. 3K). Deletion of Mage-a4 and Mage-a10 (Δ2 mice) had no effect alone and did not exaggerate the phenotype when deleted in Δ8 mice compared to Δ6, suggesting that these genes may have other functions. Analysis of testes from Mage-a Δ6 and Δ8 revealed increased p53 protein levels even in unchallenged mice (fig. S7), which suggests conservation of Mage-a gene function between human and mouse, in both germ and cancer cell types (13, 27). In addition, mutations and copy number variations in MAGE genes, including MAGE-As, have been linked to male infertility in humans (28–30).
Mage-a genes are required for robust spermatogenesis upon long-term starvation of mice
Human MAGE-As have been shown to increase glycolytic and anabolic processes while decreasing catabolic pathways in cancer cells (31, 32). The peak expression of mouse Mage-a genes occurs as germ cells switch carbon sources from blood-derived glucose to Sertoli cell–secreted lactate due to passing through the blood-testis barrier (BTB) (33, 34). In addition, we have previously shown that human MAGE-A3/6 function in metabolic regulation of cancer cells, through ubiquitination and degradation of a key energy sensor, adenosine monophosphate–activated protein kinase (AMPK), and an inhibitor of glycolysis, fructose-1,6-bisphosphatase 1 (FBP1) (31, 32). Thus, Mage-a genes may have specifically evolved to help cope with nutrient stress conditions during this change, especially when nutrients are limited. Therefore, we analyzed whether Mage-a KO mice may be hypersensitive to long-term starvation that mimics famine in nature. We exposed wild-type or Mage-a Δ6 mice to reduced food intake over a course of 100 days that resulted in dramatic weight loss (20%) and decreased serum glucose levels in both wild-type and Mage-a Δ6 mice (Fig. 4, A to C). In comparison to wild-type mice, Mage-a Δ6 mice had reduced testis size upon starvation (Fig. 4D). Histological analysis of the testis from these mice revealed an increase in the number of damaged seminiferous tubules in Mage-a Δ6 mice, compared to littermate control, indicating defects in spermatogenesis (Fig. 4, E and F). Furthermore, Mage-a Δ6 mice had reduced sperm counts upon fasting compared to littermate control mice (Fig. 4G). These findings in combination suggest that Mage-a genes are not required for spermatogenesis and sperm production in mice under normal laboratory conditions but become important upon environmental stress, such as nutrient limitation and genotoxic stress.
Fig. 4. Mage-a genes are required for robust spermatogenesis upon long-term starvation of mice.
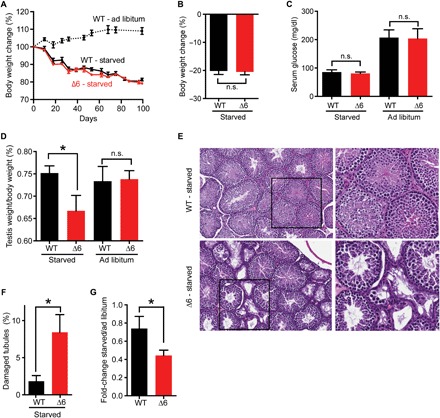
(A and B) Long-term starvation of mice (n = 5 to 6) resulted in a 20% body weight decrease. (C) Serum glucose was measured after starvation of ad libitum feeding of wild-type or Mage-a Δ6 mice (n = 3 to 9). (D to G) Starvation of Mage-a Δ6 mice decreases testis size (n = 7 to 13) (D), increases seminiferous tubule damage (>200 tubules analyzed) (E and F), and decreases sperm count (n = 7 to 12) (G). Data are means ± SEM. P values were determined by Student’s t test, *P < 0.05.
MAGE-As promote resistance to a glycolysis inhibitor in human cancer and mouse spermatogonia stem cells
Consistent with a conserved function of Mage-a genes in normal physiological and disease states, expression of human MAGE-A6 in MIA PaCa-2 cancer cells (Fig. 5A) supported increased growth compared to green fluorescent protein (GFP) control cells upon glycolysis inhibition by 2-deoxyglucose (2-DG; Fig. 5B) but had no effect in the absence of 2-DG (Fig. 5C). This function is conserved in mouse SSCs, as SSCs derived from Mage-a Δ8 mice showed increased sensitivity to 2-DG treatment compared to SSCs from littermate wild-type mice (Fig. 5D). To further understand how MAGE-As function to regulate metabolic pathways, we performed unbiased, nontargeted metabolomic analysis on MIA PaCa-2 cells expressing GFP or MAGE-A6, with or without 2-DG treatment. Principal components analysis revealed clear differences in metabolic profiles of MAGE-A6 cells compared to GFP control (Fig. 5E). Examination of those metabolites whose levels significantly changed in 2-DG–treated MAGE-A6 cells revealed increased levels of several fatty acids (Fig. 5F). Several of these are products of AMPK-regulated lipases (35). Consistent with alteration in fatty acid metabolism being important for MAGE-A6–induced cell growth upon 2-DG treatment, the carnitine palmitoyltransferase inhibitor etomoxir that inhibits fatty acid oxidation (36) reversed MAGE-A6–induced cell growth under 2-DG culture conditions (Fig. 5G) but had no effect in the absence of 2-DG (Fig. 5H). These results suggest that upon glycolysis inhibition, either by 2-DG or naturally in germ cells upon passing through the BTB, MAGE-As facilitate fuel switching to promote fatty acid oxidation and drive cell growth through production of ATP (adenosine triphosphate) and NADPH (reduced form of nicotinamide adenine dinucleotide phosphate) (37). These findings are also highly applicable in cancer cells, as expression analysis of all 185 testis-specific transcripts across 13 cancer types comprising 5532 human tumors in The Cancer Genome Atlas (TCGA) revealed that only 14 testis-specific transcripts had robust and frequent pan-cancer expression, with 8 belonging to the MAGE gene family (Fig. 5I). Thus, the MAGE gene family may have evolved specifically in eutherian mammals to protect the male germline during times of stress, such as famine, and cancer cells frequently hijack these genes to promote robust cell growth in the face of metabolic stress.
Fig. 5. MAGE-A genes promote resistance to a glycolysis inhibitor in human cancer and mouse spermatogonia stem cells.
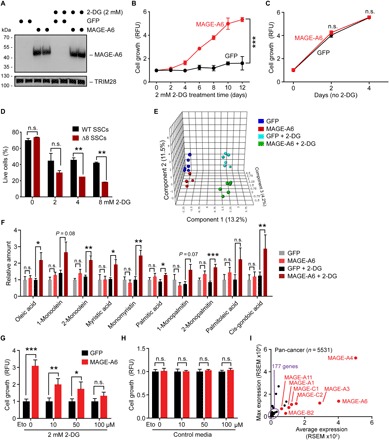
(A) Expression of human MAGE-A6 in MIA PaCa-2 cancer cells in complete media or treated with 2 mM 2-DG for 4 days. (B and C) Human MAGE-A6 expression promotes sustained growth of MIA PaCa-2 cancer cells treated with 2 mM 2-DG glycolysis inhibitor (B), but not without (C). (D) Mage-a Δ8 SSCs are more sensitive to 2-DG than littermate control wild-type SSCs. Cells were treated with indicated doses of 2-DG for 16 hours in the absence of feeder cells before the percentage of live cells was determined by annexin-V/4′,6-diamidino-2-phenylindole staining and flow cytometry analysis. (E and F) Metabolomic analysis of MAGE-A6 expressing MIA PaCa-2 cells grown in standard media or 2 mM 2-DG for 2 to 10 days. Nontargeted metabolomics was performed by the National Institutes of Health (NIH) West Coast Metabolomics Core (n = 6). Principal components analysis (E) and relative quantities of the indicated fatty acids (F) are shown. (G and H) The carnitine palmitoyltransferase inhibitor etomoxir (Eto) reverses MAGE-A6–induced growth in the presence of 2-DG after 7 days (G) but has no effect in cells grown in complete medium without 2-DG (H). (I) Pan-cancer gene expression analysis reveals that testis-specific MAGE genes are frequently expressed in tumors (n = 5532). Data are means ± SEM from n = 3 independent experiments. P values determined by Student’s t test, *P < 0.05, **P < 0.01, ***P <0.001.
DISCUSSION
A critical step in the evolution of mammals was trading reproductive capacity for brain size. Thus, protection of germ cells in times of stress is of utmost importance for mammals. However, the specific mechanisms that have evolved to do so are not understood. Here, we have identified testis-restricted genes that evolved specifically in placental mammals, including the MAGE family of E3 ubiquitin ligase regulators (Fig. 1) (12). Approximately 40 MAGE genes have been identified in each of the human and mouse genomes, two-thirds of which are classified as type I and the remainder are classified as type II (fig. S1, A to C). Type I MAGEs (MAGE-A, MAGE-B, and MAGE-C) were initially discovered as CTAs, whereas type II MAGEs are not typically associated with cancer but are implicated in several genetic disorders (12).
Since their discovery, MAGEs have garnered a lot of interest as targets for cancer immunotherapy, but their physiological functions remained understudied. Here, we systematically examined the specific location and timing of expression for the entire MAGE gene family in human and mouse. This large searchable database is publicly accessible at http://MAGE.stjude.org. We found that MAGE genes share similar expression patterns in both species (Fig. 1, B and C, and figs. S1 to S3), with type I genes being restricted or highly enriched in the testis, whereas type II MAGE genes are broadly expressed and enriched in the brain. Consistent with these findings, previous studies have noted that eutherian-specific genes are often enriched for expression in brain and testis (38).
To understand the function of type I MAGEs in the testis, we determined where and when each MAGE gene is expressed. We found that the majority of testis-restricted or enriched type I MAGEs are expressed in germ cells, in contrast to type II genes that are in somatic cells of the testis (Fig. 1, E and F, and fig. S4). However, different subsets of MAGE genes are expressed at distinct stages of male germ cell differentiation, including undifferentiated and differentiating spermatogonia, spermatocytes, and haploid spermatids. Given their molecular function in ubiquitination, our data suggest that MAGEs may work as molecular switches to fine-tune the transitions through the spermatogenesis pathway.
A set of MAGEs that particularly drew our attention were the Mage-a genes (Fig. 1F) that are expressed from undifferentiated spermatogonia to prepachytene spermatocytes. These cell populations are the most critical for ensuring robust and adequate sperm production. In the mouse genome, there are eight protein-coding Mage-a genes, Mage-a1, Mage-a2, Mage-a3, Mage-a4, Mage-a5, Mage-a6, Mage-a8, and Mage-a10. Mage-a1, Mage-a2, Mage-a3, Mage-a5, Mage-a6, and Mage-a8 proteins are more than 90% homologous (fig. S6A), and genes encoding these proteins are located in tandem on the X chromosome without intervening genes (fig. S1). Mage-a4 and Mage-a10 are more distinct members of the Mage-a subfamily and are located on the other arm of the X chromosome. Consistent with potentially redundant functions, we found that all eight Mage-a genes share similar expression profiles (Figs. 1, 2, A to C, and 3, and fig. S4), with the peak of expression just before germ cells cross the BTB in prepachytene spermatocytes.
Spermatogenesis is highly sensitive to systemic genotoxic stress, such as that often associated with chemotherapy. Chemotherapy induces massive death of spermatogonia and spermatocytes, causing near-total germ cell depletion. This leads to an interval of azoospermia and infertility, followed by a gradual recovery of spermatogenesis and restoration of fertility by the remaining stem cells (25). Our results suggest that Mage-a1, Mage-a2, Mage-a3, Mage-a5, Mage-a6, and Mage-a8 are critical for protection of the male germline against genotoxic stress. The precise mechanism(s) by which these Mage-a proteins protect against genotoxic stress is (are) not completely known. We and others have shown that Mage-a KO mice have increased p53 protein levels basally (fig. S7) or upon genotoxic stress (26). Thus, increased p53 protein levels in germ cells of Mage-a KO mice may sensitize these cells to genotoxic stress. These findings have potential clinical implications as polymorphisms in human MAGE-A genes might represent previously unknown markers for identifying patients (both adult and children) who are at increased risk of impaired spermatogenesis after chemotherapy.
In cancer cells, MAGE-A-TRIM28 E3 ubiquitin ligase complexes act as oncogenes by inhibiting critical cellular stress responses and metabolic pathways through ubiquitination and degradation of p53, AMPK, and FBP1 (13, 31, 32). AMPK is a major energy sensor that is activated under low cellular energy conditions and acts to restore metabolic homeostasis by down-regulating anabolic biosynthetic processes, such as mammalian target of rapamycin (mTOR) signaling and protein and lipid biosynthesis, while up-regulating catabolic ATP-restoring processes (39). We have found that in the testis, the expression of Mage-a genes coincides with the metabolic adaptation of developing male germ cells. Mage-a genes are expressed in spermatogonia, and their expression is increased upon RA-stimulated differentiation (Figs. 1, E and F, and 2, A and C, and figs. S4, S5, and S6B). Our results suggest that Mage-a genes are specifically expressed at this stage to alter metabolic programs through AMPK inhibition during this critical stage of spermatogenesis. For example, spermatogonial expansion requires the active synthesis of nucleic acids, proteins, and lipids, all of which are switched off by AMPK activation (3, 33, 39). Furthermore, RA-induced differentiation of spermatogonia requires active mTOR signaling, which is also inhibited by AMPK (40, 41). Male germ cells also undergo metabolic switching from glycolysis in spermatogonia that use glucose from the blood (34) to oxidative phosphorylation in spermatocytes that are completely dependent on lactate/pyruvate provided by Sertoli cells after they cross the BTB (42). Our data suggest that Mage-a genes help differentiating germ cells to cope with metabolic stress and prevent overactivation of AMPK, particularly in nutrient-poor conditions. In addition, repression of the glycolysis inhibitor FBP1 by Mage-a may maximize glycolysis and ATP output immediately before cells transverse the BTB and enter a new metabolic environment no longer supported by glycolysis. We propose that Mage-a expression turns on to maximize ATP stores (FBP1 degradation) before energy crisis and silence the “alarm” (AMPK) to allow cells time to adapt to their new carbon source.
Spermatogenesis in adult males of most species is remarkably resistant to nutritional stress, suggesting that protective mechanisms evolved that optimize spermatogenesis under fluctuating metabolic conditions. Our data suggest that the evolution of Mage-a genes might represent one such adaptation. Although Mage-a KO mice do not show obvious spermatogenesis defects in normal well-fed laboratory conditions, long-term fasting increased testicular damage and compromised spermatogenesis (Fig. 4). Similar to germ cells, cancer cells also often encounter metabolic stress when they outstrip their local nutrient supply (37). We found that MAGE-A genes are frequently expressed in cancer and that MAGE-A6 confers resistance to metabolic stress induced by the glycolysis inhibitor 2-DG in MIA PaCa-2 cells (Fig. 5). Conversely, spermatogonial cells isolated from Mage-a KO testes are more susceptible to 2-DG, further confirming the role of Mage-a proteins in preventing the detrimental effects of metabolic stress. These effects may in part be due to alterations in fatty acid metabolism, an AMPK-regulated process, as inhibition of fatty acid oxidation reversed the growth phenotype of MAGE-A6 in 2-DG–treated MIA PaCa-2 cells (Fig. 5).
In summary, our work demonstrates that the MAGE gene family evolved specifically in eutherian mammals to protect the male germline during times of stress, such as famine, and cancer cells frequently hijack these genes to promote robust cell growth in the face of metabolic stress.
MATERIALS AND METHODS
Identification of testis-restricted transcripts and aberrant expression in tumors
To identify testis-restricted transcript, we downloaded preanalyzed global gene expression data from the GTEx Consortium (16). Normalized expression levels [transcripts per million reads (TPM)] of 18,059 transcripts in all nonreproductive tissues (42) and testis were analyzed. To stringently exclude transcripts with expression in nonreproductive tissues, we used a 0.5-TPM expression value as a cutoff. The number of nonreproductive tissues with >0.5 TPM expression for each transcript was plotted against the log2 TPM expression in testis. Transcripts with robust expression in testis (>10 TPM) and <0.5 TPM in all other tissue were considered testis-restricted. One hundred eighty-five transcripts were identified by these filtering criteria. Note that the number of testis-specific genes is likely higher due to the GTEx data analysis that discards multimapping reads that could prevent discovery of genes from multicopy gene families. To identify which of the 185 transcripts are eutherian specific, we used OrthoDB (17) to manually curate those without orthologs outside of eutherian mammals.
To determine which of the 185 testis-restricted genes are aberrantly expressed in cancer (CTAs), we queried TCGA using cBioPortal (43) for expression in breast invasive carcinoma (n = 1105), cervical squamous cell carcinoma and endocervical adenocarcinoma (n = 306), colon adenocarcinoma (n = 382), esophageal carcinoma (n = 185), head and neck squamous cell carcinoma (n = 522), liver hepatocellular carcinoma (n = 374), lung adenocarcinoma (n = 523), lung squamous cell carcinoma (n = 501), ovarian serous cystadenocarcinoma (n = 307), sarcoma (n = 263), skin cutaneous melanoma (n = 472), stomach adenocarcinoma (n = 415), and uterine corpus endometrial carcinoma (n = 177) datasets. The average and maximal RNA-Seq by Expectation Maximization (RSEM) expression value were calculated across all 13 tumor types (n = 5532).
Animals and tissue collection
Human tissues were obtained from commercially available sources—Clontech Laboratories (bone marrow and fetal liver) and Agilent Technologies (adrenal, uterus, umbilical cord, and breast)—and remaining tissues were obtained from Ambion FirstChoice Human Total RNA Survey Panel. All procedures and use of mice were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital. For adult mouse expression profiling, C57BL/6 and BALB/c mice were purchased from the University of Texas (UT) Southwestern mouse breeding core facility, which sources its colony breeders from the Jackson Laboratory. All mice were maintained on 2916 Global Diet (Harlan Teklad). Male and female mice (n = 6) at 7 to 8 weeks of age were euthanized at lights on by isoflurane inhalation. Mice were exsanguinated via vena cava, and tissues were collected and snap-frozen in liquid nitrogen. Except for female-reproductive tissues, all tissues were from male mice. Tissues were isolated from appropriate anatomical locations according to established laboratory methods as previously described with slight modifications. Briefly, brown adipose tissue was collected from the dorsal interscapular region and surrounding white adipose or connective tissue was removed. White adipose tissue was collected from the inguinal (referred to as subcutaneous white adipose) and epididymal (referred to as epididymal white adipose) area. Skeletal muscle (quadriceps) was isolated from both femurs, and the remaining bone was flushed with RNAlater (Life Technologies) to remove and collect bone marrow. The enteric tract, including duodenum, jejunum, ileum, and proximal and distal colon, was sectioned and flushed with phosphate-buffered saline (PBS). For duodenum, jejunum, and ileum, the intestinal mucosa was scraped and collected separately (referred to as duodenum, jejunum, and ileum) from the remaining tissue (referred to as duodenum, jejunum, and ileum musculature). The brain was sectioned into pituitary, hypothalamus, hippocampus, brainstem, cerebrum, cerebellum, olfactory bulb, and corpus striatum, which was the remaining brain tissue after the other sections had been removed. Eyes were sectioned into eye lens and retina (remaining tissue after eye lens removal). Both uterine horns were collected. Mammary gland was collected from a lactating mouse 10 days after birth. Pancreas was collected directly into the RNA Stat-60 reagent and prepared immediately because of its high ribonuclease (RNase) content. The rest of the snap-frozen tissues were stored at −80°C until RNA extraction.
For embryonic tissue panel, Institute of Cancer Research mouse embryos were taken at embryonic day 15 (E15), E16, E17, E18, and E19. Placenta and fetal organs were collected (heart, lung, brain, adrenal, liver, kidney, colon, and gonads). Except for ovary, all other organs were from male embryos. Embryonic stem cells were from 129/SvEvTac mice and prepared by the Transgenic Technology Center at UT Southwestern Medical Center.
To study transcript profiles during spermatogenesis, testes were collected from C57BL/6 mice at P5, P7, P10, P14, P21, P28, P35, P42, and P56. Populations of cells highly enriched for specific spermatogenic cell types (>80% by microscopic evaluation) and Sertoli cells (>30%) were prepared from C57BL/6 mice using the STA-PUT method based on sedimentation velocity at unit gravity (44). Type A spermatogonia and Sertoli cells were prepared from mice at P6; spermatogonia A and B, at P9; premeiotic spermatocytes, at P12; meiotic pachytene spermatocytes, at P21; and postmeiotic round spermatids, at P35. Fractions with expected cell type and high purity were pooled and used for RNA extraction. Genes known to be expressed in particular cell types were used as positive-control markers for the fractionation technique (PLZF for spermatogonia, DAZL for pre-meiotic spermatocytes, SYCP3 for pachytene spermatocytes, ACRV1 and PRM1 for post-meiotic round spermatids, and RHOX5 for Sertoli cells). To confirm germ cell–specific expression, we used testes from KitlSl/KitlSl-d (steel) mice that lack most differentiated germ cells (stock no. 100401, the Jackson Laboratory).
RNA preparation and cDNA synthesis
RNA was extracted using the RNA Stat-60 reagent (Tel-Test) according to the manufacturer’s directions. Skeletal muscle, bone, and skin were crushed into a powder using a Bessmann pulverizer (Fisher Scientific). To minimize organ lobe-to-lobe/part-to-part differences in mRNA expression, whole liver, kidney, and testis were crushed and the resulting powder was homogenized. Total RNA was pooled in equal quantities for each tissue (n = 6). Genomic DNA contamination was eliminated by deoxyribonuclease (DNase) I (Roche) treatment in 4.5 mM MgCl2. cDNA for RT-QPCR assays was prepared from 4 μg of DNased RNA using a High-Capacity cDNA Reverse Transcription kit (Life Technologies) in 100 μl final volume. Following cDNA synthesis, RNase-free water was added to increase the sample volume to 300 μl.
Primer design and RT-QPCR analysis
Gene expression levels were measured by RT-QPCR in triplicate wells of a 384-well reaction plate with 10 ng of cDNA per well on an Applied Biosystems 7900HT with SYBR Green chemistry. Control reactions were performed without RT (−RT) to ensure that amplifications were from RNA and not contaminating genomic DNA. Specific primers were designed for every gene, except for human MAGE-A2/A2B, A3/A6, A9/A9B, and D4/D4B and mouse MAGE-B1/B2/B3, B5/5L, and B4/B10. In these cases, genes are too similar for specific primers and thus primers that detect both genes were used. Primers were designed using Roche Universal ProbeLibrary Assay Design Center and used at 150 nM concentration. Primers are listed in table S3. For mouse MAGE-A3, a TaqMan predesigned assay was used (MM00850994_SH). For primer validation and efficiency determination, standard cDNA generated from mouse/human testis RNA was used. PCR efficiencies were calculated from the slope of the resulting standard curves. RT-QPCR data were analyzed by ABI instrument software SDS2.1. Baseline values of amplification plots were set automatically, and threshold values were kept constant to obtain normalized cycle time and linear regression data. Normalized mRNA levels are expressed as arbitrary units (AU) and were obtained by dividing the averaged, efficiency-corrected values for mRNA expression by that for 18S ribosomal RNA and multiplying the quotient by 1 × 105 for scaling purposes. Expression levels were defined as absent if the cycle time value was more than 34, low if the level was below 0.02 AU, medium if the level was between 0.02 and 0.2 AU, high if the level was between 0.2 and 1 AU, and very high when value was equal to or greater than 1 AU.
Heatmap and hierarchical clustering
The heatmap figures were generated using the statistical analysis software R along with the following R packages: ggplot2 ver.1.0.0, reshape ver.1.4, and NMF ver.0.20.5. Hierarchical clustering of the genes and tissues was done by first computing the similarity distances between expression values using the standard Euclidean method where the distance between two vectors is calculated as the square of the difference between the two vectors. The cluster linkage was then computed using the complete linkage method where individual clusters are sequentially combined into larger clusters. For human and mouse adult and embryonic datasets, gene expression values were log-scaled for better visual representation. Other datasets were scaled by gene where the sum of expression values for each gene was set to one. The statistical software package R was also used to generate the correlation plots. Correlation coefficients were computed using the Pearson correlation method, and the figures were generated using the standard plot function in R.
In situ hybridization
The fragment of Mage-a (recognizing Mage-a1, Mage-a2, Mage-a3, Mage-a4, Mage-a5, Mage-a6, and Mage-a8) was amplified using PCR primers 5′-CAGGATCCCCATCAAAGGATCAGGGGGC-3′ and 5′-CACTCGAGAGATGAGCTTCCTGGGGTCT-3′ and subcloned into pBluescript II KS (+/−) vector (Stratagene). S35-labeled antisense probes were prepared after linearization with Not I and transcription with T3 polymerase by in vitro transcription using the Maxiscript kit (Ambion). Sense probes were prepared after linearization of the plasmid with Bam HI and transcribed with T7 polymerase. Radioisotopic in situ hybridization was performed on paraffin-embedded sections of P13 and adult C57BL/6 mouse testis. Autoradiographic exposure ranged from 14 to 28 days at 4°C. Epithelial stages were identified by evaluation of hematoxylin-stained serial sections as previously described (45).
Mage-a antibody generation
Glutathione S-transferase–Mage-a3 (full length) was produced in BL21(DE3) cells by overnight induction at 16ºC with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Protein was purified from bacterial lysates with glutathione Sepharose (GE Amersham) and eluted with 10 mM glutathione. Purified recombinant protein was sent to Cocalico Biologicals Inc. for polyclonal antibody production. Crude antibody serum was affinity-purified against full-length recombinant Mage-a3. Antibody is predicted to recognize Mage-a1/Mage-a2/Mage-a3/Mage-a5/Mage-a6/Mage-a8 proteins but not Mage-a4 and Mage-a10.
Immunoblotting
Total testis lysates were prepared by homogenizing frozen testis tissue in radioimmunoprecipitation assay buffer [50 mM tris, 150 mM NaCl, 0.1% SDS (w/v), 0.5% sodium deoxycholate (w/v), 1% Nonidet P-40 (v/v), and protease inhibitor cocktail (Roche)]. Protein concentration in lysates was determined by bicinchoninic acid assay (Pierce); 30 μg of protein was resolved by SDS–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Bio-Rad). The protein blots were incubated with anti–Mage-a (1:1000) and anti-p53 (1:1000, Sc-126) antibodies. REVERT Total Protein Stain (LI-COR) of membranes was used to ensure equal loading.
Primary cultures of spermatogonia and RA treatment
Primary cultures of undifferentiated spermatogonia were generated from the double-transgenic Id4-eGFP;Rosa26-LacZ hybrid mice that express the LacZ transgene in all germ cells but express the enhanced GFP (EGFP) transgene only in ID4+ SSCs (21, 34). Primary cultures were established from the magnetic-activated cell–sorted THY1+ fraction of testis homogenates of P6 to P8 mice (46). Spermatogonial cultures were maintained on mitotically inactivated Sandos Inbred Mice (SIM) mouse embryo–derived thioguanine- and ouabain-resistant feeder monolayers (STOs) in mouse serum-free medium (mSFM), devoid of fatty acids and supplemented with the growth factors GDNF (glial cell line–derived neurotrophic factor; 20 ng/ml; PeproTech, NJ, USA) and FGF2 (1 ng/ml; PeproTech). Cultures were kept under glycolysis-optimized conditions in humidified incubators at 37°C, 10% O2, and 5% CO2 in air (34). Culture medium was replaced every second day, and passaging onto fresh feeders was performed every 6 to 8 days. For treatment of cultured spermatogonia with all-trans RA (0.5 μM) or dimethyl sulfoxide (DMSO; vehicle), spermatogonial clumps were separated from feeders by gentle pipetting, single-cell suspension was generated by trypsin-EDTA digestion, and cells were plated on STOs. After 2 to 3 days in culture, spermatogonial cells were exposed to all-trans RA and 24 to 48 hours later collected from feeders for RNA preparation and gene expression analysis.
siRNA transfection cultures, flow cytometry, and transplantation analyses
Cultured spermatogonial clumps were separated from feeders by gentle pipetting, single-cell suspension was generated by trypsin-EDTA digestion, and 1 × 105 or 2 × 104 cells were plated in 24- or 96-well plates, respectively, without STO feeder cells in mSFM with GDNF and FGF2. Cells were transfected with either nontargeting control (D-001810-10-05, Dharmacon) or Mage-specific predesigned siRNA oligonucleotides (Sigma, Table S3) by Lipofectamine 3000 (Invitrogen) as follows: 2 μl of Lipofectamine 3000 in 100 μl of OptiMem and 75 pmol siRNA in 100 μl of OptiMem per 1 × 105 cells. When more than one siRNA were available to target the same gene, a pool of two or three siRNAs was used. Sixteen hours after transfection, cells were washed with Hanks’ balanced salt solution and fresh mSFM with GDNF and FGF2 was added. To assess GFP expression level, cells were analyzed by flow cytometry 6 days after transfection. Single-cell suspensions were generated by trypsin-EDTA digestion as described previously and analyzed using an Attune NxT Flow Cytometer (Thermo Scientific, MA, USA). Identification and gating of the ID4-eGFPbright and ID4-eGFPdim populations from cultures were done as described previously (21). To compare the regenerative capacity of spermatogonial culture populations after siRNA transfection, transplantation analysis was performed (46). Six days after siRNA transfection, spermatogonial clumps were separated from feeders by gentle pipetting, single-cell suspension was generated by trypsin-EDTA digestion, and cells were suspended in mouse serum-free medium at 1 × 106 cells/ml. Ten microliters (10,000 cells) was microinjected into each recipient testis. Recipient testes were evaluated for colonies of donor-derived spermatogenesis 2 months later.
Generation of Mage-a KO mouse models
Mouse lines were generated by injecting sgRNAs and Cas9 protein in the pronucleus and cytoplasm of C57BL/6 zygote (see Fig. 3A). Single-guide RNA (sgRNA) was prepared by in vitro transcription (MEGAshortscript T7 Kit, Ambion) and purified by the MEGAclear Transcription Clean-Up Kit (Ambion). Δ6 Mage-a KO mice were generated by injecting gRNA #1 (tgaggactctggggaggact) and #2 (catctctgaggagtttgtgc). The progeny was screened for deletion of ~170 kb (encompassing Mage-a1, Mage-a2, Mage-a3, Mage-a5, Mage-a6, and Mage-a8) by PCR (aDEL-F: gtcataatggctgactcccgt, aDEL-R: accacccagctctcaaataagga). Δ2 Mage-a KO mouse (indel in Mage-a4 and Mage-a10) was generated by targeting Mage-a4 (gRNA #3: tgaggactctggggaggact) and Mage-a10 (gRNA #4: tgcctagaccccggaagcgt). The progeny was screened for frameshift mutations by Cel-1 assay and Sanger sequencing of the PCR products [a10-F: catgcacacttctggcaac and a10-R: cacagccctcctctgagttg, 470 base pairs (bp); a4-F: tcttcctttccccaccaggtg and A4-R: ttgccaattcatataac, 394 bp]. To generate Δ8 Mage-a KO mice, we crossed a Δ2 male with a Δ6 female, screened the female progeny for double mutation Δ2/Δ6, and mated the Δ2/Δ6 heterozygous females with a wild-type male. The progeny were screened for the presence of both mutations, Δ2 and Δ6, which was a result of crossing over between the two X chromosomes and referred to as Δ8. To generate littermate males for experiments, Δ8 heterozygous females were mated with a wild-type male.
Animals were genotyped as follows: Tail snips (1 to 2 mm) were collected at weaning (~21 days old) and again when animals were euthanized for organ collection. Genomic DNA (gDNA) was prepared by incubating tails in 200 μl of 50 mM NaOH at 95°C for 35 min, allowed to cool for 3 min, and neutralized by the addition of 20 μl of 1 M tris (pH 8.0). PCR was performed using the KAPA2G Robust HotStart PCR kit (no. KK5518, KAPA Biosystems), following the manufacturer’s protocol. Buffer A, enhancer, and 1 μl of gDNA were used. Primers used were as follows: for Mage-a1/Mage-a2/Mage-a3/Mage-a5/Mage-a6/Mage-a8 wild-type allele, forward: AGTTGTGAGTGGAGCTCTATGG; reverse: CTATCCAGGACACAGCCACAGAAT; annealing temperature, 65°C; product size, 808 bp; for Mage-a1/Mage-a2/Mage-a3/Mage-a5/Mage-a6/Mage-a8 mutant allele, forward: GTCATAATGGCTGACTCCCGT; reverse: ACCACCCAGCTCTCAAATAAGGA; annealing temperature, 65°C; product size, 500 bp; for Mage-a4 wild-type, forward: ACAGGAAAGTCACAATGGAAAA; reverse: CTCTTTGAGGACTCTGGGGA; annealing temperature, 60°C; product size, 189 bp; for Mage-a4 mutant, forward: ACAGGAAAGTCACAATGGAAAA; reverse: CTCTTTGAGGACTCTGGTAT; annealing temperature, 60°C; product size, 178 bp; for Mage-a10 wild type, forward: CATGCACACTTCTGGCAAC; reverse: TCAACCATACATCTCCTACG; annealing temperature, 60°C; product size, 321 bp; and for Mage-a10 mutant, forward: CATGCACACTTCTGGCAAC; reverse: TCAACCATACATCTCCTTCC; annealing temperature, 60°C; product size, 316 bp.
Tissue weights and fertility evaluation
To assess the effect of Mage-a depletion on organ weights, wild-type and Δ2/Δ6/Δ8 KO mice were euthanized and organ weights were measured immediately after dissection. To test the fertility in males, long-term and short-term mating tests were performed. For long-term tests, wild-type and KO males were housed individually together with a wild-type female mouse for 5 months, and the number of pregnancies, number of pups, and the interval between births were recorded for each mating pair. For short-term tests, each male was set with one female at the time until plugged and then the female was removed from the male cage and the successful pregnancies and number of pups born were recorded for each mating pair. When several sequential matings were set, a new female was placed in a male cage after the previous female was plugged and removed. In all tests, four to seven males per genotype were used.
Busulfan recovery
To evaluate spermatogenesis recovery after partial germ cell depletion, 5-month-old wild-type and Δ2, Δ6, and Δ8 Mage-a KO mice were intraperitoneally injected with busulfan (20 mg/kg; Sigma) dissolved in 1:1 volume ratio of DMSO and water. Eight weeks after treatment, mice were euthanized; testes were weighed, fixed in 4% paraformaldehyde (PFA) for 24 hours, and paraffin-embedded; and hematoxylin and eosin (H&E)–stained sections were analyzed for percentage of damaged tubules. To evaluate fertility after busulfan treatment, 8 weeks after treatment, four wild-type and four Δ8 mice were subjected to four sequential short-term mating tests as described above.
Long-term nutrient deprivation (starvation) and spermatogenesis
To assess the role of Mage-a genes in preserving male fertility during long-term nutrient deprivation, we subjected wild-type and KO mice to a ~70% calorie restriction feeding paradigm, referred to as starvation. Wild-type and KO mice were housed individually, and after 1 week of acclimatization, food intake was determined. Mice were then fed ad libitum (control) or offered daily ~70% of food intake in a petri dish. To obtain and maintain similar body weight decrease (~20%) throughout the starving period, body weights were measured weekly and the amount of food offered was individually corrected accordingly. To prevent dehydration, an additional water bottle was offered. Mice were kept under starvation conditions for 80 to 100 days. Mice were then euthanized; testes were weighed, fixed in 4% PFA for 24 hours, and paraffin-embedded; and H&E-stained sections were analyzed for percentage of damaged tubules. Sperm was prepared from cauda epididymis. Cauda epididymal sperm were allowed to swim out and incubated for 15 min at 37°C in DPBS [Dulbecco’s PBS with 0.1% fetal bovine serum, 10 mM Hepes, 10 mM sodium pyruvate, glucose (1 mg/ml), and penicillin/streptomycin (1 mg/ml)]. Sperm number was quantified using a computer-assisted semen analysis system (Hamilton Thorne Research) and by manual counting using a hemocytometer.
Immunohistochemistry and immunofluorescence
Testes were fixed for 24 hours at 4°C in 0.1 M sodium phosphate buffer (pH 7.2) containing 4% (w/v) PFA. Fixed testes were paraffin-embedded for H&E staining and immunohistochemistry (IHC). For immunofluorescence (IF), fixed testes were equilibrated through a 10, 18, and 25% sucrose (w/v, dissolved in 1× PBS) gradient by sequential overnight incubations at 4°C. Once equilibrated to 25% sucrose, testes were embedded in tissue freezing medium (Electron Microscopy Sciences) and frozen using a Shandon Lipshaw cryobath. IHC-based labeling was performed after deparaffinization using a DISCOVERY XT autostainer (Ventana Medical Systems) with antimouse Mage-a3 antibody (1:1000). All slides were counterstained with hematoxylin. Bright-field images were taken with an upright Eclipse Ni (Nikon) or constructed from digitized images using Aperio ImageScope (Leica Biosystems). The percentage of damaged tubules showing vacuolization and reduced germ cell layers (<4) was determined in a blinded manner in which >200 tubules from 2 to 6 mice per genotype per experiment were analyzed.
IF was performed on 8-μm cryosections. Before labeling, sections were equilibrated in air to room temperature for 15 min, hydrated for 10 min at room temperature, heat-treated at 80°C for 8 min in 10 mM sodium citrate (pH 6.0), and then incubated for 1 hour at room temperature in blocking buffer [Roche Blocking Reagent (1% v/v) diluted in 0.1 M sodium phosphate buffer, containing Triton X-100 (0.1% v/v)]. Sections were then treated for 18 to 24 hours at room temperature with antibody diluted in blocking buffer. Antibodies used were as follows: anti-Mage-a (production described above), rabbit polyclonal (1:500), anti-Stra8 (no. ab49602, Abcam), rabbit polyclonal (1:250), and anti-c-Kit (no. 3074, Cell Signaling Technology) rabbit monoclonal (1:400). After treatment with primary antibodies, sections were washed three times for 10 min per wash in PBS and then incubated for 40 min at room temperature with Alexa Fluor 568 secondary antibodies (Molecular Probes) diluted to 4 μg/ml in PBS containing Hoechst 33342 (5 μg/ml; Molecular Probes). After treatment with secondary antibodies, sections were washed three times in PBS and cover-slipped for viewing using Fluoro-Gel mounting medium (Electron Microscopy Sciences).
MIA Paca-2 cells, 2-DG treatment, viability assays, and metabolomics
MIA PaCa-2 cancer cells stably expressing GFP or human MAGE-A6 (provided by K. Scott, Baylor College of Medicine) were cultured in Dulbecco’s modified Eagle’s medium high glucose, 10% FBS, and 2.5% horse serum. Cells were plated in a 96-well plate, 1 × 104 cells per well, and treated with 2 mM 2-DG glycolysis inhibitor (Sigma) for 12 days. Viability was assessed using alamarBlue viability reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were incubated with 10% reagent in complete medium for 4 hours, and increased fluorescence was detected using an excitation between 530 and 560 nm and an emission at 590 nm.
For nontargeted metabolomics analysis, MIA-PaCa-2 cells stably expressing GFP or MAGE-A6 were grown in standard media or 2 mM 2-DG for 2 to 10 days. Nontargeted metabolomics was performed by the NIH West Coast Metabolomics Center (n = 6 for each group) (47). Data were normalized and analyzed by MetaboAnalyst (48).
Supplementary Material
Acknowledgments
We thank D. Alford for help in creating the MAGE expression web resource. We are grateful to M. Lu for performing IHC, to S. Savage and M. Straign for help with dissections and sperm analyses, and to C. Rolle for animal care. We thank K. Scott from the Baylor College of Medicine for providing MIA Paca2 cells expressing GFP or MAGE-A6. We also thank Potts laboratory members and Charles Rock for helpful discussions. Funding: This work was supported by the St. Jude Children’s Research Hospital (P.R.P.), NIH/NICHD grant R01HD061665 (J.M.O.), and NIH T32 Pharmacology training grant GM007062 (A.L.B). Author contributions: P.R.P. and K.F.T. conceptualized the study and designed experiments. K.F.T., M.C.M., and A.L.B. performed tissue gene expression analysis. M.J.O., T.L., J.M.O., and K.F.T. performed spermatogonia experiments. J.K., R.R., H.T., E.B., M.M.K., and K.F.T. performed experiments and analyzed data. M.S.K. designed software. J.P.C. and S.M.P.-M. performed gene editing. K.F.T. and P.R.P. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/5/eaav4832/DC1
Table S1. Expression (TPM) of testis-specific genes using the GTex dataset.
Table S2. Ortholog analysis of testis-specific genes using OrthoDB v9.1.
Table S3. Primers and siRNAs for MAGE genes.
Fig. S1. Human MAGE gene expression analysis.
Fig. S2. Mouse anatomical MAGE gene expression map in an adult organism.
Fig. S3. Tissue expression map of mouse MAGE genes during embryonic development and in embryonic stem cells.
Fig. S4. MAGE gene expression in mouse testis.
Fig. S5. Validation of SCC cultures.
Fig. S6. Phenotyping of Mage-a KO mice.
Fig. S7. p53 protein levels are increased in Mage-a KO mouse testis.
REFERENCES AND NOTES
- 1.D. Lack, in Evolution as a Process, J. Huxley, A. C. Hardy, E. B. Ford, Eds. (George Allen and Unwin, 1954), pp. 143–156. [Google Scholar]
- 2.de Rooij D. G., The nature and dynamics of spermatogonial stem cells. Development 144, 3022–3030 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Griswold M. D., Spermatogenesis: The commitment to meiosis. Physiol. Rev. 96, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oatley J. M., Brinster R. L., Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24, 263–286 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlatt S., Ehmcke J., Regulation of spermatogenesis: An evolutionary biologist's perspective. Semin. Cell Dev. Biol. 29, 2–16 (2014). [DOI] [PubMed] [Google Scholar]
- 6.White T. C., The importance of a relative shortage of food in animal ecology. Oecologia 33, 71–86 (1978). [DOI] [PubMed] [Google Scholar]
- 7.Soubry A., Hoyo C., Jirtle R. L., Murphy S. K., A paternal environmental legacy: Evidence for epigenetic inheritance through the male germ line. Bioessays 36, 359–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maher G. J., Ralph H. K., Ding Z., Koelling N., Mlcochova H., Giannoulatou E., Dhami P., Paul D. S., Stricker S. H., Beck S., McVean G., Wilkie A. O. M., Goriely A., Selfish mutations dysregulating RAS-MAPK signaling are pervasive in aged human testes. Genome Res. 28, 1779–1790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee T.-L., Pang A. L.-Y., Rennert O. M., Chan W.-Y., Genomic landscape of developing male germ cells. Birth Defects Res. C Embryo Today 87, 43–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng K., Yang F., Wang P. J., Regulation of male fertility by X-linked genes. J. Androl. 31, 79–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P. J., McCarrey J. R., Yang F., Page D. C., An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27, 422–426 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Lee A. K., Potts P. R., A comprehensive guide to the MAGE family of ubiquitin ligases. J. Mol. Biol. 429, 1114–1142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle J. M., Gao J., Wang J., Yang M., Potts P. R., MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell 39, 963–974 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weon J. L., Yang S. W., Potts P. R., Cytosolic iron-sulfur assembly is evolutionarily tuned by a cancer-amplified ubiquitin ligase. Mol. Cell 69, 113–125.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Weon J. L., Potts P. R., The MAGE protein family and cancer. Curr. Opin. Cell Biol. 37, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melé M., Ferreira P. G., Reverter F., DeLuca D. S., Monlong J., Sammeth M., Young T. R., Goldmann J. M., Pervouchine D. D., Sullivan T. J., Johnson R., Segrè A. V., Djebali S., Niarchou A.; The GTEx Consortium, Wright F. A., Lappalainen T., Calvo M., Getz G., Dermitzakis E. T., Ardlie K. G., Guigó R., Human genomics. The human transcriptome across tissues and individuals. Science 348, 660–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zdobnov E. M., Tegenfeldt F., Kuznetsov D., Waterhouse R. M., Simão F. A., Ioannidis P., Seppey M., Loetscher A., Kriventseva E. V., OrthoDB v9.1: Cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45, D744–D749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellve A. R., Cavicchia J. C., Millette C. F., O’Brien D. A., Bhatnagar Y. M., Dym M., Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 74, 68–85 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endo T., Freinkman E., de Rooij D. G., Page D. C., Periodic production of retinoic acid by meiotic and somatic cells coordinates four transitions in mouse spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 114, E10132–E10141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed E. A., de Rooij D. G., Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 558, 263–277 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Chan F., Oatley M. J., Kaucher A. V., Yang Q.-E., Bieberich C. J., Shashikant C. S., Oatley J. M., Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 28, 1351–1362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Q.-E., Gwost I., Oatley M. J., Oatley J. M., Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol. Reprod. 89, 113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oatley M. J., Kaucher A. V., Racicot K. E., Oatley J. M., Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol. Reprod. 85, 347–356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinster R. L., Zimmermann J. W., Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. U.S.A. 91, 11298–11302 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bucci L. R., Meistrich M. L., Effects of busulfan on murine spermatogenesis: Cytotoxicity, sterility, sperm abnormalities, and dominant lethal mutations. Mutat. Res. 176, 259–268 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Hou S., Xian L., Shi P., Li C., Lin Z., Gao X., The Magea gene cluster regulates male germ cell apoptosis without affecting the fertility in mice. Sci. Rep. 6, 26735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladelfa M. F., Peche L. Y., Toledo M. F., Laiseca J. E., Schneider C., Monte M., Tumor-specific MAGE proteins as regulators of p53 function. Cancer Lett. 325, 11–17 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Krausz C., Giachini C., Giacco D. L., Daguin F., Chianese C., Ars E., Ruiz-Castane E., Forti G., Rossi E., High resolution X chromosome-specific array-CGH detects new CNVs in infertile males. PLOS ONE 7, e44887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Y., Xu J., Yang X., Liu Y., Ma Y., Yang D., Dong Q., Yang Y., Evidence for the involvement of the proximal copy of the MAGEA9 gene in Xq28-linked CNV67 specific to spermatogenic failure. Biol. Reprod. 96, 610–616 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Okutman O., Muller J., Skory V., Garnier J. M., Gaucherot A., Baert Y., Lamour V., Serdarogullari M., Gultomruk M., Röpke A., Kliesch S., Herbepin V., Aknin I., Benkhalifa M., Teletin M., Bakircioglu E., Goossens E., Charlet-Berguerand N., Bahceci M., Tüttelmann F., Viville S., A no-stop mutation in MAGEB4 is a possible cause of rare X-linked azoospermia and oligozoospermia in a consanguineous Turkish family. J. Assist. Reprod. Genet. 34, 683–694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin X., Pan Y., Wang L., Zhang L., Ravichandran R., Potts P. R., Jiang J., Wu H., Huang H., MAGE-TRIM28 complex promotes the Warburg effect and hepatocellular carcinoma progression by targeting FBP1 for degradation. Oncogene 6, e312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pineda C. T., Ramanathan S., Tacer K. F., Weon J. L., Potts M. B., Ou Y.-H., White M. A., Potts P. R., Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell 160, 715–728 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rato L., Alves M. G., Socorro S., Duarte A. I., Cavaco J. E., Oliveira P. F., Metabolic regulation is important for spermatogenesis. Nat. Rev. Urol. 9, 330–338 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Helsel A. R., Oatley M. J., Oatley J. M., Glycolysis-optimized conditions enhance maintenance of regenerative integrity in mouse spermatogonial stem cells during long-term culture. Stem Cell Rep. 8, 1430–1441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garton A. J., Campbell D. G., Carling D., Hardie D. G., Colbran R. J., Yeaman S. J., Phosphorylation of bovine hormone-sensitive lipase by the AMP-activated protein kinase. A possible antilipolytic mechanism. Eur. J. Biochem. 179, 249–254 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Qu Q., Zeng F., Liu X., Wang Q. J., Deng F., Fatty acid oxidation and carnitine palmitoyltransferase I: Emerging therapeutic targets in cancer. Cell Death Dis. 7, e2226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carracedo A., Cantley L. C., Pandolfi P. P., Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 13, 227–232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunwell T. L., Paps J., Holland P. W. H., Novel and divergent genes in the evolution of placental mammals. Proc. Biol. Sci. 284, 20171357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardie D. G., AMPK—Sensing energy while talking to other signaling pathways. Cell Metab. 20, 939–952 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobbs R. M., Seandel M., Falciatori I., Rafii S., Pandolfi P. P., Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell 142, 468–479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busada J. T., Geyer C. B., The role of retinoic acid (RA) in spermatogonial differentiation. Biol. Reprod. 94, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira S. L., Rodrigues A. S., Sousa M. I., Correia M., Perestrelo T., Ramalho-Santos J., From gametogenesis and stem cells to cancer: Common metabolic themes. Hum. Reprod. Update 20, 924–943 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., Schultz N., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellvé A. R., Millette C. F., Bhatnagar Y. M., O’Brien D. A., Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J. Histochem. Cytochem. 25, 480–494 (1977). [DOI] [PubMed] [Google Scholar]
- 45.Meistrich M. L., Hess R. A., Assessment of spermatogenesis through staging of seminiferous tubules. Methods Mol. Biol. 927, 299–307 (2013). [DOI] [PubMed] [Google Scholar]
- 46.J. M. Oatley, R. L. Brinster, Spermatogonial stem cells, in Methods in Enzymology, K. Irina, L. Robert, Eds. (Academic Press, 2006), vol. 419, pp. 259–282. [DOI] [PubMed] [Google Scholar]
- 47.Fiehn O., Metabolomics by gas chromatography–mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 114, 30.4.1–30.4.2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S., Xia J., MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/5/eaav4832/DC1
Table S1. Expression (TPM) of testis-specific genes using the GTex dataset.
Table S2. Ortholog analysis of testis-specific genes using OrthoDB v9.1.
Table S3. Primers and siRNAs for MAGE genes.
Fig. S1. Human MAGE gene expression analysis.
Fig. S2. Mouse anatomical MAGE gene expression map in an adult organism.
Fig. S3. Tissue expression map of mouse MAGE genes during embryonic development and in embryonic stem cells.
Fig. S4. MAGE gene expression in mouse testis.
Fig. S5. Validation of SCC cultures.
Fig. S6. Phenotyping of Mage-a KO mice.
Fig. S7. p53 protein levels are increased in Mage-a KO mouse testis.


