Abstract
IKAROS (encoded by IKZF1) is an important hematopoietic transcription factor critical for early B cell differentiation, with major defects known to lead to low B cell numbers and hypogammaglobulinemia. More perplexing is the link between IKZF1 variants and autoimmunity, including polymorphisms associated with susceptibility to SLE, and recently, rare variants driving monogenic autoimmunity. We identified a novel p.L188V mutation in IKZF1 in the index patient and her father and found this mutation to lead to loss of DNA binding. Peripheral B cells lacking a full complement of IKAROS function show upregulation of molecules accentuating B cell activation, while CD22, a key negative feedback circuit, is suppressed. The resulting hyperresponsiveness of peripheral B cells, in combination with elevated follicular helper T cell (Tfh) numbers, provides a putative mechanistic explanation for the association of IKZF1 variants with the emergence of autoimmune manifestations in this kindred.
Keywords: IKAROS, IKZF1, systemic lupus erythematosus, autoimmunity, B cells, CD22
Capsule summary
Mutation in IKZF1, a hematopoietic transcription factor, gives rise to autoimmunity in a novel kindred. Our study identifies a link between mutant IKAROS and decreased CD22 expression driving the hyperresponsive B cell phenotype.
To the Editor:
Ikaros is the founding member of a zinc finger transcription factor family, with crucial functions in murine hemato-lymphoid development, differentiation and homeostasis (1). This function was illustrated first in mice, with the complete arrest of B cell development in Ikaros-null mice and later by the generation of transgenic mice with dominant-negative Ikaros or reduced expression of Ikaros (1,2). Ikaros exerts transcriptional repression of target genes through DNA binding. Distinct isoforms of Ikaros have a common C-terminal dimerization domain but differ in the composition of their N-terminal DNA binding domain. Isoforms that do not bind DNA but retain dimerization potential are dominant negative. IKAROS function is conserved across species, with Kuehn et al. identifying novel heterozygous IKZF1 mutations as the cause of an autosomal dominant form of common variable immunodeficiency (CVID), characterized by hypogammaglobulinemia and a progressive loss of B cells (3). The heterozygous genotype in patients resulted in a less severe phenotype than the complete Ikaros-deficiency in mice, suggesting a partial retention of function in the human CVID context (1). Additional germline mutations were recently identified in patients with early onset hypogammaglobulinemia and autoimmunity, including one patient with SLE (4,5). While the impact of IKAROS deficiency on early B cell differentiation has been studied (3–5), the mechanism of action giving rise to autoimmunity remains undefined.
The index patient, P2, born to non-consanguineous parents, was diagnosed with juvenile-onset systemic lupus at the age of 12 years. She presented with asthenia, weight loss, a lingual ulcer, hepatosplenomegaly and hair loss. Blood analysis showed pancytopenia, systemic inflammation, complement activation, isolated IgA deficiency, highly increased levels of anti-nuclear and anti-dsDNA antibodies and adrenal failure resulting from bilateral venous thrombosis due to antiphospholipid syndrome (Table E1). Except for a recent pneumococcal pleuropneumonia, her medical history was unremarkable. Immunological work-up revealed low B cell numbers and low-to-absent antibody titers for rubella, mumps and hepatitis B virus despite immunization (Table E1). After initiation of steroids (with hydroxychloroquine and anticoagulation), a progressive and persistent decline in B cell numbers and levels of all immunoglobulin isotypes was noted. Subcutaneous IVIG was started and no notable infections have occurred since. Further immunological analysis revealed low IgM values and the presence of lupus anticoagulant and anti-nuclear antibodies in her self-reported healthy father (P1) (Table E1). Genetic variants detected through whole exome sequencing of the index patient and her parents were filtered for rare coding mutations predicted to be damaging. Possible de novo candidates were found unlikely to be biologically relevant directing our focus on the 48 variants shared by both P2 and her father (P1). Based on available literature a novel heterozygous mutation in IKZF1 (NM_006060), c.C562G;p.Leu188Val, was noted as the most plausible pathogenic candidate and confirmed by Sanger sequencing (Fig. 1A). The mutation is located in a highly conserved amino acid in the zinc finger 3 domain of IKAROS (Fig. 1B), necessary for DNA binding. A direct relation between this novel IKZF1 mutation and juvenile-onset SLE was considered highly probable based on the association of common IKZF1 polymorphisms with adult-onset SLE, the development of lupus-like symptoms in DN-Ikaros transgenic mice (6) and the recent report of a splicing IKZF1 mutation with incomplete penetrance in a family with juvenile-onset SLE and progressive hypogammaglobulinemia (4).
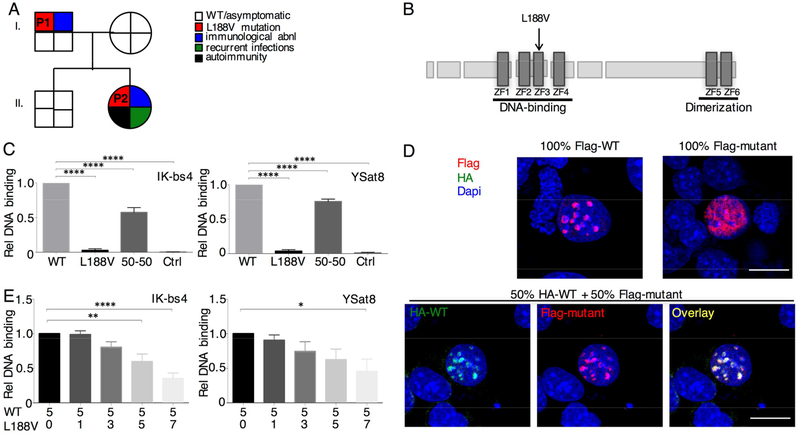
Fig. 1. Heterozygous mutation in IKZF1 is a strong driver of autoimmunity through loss of DNA binding capacity.
(A) Segregation of the p.L188V mutant allele and the clinical and immunological phenotype. P1 and P2 are indicated. (B) Schematic representation of the IKAROS protein. Zinc-fingers (ZF) are shown as dark rectangles. The p.L188V mutation is marked by a black arrow. (C) EMSAs were performed using two different probes: IK-bs4 and ΥSat8 as indicated. Quantification data were normalized using the binding of the extracts from WT transfected cells as 100% (6 independent experiments). (D) NIH3T3 cells transfected with Flag-mutant, Flag-WT and/or HA-WT as indicated were stained with anti-Flag antibody +/− an anti-HA antibody followed by fluorochrome-conjugated secondary antibodies. Representative of 4 independent confocal experiments. Scale bar 20μm. (E) IK-bs4 and YSat8 EMSAs, where an increasing amount of L188V mutant was transfected with an equal amount of WT IKAROS. Quantified data were normalized using the binding of the extracts from WT transfected cells set as 100% (4 independent experiments). Statistics were performed using one-way ANOVA. Mean±SEM. **** = p<0.0001, ** = p<0.01, * = p<0.05.
Preserved IKAROS mRNA (Fig. E1A) and protein (Fig. E1B–C) expression in primary B cells and EBV-transformed B cells respectively indicated a functional rather than a stability defect. Using electrophoretic mobility-shift assay (EMSA) with two different validated probes (Ik-bs4 and ySat8), nuclear enriched fractions from transfected HEK293 cells (Fig. E2A–B) demonstrated that homodimers of mutant IKAROS are incapable of binding IKAROS consensus sequence (Fig. 1C, Fig. E2C lanes 6 and 7). Confocal microscopic analysis of transfected NIH-3T3 cells with Flag and/or HA-tagged expression vectors, demonstrated characteristic punctate staining for wild-type (WT) in accordance with pericentromeric heterochromatin colocalization (Fig. 1D). However p.L188V Ik-1 exhibited a diffuse nuclear staining pattern which could be rescued by cotransfection with WT. To determine if mutant IKAROS participated in this restored DNA binding or was merely present as part of a multimeric complex, we titrated mutant IKAROS into cells with a fixed amount of WT IKAROS (Fig. 1E, Fig. E2D). This was not performed for previously reported IKZF1 mutations (3–5). Similar results were observed when the two vectors were titrated using different promoters (Fig. E3). Together, these data demonstrate that p.L188V IKAROS, despite retaining nuclear localization and capacity to integrate into IKAROS complexes, can cause a decrease in overall DNA binding.
Detailed immune profiling revealed total B cell numbers were slightly low in P2 and normal for P1 (Fig. E4A), but common defects in B cell differentiation were apparent with low KRECs (Fig. E4B), reduced naïve B cells (Fig. E4C–D) and markedly increased memory IgM B cells (Fig. E4E). Analysis of the BCR repertoire of naïve B-cells (Fig. E5A–C) was unremarkable except for a decrease in repertoire diversity in P2 (Fig. E5D), reminiscent of other conditions with low B cells and autoimmunity such as Omenn syndrome. No consistent difference was observed in class switch recombination (CSR) or somatic hyper-mutation (only reduced in P2)(Fig. E5E–F), contrary to the reported role of CSR in mice (2). Despite normal total T cell numbers (Table E1), there was a reversed CD4/CD8 ratio (Fig. E4H–I) mimicking previous observations in both SLE and CVID. A profound increase was observed in the circulating follicular T cells, both within the follicular helper Tfh (Fig. E4M) and follicular regulatory Tfr (Fig. E4N) CD4+ T cell subsets. The capacity of Tfh to support antibody generation and the association of Tfh responses with SLE and the lupus-like disease that occurs in Roquinsan/san mice due to exhuberant Tfh responses indicates this follicular differentiation likely contributes to the immunological dysregulation identified in the IKZF1 p.L188V patients.
Mutant IKAROS patients with adequate B cell generation but autoimmunity provide a unique opportunity to investigate the impact of IKAROS on peripheral B cells. Using a CFSE-based proliferation assay or 24-hour stimulation followed by Ki67 staining on total B cells no difference in proliferative potential was seen, unlike observations made in mice (Fig. E6A–D) (2). Furthermore, we observed an increasing MFI for CD19, an activating co-receptor (Fig. 2A), corresponding with upregulated CD19 expression in unstimulated IkL/L cells reported by Heizmann et al (7). These data suggested that particular molecular pathways may be disturbed in IKZF1 p.L188V B cells and led us to investigate the activation response of mutant B cells. First, we measured the response of the Erk pathway as naïve Ikaros-deficient B cells show constitutive phosphorylation of Erk resulting in hyper-proliferation (2). Following stimulation with anti-IgM mix, primary and EBV transformed B cells bearing the IKZF1 p.L188V mutation demonstrated persistent elevation of Erk phosphorylation, already at baseline (Fig. 2B–C, Fig. E7) with a prolonged activation at high doses. Second, we stimulated PBMCs in vitro for 24 hours with anti-IgM mix and anti-IgM mix plus anti-CD40L. At baseline, B cells of both P1 and P2 displayed elevated CD69 expression which responded in a normal manner to stimulation (Fig. 2D). Interestingly, CD22 expression in patient B cells was diminished at baseline despite the over-representation of memory B cells, reported to have higher CD22 expression in healthy individuals (8) (Fig. 2E). CD22 expression increased upon stimulation, but levels in mutant IKAROS individuals remained decreased compared to healthy controls (HC) (Fig. 2E). Furthermore, CD22 mRNA expression was reduced on mutant IKAROS primary B cells (p-value 0.14) (Fig. 2F). IKAROS p.L188V mutation thus results in hyperresponsive B cells with upregulated CD69 expression and downregulated CD22 expression at baseline, in line with transcriptome profiling of hyperactivated unstimulated IkL/L follicular B cells which showed increased CD19 and decreased CD22 expression.
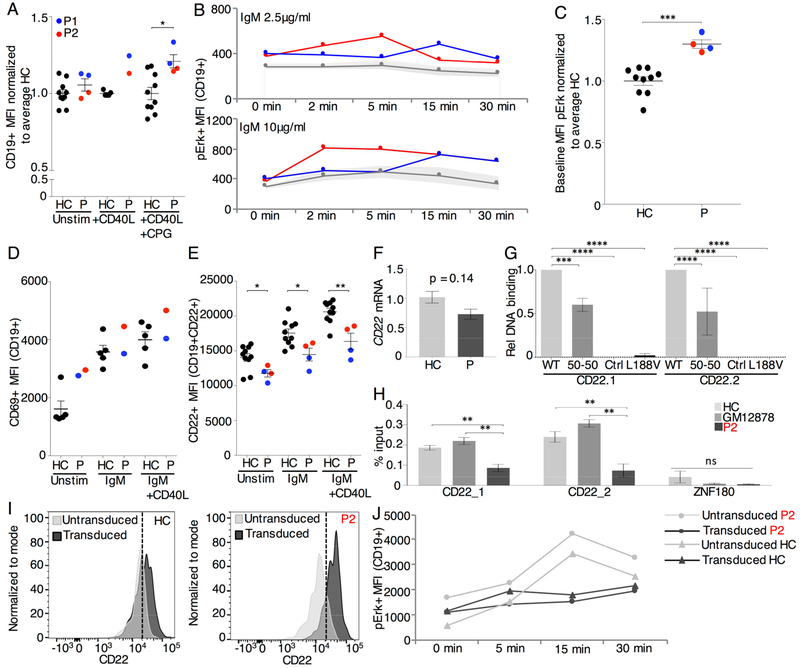
Fig. 2. Hyperresponsive phenotype of mutant IKAROS B cells is rescued by CD22 overexpression.
(A) CD19+ MFI on B cells from the CFSE assay in Fig. E6. (B) PBMCs stimulated with 2.5μg/ml or 10μg/ml anti-IgM-IgA-IgG were stained for pErk. Shaded area marks one standard deviation above and below the mean of the HC (2 repeats). (C) Baseline MFI of pErk in primary B cells for patients versus HC normalized to the average of HC. (D) PBMCs stimulated with anti-IgM-IgA-IgG 10μg/ml alone or or anti-IgM-IgA-IgG 10μg/ml + CD40L 1μg/ml for 24hrs were stained for CD69 (1 repeat) and (E) CD22 (2 repeat). (F) CD22 mRNA expression normalized to GAPDH and Actin for P and seven HC for 3 repeats. (G) Relative quantification of DNA binding on CD22 specific EMSA probes, normalized to WT binding (3 repeats). (H) ChIP-qPCR of IKAROS in EBV transformed B cells of P2, HC and GM12878 human B lymphocytes for CD22 and ZNF180 (not bound by IKAROS) (3 repeats). (I) CD22 MFI on CD19+ primary B cells after transduction with hCD22 for P2 and HC. (J) pErk MFI on hCD22 transduced CD19+ primary B cells after stimulation with 10μg/ml anti-IgM-IgA-IgG (1 repeat). Mean±SEM. **** = p<0.0001, *** = p<0.001 ** = p<0.01, * = p<0.05.
In murine studies, reductions of Ikaros in IkL/L mice result in mature B cells that exhibit a lower activation threshold (2) while B cells from CD22-deficient mice demonstrate increased calcium fluxes and cell proliferation with subsequent production of autoantibodies (9). Analysis of available ChIP-seq data indicates that both CD22 and CD19 genes are bound by IKAROS in pre-B ALL cells and in mature B-lymphocytes (GSE58825 and ENCODE data GSE92125, respectively) (Fig. E8A, data not shown). To evaluate a direct link between IKAROS and CD22 we repeated EMSAs with CD22 specific probes which demonstrated DNA binding ability for WT IKAROS but abrogated DNA binding for mutant IKAROS (Fig. 2G, Fig. E8B). Reduced endogenous IKAROS binding to CD22 was confirmed true for patient EBV-transformed B cells using ChIP-qPCR (Fig. 2H, Fig. E8C) demonstrating a cross-species conservation of the IKAROS-CD22 interaction. CD22 overexpression in primary B cells using lentiviral transduction rescued the hyperresponsive B cell phenotype as demonstrated by a post-transduction decrease in phosphorylated Erk to healthy control levels both at baseline and after stimulation (Fig. 2I–J). We propose mutant IKAROS B cells are reprogrammed to a hyperactivated state at baseline with depressed CD22 expression contributing to a lower threshold for activation.
Methods
Written informed consent was obtained from all participants and the study was approved by the Ethics Committee of University Hospitals Leuven. The index (P2) and her father (P1) are of European Belgian descent. Unless otherwise stated healthy controls included the mother and brother of the affected kindred, young adults aged 25–28 years and a male of 47years (to match the age of P1).
Genetic analysis
Whole exome sequencing was performed as previously described on the index patient and her self-reported healthy parents(1). Only coding nonsynonymous variants with genotype quality > 60, CADD score > 10(2), gene damage index score of <12,405 (3) and mean allele frequency of <0.005 in NHLBI GO Exome Sequencing Project 6500, 1000 Genomes Project (October 2014) or in The Exome Aggregation Consortium database were considered. Gel purified products (Macherey-Nagel) amplified with IKZF1_exon5 primers (see below) were sent to LGC Genomics Facility in Berlin, Germany.
Reverse transcriptase-polymerase chain reaction
Total RNA was isolated from blood mononuclear cells (PBMCs) using TRIzol® reagent (Ambion, Life Technologies) and for primary B cells the aqueous phase was transferred to the Zymo Research RNA Clean & Concentrator-5 Kit. Complementary DNA was synthesized from RNA using the GoScript™ Reverse Transcription System (Promega). Real time quantitative PCR was performed on a StepOnePlus real-time PCR system (ABI). cDNA was added to Fast SYBR Green Master Mix (Applied Biosystems) supplemented with gene-specific primers. Analysis was performed with the efficiency corrected 2−ΔΔCT method, and quantifications were normalized to the average of the housekeeping genes. Experiments were performed in triplicate and repeated twice for IKZF1 expression and performed in duplicate and repeated 3 times for CD22 expression. For CD22 expression PrimeTime® qPCR primers were ordered from Integrated DNA Technologies.
Western blotting
Cell pellets from EBV clones, generated from fresh PBMCs(4), were solubilized in lysis buffer (50mM Tris-HCl pH 7,5, 100mM NaCl, 5% glycerol, 2mM DTT and 1% Triton-X, protease inhibitor (Pierce) and phosphatase inhibitor (Roche)) and sonicated. Fifty μg of lysate were separated on 4–12% Bis-Tris acrylamide gels (NuPAGE Precast Gel System, Life Technologies) and blotted on a PVDF membrane (GE Healthcare). For nuclear extracts 5μg was loaded. After blotting, membranes were incubated using specific antibodies; r α-IKAROS (Cell Signaling, D10E5), m α-vinculine (Sigma-Aldrich, V9264), m α-histone H1 (Santa Cruz, sc-393358), m α-Flag (Cell Signaling, 9A3), m α-HA (Eurogentec, 16B12), m α-GAPDH (Invitrogen, MA5–15738), Proteins were revealed using western Lightning Plus-ECL (Perkin Elmer) or ECL prime (GE Healthcare). All blots were acquired with the G:box Chemi-XRQ. Quantification was performed using AIDA software (Raytest, version 5.0).
Electrophoretic mobilitiy shift assay (EMSA)
Flag-tagged and HA-tagged human wild-type and L188V mutant IKAROS (IKZF1; NM_006060) cDNA were each cloned downstream of the CAG promoter. The SV40 promoter was cloned into the HA-tagged human wild-type IKAROS expression plasmid using the In-Fusion HD cloning kit (Clontech). 7μg of flag-tagged full-length WT IKAROS, mutant L188V IKAROS, p.173–253del mutant (control) were used for transfections in HEK293T cells using GeneJuice® transfection reagent (Novagen). After 24 hours, nuclear enriched fractions were prepared, as adapted from Denayer et al, 2010. Double stranded oligonucleotides(5), IKAROS consensus-binding sequence (Ik-bs4) and γ satellite from human chromosome 8 (γSat8), were radioactively labeled with [α−32P] dCTP by a fill-in reaction by the Klenow fragment of DNA polymerase (6). Nuclear extracts were incubated with 1 μl of radiolabeled probe (20000cpm/μl). Sample conditions for separation by electrophoresis are 10 mM Hepes pH 7.9, 2.5 mM MgCl2, 0.05 mM EDTA, 8% glycerol, 0.05% Triton X-100, 1 mM DTT and 50 ng/μl poly(dIdC) and supershift was induced by adding undiluted Flag antibody (Sigma-Aldrich, F7425). Films were developed with the CURIX 60 (AGFA Healthcare).
Measurement of DNA-binding capacity by confocal microscopy
HA/Flag-tagged human wild-type and Flag-tagged L188V mutant IKAROS plasmids were transfected in NIH3T3 cells using Lipofectamine 3000 Reagent (Invitrogen). Cells were stained with mouse anti-HA monoclonal antibody (BIOT-101L050; Eurogentec) and/or rabbit anti-Flag polyclonal affinity antibody (F7425; Sigma Aldrich), DAPI (D1306; Molecular Probes) and either Streptavidin-Alexa Fluor 488 (S32354; Molecular Probes) or Alexa Fluor 555-donkey anti-rabbit (A31572; Molecular Probes). Images were collected on an LSM 510 Meta confocal microscope (Ziess) with a 60X immersion objective.
Flow cytometry
Blood from controls and patients (now aged 19 years and 49 years) was harvested at the same time, all PBMCs were isolated using lymphocyte separation medium (LSM, MP Biomedicals) and used fresh for analysis. However when age-matched controls were needed samples were frozen and stained within one month. For activation studies PBMCs were stimulated in culture for 24hr at 37°C in a 5% CO2 incubator with 10μg/ml anti-IgM mix (AffiniPure Goat Anti-Human IgA+IgG+IgM (H+L), Jackson Immunoresearch Lab) or 10μg/ml anti-IgM mix + 1μg/ml recombinant human Soluble CD-40 Ligand (CD40L, ProSpec). For intracellular phospho-staining PBMCs were stimulated at 37*0 in fetal bovine serum with 2.5μg/ml anti-IgM mix or 10μg/ml anti-IgM mix for 0min, 2min, 15min or 30min, as indicated. Staining with appropriate fluorophore-conjugated antibodies was performed with the PERFIX EXPOSE kit (Beckman Coulter). Unless otherwise stated, all antibodies used were from eBioscience and include the following (clones are enclosed in brackets): IgM PE (SA-DA4), CD40 PE-Cy7 (5C3), CD38 APC (HIT2), CD27 AF-700 (O323), IgD APC-Cy7 (BioLegend, IA6–2), CD24 BV421 (BioLegend, ML5), CD19 BV510 (BioLegend, HIB19), CD25 AF488 (Biolegend, BC96), CD25 (BD, BB515), CXCR5 PE (BioLegend, J252D4), CD4 PE-eFluor 610 (RPA-T4), IL7Ra (CD127) Biotin PerCPcy5.5 (eBioRDR5), CD28 PE-Cy7 (CD28.2), Foxp3 AF647 APC (BioLegend, 206D), CD8a AF700 (BioLegend, RPA-T8), CD45RA APC-Cy7 (HI 100), PD-1 BV421 (BioLegend, EH12.2H7), Zombie Aqua™ (Biolegend), CD4-EF450 (RPA-T4), CD8 V500 (BD, RPA-T8), CD45RA PeCy7 (HI100), CCR7 PE (3D12), CD62L APC-EF780 (DREG-56),CD19 EF450 (HIB19), CD14 BV510 (BioLegend, M5E2), CD22 PerCyP EF710 (eBio4KB128), CD40 Pe-Cy7 (5C3), CD27 FITC (O323), CD69 PeCy7 (FN50), HLA-DR APC-Cy7 (LN3), pErk APC (T202), CD19 PE (SJ25C1), CD27 EF450 (O323), Alexa Fluor® 647 Mouse IgG1 κ Isotype control (BioLegend, MPOC-21), GFP (Biolegend, FM264G), ERK1/ERK2 (Invitrogen, K.913.4), F(ab’)2-goat anti-rabbit IgG secondary antibody AF-647 (Invitrogen).
TREC and KREC
Droplet digital PCR (Bio-Rad, Hercules, CA) with DNA detection assays (Life Technologies) for cjKREC (coding joint κ-deleting recombination excision circles), cjTREC (coding joint T-cell receptor excision circles) (both custom made, sequences available on request) and copy number reference assay RNAseP were performed using 250ng of restriction digested (EcoRI, New England Biolabs) gDNA according to the manufacturer’s instructions. Relative quantity of cjKREC and cjTREC per (RNAseP/2)*100,000 cells was measured with QuantaSoft v1.4 (Bio-Rad).
B cell antigen receptor analysis using next generation sequencing
For the analysis of the naïve B-cell receptor repertoire, CD19+ CD27- IgD+ CD3- naïve B-cells were FACS sorted. DNA was isolated using direct lysis(7) and IGH rearrangements were amplified and sequenced using Roche 454 sequencing as previously described(8,9). In short, IGH rearrangements were amplified from a multiplex PCR using the forward VH1–6 FR1 and reverse JH consensus BIOMED-2 primers(10). These PCR products were purified and sequenced using Roche 454 sequencing as previously described(8,9). In short, PCR products were purified by gel extraction (Qiagen, Valencia, CA) and Agencourt AMPure XP beads (Beckman Coulter, Fullerton, CA). Subsequently, the concentration of the PCR product was measured using the Quant-it Picogreen dsDNA assay (Invitrogen, Carlsbad, CA). The purified PCR products were sequenced on the 454 GS junior instrument using the Lib-A kit according the manufacturer’s recommendations. For analysis sequences were demultiplexed based on their multiplex identifier sequence and 40 nucleotides trimmed from both sides to remove the primer sequence using ARGalaxy (11,12). Fasta files were analyzed in IMGT/High-V-Quest (selection of IMGT reference directory set: F+ORF+in-frame P with all alleles, search for insertions and deletions: yes, parameters for IMGT/Junction Analysis: default)(13). Subsequently unique sequences were selected based upon V and J gene usage and CDR3 amino acid sequence, and analyzed for V and J gene usage, CDR3 length and junction characteristics using ARGalaxy(12). For the healthy control data, previously published controls were used (European Nucleotide Archive (ENA) project number PRJEB15348)(9,14). For analysis of SHM frequency and CSR RNA was extracted from total PBMC’s using RNeasy Plus Kits (Qiagen) and transcribed into cDNA. IGH transcripts were amplified from 5μl cDNA per reaction in a multiplex PCR using the forward VH1–6 FR1 (BIOMED-2) primers(10) and the CgCH1(15) or the IGHA(16) reverse primer reverse primer. PCR products were purified and sequenced as described above only now using the Lib-A V2 454 sequencing kit. Sequences were analyzed using IMGT and ARGalaxy (11–13). Only unique productive sequences, that were complete, without ambiguous bases, with a subclass that could be defined, were included in the analysis. Previously published aged matched healthy control data were used (European Nucleotide Archive (ENA) project number PRJEB15348)(14).
Proliferation assay
B cells were negatively selected using EasySep™Human B Cell Enrichment Kit (Stem cell technologies) and labeled with CFSE (final working solution 1μM) using the CellTrace™ CFSE Cell Proliferation Kit (Life Technologies). Primary B cells were stimulated with 10ug/ml anti-IgM mix (AffiniPure Goat Anti-Human IgA+IgG+IgM (H+L), Jackson Immunoresearch Lab) + 1μg/ml recombinant human Soluble CD-40 Ligand (CD40L, ProSpec) + 1μg/ml Class B CpG oligonucleotide (Invivogen) for 5 days. Data was analyzed for proliferation index with FlowJo (Version 9).
Chromatin immunoprecipitation (ChIP)
Analysis of ChIP-seq data was performed on available data (ENCODE data GSE92125) (17). Briefly, raw reads were mapped to hg38 and significantly enriched peaks were called against the corresponding input control using MACS2. Signal tracks were produced for visualization using the Integrated genome browser. Chromatin Immunoprecipitation (ChIP) was performed essentially according to (PMID:20680851). Briefly, lymphoblast samples were grown in RPMI +10% FBS and fixed with 1 % formaldehyde for 10 minutes and then fixation was quenched with 1.25M glycine. Samples were then lysed and sonicated for 18 minutes using a Covaris S2. 2ug of anti-IKAROS antibody (Genetex; GTX129438) or control antibody (Genetex; GTX35035) were used with 30ug of input sonicated chromatin. The precipitated DNA products were then analyzed by qPCR and compared to unenriched input DNA samples. Sequences for primers used for ChIP-qPCR are listed under ‘Primer and probe sequences’.
Lentiviral transduction
Custom gene expression lentiviral vector for EGFP-EF1A-hIKZF1 (NM_006060.5) was constructed and packaged by Cyagen Biosciences. The lentiviral vector EGFP-EF1A-hCD22 (NM_001771.3]) was constructed by Cyagen Biosciences but packaged with a specialized baboon retroviral envelope (BaEV) for human B-cell transduction as previously described by the Leuven Viral Vector Core (18). After 24 hours of pre-stimulation with 10ug/ml anti-IgM mix + 1μg/ml recombinant human Soluble CD-40 Ligand (CD40L, ProSpec) + 1μg/ml Class B CpG oligonucleotide, primary human B cells were transduced by spin inoculation at 900g for 90 minutes with polybrene 5ug/ml. B cells were cultured in stimulation media for a further 24 hours and subsequently acquired on day 4 for downstream analysis. Transduction of EBV transformed B cells was performed similarly but without stimulation.
Statistics
Statistics were performed using an unpaired t test or one-way ANOVA with Sidak’s multiple comparison test.
Primer and probe sequences
| Name | Purpose | Forward primer 5’ – 3’ | Reverse primer 5’ – 3’ |
|---|---|---|---|
| IKZF1_exon5 | Sanger | TCCCCACGCTGAGTTTAGTTC | CAGGGTTAGCCAGCAAGGAC |
| IKZF1 | qPCR | AACGTCGCCAAACGTAAGAG | CCATCACGTGGGACTTCATCA |
| BETA-ACTIN | qPCR | CTGGGACGACATGGAGAAAA | AAGGAAGGCTGGAAGAGTGC |
| GAPDH | qPCR | AGAAGGCTGGGGCTCATTTG | GCATCAGCAGAGGGGGCAGA |
| HPRT | qPCR | GTAGCCCTCTGTGTGCTCAAGG | GGCTTATATCCAACACTTCGTGGGG |
| Ik-bs4 | EMSA | tcgagTGACAGGGAATACACATTCCCAAAAGCg | |
| YSat8 | EMSA | tcgagTATGGCGAGGAAAACTGAAAAAGGTGGAAAATTTAGAAATGTg | |
| CD22 | qPCR | GAGATGCATGGTGTCGTGT | CTCCTTTTGCTCTCAGATGCT |
| CD22.1 | EMSA | tcgagGTATTTCTCCCACTATCTTCTTTAGTCTTTAGAAAGCg | |
| CD22.2 | EMSA | tcgagTCTGGAAAACCCAATGGCTCCCCAGGGCTTCGCTCTTGCTGCAGGAGGGGAAACg | |
| CD22_1 | ChIP-qPCR | CCAGAAGATGAGGGGATTCA | AGCCCATCCAGTGTCAATGT |
| CD22_2 | ChIP-qPCR | GGGAAGGAAATGGAGTCACA | TGTCTGCAAAGCCAGTATGC |
| ZNF180 | ChIP-qPCR | TGATGCACAATAAGTCGAGCA | TGCAGTCAATGTGGGAAGTC |
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine guanine phosphoribosyltransferase (HPRT), fms like tyrosine kinase 3 (FLT3), DNA Nucleotidylexotransferase (DNTT), Zinc Finger Protein 180 (ZNF180)
Supplementary Material
Disclosure of potential conflict of interest:
This work was supported by the ERC grant IMMUNO and the KUL (GOA). EVN is an FWO fellow (1S22716N). SH-B is an FWO fellow (1272517N). Part of this work was supported by the Dutch Organisation for Scientific Research (NWO/ZonMw VIDI grant 91712323 to M. van der Burg, VENI grant 91616058 to P.A. van Schouwenburg) and by an Institutional Research Grant from the American Cancer Society (14–196-01 to S.F.). H.S. is an Arthritis National Research Foundation (ANRF) Scholar and a Hellman Fellow. The other authors have declared that no conflict of interest exists.
We thank Hannah Van Hove (KUL), John Barber (KUL), Teresa Prezzemolo (VIB), Emanuela Pasciuto (KUL) and Klara Mallants (KUL) for technical assistance as well as the patients and their family who participated in this study.
Erika Van Nieuwenhove, MD1,2,3
Josselyn E. Garcia-Perez, PhD1,2
Christine Helsen, PhD4
Princess D. Rodriguez, MSc5
Pauline A. van Schouwenburg, PhD6
James Dooley, PhD1,2
Susan Schlenner, PhD1,2
Mirjam van der Burg, MD, PhD6
Els Verhoeyen, PhD7,8
Rik Gijsbers, PhD9,10
Seth Frietze, PhD5
Hilde Schjerven, PhD11
Isabelle Meyts, MD, PhD1,3
Frank Claessens, PhD4
Stephanie Humblet-Baron, MD, PhD1,2*
Carine Wouters, MD, PhD1,3*
Adrian Liston, PhD1,2*
From 1 KUL - University of Leuven, Department of Microbiology and Immunology, Leuven, Belgium, 2 VIB Center for Brain and Disease Research, Leuven, Belgium, 3 University Hospitals Leuven, Leuven, Belgium, 4 KUL - University of Leuven, Department of Cellular and Molecular Medicine, Leuven, Belgium, 5 Department of Medical Laboratory and Radiation Science, University of Vermont, Burlington, VT 05405, USA, 6 Department of Immunology, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands, 7 CIRI - International Center for Infectiology Research,Team EVIR, Inserm, U1111, Université Claude Bernard Lyon 1, CNRS, UMR5308, Ecole Normale Supérieure de Lyon, Univ Lyon, F-69007, Lyon, France,8 Université Côte d’Azur, INSERM, C3M, Nice, France, 9 Laboratory for Viral Vector Technology and Gene Therapy, Department of Pharmaceutical and Pharmacological Sciences, KU Leuven, Leuven, Belgium, 10 Leuven Viral Vector Core, Leuven, Belgium, 11 Department of Laboratory Medicine, University of California, San Francisco, CA 94143, USA. adrian.liston@vib.be and carine.wouters@uzleuven.be.
* These authors contributed equally to the work
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997; 15(1): 155–176. [DOI] [PubMed] [Google Scholar]
- 2.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32(3):720–730. [DOI] [PubMed] [Google Scholar]
- 3.Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, et al. Loss of B Cells in patients with heterozygous mutations in IKAROS. N Engl J Med. 2016;374(11):1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshino A, Okada S, Yoshida K, Nishida N, Okuno Y, Ueno H, et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J Allergy Clin Immunol. 2016:1–9. [DOI] [PubMed] [Google Scholar]
- 5.Bogaert D, Kuehn H, Bonroy C, Calvo KR, Dehoorne J, Vanlander AV, et al. A novel IKAROS haploinsufficiency kindred with unexpectedly late and variable B-cell maturation defects. J Allergy Clin Immunol. 2018. January;141 (1):432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wojcik H, Griffiths E, Staggs S, Hagman J, Winandy S. Expression of a non-DNA-binding Ikaros isoform exclusively in B cells leads to autoimmunity but not leukemogenesis. Eur J Immunol. 2007;37(4):1022–1032. [DOI] [PubMed] [Google Scholar]
- 7.Heizmann B, Sellars M, Macias-Garcia A, Chan S, Kastner P. Ikaros limits follicular B cell activation by regulating B cell receptor signaling pathways. Biochem Biophys Res Commun. 2016;470(3):714–720. [DOI] [PubMed] [Google Scholar]
- 8.Dörken B, Moldenhauer G, Pezzutto A, Schwartz R, Feller A, Kiesel S, et al. HD39 (B3), a B lineage-restricted antigen whose cell surface expression is limited to resting and activated human B lymphocytes. J Immunol. 1986; 136:4470–4479. [PubMed] [Google Scholar]
- 9.O’Keefe TL, Williams GT, Davies SL, Neuberger MS. Hyperresponsive B cells in CD22-deficient mice. Science.1996;November(1:274(5288)):798–801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.