Abstract
Mounting evidence suggests that Type 3 Secretions Systems (T3SS) are widespread among Vibrio species, and are present in strains isolated from diverse sources such as human clinical infections, environmental reservoirs, and diseased marine life. Experiments evaluating V. parahaemolyticus and V. cholerae T3SS mediated virulence suggest that Vibrio T3SS pathogenicity islands have a tripartite composition. A conserved “core” region encodes functions essential for colonization and disease in vivo, including modulation of innate immune signaling pathways and actin dynamics, whereas regions flanking core sequences are variable among strains and encode effector proteins performing a diverse array of activities. Characterizing novel functions associated with Vibrio-specific effectors is therefore essential for understanding how vibrios employ T3SS mechanisms to cause disease in a broad range of hosts and how T3SS island composition potentially defines species specific disease.
INTRODUCTION
Vibrio T3SS identification.
Despite widespread identification in many bacterial genera that began in the mid-1980’s with studies on pathogenic Yersinia species, Type 3 Secretion Systems (T3SSs) were not recognized as virulence mechanisms in pathogenic Vibrio species until the completed genome sequence of an O3:K6 serotype V. parahaemolyticus strain, RIMD 2210633, was reported in 2003 [1]. Rapid identification of T3SSs in other Vibrio species followed, and in 2005, genomic sequencing identified the first V. cholerae T3SS in AM-19226, a clinically isolated non-O1/non-O139 serogroup, cholera toxin negative strain that causes non-epidemic cholera [2]. More recently, molecular methods combined with annotation of genomic sequence data expanded the list of T3SS-positive Vibrio species [3-10]. T3SS association with pathogenic Vibrio species is easily explained within the context of a virulence mechanism, but knowing that Vibrio species often interact with multiple hosts (and not always as pathogens) raises the question of whether T3SSs can promote a more symbiotic relationship or alternatively, an advantage in the environmental niche. In either case, researchers now face the challenge of identifying and characterizing novel, often Vibrio-specific effector proteins in an effort to mechanistically understand T3SS mediated interactions with a wide variety of eukaryotic hosts.
Two T3SSs.
Most Vibrio spp. encode one T3SS, but V. parahaemolyticus strains can carry one on each chromosome, respectively termed T3SS1 and T3SS2 [1]. T3SS1 and 2 gene organization and content differs; each T3SS is assembled from distinct proteins and functions independently [3]. T3SS1 is nearly ubiquitous among V. parahaemolyticus strains, is most similar to the Yersinia Ysc T3SS in sequence and synteny, and is associated with mammalian cell cytotoxicity in vitro [1]. Multiple lines of evidence support an ancestral origin, and Vibrio species pathogenic for non-human hosts (e.g. V. alginolyticus, V. campbellii, V. caribbenthicus, V. harveyi, and V. tubiashii) typically encode T3SS1 [1,3,11,12].
In contrast, V. parahaemolyticus T3SS2 is encoded on a genomic pathogenicity island and appears restricted to pandemic O3:K6 serotype isolates and related, pathogenic serovariant strains. Historically, pathogenic strains were identified by a hemolytic property known as the Kanagawa phenomenon, encoded by tdh or trh genes. We now know that the tdh and trh loci are typically found within the T3SS2 genomic island, although the protein products are secreted by another mechanism and are not T3SS substrates [1,3,13].
A subset of V. cholerae non-O1/non-O139 serogroup strains, which can cause sporadic cholera but do not cause epidemic disease, encode T3SS2. The vast majority of T3SS-positive strains lack the major, canonical virulence factors associated with epidemic strains (i.e. toxin co-regulated pilus and cholera toxin), employing the T3SS mediated pathogenic mechanisms instead [2,14-16]. For both V. cholerae and V. parahaemolyticus, experimental evidence using animal models of infection indicates that T3SS2 is required for colonization and disease [17-20].
V. mimicus and V. anguillarum strains pathogenic for humans can also encode a T3SS2 [6,7]. Regarding other species, the use of increasingly sophisticated phylogenetic methods to redefine evolutionary relationships combined with our expanding knowledge of pangenomes results in a fluid understanding of pathogenic mechanisms associated with a particular species [10,21-23]. What follows next therefore summarizes our current knowledge of T3SS effector protein functions and species associations, based largely on experiments in V. parahaemolyticus and V. cholerae.
TYPE THREE SECRETION SYSTEM 1 (T3SS1)
Based on genetic content and organization, the ~40 kb V. parahaemolyticus T3SS1 locus is similar to that found in other Vibrio species. Effector proteins characterized thus far appear to functionally converge at the level of the host membrane, and in some cases, with multiple activities attributed to single effectors. For example, VopQ (also known as VepA) reported activities include induction of autophagy in vitro, activation of the p38, JNK, and ERK Mitogen-Activated Protein Kinase (MAPK) pathways, and host cell lysosome rupture via interaction with the V0 domain of the V-ATPase that forms gated channels [24-26]. VopS encodes a bacterial phosphoinositide-binding (PIB) domain, and PIP2 ligand binding directs effector folding and targeting to the host plasma membrane where VopS mediates actin reorganization by AMPylation of Rho family GTPases, resulting in cytoskeletal collapse and cell rounding [27-29]. VopR also encodes a PIB domain and is localized to the plasma membrane, although its functions remain to be fully elucidated [29]. Interestingly, an effector encoded outside the T3SS1 island displays phosphatidylinositol phosphatase activity, leading Orth and colleagues to propose that other effector activities are influenced by depleting PIP2 from the host cell membrane [30,31].
Although primarily a pathogen of marine life, V. alginolyticus causes T3SS1 dependent cytotoxicity in both fish and mammalian cell lines [32,33]. Interestingly, apoptotic features were present in fish cell lines, whereas mammalian cells appeared to undergo autophagy. Two identified effectors, Val1686 and Val1680, are VopS and VopQ orthologues. Like VopS, Val1686 induces cell rounding, but is also sufficient to trigger apoptosis in infected fish cells. Unlike VopQ, Val1680 does not induce autophagy in fish cells, but it does contribute to T3SS-induced LDH release by an unidentified mechanism [34].
Speculation that T3SS1 is important for survival in the aquatic environment is supported by its presence in both environmental and clinical strains of V. parahaemolyticus and evidence indicating an ancestral origin, consistent with the theory proposed by Zhang et al. that T3SS1-mediated cytotoxicity provides a mechanism to supply nutrients in a nutrient-poor environmental reservoir [35]. Additionally, T3SS1 is not ubiquitously found in V. harveyi strains, and T3SS1 presence was not associated with pathogenicity in a shrimp model even though V. harveyi is documented as a significant marine pathogen, particularly of shrimp [10,36]. A definitive association between T3SS1-mediated phenotypes and human/marine-life infection thus awaits additional molecular characterizations in model systems.
TYPE THREE SECRETION SYSTEM 2 (T3SS2)
T3SS2 clade classification and defining the core region.
The V. cholerae and V. parahaemolyticus T3SS2 gene clusters are more similar to each other in content and synteny than they are to T3SSs from other species. In addition, the Vibrio T3SS structural machinery components do not collectively align with a single T3SS family classification [37]. Rather, structural protein orthologues from each of the three families (Inv-Mxi-Spa, Ysc, and Ssa-Esc) are represented in the Vibrio gene clusters, although some components await definitive identification (Table 1). Given that the T3SS2 has been experimentally shown to function in both V. cholerae and V. parahaemolyticus, the prevailing opinion is that such proteins exist, but are encoded by novel sequences [2,3,17,23].
Table 1.
T3SS Nomenclature. Although a universal nomenclature remains elusive (even between Vibrio species), Table 1 provides a reference to itemize T3SS components. Note that Vibrio ORFs encoding the needle filament, the needle length control protein, pilotin, and inner rod were not identified by initial sequence annotation and remain unidentified.
| Predicted Function |
Yersinia spp. | Salmonella SPI-1 |
V. parahaemolyticus T3SS2 |
V. cholerae T3SS2 |
|---|---|---|---|---|
| IM ring | LcrD/YscV | InvA | VscV2 | VcsV2 |
| IM ring | YscU | SpaS | VscU2 | VcsU2 |
| IM ring | YscR | InvL/SpaP | VscR2 | VcsR2 |
| IM ring | YscT | InvN/SpaR | VscT2 | VcsT2 |
| IM ring | YscS | SpaQ | VscS2 | VcsS2 |
| IM ring | YscD | PrgH | - | VopH? |
| Periplasmic ring | YscJ | PrgK | VscJ2 | VcsJ2 |
| Inner Rod | YscI | PrgJ | - | - |
| ATPase | YscN | InvC/SpaL | VscN2 | VcsN2 |
| Cytoplasmic ring | YscQ | InvK/SpaO | VscQ2 | VcsQ2 |
| Complex with ATPase | YscL | - | - | - |
| Secretin (OM ring) | YscC | MxiD | VscC2 | VcsC2 |
| Pilotin | YscW | MxiM | - | - |
| Needle | YscF | PrgI | - | - |
| Needle length determinant | YscP | InvJ | - | - |
| Hydrophilic translocator | LcrV | SipD | VopW | VopW |
| Translocon | YopB | SipB | VopB2 | VspD2 |
| Translocon | YopD | SipC | VopD2 | VspB2 |
SPI-1, Salmonella pathogenicity island 1; IM, inner membrane OM, outer membrane
Despite lacking sequence similarity to other known T3SS hydrophilic translocators, VopW was experimentally identified by Zhou et al. as a third translocon component essential for effector translocation in V. parahaemolyticus [38,39]. vopW sequences are present in all V. cholerae and V. parahaemolyticus T3SS2 islands, although gene location, number, and sequence identity are variable. Somewhat paradoxically, VopW was identified as a translocated effector protein in both V. parahaemolyticus and V. cholerae [38,40]. However, the collective results are consistent with reports from other systems of effector proteins having dual structural/effector function or T3SS-independent entry [41,42].
T3SS2 has been categorized into two clades, alpha (α) and beta (β), based on the sequences of genes encoding structural components and collective observations suggesting independent acquisition events by ancestral clones [23,43,44]. T3SS2 sequence comparisons that also combine genomic island organization indicate that V. parahaemolyticus and V. cholerae T3SS2α (e.g. RIMD 2210633 and AM-19226) are more similar to each other than T3SS2α and T3SS2β of the same species (e.g. V. cholerae strains AM-19226 and 1587). V. anguillarum and V. mimicus T3SS2 clade classifications have remained elusive, largely due to insufficient sequence data and/or hybrid characteristics [6].
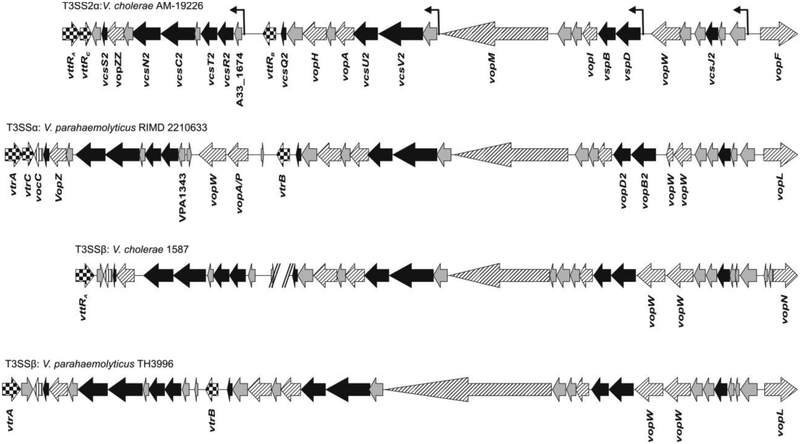
Seven proteins identified as V. cholerae effectors are encoded within and immediately adjacent to the cluster of operons encoding structural apparatus proteins (Vops, Figure 1). Based on sequence similarity, synteny and/or evidence of translocation, each has a V. parahaemolyticus ortholog. For both species, experiments demonstrated that effectors encoded within the structural apparatus cluster are essential in vivo for colonization or disease related phenotypes (described below). Effector proteins are also encoded within mosaic “flanking regions” that lie 5’ and 3’ adjacent to the structural gene operon cluster, but associated phenotypes are less dramatic or remain unknown.
Figure 1.
Genetic organization of representative V. cholerae and V. parahaemolyticus T3SS2 island core regions. T3SS2αclade islands are depicted for V. cholerae strain AM-19226 and V. parahaemolyticus strain RIMD 2210633. T3SS2βclade islands are depicted for V. cholerae strain 1587 and V. parahaemolyticus strain TH3996. Black arrows designate structural apparatus genes, checkered arrows designate transcriptional regulator genes, diagonally lined arrows designate effector protein genes, vertically lined arrows designate chaperone genes, and gray arrows designate genes predicted to encode hypothetical or conserved hypothetical proteins. In addition to vopW position and copy number, differences include a variable length sequence between vcsR2 and vttRb, encoding one of two ToxR-like T3SS transcriptional regulators. vopW is denoted as an effector protein gene here, although its role as a structural component has been described and is discussed in the text. Double hatch lines indicate the end of a contig in the NCBI sequence. Bent arrows above the genes indicate putative promoters for the three major operons, named for the genes encoding the 11 identified structural proteins in the core region: 1) vcsRTCNS2, 2) vcsVUQ2, and 3) vcsJ2, vspD, and vspB [45,82]. Transcriptional reporter fusion data combined with transcriptome analyses suggest the presence of a fourth promoter upstream of vspD (downstream of vopW). 5’ and 3’ genes and regulatory genes are labeled in all four strains for positional reference. Otherwise, only genes differing in position among the four strains are labeled.
We thus conclude that a Vibrio-specific “core” region can be defined within the T3SS pathogenicity island, having the following properties: 1) gene content and position well conserved between species, 2) encoding proteins essential for T3SS structural apparatus function, 3) including effector functions necessary and potentially sufficient for pathogenic mechanisms, such as colonization, and 4) encoding transcriptional regulatory proteins required for T3SS expression. Based on such criteria, the core region (using strain AM-19226 as a reference) is bounded 5’ by vttRa, encoding one of two essential, ToxR-like transmembrane transcriptional regulators, and 3’ by vopF [45]. Shared features between V. cholerae and V. parahaemolyticus thus raises the interesting possibility of a common T3SS mediated mechanism of Vibrio colonization orchestrated by orthologous, core encoded effector proteins.
The clinical spectrum of Vibrio T3SS2-associated human disease
V. parahaemolyticus and V. cholerae primarily cause gastroenteritis, though clinical manifestations of disease are host variable and are also influenced by species and serogroup differences: V. parahaemolyticus typically induces an inflammatory diarrhea, whereas epidemic, O1/O139 serogroup V. cholerae infection is characterized by secretory diarrhea with no damage to the intestinal epithelium [46,47]. Infection by cholera toxin negative, non-O1/non-O139 serogroup V. cholerae strains is historically considered clinically indistinguishable from epidemic strains, but a subset of cases presents with a mild inflammatory component [47,48]. V. mimicus can cause acute gastroenteritis and otitis media after exposure to seawater or contaminated seafood. V. anguillarum is largely a pathogen of crustaceans and bivalves, but along with V. hollisae, is associated with wound infections and can cause severe illness in immunocompromised individuals [21].
It is therefore interesting to note that the inflammatory component of Vibrio spp. associated gastroenteritis has been linked to T3SS2 presence and the causality borne out by experiments recapitulating disease using an orogastrically inoculated infant rabbit model [18]. The intestinal epithelium remains intact during infection by cholera toxin-positive O1 serogroup strains, but infection by T3SS2α-positive strains results in both diarrhea and an altered epithelial cell architecture, with V. parahaemolyticus infection causing increased inflammation and disruption compared to less dramatic damage observed by T3SS-positive V. cholerae strain infection [18,49,50]. One interpretation of the differing T3SS-related pathologies is that common effector proteins and mechanisms are used to colonize and establish an infection, but that clinical variations result from a combination of host factors and species-specific effector proteins or effector alleles.
Core encoded effectors.
The prototype T3SS2α V. cholerae strain, AM-19226, encodes at least 13 translocated proteins. Seven are found within the core region of V. cholerae and are shared with V. parahaemolyticus (differing nomenclature is indicated in parentheses): VopZZ(VopZ), VopH, VopA, VopM (VopV), VopI, VopW, and VopF, (VopN/ VopL) (Figure 1) [40] (Dziejman laboratory, unpublished data). Variable amino acid sequence conservation (28-49% identity and 42-65% similarity) suggests that Vops have strain/species specific attributes while retaining structural conservation to carry out a subset of conserved functions. Whether all proteins detected as present in host cells and translocated in a T3SS-dependent manner function solely as effectors remains to be determined, since bioinformatic and experimental data suggest functions consistent with “missing” secretory apparatus components [19,51].
VopF/N/L all possess Wiscott-Aldrich homology 2 (WH2) domains, which promote mammalian cell actin rearrangement [17,52-55]. All three proteins nucleate actin in vitro, but the phenotypes associated with infection or transfection of mammalian cells differ in that VopF induces actin-rich protrusions, whereas VopN and VopL form actin stress fibers/non-functional actin filaments. V. cholerae vopF is required for wild-type levels of colonization in the infant mouse model, consistent with a role for modulating host cell cytoskeletal dynamics during the early stages of infection. A recent report documented an association between VopL actin dysregulation and limited ROS production resulting from halted assembly of the NADPH oxidase complex at the plasma membrane during V. parahaemolyticus infection [56].
VopV/VopM also display actin reorganization activities, and are essential for in vivo colonization. Biophysical analyses and in vivo results demonstrate that VopV binds actin via ~400bp repeated sequences and interacts with filamin via C-terminal sequences. The resulting cytoskeletal rearrangements play a critical role in brush border effacement and remodel the epithelial cell surface to promote attachment [57-59]. Thus, modulation of actin dynamics clearly plays an important role in Vibrio T3SS pathogenesis, although it is often challenging to determine the pathogenic outcome of cytoskeletal alterations and direct activities vs. global effects resulting from rearrangements, and the precise mechanisms of bacterial adherence to host tissues remain unclear.
The V. parahaemolyticus VopZ effector protein (not to be confused with the V. cholerae effector VopZ, function unknown) is a bifunctional protein important for colonization and intestinal fluid accumulation in the infant rabbit model [60]. Investigators also identified N-terminal domain sequences required for inhibiting TAK1 kinase activation and thus interfering with the NFκB and MAPK signaling pathways. The results thus suggest that like other T3SS positive bacteria, vibrios can modulate the immune response during infection. In V. cholerae strain AM-19226, the VopZZ effector (a VopZ homolog) is absolutely required to cause cytotoxicity in vitro and colonization in vivo, although molecular activities remain to be uncovered and are difficult to reliably predict based on sequence similarity (Dziejman laboratory, unpublished data).
Regions flanking the structural core are mosaic and encode diverse proteins.
Notable T3SS2 genomic island diversity in terms of size (~47kb-~100kb) and genetic content is conferred by 5’ and 3’ flanking regions, which carry sequence remnants consistent with lateral acquisition events [1,2]. The V. cholerae 5’ genomic island flanking sequences encode the VopE and VopX effector proteins [40]. Although VopE is not required for infant mouse colonization, a conserved Rho GTPase-activating domain is responsible for an activity that interferes with mitochondrial dynamics and innate immune responses that utilize mitochondria as a signaling platform [18,40,61-63]. VopX is dispensable for colonization in the infant mouse model, but mediates a cell growth defect in S. cerevisiae by interacting with components of the cell wall integrity (CWI) MAPK pathway, similar to results observed when VopE is expressed in yeast [64,65]. In place of VopX, some strains encode an effector similar to Shigella OspB, which has been shown to modulate the host inflammatory response [66].
The 5’ and 3’ flanking regions in V. parahaemolyticus are more variable and can encode VopO, VopT, VopC, VopA/P and/or VopG, although vopT is not present in T3SS2 stains. The unique VopO effector has no known homologues, but is critical for host cell stress fiber formation and epithelial barrier disruption in vitro [67]. VopT functions as an ADP-ribosyltransferase that targets the mammalian small G protein, Ras, and plays a role in in vitro cytotoxicity of Caco2 and HCT-8 cells [68]. VopA/P (a YopJ homolog, independent from the VopA encoded within the core region) is an acetyltransferase that inactivates MAP Kinase proteins through acetylation [69,70]. Although widely distributed in V. cholerae and V. parahaemolyticus, VopG (function unknown) is not required for V. cholerae infant mouse colonization and combined with the variable location, led to exclusion as a core cluster effector.
The ability of V. parahaemolyticus strain RIMD 2210633 (T3SS2α) to invade HeLa and Caco2 cells as well as HeLa cell invasion by T3SS2β V. cholerae strain 1587 has been attributed to VopC effector protein activity [71-73]. Limited Vibrio species invasion was documented more than 30 years ago prior to T3SS identification, and although the T3SS status of all strains in earlier studies is unknown, it is interesting to note that strain 1587 was among the strains analyzed [74-78]. However, current data indicate that the presence of a T3SS cannot be strictly correlated with an invasive phenotype, and VopC is not required for V. parahaemolyticus colonization or fluid accumulation in the infant rabbit model, nor is it present in V. cholerae T3SSα clade strains [73]. Furthermore, in vivo imaging data from infant rabbit model studies and recently published evidence of intracellular K+ levels in target cells serving as a signal to switch secretion from middle (translocator) to late (effector) substrates strongly support the predominantly extracellular nature of the vibrio-host relationship during gastrointestinal infection [49,50,79].
The V. cholerae mosaic 3’ region (~6.4 kb-17 kb) is comprised of sequences that lie downstream of VopF/N/L. Most V. cholerae strains encode four effector proteins in the 3’ region: VopG , VopK , VopY , and VopZ (which is not a V. parahaemolyticus homolog despite the same name) [40], although VopY is annotated in a limited number of strains. In AM-19226, no 3’ encoded effector is required for infant mouse colonization, but in an infant rabbit model of infection, moderate reductions in the incidence and severity of diarrhea as well as a slight decrease in colonization is observed during infection with VopK or VopY deletion strains [18]. In yeast, VopK toxicity is dependent on residues in the C-terminal domain postulated to comprise an MCF1-SHE serine peptidase domain, although peptidase activity was not detected and motif conservation is imprecise [80].
Conclusions
As we continue to recognize and catalog both similarities and differences, we begin to uncover how Vibrios have diversified T3SS functions to suit specific roles, adaptations, or environments. Thus, it seems likely that effector proteins present in the conserved core region of all T3SS2 islands dictate common mechanisms employed during infection, such as colonization, whereas the mosaic regions encode unique sets of effector proteins that may dictate specific characteristics of infection. Although T3SS1 has been difficult to definitively associate with disease in non-human pathogens, discovering T3SS1 encoded effector associated phenotypes has provided insight into how Vibrio encoded effectors interface with host proteins and pathways. As both V. cholerae and V. parahaemolyticus are considered to be environmental organisms that can act as human pathogens, we must consider whether the T3SS2 provides beneficial phenotypes in the natural aquatic reservoir. Matz et al. demonstrated that T3SS2 promotes V. parahaemolyticus survival during co-culture with a marine flagellate, which correlates with flagellate killing, and ciliates and amoeba were also susceptible to T3SS2-mediated killing [81]. Further studies examining the molecular mechanisms of effector protein function will help to elucidate how these proteins collectively or individually contribute to bacterial fitness and survival in the environment. Indeed, many challenges remain: to identify which effectors are both necessary and sufficient for colonization, to identify effectors specific for activity in the human host during disease, to determine whether effectors are required for a particular niche or lifestyle, and to elucidate effector protein functions, be they unique or redundant within a strain or across T3SS clades and species.
Highlights.
Vibrio spp. can encode one or two unique T3SSs
An environmental role for the T3SS remains largely unknown
Type 3 Secretion System 2 genomic islands in Vibrio species have conserved features
Effectors encoded in the conserved, core gene cluster are critical for colonization
Variable sequence flanking regions encoded effectors are clade and species-specific
ACKNOWLEDGEMENTS
We are grateful to the members of the Dziejman Lab and to Marty Pavelka for critically reading the manuscript, and especially to Chris Seward for his expert assistance with the figure. The Dziejman Lab acknowledges current funding from NIH/NIAID AI126005-01A1 to M.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author declarations of interest: none
The authors declare no conflict of interest.
References:
- 1.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, et al. : Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 2003, 361:743–749. [DOI] [PubMed] [Google Scholar]
- 2.Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, et al. : Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci U S A 2005, 102:3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T: Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun 2004, 72:6659–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henke JM, Bassler BL: Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol 2004, 186:3794–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Persson OP, Pinhassi J, Riemann L, Marklund BI, Rhen M, Normark S, Gonzalez JM, Hagstrom A: High abundance of virulence gene homologues in marine bacteria. Environ Microbiol 2009, 11:1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okada N, Matsuda S, Matsuyama J, Park KS, de Los Reyes C, Kogure K, Honda T, Iida T: Presence of genes for type III secretion system 2 in Vibrio mimicus strains. BMC Microbiol 2010, 10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naka H, Dias GM, Thompson CC, Dubay C, Thompson FL, Crosa JH: Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect Immun 2011, 79:2889–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, Monday SR, Allard MW, Strain EA, Whittaker P, Naum M, McCarthy PJ, Lopez JV, Fischer M, Brown EW: Vibrio caribbeanicus sp. nov., isolated from the marine sponge Scleritoderma cyanea. Int J Syst Evol Microbiol 2012, 62:1736–1743. [DOI] [PubMed] [Google Scholar]
- 9.Dias GM, Thompson CC, Fishman B, Naka H, Haygood MG, Crosa JH, Thompson FL: Genome sequence of the marine bacterium Vibrio campbellii DS40M4, isolated from open ocean water. J Bacteriol 2012, 194:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin B, Wang Z, Malanoski AP, O'Grady EA, Wimpee CF, Vuddhakul V, Alves N Jr, Thompson FL, Gomez-Gil B, Vora GJ: Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep 2010, 2:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espejo RT, Garcia K, Plaza N: Insight Into the Origin and Evolution of the Vibrio parahaemolyticus Pandemic Strain. Front Microbiol 2017, 8:1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Austin B: Taxonomy of bacterial fish pathogens. Vet Res 2011, 42:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izutsu K, Kurokawa K, Tashiro K, Kuhara S, Hayashi T, Honda T, Iida T: Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect Immun 2008, 76:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Octavia S, Salim A, Kurniawan J, Lam C, Leung Q, Ahsan S, Reeves PR, Nair GB, Lan R: Population structure and evolution of non-O1/non-O139 Vibrio cholerae by multilocus sequence typing. PLoS One 2013, 8:e65342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta D, Chowdhury G, Pazhani GP, Guin S, Dutta S, Ghosh S, Rajendran K, Nandy RK, Mukhopadhyay AK, Bhattacharya MK, et al. : Vibrio cholerae non-O1, non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg Infect Dis 2013, 19:464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasan NA, Ceccarelli D, Grim CJ, Taviani E, Choi J, Sadique A, Alam M, Siddique AK, Sack RB, Huq A, et al. : Distribution of virulence genes in clinical and environmental Vibrio cholerae strains in Bangladesh. Appl Environ Microbiol 2013, 79:5782–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ: A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 2007, 1:95–107. [DOI] [PubMed] [Google Scholar]
- 18.Shin OS, Tam VC, Suzuki M, Ritchie JM, Bronson RT, Waldor MK, Mekalanos JJ: Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. MBio 2011, 2:e00106–00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller KA, Chaand M, Gregoire S, Yoshida T, Beck LA, Ivanov AI, Dziejman M: Characterization of V. cholerae T3SS-dependent cytotoxicity in cultured intestinal epithelial cells. Cell Microbiol 2016, 18:1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel Zur Wiesch P, Zhou X, Davis BM, Waldor MK: Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Natl Acad Sci U S A 2016, 113:6283–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson FL, Iida T, Swings J: Biodiversity of vibrios. Microbiol Mol Biol Rev 2004, 68:403–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin H, Yu M, Wang X, Zhang XH: Comparative genomic analysis reveals the evolution and environmental adaptation strategies of vibrios. BMC Genomics 2018, 19:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada N, Iida T, Park KS, Goto N, Yasunaga T, Hiyoshi H, Matsuda S, Kodama T, Honda T: Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun 2009, 77:904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimohata T, Nakano M, Lian X, Shigeyama T, Iba H, Hamamoto A, Yoshida M, Harada N, Yamamoto H, Yamato M, et al. : Vibrio parahaemolyticus infection induces modulation of IL-8 secretion through dual pathway via VP1680 in Caco-2 cells. J Infect Dis 2011, 203:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuda S, Okada N, Kodama T, Honda T, Iida T: A cytotoxic type III secretion effector of Vibrio parahaemolyticus targets vacuolar H+-ATPase subunit c and ruptures host cell lysosomes. PLoS Pathog 2012, 8:e1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matlawska-Wasowska K, Finn R, Mustel A, O'Byrne CP, Baird AW, Coffey ET, Boyd A: The Vibrio parahaemolyticus Type III Secretion Systems manipulate host cell MAPK for critical steps in pathogenesis. BMC Microbiol 2010, 10:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharjee RN, Park KS, Chen X, Iida T, Honda T, Takeuchi O, Akira S: Translocation of VP1686 upregulates RhoB and accelerates phagocytic activity of macrophage through actin remodeling. J Microbiol Biotechnol 2008, 18:171–175. [PubMed] [Google Scholar]
- 28.Casselli T, Lynch T, Southward CM, Jones BW, DeVinney R: Vibrio parahaemolyticus inhibition of Rho family GTPase activation requires a functional chromosome I type III secretion system. Infect Immun 2008, 76:2202–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salomon D, Guo Y, Kinch LN, Grishin NV, Gardner KH, Orth K: Effectors of animal and plant pathogens use a common domain to bind host phosphoinositides. Nat Commun 2013, 4:2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono T, Park KS, Ueta M, Iida T, Honda T: Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect Immun 2006, 74:1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broberg CA, Zhang L, Gonzalez H, Laskowski-Arce MA, Orth K: A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science 2010, 329:1660–1662. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Z, Chen C, Hu CQ, Ren CH, Zhao JJ, Zhang LP, Jiang X, Luo P, Wang QB: The type III secretion system of Vibrio alginolyticus induces rapid apoptosis, cell rounding and osmotic lysis of fish cells. Microbiology 2010, 156:2864–2872. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Z, Zhang L, Ren C, Zhao J, Chen C, Jiang X, Luo P, Hu CQ: Autophagy is induced by the type III secretion system of Vibrio alginolyticus in several mammalian cell lines. Arch Microbiol 2011, 193:53–61. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Z, Liu J, Deng Y, Huang W, Ren C, Call DR, Hu C: The Vibrio alginolyticus T3SS effectors, Val1686 and Val1680, induce cell rounding, apoptosis and lysis of fish epithelial cells. Virulence 2018, 9:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Orth K: Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol 2013, 16:70–77. [DOI] [PubMed] [Google Scholar]
- 36.Rattanama P, Srinitiwarawong K, Thompson JR, Pomwised R, Supamattaya K, Vuddhakul V: Shrimp pathogenicity, hemolysis, and the presence of hemolysin and TTSS genes in Vibrio harveyi isolated from Thailand. Dis Aquat Organ 2009, 86:113–122. [DOI] [PubMed] [Google Scholar]
- 37.Burkinshaw BJ, Strynadka NC: Assembly and structure of the T3SS. Biochim Biophys Acta 2014, 1843:1649–1663. [DOI] [PubMed] [Google Scholar]
- 38.Zhou X, Ritchie JM, Hiyoshi H, Iida T, Davis BM, Waldor MK, Kodama T: The hydrophilic translocator for Vibrio parahaemolyticus, T3SS2, is also translocated. Infect Immun 2012, 80:2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodama T, Hiyoshi H, Gotoh K, Akeda Y, Matsuda S, Park KS, Cantarelli VV, Iida T, Honda T: Identification of two translocon proteins of Vibrio parahaemolyticus type III secretion system 2. Infect Immun 2008, 76:4282–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam A, Miller KA, Chaand M, Butler JS, Dziejman M: Identification of Vibrio cholera type III secretion system effector proteins. Infect Immun 2011, 79:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fields KA, Straley SC: LcrV of Yersinia pestis enters infected eukaryotic cells by a virulence plasmid-independent mechanism. Infect Immun 1999, 67:4801–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchrieser C, Glaser P, Rusniok C, Nedjari H, D'Hauteville H, Kunst F, Sansonetti P, Parsot C: The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 2000, 38:760–771. [DOI] [PubMed] [Google Scholar]
- 43.Murphy RA, Boyd EF: Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol 2008, 190:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morita M, Yamamoto S, Hiyoshi H, Kodama T, Okura M, Arakawa E, Alam M, Ohnishi M, Izumiya H, Watanabe H: Horizontal gene transfer of a genetic island encoding a type III secretion system distributed in Vibrio cholerae. Microbiol Immunol 2013, 57:334–339. [DOI] [PubMed] [Google Scholar]
- 45.Alam A, Tam V, Hamilton E, Dziejman M: vttRA and vttRB Encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain. Infect Immun 2010, 78:2554–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qadri F, Alam MS, Nishibuchi M, Rahman T, Alam NH, Chisti J, Kondo S, Sugiyama J, Bhuiyan NA, Mathan MM, et al. : Adaptive and inflammatory immune responses in patients infected with strains of Vibrio parahaemolyticus. J Infect Dis 2003, 187:1085–1096. [DOI] [PubMed] [Google Scholar]
- 47.Nelson EJ, Harris JB, Morris JG Jr., Calderwood SB, Camilli A: Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol 2009, 7:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris JG Jr.: Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin Infect Dis 2003, 37:272–280. [DOI] [PubMed] [Google Scholar]
- 49.Ritchie JM, Rui H, Zhou X, Iida T, Kodoma T, Ito S, Davis BM, Bronson RT, Waldor MK: Inflammation and Disintegration of Intestinal Villi in an Experimental Model for Vibrio parahaemolyticus-Induced Diarrhea. PLoS Pathog 2012, 8:e1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie JM, Rui H, Bronson RT, Waldor MK: Back to the future: studying cholera pathogenesis using infant rabbits. MBio 2010, 1:e00047–00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaand M, Miller KA, Sofia MK, Schlesener C, Weaver JW, Sood V, Dziejman M: Type 3 Secretion System Island Encoded Proteins Required for Colonization by Non-O1/non-O139 Serogroup V. cholerae. Infect Immun 2015, 83:2862–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tam VC, Suzuki M, Coughlin M, Saslowsky D, Biswas K, Lencer WI, Faruque SM, Mekalanos JJ: Functional analysis of VopF activity required for colonization in Vibrio cholerae. MBio 2010, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, Rosen MK, Orth K: Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci U S A 2007, 104:17117–17122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Avvaru BS, Pernier J, Carlier MF: Dimeric WH2 repeats of VopF sequester actin monomers into non-nucleating linear string conformations: An X-ray scattering study. J Struct Biol 2015, 190:192–199. [DOI] [PubMed] [Google Scholar]
- 55.Pernier J, Orban J, Avvaru BS, Jegou A, Romet-Lemonne G, Guichard B, Carlier MF: Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat Struct Mol Biol 2013, 20:1069–1076. [DOI] [PubMed] [Google Scholar]
- 56.de Souza Santos M, Salomon D, Orth K: T3SS effector VopL inhibits the host ROS response, promoting the intracellular survival of Vibrio parahaemolyticus. PLoS Pathog 2017, 13:e1006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiyoshi H, Kodama T, Saito K, Gotoh K, Matsuda S, Akeda Y, Honda T, Iida T: VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe 2011, 10:401–409. [DOI] [PubMed] [Google Scholar]
- 58.Zhou X, Massol RH, Nakamura F, Chen X, Gewurz BE, Davis BM, Lencer WI, Waldor MK: Remodeling of the intestinal brush border underlies adhesion and virulence of an enteric pathogen. MBio 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimura M, Fujii T, Hiyoshi H, Makino F, Inoue H, Motooka D, Kodama T, Ohkubo T, Kobayashi Y, Nakamura S, et al. : A repeat unit of Vibrio diarrheal T3S effector subverts cytoskeletal actin homeostasis via binding to interstrand region of actin filaments. Sci Rep 2015, 5:10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou X, Gewurz BE, Ritchie JM, Takasaki K, Greenfeld H, Kieff E, Davis BM, Waldor MK: A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Rep 2013, 3:1690–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aepfelbacher M, Roppenser B, Hentschke M, Ruckdeschel K: Activity modulation of the bacterial Rho GAP YopE: an inspiration for the investigation of mammalian Rho GAPs. Eur J Cell Biol 2011, 90:951–954. [DOI] [PubMed] [Google Scholar]
- 62.Von Pawel-Rammingen U, Telepnev MV, Schmidt G, Aktories K, Wolf-Watz H, Rosqvist R: GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol Microbiol 2000, 36:737–748. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki M, Danilchanka O, Mekalanos JJ: Vibrio cholerae T3SS effector VopE modulates mitochondrial dynamics and innate immune signaling by targeting Miro GTPases. Cell Host Microbe 2014, 16:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seward CH, Manzella A, Alam A, Butler JS, Dziejman M: Using S. cerevisiae as a Model System to Investigate V. cholerae VopX-Host Cell Protein Interactions and Phenotypes. Toxins (Basel) 2015, 7:4099–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bankapalli LK, Mishra RC, Raychaudhuri S: VopE, a Vibrio cholerae Type III Effector, Attenuates the Activation of CWI-MAPK Pathway in Yeast Model System. Front Cell Infect Microbiol 2017, 7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zurawski DV, Mumy KL, Faherty CS, McCormick BA, Maurelli AT: Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol Microbiol 2009, 71:350–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hiyoshi H, Okada R, Matsuda S, Gotoh K, Akeda Y, Iida T, Kodama T: Interaction between the type III effector VopO and GEF-H1 activates the RhoA-ROCK pathway. PLoS Pathog 2015, 11:e1004694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kodama T, Rokuda M, Park KS, Cantarelli VV, Matsuda S, Iida T, Honda T: Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol 2007, 9:2598–2609. [DOI] [PubMed] [Google Scholar]
- 69.Trosky JE, Li Y, Mukherjee S, Keitany G, Ball H, Orth K: VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem 2007, 282:34299–34305. [DOI] [PubMed] [Google Scholar]
- 70.Trosky JE, Mukherjee S, Burdette DL, Roberts M, McCarter L, Siegel RM, Orth K: Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J Biol Chem 2004, 279:51953–51957. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L, Krachler AM, Broberg CA, Li Y, Mirzaei H, Gilpin CJ, Orth K: Type III effector VopC mediates invasion for Vibrio species. Cell Rep 2012, 1:453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Souza Santos M, Orth K: Intracellular Vibrio parahaemolyticus escapes the vacuole and establishes a replicative niche in the cytosol of epithelial cells. MBio 2014, 5:e01506–01514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okada R, Zhou X, Hiyoshi H, Matsuda S, Chen X, Akeda Y, Kashimoto T, Davis BM, Iida T, Waldor MK, et al. : The Vibrio parahaemolyticus effector VopC mediates Cdc42-dependent invasion of cultured cells but is not required for pathogenicity in an animal model of infection. Cell Microbiol 2014, 16:938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dalsgaard A, Albert MJ, Taylor DN, Shimada T, Meza R, Serichantalergs O, Echeverria P: Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol 1995, 33:2715–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akeda Y, Nagayama K, Yamamoto K, Honda T: Invasive phenotype of Vibrio parahaemolyticus. J Infect Dis 1997, 176:822–824. [DOI] [PubMed] [Google Scholar]
- 76.Boutin BK, Townsend SF, Scarpino PV, Twedt RM: Demonstration of invasiveness of Vibrio parahaemolyticus in adult rabbits by immunofluorescence. Appl Environ Microbiol 1979, 37:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russell RG, Tall BD, Morris JG Jr.,: Non-O1 Vibrio cholerae intestinal pathology and invasion in the removable intestinal tie adult rabbit diarrhea model. Infect Immun 1992, 60:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chattarjee BD, Mukherjee A, Sanyal SN: Rabbit ileal loop invasion of Vibrio parahaemolyticus. Indian J Pathol Microbiol 1982, 25:213–218. [PubMed] [Google Scholar]
- 79.Tandhavanant S, Matsuda S, Hiyoshi H, Iida T, Kodama T: Vibrio parahaemolyticus Senses Intracellular K(+) To Translocate Type III Secretion System 2 Effectors Effectively. MBio 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bankapalli LK, Mishra RC, Singh B, Raychaudhuri S: Identification of Critical Amino Acids Conferring Lethality in VopK, a Type III Effector Protein of Vibrio cholerae: Lessons from Yeast Model System. PLoS One 2015, 10:e0141038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matz C, Nouri B, McCarter L, Martinez-Urtaza J: Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS One 2011, 6:e20275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaand M, Dziejman M: Vibrio cholerae VttRA and VttRB Regulatory Influences Extend beyond the Type 3 Secretion System Genomic Island. J Bacteriol 2013, 195:2424–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]