Abstract
Glial cell line Derived Neurotrophic Factor (GDNF) is a protein that is required for the development and survival of enteric, sympathetic, and catecholaminergic neurons. We previously reported that GDNF is protective against high fat diet (HFD)-induced hepatic steatosis in mice through suppression of hepatic expression of peroxisome proliferator activated receptor- γ (PPAR-γ) and genes encoding enzymes involved in de novo lipogenesis. We also reported that transgenic overexpression of GDNF in mice prevented the HFD-induced liver accumulation of the autophagy cargo-associated protein p62/sequestosome 1 (p62/SQSTM1) characteristic of impaired autophagy. Here we investigated the effects of GDNF on hepatic autophagy in response to increased fat load, and on hepatocyte mitochondrial fatty acid β-oxidation and cell survival. GDNF not only prevented the reductions in the liver levels of some key autophagy-related proteins, including Atg5, Atg7, Beclin-1 and LC3A/B-II, seen in HFD-fed control mice, but enhanced their levels after 12-weeks of HFD-feeding. In vitro, GDNF accelerated autophagic cargo clearance in primary mouse hepatocytes and a rat hepatocyte cell line, and reduced the phosphorylation of the mTOR complex downstream target p70S6 kinase similar to the autophagy activator rapamycin. GDNF also enhanced mitochondrial fatty acid β-oxidation in primary mouse and rat hepatocytes, and protected against palmitate-induced lipotoxicity. Conclusion: We demonstrate a novel role for GDNF in enhancing hepatic autophagy and in potentiating mitochondrial function and fatty acid oxidation. Our studies show that GDNF and its receptor agonists could be useful for enhancing hepatocyte survival and protecting against fatty acid-induced hepatic lipotoxicity.
Keywords: autophagic flux, GDNF transgenic, mice, respiration, survival
Nonalcoholic fatty liver disease (NAFLD) has become a major health concern globally due to the worldwide obesity epidemic. NAFLD is a complex disease which in its simplest form is characterized by excessive accumulation of triglyceride (TG) in hepatocytes due to increased uptake of free fatty acids (FFA), increased fatty acid (FA) synthesis through de novo lipogenesis, and reduced fatty acid clearance (1). This simple steatosis can progress to the more severe nonalcoholic steatohepatitis (NASH) which is characterized by steatosis, hepatocellular injury, inflammation and liver fibrosis (2).
Autophagy is a cell survival pathway in which intracellular components are degraded in lysosomes to release substrates which can be used for energy production and the synthesis of new macromolecules (3). Macroautophagy has been shown to play an important role in lipid homeostasis (lipophagy) and mitochondrial quality control (mitophagy) in hepatocytes (4, 5). The dysregulation of hepatic autophagy seen in animal models of obesity and in some NAFLD patients, as well as the reduction in TG accumulation observed in response to the induction of liver autophagy, suggests that hepatic autophagy could be a useful target in the treatment of NAFLD (6–10).
We recently reported that glial cell line derived neurotrophic factor (GDNF), a factor required for the survival of sympathetic, catecholaminergic and enteric neurons, is protective against high fat diet (HFD)-induced hepatic steatosis in mice. The mechanism of this effect is in part through suppression of hepatic expression of peroxisome proliferator activated receptor-γ (PPAR-γ) and its downstream targets as well as suppression of gene expression of fatty acid synthase (Fasn) and stearoyl-CoA desaturase (Scd1) which encode enzymes involved in de novo lipogenesis (11). Interestingly, we also found that liver p62/sequestosome 1 (p62/SQSTM1) levels were highly elevated in HFD-fed CF-1 mice, but significantly decreased in GDNF overexpressing mice. This protein is a key autophagic clearance protein which recognizes and delivers ubiquitinated protein cargo to the autophagosome for downstream degradation in lysosomes, and increased levels of p62/SQSTM1 are often associated with inhibition of autophagy while decreased levels of the protein are associated with autophagy activation (12–15). The finding that GDNF overexpression resulted in reductions in liver p62/SQSTM1 levels HFD-fed mice thus suggested that GDNF might up regulate liver autophagy in obesity.
In the current study we investigated the effects of GDNF on the hepatic autophagic pathway and the role of GDNF in modulating hepatocyte mitochondrial function, fatty acid oxidation and cell survival. We show that GDNF protects against HFD-induced suppression of liver autophagy and enhances mitochondrial fatty acid oxidation in hepatocytes. We also show that GDNF protects against palmitate-induced toxicity in hepatocytes. Our findings demonstrate an important ability of GDNF to regulate autophagy and lipid metabolism in hepatocytes.
Experimental Procedures
Animals
Animal studies were conducted in male and female CF-1 (control) mice and GDNF transgenic (Tg) littermates (on a CF-1 background). GDNF transgenic mice overexpress GDNF in cells expressing glial fibrillary acidic protein (GFAP) which are found in several tissues including the liver (Supplementary Fig. 1) (16). The generation of these mice has been previously described (17). The mice were used at 5–6 weeks of age and were maintained on a 12 h light-dark cycle in a temperature controlled barrier facility with free access to food and water. They were fed a regular diet (RD) (2018SX; Teklad Global 18% Protein Extruded Rodent Diet, Harlan Laboratories, Madison, WI) that contained 6.2% fat by weight or a HFD (TD.06414, Harlan) that contained 34.3% fat by weight. The complete composition of the diets has previously been reported (16). All animal studies were approved by the Atlanta Veteran Affairs Medical Center Institutional Animal Care and Use Committee.
Cell Culture
RALA255–10G rat hepatocytes infected with empty lentiviral vector alone (VEC cells) or lentivirus expressing shRNA to Atg5 (siAtg5 cells) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) without pyruvate as previously described (4, 18, 19). The human hepatoma-derived HepG2 cell line (20) was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Minimum Essential Medium (ATCC) supplemented with fetal bovine serum (10%) according to recommended procedure. Primary mouse hepatocytes were isolated as previously described (21) using Gibco (Life Technologies Corp, Grand Island, NY, USA) hepatocyte isolation and culture media. Stock (6 mM) palmitate (Sigma-Aldrich, St. Louis, MO) conjugated to fatty acid-free bovine serum albumin (BSA) (Sigma-Aldrich) was prepared as previously described (22) and used at final concentration of 0.1–0.3 mM. Recombinant human and mouse GDNF (Shenandoah Biotechnology, Warwick, PA) were used at a final concentration of 100–200 ng/ml. Medium for all the cell types were replaced with fresh media every 48 h.
Oxygen consumption rate measurement
Primary mouse hepatocytes, RALA255–10G cells and HepG2 were seeded in 96-well XFe96 cell culture microplates (Agilent Technologies Seahorse Bioscience, Lexington, MA, USA) and cultured for 24 h to six days in complete medium supplemented with or without GDNF and palmitate (0.1–0.2 mM). The β-adrenergic receptor antagonist SR 59230A (Sigma-Aldrich) was added (10 µM) to the appropriate wells during the last 4 days of culture while rapamycin (Calbiochem EMD Chemicals, Darmstadt, Germany) was added (20 nM) during the last 24 h hours of culture. The culture media were replaced every 48 h with fresh media supplemented with or without the test agents. Before the start of the respiration assay, the cells were washed twice with fatty acid oxidation (FAO) assay medium (111 mM NaCl, 4.7 mM KCl, 1.25 mM CaCl2, 2 mM MgSO4, 1.2 mM NaH2PO4, 2.5 mM glucose, 0.5 mM carnitine, and 5 mM HEPES) and incubated in the same medium for 45 min at 37°C in a non-CO2 incubator. Just before the start of the assay GDNF (100 ng/ml), palmitate (1 mM), rapamycin (20 nM) and SR 59230A (10 µM) were added to the appropriate wells. Mitochondrial bioenergetic function and fatty acid oxidation were then assessed by measuring the oxygen consumption rates (OCR) using a Seahorse XFe96 Extracellular Flux Analyzer (Seahorse Bioscience) before and after the addition of 1.0 mM oligomycin (Seahorse Bioscience), 1.0 mM Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) (Seahorse Bioscience) and 0.5 mM rotenone/antimycin A (Seahorse Bioscience) (23, 24). Oxygen consumption rates were normalized to protein.
Cellular toxicity Assay
To assess the toxicity of palmitate, RALA255–10G cells and HepG2 cells were plated at a density of 5 × 103 cells/well in 96-well plates. After 48 h in regular cell culture medium, the medium was replaced with fresh culture medium supplemented with or without GDNF (200 ng/ml) and palmitate (0.2 mM and 0.25 mM). Cell death was assessed after 24h and 48h using the CellTox Green Cytotoxicity Assay kit (Promega, Madison, WI). Fluorescence was measured at 485 nmEx/ 528 nmEm and the percentage of cell death determined by dividing the fluorescence of the GDNF and palmitate treated groups by the fluorescence of the vehicle treated control group, multiplying by a hundred and then subtracting the result from 100.
Western blotting
Western blotting was performed as previously described (11) using rabbit primary antibodies to Atg5 (D5F5U), Atg7, Beclin-1, cleaved caspase-3 (Asp175) LC3A/B (D3U4C), total and phospho-mTOR (Ser 2448), total and phospho-p70SK6 (Thr389), total and phospho-4E-BP1 (Ser65) (Cell Signaling Technologies), and p62/SQSTM1 (Sigma-Aldrich) diluted 1:1000. Mouse monoclonal primary antibodies to α-tubulin (DM1A) (Cell Signaling Technologies, Danvers, MA) and β-actin (A5441, clone AC-15) (Sigma-Aldrich) were diluted, respectively, 1:1000 and 1:5000 before use. Horseradish peroxidase conjugated anti-mouse and anti-rabbit IgG (Cell Signaling Technologies) secondary antibodies were used at 1:2,000 dilution. A semi quantitative measurement of band intensity was performed using the Carestream Molecular Imaging Software (Carestream Molecular Imaging, New Haven, CT).
Assessment of Autophagic Flux
To assess autophagic flux, primary mouse hepatocytes and RALA255–10G rat hepatocytes were cultured for 1 h in cell culture medium supplemented with or without the lysosomal inhibitors ammonium chloride (20 mM) (Sigma-Aldrich) and leupeptin (100 μM) (Roche Diagnostics, Mannheim, Germany) followed by 4 h in the presence or absence of the inhibitors, GDNF (200 ng/ml) and palmitate (0.2 mM). Protein were isolated and subjected to Western blotting for LC3A/B-II. Autophagic flow was then determined as previously described (4, 25) by calculating the ratio of the LC3A/B-II band intensity value in the presence of the lysosomal inhibitors (normalized to its β-actin levels) to the LC3A/B-II band intensity value for the same treatment in the absence of the inhibitors.
Immunofluorescence staining
Liver sections (8 μm thick) were obtained from mice liver tissues previously frozen in Tissue-Tek O. C. T. compound (Sakura Finetek, Torrance, CA, U.S.A.) and stained with antibodies to LC3A/B (Cell Signaling Technologies) and p62/SQSTM1 (Sigma) as previously described (11) and counterstained for lipid droplets using BODIPY 493/503 (Life Technologies) according to suggested procedure.
Statistical Analysis
Statistical analyses were conducted using the GraphPad Prism software version 3.00 for Windows (GraphPad Software, San Diego, CA). Data were tested for normality and subjected to unpaired t test or one-way ANOVA with Tukey post test.
Results
GDNF transgenic mice are protected against high fat diet-induced impairment of liver autophagy
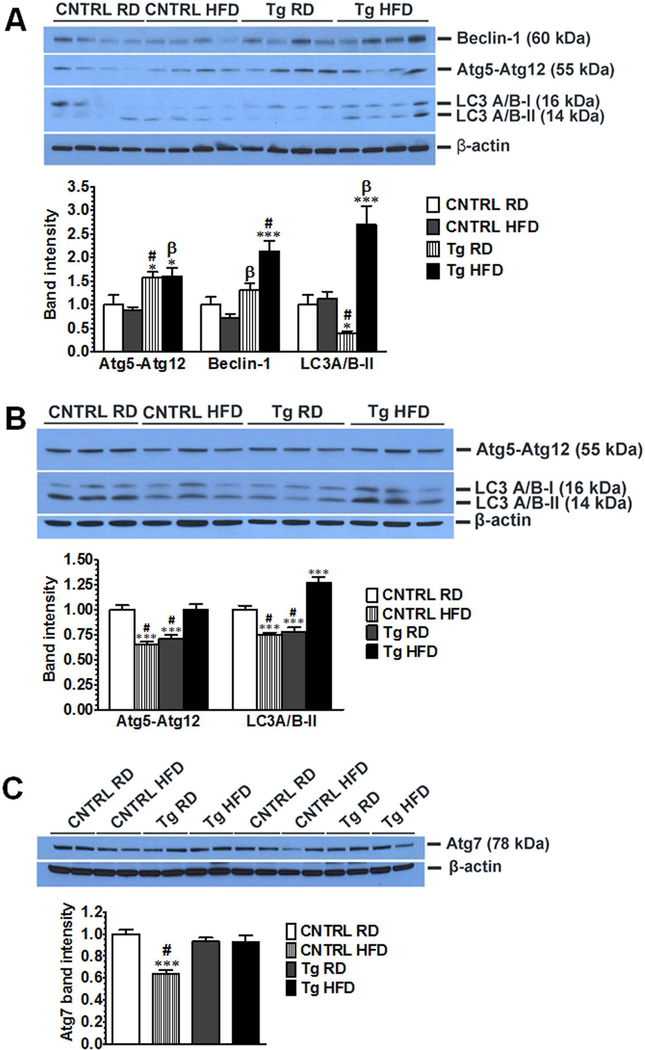
To investigate the ability of GDNF to regulate hepatocyte autophagy in response to increased fat load, we first assessed the effects of GDNF overexpression on liver levels of key components of the autophagy pathway in a dietary mouse model of obesity. GDNF Tg (Tg) mice fed the HFD for 12 weeks had normal liver triglyceride levels, serum alanine aminotransferase (ALT) levels and low to moderate hepatic steatosis while control (CNTRL) mice fed the HFD had elevated liver triglyceride levels and serum ALT levels and increased hepatic steatosis (16) (Supplementary Fig. 2). We also observed that 12 weeks of HFD feeding resulted in significant increases in liver Atg5, Beclin-1, and LC3A/B-II levels in male and female GDNF Tg mice, and decreases in liver Atg5, Atg7 and LC3A/B-II levels in female CNTRL mice (Fig. 1A–C). These data thus suggested an increase in hepatic autophagy geared towards protecting against steatosis and lipotoxicity in HFD-fed GDNF Tg mice.
FIG. 1.
GDNF prevents high fat diet-induced reduction in the levels of components of the autophagic pathway in mouse liver. Western blot analysis of (A) Beclin-1, Atg5, and LC3A/B levels in liver from male control (CNTRL) and GDNF transgenic (Tg) mice fed a RD or HFD for 12 weeks. β-actin was used as a loading control. Histogram shows fold change relative to RD-fed control mice of band intensities per β-actin band intensities. Plotted are means + SEM (***, P<0.001; *, P<0.05 relative to CNTRL RD; #, P<0.001; β, P<0.01 relative to CNTRL HFD. n = 4 mice in each group). (B) Western blot analysis of Atg5 and LC3A/B protein levels in liver from female control (CNTRL) and GDNF transgenic (Tg) mice fed a RD or HFD for 12 weeks. Plotted are means + SEM (***, P<0.001 relative to CNTRL RD; #, P<0.001 relative to Tg HFD. n = 3 mice per group). (C) Western blot analysis of Atg7 protein levels in liver from female control (CNTRL) and GDNF transgenic (Tg) mice fed a RD or HFD for 12 weeks. Plotted are means + SEM (***, P<0.001 relative to CNTRL RD; #, P<0.001 relative to Tg HFD. n = 4 mice per group).
GDNF enhances autophagy in vitro
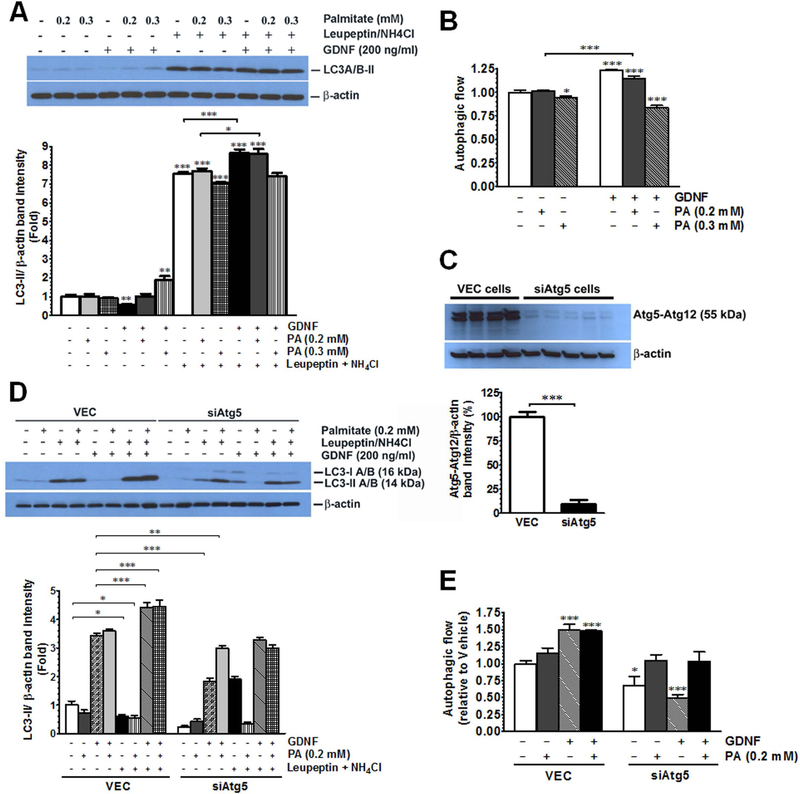
While an increase in LC3 levels can be an indicator of autophagy induction, a decrease in cellular LC3-II levels is also known to result from increased autophagic flux (turnover) (15). To examine if GDNF could enhance autophagic flux in response to exposure to saturated fatty acids, we first pretreated isolated mouse hepatocytes for 1 h with or without the lysosomal inhibitors ammonium chloride and leupeptin to block autophagic cargo clearance. We then exposed the cells to GDNF (200 ng/ml) and palmitate (0.2–0.3 mM) for 4 h in the presence or absence of the inhibitors and then isolated protein to assess autophagic flow. Hepatocytes exposed to GDNF or to 0.2 mM palmitate in the presence of GDNF had significantly higher autophagic flow than cells exposed either to vehicle or palmitate alone (Fig. 2A and B). To verify that the observed GDNF effects were direct effects on autophagy we compared changes in autophagic flux between VEC RALA255–10G rat hepatocytes (control) which have an intact autophagy machinery and siAtg5 RALA255–10G hepatocytes which have impaired autophagy due to knockdown of Atg5 expression (Fig. 2C). Controls cells exposed to GDNF alone or palmitate in the presence of GDNF exhibited increased autophagic flow when compared to cells vehicle or palmitate alone (Fig. 2D and E). These effects of GDNF were, however, absent in siAtg5 RALA255–10G rat hepatocytes (Fig. 2D and E). The ability of GDNF to enhance autophagy flux was also evident in VEC RALA255–10G hepatocytes, but not siAtg5 RALA255–10G hepatocytes, exposed to palmitate for 6 days (supplementary Fig. 3A and B). GDNF also significantly decreased p62/SQSTM1 levels in VEC cells exposed to palmitate for 6 days while lysosomal inhibitors induced similar increases as those seen in cells exposed to vehicle alone (Supplementary Fig. 3C) thus providing further support that GDNF can stimulate autophagy in response to a lipid challenge.
FIG. 2.
GDNF enhances autophagic flux in rat hepatocytes and primary mouse primary mouse hepatocytes in vitro (A) Western blot analysis of LC3 levels in VEC (empty vector-infected) and siAtg5 (Atg5 siRNA infected) RALA255–10G rat hepatocytes pre-cultured for 1 h in medium supplemented with or without the lysosomal inhibitors leupeptin and ammonium chloride followed by 4 h in medium supplemented with or without GDNF and palmitate (PA), and (B) plot of autophagic flow for GDNF and PA treated cells relative to cells treated with vehicle only. Plotted are means + SEM (***, P<0.001; *, P<0.05. n = 3). (C) Western blot comparing Atg5 protein expression levels between VEC and siAtg5 RALA255–10G rat hepatocytes. Plotted are means + SEM (***, P<0.001. n= 4–5). (D) Western blot analysis of LC3 levels in primary mouse hepatocytes pre-cultured for 1h in medium supplemented with or without the lysosomal inhibitors leupeptin and ammonium chloride followed by 4h in medium supplemented with or without GDNF and palmitate (PA), and (E) plot of autophagic flow for GDNF and PA treated cells relative to cells treated with vehicle only. Plotted are means + SEM (***, P<0.001; *, P<0.05. n = 3).
GDNF suppresses mTOR signaling in hepatocytes and enhances cellular respiration similar to the autophagy inducer rapamycin
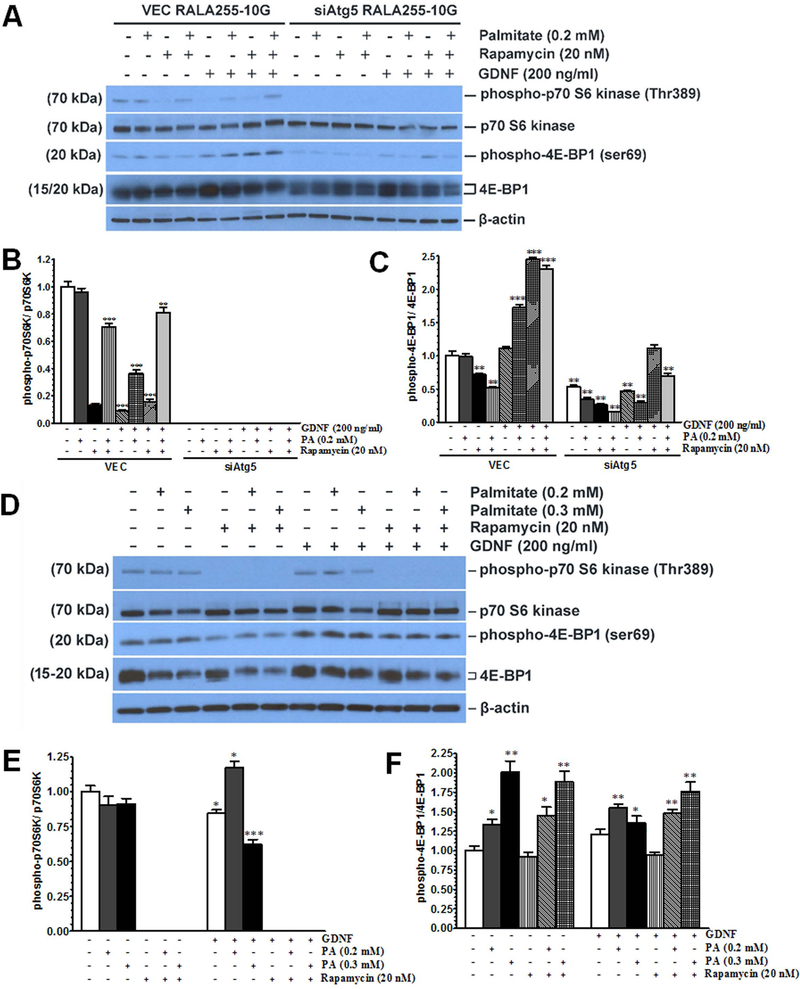
Autophagy plays an important role in fat metabolism by breaking down lipid droplets into free fatty acids which are then transported to the mitochondria for β-oxidation to release energy (4). The mechanistic target of rapamycin (mTOR) signaling pathway regulates cellular anabolic processes, such as lipid synthesis, as well as catabolic processes, such as autophagy, breakdown of intracellular triglycerides, and fatty acid β-oxidation (26). Anabolic signals activate the mTOR complex 1 (mTORC1) through phosphorylation of its serine 2448 residue while catabolic signals inhibit this phosphorylation (27). To understand the role of GDNF in modulating the mTOR signaling pathway in response to saturated fatty acid challenge, we first compared the effects of GDNF on mTOR phosphorylation to those of the mTOR inhibitor rapamycin and the lipolysis-inducing β3-adrenergic receptor agonist CL316,243. HepG2 cells cultured for 48 h in medium supplemented with GDNF alone showed similar decreases in mTOR phosphorylation as cells exposed to rapamycin or CL316,243 alone during the last 24 h of culture (Supplementary Fig. 4A and B). In culture medium supplemented with palmitate, GDNF was also able to blunt palmitate-induced increase in mTOR phosphorylation (Supplementary Fig. 4 A and B). GDNF, however, had no additive effect on rapamycin-mediated mTOR phosphorylation (Supplementary Fig. 4A). We next performed experiments in rat hepatocytes and primary mouse hepatocytes to determine the effects of GDNF on downstream targets of mTOR and the possible link to the autophagic pathway. Exposure (48 h) of control rat hepatocytes (VEC) with an intact autophagic machinery to GDNF alone or GDNF in combination with palmitate decreased the phosphorylation of p70S6 kinase on threonine 389 significantly similar to rapamycin, and enhanced the phosphorylation of 4E-BP1 on serine 69 (Fig. 3A–C). However, in rat hepatocytes in which autophagy was impaired due to a knockdown of Atg5 (siAtg5), the phosphorylation of p70S6K was blocked completely while the phosphorylation of 4E-BP1 was significantly decreased (Fig. 3A–C). Similarly, GDNF either alone or in the presence of palmitate (0.3 mM) significantly decreased p70S7K phosphorylation in primary mouse hepatocytes, but had no significant effects on 4E-BP1 phosphorylation (Fig. 3D–F). This suggested that p70S6K might be a critical link between the autophagic pathway and the mTOR signaling pathway.
FIG. 3.
GDNF blocks the phosphorylation of p70S6 kinase in hepatocytes. (A-C) Western blot analysis of phospho-p70S6K (Thr389) and phospho-4E-BP1 (ser69) levels in RALA255–10G rat hepatocytes cultured for 48h in medium supplemented with or without GDNF (200 ng/ml) and palmitate (0.2 mM) and during the last 24h with or without rapamycin (20 nM). Plotted are means + SEM (***, P<0.001; **, P<0.01. n = 3). (D-F) Western blot analysis of phospho-p70S6K (Thr389) and phospho-4E-BP1 (ser69 levels in primary mouse hepatocytes cultured for 48 h in medium supplemented with or without GDNF (200 ng/ml) and palmitate (0.2 mM) and during the last 24 h with or without rapamycin (20 nM). Plotted are means + SEM (***, P<0.001; **, P<0.01. n = 3).
The fact that GDNF promotes mTOR dephosphorylation similar to rapamycin suggests that GDNF could inactivate mTOR and promote catabolic processes that favor the breakdown of cellular fuels. To test if GDNF could activate mitochondrial respiration and fatty acid β-oxidation in hepatocytes similar to the autophagy activator rapamycin (28, 29), we first cultured HepG2 cells for 6 days in medium supplemented with or without GDNF and rapamycin during the last 24 h. Mitochondrial respiration was then assessed by measuring oxygen consumption rates (OCR) before and after addition of the electron transport chain inhibitors oligomycin, FCCP, rotenone and actinomycin A. GDNF significantly enhanced basal oxygen consumption rate and spare capacity similar to rapamycin (Supplementary Fig. 5A and B), thus confirming its ability to enhance cellular respiration.
GDNF enhances mitochondrial fatty acid β-oxidation
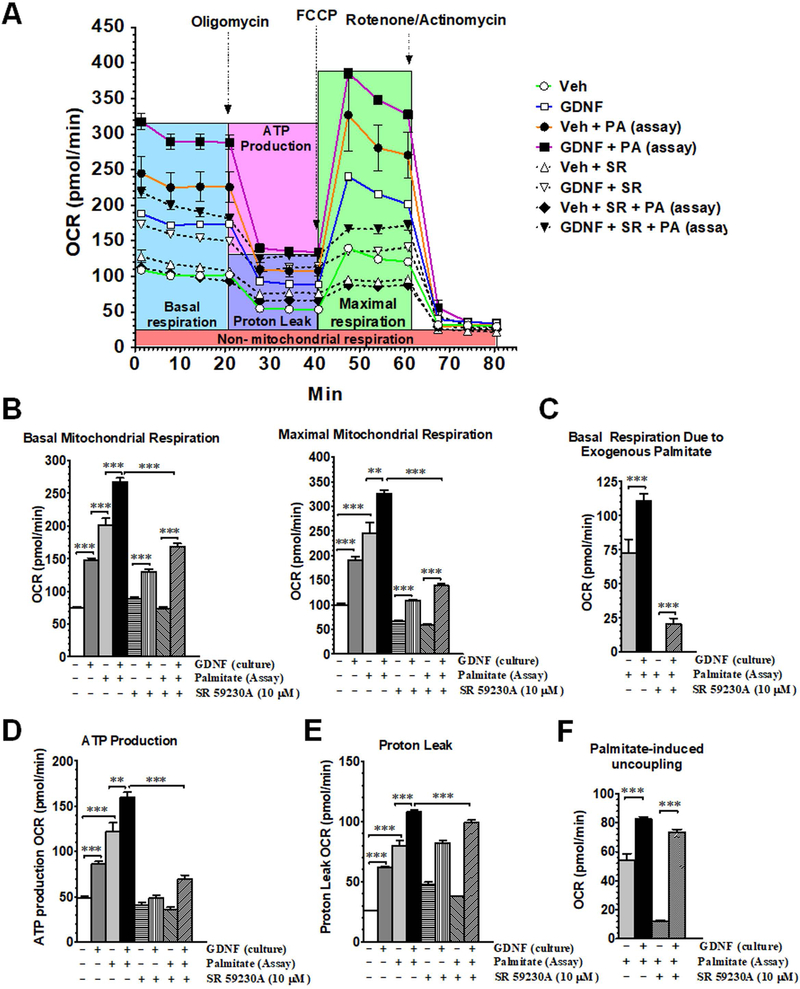
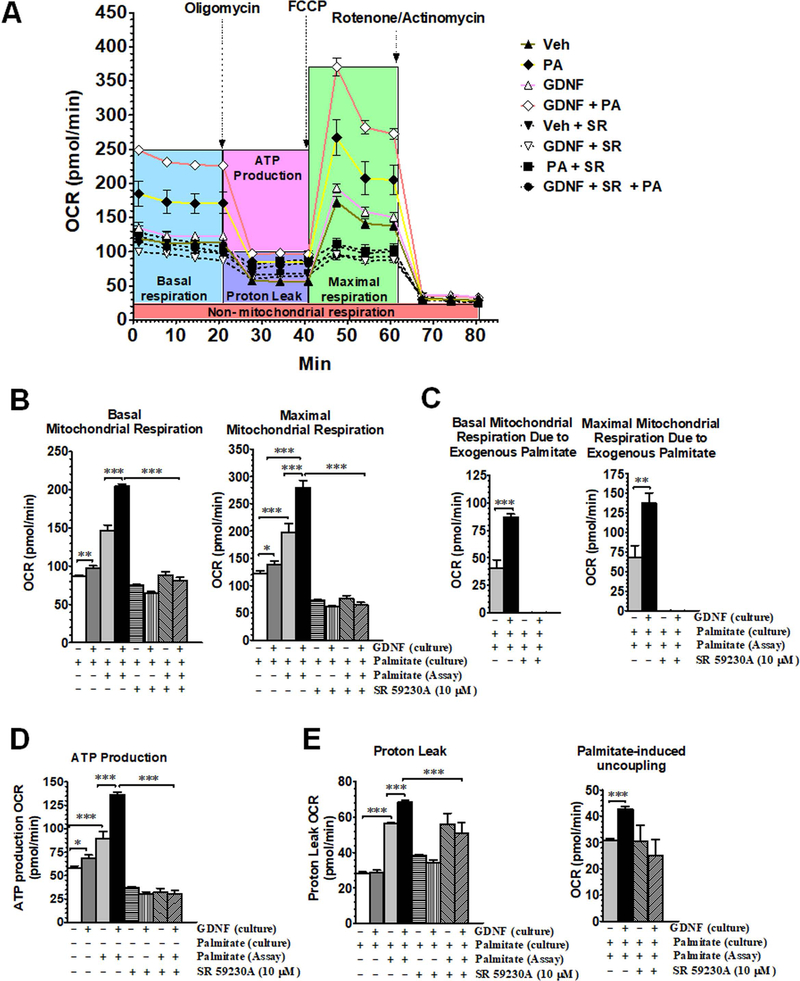
We next assessed the ability of GDNF to enhance mitochondrial fatty acid β-oxidation in response to palmitate challenge. We first cultured HepG2 cells for 6 days in palmitate-free cell culture medium supplemented with or without GDNF and then exposed them to palmitate during the assay of mitochondrial respiration to assess the ability of GDNF to enhance mitochondrial respiration in response to acute exposure to a saturated fatty acid. HepG2 cells cultured in the presence of GDNF had higher basal and maximal mitochondrial respiration relative to controls and had even higher OCRs when exposed to exogenous palmitate (Fig. 4A–C). Cells cultured in the presence of GDNF also had higher ATP synthesis-linked basal respiration and higher proton leak and palmitate-induced mitochondrial uncoupling than cells cultured in the absence of GDNF (Fig. 4D–F). The β-oxidation enhancing effects of GDNF were, however, lost when the cells were exposed to the β-adrenergic antagonist SR 50230A during the last 4 days of culture (Fig. 4B–E) suggesting that the β-adrenergic signaling pathway might be involved in GDNF-mediated fatty acid β-oxidation in hepatocytes. Overall these results suggest an ability of GDNF to enhance mitochondrial respiration to supply ATP during times of increased energy demand, and also to increase the uncoupling of respiration from ATP production during β-oxidation which could be protective against the release of toxic reactive oxygen species.
FIG. 4.
GDNF enhances mitochondrial β-oxidation cells after acute exposure to palmitate. Mitochondrial respiration in HepG2 cells cultured for 6 days in medium supplemented with or without GDNF with or without the β3-adrenergic receptor antagonist SR 59230A (10 µM) added during the last 4 days of culture and oxygen consumption rates (OCR) assayed in absence and presence of palmitate (PA). (A) Mitochondrial respiration profile, (B) basal and maximal mitochondrial respiration, (C) basal mitochondrial respiration due to exogenous palmitate, (D) ATP production-linked mitochondrial respiration, (E) proton leak, and (F) palmitate-induced uncoupling after acute exposure to palmitate. Plotted are means + SEM (***, P<0.001; **, P<0.01. n = 5).
We next tested the effects of GDNF on mitochondrial respiration after prolonged exposure to palmitate. HepG2 cells were cultured for 6 days in cell culture medium containing palmitate (0.1 mM) with or without GDNF. We then assayed mitochondrial respiration in fatty acid oxidation assay medium supplemented with or without additional palmitate. Similar to cells cultured in culture medium without palmitate, cells cultured in the presence of GDNF in medium containing palmitate had higher basal and maximal mitochondrial respiration, higher basal and maximal respiration due to exogenous palmitate, higher ATP synthesis-linked basal respiration, and higher proton leak and palmitate-induced mitochondrial uncoupling than controls (Fig. 5A–E). The ability to metabolize PA was, however, lower in cells chronically exposed to PA than in cells cultured in the absence of PA. Also, similar to cells cultured in culture medium without palmitate, the β-oxidation enhancing effects of GDNF in cells cultured in medium containing palmitate was lost when the cells were exposed to the β-adrenergic antagonist (Fig. 5A–E).
FIG. 5.
GDNF prevents palmitate-induced suppression of mitochondrial β-oxidation. (A) Mitochondrial respiration profile, (B) basal and maximal mitochondrial respiration, (C) basal and maximal mitochondrial respiration due to exogenous palmitate, (D) ATP production-linked mitochondrial respiration, and (E) proton leak and palmitate-induced uncoupling in HepG2 cells cultured for 6 days in medium containing palmitate (0.1 mM) and supplemented with or without GDNF and during the last 4 days of culture with or without the β3-adrenergic receptor antagonist SR 59230A (10 µM). Oxygen consumption rates (OCR) were assayed in buffer (Assay) with and without GDNF, palmitate and SR 59230A. Plotted are means + SEM (***, P<0.001; **, P<0.01; *, P<0.05. n = 5).
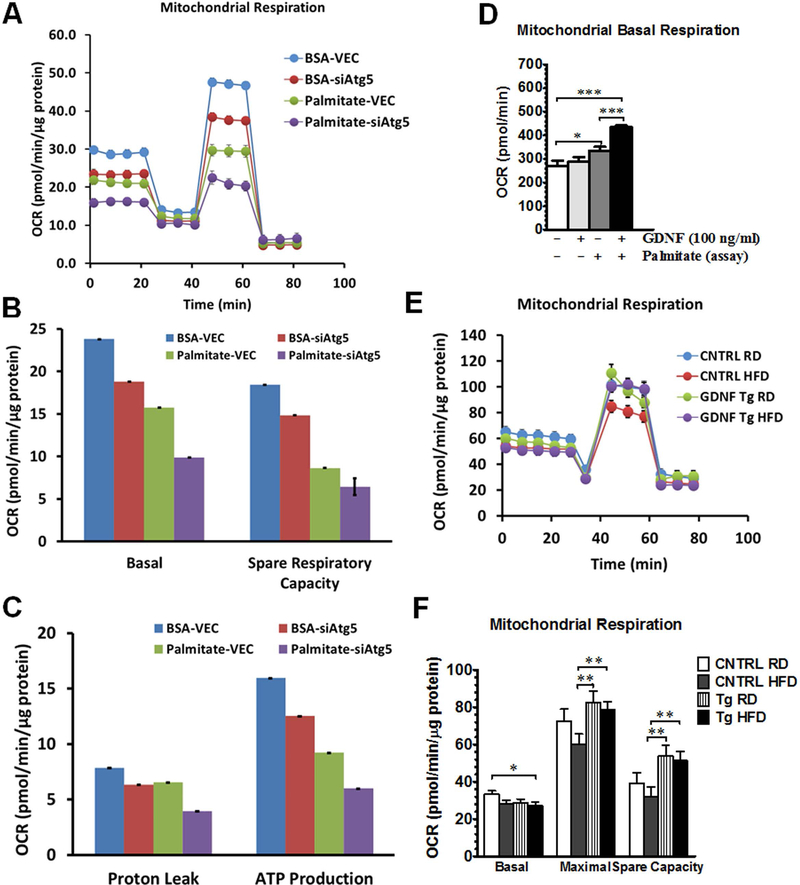
To establish the necessity of the autophagic pathway in GDNF’s ability to enhance mitochondrial fatty acid β-oxidation, we compared mitochondrial respiration in RALA255–10G rat hepatocytes with (VEC) and without an intact autophagy machinery (siAtg5). RALA255–10G rat hepatocytes were cultured for 24h in medium supplemented with or without palmitate (0.2 mM) and mitochondrial respiration assayed without additional palmitate supplementation. Overall, cells with an intact autophagy machinery had higher mitochondrial respiration profiles including basal maximal spare respiratory capacity, proton leak and ATP production than autophagy knockout cells (siAtg5) (Fig. 6A–C). In the presence of the higher dose of palmitate used (0.2 mM), there was, however, a reduction in mitochondrial respiration in both cells, and more so in siAtg5 cells than in VEC cells (Fig. 6A–C). We also examined if GDNF had similar effects on mitochondrial respiration in VEC rat hepatocytes as it did in HepG2 cells, VEC hepatocytes cultured for 3 days in the presence of GDNF and then exposed to palmitate during the assay had significantly higher basal oxygen consumption rates than cells cultured in medium alone and assayed in the presence or absence of palmitate (Fig. 6D). We also investigated if the overexpression of GDNF in the liver from GDNF Tg mice enhances their hepatocytes mitochondrial respiration. Hepatocytes isolated from GDNF Tg mice fed a regular rodent diet (RD) or a HFD for 7 days had significantly higher maximal respiration and spare respiratory capacity than control mice fed the HFD (Fig. 6 E and F).
FIG. 6.
GDNF enhances mitochondrial respiration in rat hepatocytes and primary mouse hepatocytes. Comparison of mitochondrial respiration between VEC RALA255–10G rat hepatocytes (with intact autophagy) and siAtg5 RALA255–10G hepatocytes with defective autophagy (A-C). Plotted are means + SEM. (D) Basal oxygen consumption rates (OCR) of VEC RALA255–10G rat hepatocytes cultured for 3 days in medium with or without GDNF and assayed in assay buffer with or without palmitate (Assay). Plotted are means + SEM (***, P<0.001; *, P<0.05. n = 3). (E and F) Mitochondrial respiration profiles of hepatocytes isolated from control (CNTRL) and GDNF transgenic (Tg) that had been fed on a RD or HFD for 7 days. Plotted are means + SEM (**, P<0.01; *, P<0.05. n = 3).
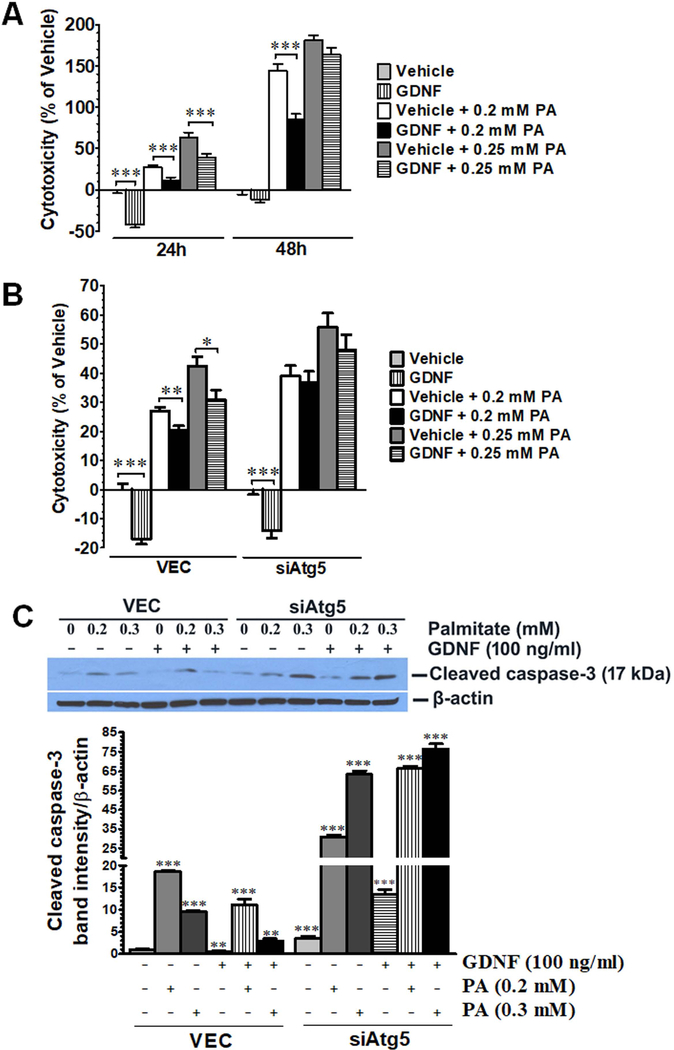
GDNF is protective against palmitate-induced cytotoxicity
In light of the observed GDNF’s ability to enhance autophagy and β-oxidation in rat hepatocytes and HepG2 cells in response to palmitate challenge, we also assessed if GDNF could protect against palmitate-mediated lipotoxicity. GDNF enhanced cell survival in both HepG2 cells and VEC-infected RALA255 hepatocytes and protected against palmitate-induced cell death in part by suppressing cleaved caspase-3 mediated apoptosis (Fig. 7A–C). The protective effects of GDNF against palmitate-induced hepatocyte cell death were, however, diminished in RALA255–10G cells with defective autophagy (Fig. 7B and C).
FIG. 7.
GDNF protects against palmitate-induced cytotoxicity in HepG2 cells and RALA255 rat hepatocytes. (A) Assessment of cytotoxicity in HepG2 cell death cultured for 24 and 48 h in medium supplemented with or without GDNF (100 ng/ml) and palmitate (PA) (0.2 and 0.25 mM). Plotted are means + SEM relative to vehicle (***, P<0.001. n = 5). (B) Assessment of cytotoxicity in VEC and siAtg5 Rala255–10G rat hepatocytes cultured for 48h of in the presence and absence of GDNF (100 ng/ml) and PA (0.2 and 0.25 mM). Plotted are means + SEM relative to vehicle (***, P<0.001 **, P<0.01; *, P<0.05. n = 4). (C) Western blot analysis of cleaved caspase-3 levels in VEC and siAtg5 hepatocytes cultured for 24 h in the presence and absence of GDNF (100 ng/ml) and PA (0.2 and 0.3 mM). Plotted are means + SEM (***, P<0.001) **, P<0.01. n = 3).
Discussion
In our previous study we demonstrated a new role for GDNF in hepatic lipid metabolism involving suppression of the transcription of proteins involved in hepatic lipid uptake and de novo lipogenesis (11). In this study we have uncovered a new mechanism involving increased autophagy and fatty acid β-oxidation by which GDNF can potentially protect hepatocytes against the harmful effects of a HFD and saturated fatty acids.
Autophagy plays an important role in hepatocyte lipid metabolism (4, 30). Defective hepatic autophagy characterized by increased p62/SQSTM1 levels and reduced Atg5, Atg7 and Beclin-1 levels similar to what we observed in HFD-fed control mice in this study and our previous study (11) has been reported in some rodent models of obesity and fatty liver disease as well as in some NAFLD patients (6, 10, 31). It is not known how much this impairment in autophagy contributes to the hepatic steatosis and the accompanying disease. It is, however, clear that defective autophagy has an impact on hepatic lipid metabolism since induction or restoration of autophagy is able to lower hepatocyte TG accumulation (7, 10).
A general indicator of defective autophagy is increased LC3-II and p62/SQSTM1 cellular levels with decreased autophagic flux (15). In this study liver GDNF overexpression was associated with increased liver LC3A/B-II, Atg5, and Beclin-1 levels in HFD-fed male mice and prevented the decreases in liver Atg5, Atg7 and LC3-II levels seen in the liver of HFD-fed female control mice. In vitro, GDNF treatment of palmitate-exposed primary mouse and rat hepatocytes resulted in greater reductions in LC3-II levels than that seen in vehicle-treated cells. These reductions were, however, not due to impairment of autophagy, but to increased autophagic clearance since treatment with lysosomal inhibitors indicated that GDNF-treated hepatocytes had higher autophagic flux than hepatocytes cultured in the absence of GDNF. The ability of GDNF to enhance autophagy was blocked in cells with defective autophagy due to a knockdown of the Atg5 gene. Acute exposure to palmitate results in increased LC3-II and p62/SQSTM1 accumulation in hepatocytes cells due to the initial induction of autophagy (32). In this study we adopted a cell treatment strategy that resembled chronic fatty acid exposure similar to that seen in obesity. Using this strategy we were able to assess change in flux rather than the initial induction of autophagy. Hence in our study treatment with palmitate induced autophagic flux which was highest in GDNF-treated cells. These findings, thus, strongly point to an ability of GDNF to modulate the rate of autophagy in hepatocytes in response to excess lipid challenge.
The mTOR kinase plays an important role in lipid metabolism with its activity being regulated based on energy demands and nutrient supply. In this study we observed the ability of GDNF to reduce the phosphorylation of mTOR and its downstream target p70S6K. However, the phosphorylation of 4E-BP1, another downstream target of mTOR, was enhanced. The seemingly opposite effects of GDNF on the two mTOR targets was possibly due to their different roles in metabolism thus requiring differential regulation (33, 34). Indeed, there is evidence showing that these factors are differentially regulated to achieve metabolic homeostasis. Choo et, al. (35) have presented data showing differential inhibition of 4E-BP1 and p70S7K by rapamycin in several cell lines with p70S6K phosphorylation being inhibited during the entire 48 h period of treatment, and 4E-BP1 phosphorylation being inhibited only during the first 3 h of exposure and coming back to normal within 6 h to in order to maintain translation.
The phosphorylation of mTOR is associated with increased activity which favors anabolic cellular processes such as lipogenesis while its dephosphorylation can result in autophagy activation, increased lipolysis and β-oxidation (26). GDNF improved general hepatocyte cellular respiration and increased mitochondrial fatty acid β-oxidation. GDNF not only enhanced mitochondrial respiration coupled to ATP production in response to palmitate exposure, but also enhanced mitochondrial uncoupling which may serve to protect the cells against palmitate-induced oxidative stress (36). GDNF protected rat hepatocytes against palmitate-induced cell death. These effects of GDNF were, however, reduced in hepatocytes with defective autophagy thus reinforcing the interdependence between autophagy and mitochondrial respiration in hepatocyte fatty acid oxidation.
Impairment of hepatic mitochondrial activity with accompanying endoplasmic reticulum stress and cellular toxicity have been observed in obesity and NAFLD as well as following exposure to saturated long chain fatty acids such as palmitic acid. ER stress also occurs with impaired autophagy (22, 37–39). This effect has been linked to increased mitochondrial production of reactive oxygen species, excessive ER-mitochondrial coupling leading to mitochondrial calcium overload, and reductions in hepatic nicotinamide adenine dinucleotide (NAD) levels (40–42). Restoration of hepatic NAD levels has been shown to increase mitochondrial β-oxidation and prevent hepatic steatosis both in vitro and in vivo (41).
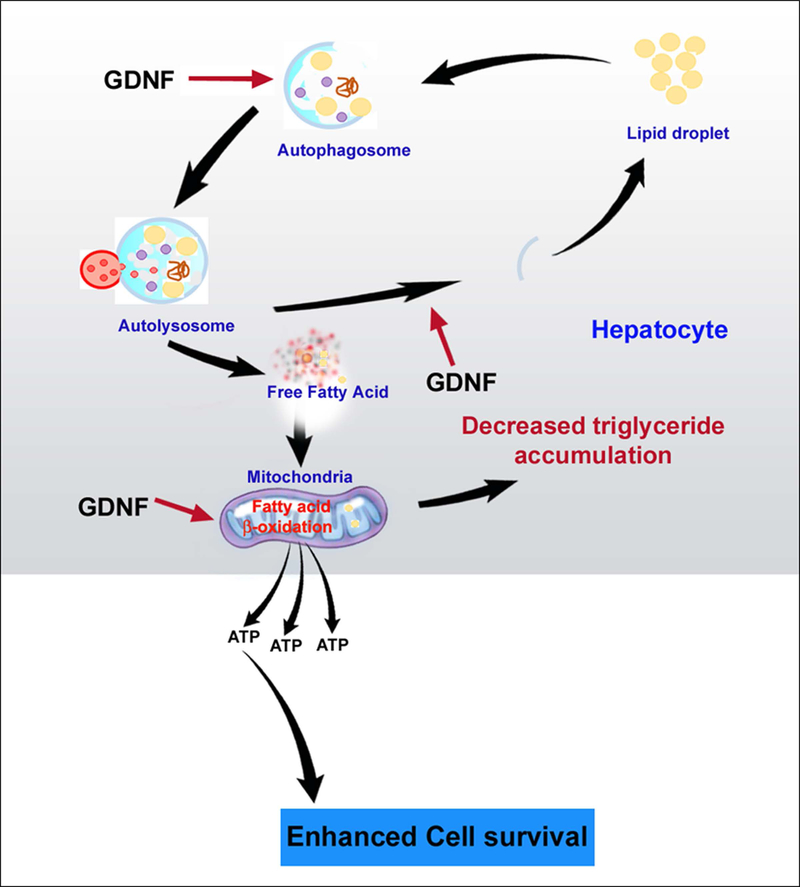
In this study GDNF was protective against palmitate-induced hepatocyte cytoxicity and cell death partly through apoptosis. As summarized in Fig. 8 we have demonstrated that GDNF can protect against obesity-induced derangement of hepatic autophagy through its ability to enhance autophagic flux. We have also demonstrated that GDNF can enhance fatty acid β-oxidation and protect against fatty acid-induced toxicity in hepatocytes. These findings open the possibility that GDNF or its agonist could be used to improve hepatic fatty acid metabolism in NAFLD patients.
FIG. 8.
Schematic representation of the proposed model of GDNF action in hepatocytes involving enhancement of lipophagy and fatty acid β-oxidation following lipid challenge. In response to increased triglyceride accumulation in the form of lipid droplets within the hepatocyte, GDNF enhances the formation of the autophagosome which grows to enclose the lipid droplet. Fusion of the autophagosome to lysosomes results in the release of lytic enzymes which breakdown the lipid droplet into free fatty acids. The autophagosome wall breaks down to release the free fatty acids into the cytoplasm from where they enter mitochondria and undergo β-oxidation.
Supplementary Material
Acknowledgments
Financial Support:
This work is supported by U.S. National Institutes of Health (NIH) grants R01DK080684 (Shanthi Srinivasan) and R01DK044234 and R01AA022601 (Mark Czaja), and VA Research and Development Merit Review Award # BX000136-08 (Shanthi Srinivasan).
List of Abbreviations:
- CNTRL
CF-1 control mice
- EM
emission
- EX
extinction
- FAO
fatty acid oxidation
- Fasn
fatty acid synthase
- FCCP
Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone
- FFA
free fatty acids
- GDNF
Glial cell line Derived Neurotrophic Factor
- GFAP
glial fibrillary acidic protein
- HFD
high fat diet
- NAFLD
Nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- OCR
oxygen consumption rate
- p62/SQSTM1
p62/sequestosome 1
- PPAR-γ
peroxisome proliferator activated receptor-γ
- RD
regular diet
- Scd1
stearoyl-CoA desaturase
- TG
triglyceride
- FA
fatty acid
- Tg
GDNF transgenic mice
References
- 1.Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology 2010;51:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012;55:2005–2023. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol 2014;21:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, et al. Autophagy regulates lipid metabolism. Nature 2009;458:1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madrigal-Matute J, Cuervo AM. Regulation of Liver Metabolism by Autophagy. Gastroenterology 2016;150:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Rodriguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, Vargas-Castrillon J, et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis 2014;5:e1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park HW, Park H, Semple IA, Jang I, Ro SH, Kim M, Cazares VA, et al. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat Commun 2014;5:4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, Privalsky ML, et al. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. J Clin Invest 2012;122:2428–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czaja MJ. Function of Autophagy in Nonalcoholic Fatty Liver Disease. Dig Dis Sci 2016;61:1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 2010;11:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mwangi SM, Peng S, Nezami BG, Thorn N, Farris AB 3rd, Jain S, Laroui H, et al. Glial cell line-derived neurotrophic factor protects against high-fat diet-induced hepatic steatosis by suppressing hepatic PPAR-gamma expression. Am J Physiol Gastrointest Liver Physiol 2016;310:G103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, et al. p62/SQSTM1 Binds Directly to Atg8/LC3 to Facilitate Degradation of Ubiquitinated Protein Aggregates by Autophagy. Journal of Biological Chemistry 2007;282:24131–24145. [DOI] [PubMed] [Google Scholar]
- 13.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS Journal 2015;282:4672–4678. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell 2011;44:279–289. [DOI] [PubMed] [Google Scholar]
- 15.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mwangi SM, Nezami BG, Obukwelu B, Anitha M, Marri S, Fu P, Epperson MF, et al. Glial Cell Line-Derived Neurotrophic Factor Protects Against High Fat Diet-Induced Obesity. Am J Physiol Gastrointest Liver Physiol 2014;306:G515–G525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Z, Alam S, Oppenheim RW, Prevette DM, Evenson A, Parsadanian A. Overexpression of glial cell line-derived neurotrophic factor in the CNS rescues motoneurons from programmed cell death and promotes their long-term survival following axotomy. Experimental Neurology 2004;190:356–372. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Schattenberg JM, Rigoli RM, Storz P, Czaja MJ. Hepatocyte Resistance to Oxidative Stress Is Dependent on Protein Kinase C-mediated Down-regulation of c-Jun/AP-1. Journal of Biological Chemistry 2004;279:31089–31097. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology 2010;52:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980;209:497–499. [DOI] [PubMed] [Google Scholar]
- 21.Akie TE, Cooper MP. Determination of Fatty Acid Oxidation and Lipogenesis in Mouse Primary Hepatocytes. J Vis Exp 2015:e52982. [DOI] [PMC free article] [PubMed]
- 22.Egnatchik RA, Leamy AK, Jacobson DA, Shiota M, Young JD. ER calcium release promotes mitochondrial dysfunction and hepatic cell lipotoxicity in response to palmitate overload. Mol Metab 2014;3:544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers GW, Nadanaciva S, Swiss R, Divakaruni AS, Will Y. Assessment of fatty acid beta oxidation in cells and isolated mitochondria. Curr Protoc Toxicol 2014;60:25 23 21–19. [DOI] [PubMed] [Google Scholar]
- 24.Hill Bradford G, Benavides Gloria A, Lancaster Jack R, Ballinger S, Dell’Italia L, Zhang J, Darley-Usmar Victor M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. In: Biological Chemistry; 2012. p. 1485–1512. [DOI] [PMC free article] [PubMed]
- 25.Tanaka S, Hikita H, Tatsumi T, Sakamori R, Nozaki Y, Sakane S, Shiode Y, et al. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology 2016;64:1994–2014. [DOI] [PubMed] [Google Scholar]
- 26.Caron A, Richard D, Laplante M. The Roles of mTOR Complexes in Lipid Metabolism. Annual Review of Nutrition 2015;35:321–348. [DOI] [PubMed] [Google Scholar]
- 27.Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J 1999;344 Pt 2:427–431. [PMC free article] [PubMed] [Google Scholar]
- 28.Brown NF, Stefanovic-Racic M, Sipula IJ, Perdomo G. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism - Clinical and Experimental 2007;56:1500–1507. [DOI] [PubMed] [Google Scholar]
- 29.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR Signaling Extends Chronological Life Span via Increased Respiration and Upregulation of Mitochondrial Gene Expression. Cell Metabolism 2007;5:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ 2013;20:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H-Y, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. The Journal of biological chemistry 2009;284:31484–31492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan SH, Shui G, Zhou J, Li JJE, Bay B-H, Wenk MR, Shen H-M. Induction of Autophagy by Palmitic Acid via Protein Kinase C-mediated Signaling Pathway Independent of mTOR (Mammalian Target of Rapamycin). Journal of Biological Chemistry 2012;287:14364–14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita M, Gravel S-P, Chénard V, Sikström K, Zheng L, Alain T, Gandin V, et al. mTORC1 Controls Mitochondrial Activity and Biogenesis through 4E-BP-Dependent Translational Regulation. Cell Metabolism 2013;18:698–711. [DOI] [PubMed] [Google Scholar]
- 34.Bae EJ, Xu J, Oh DY, Bandyopadhyay G, Lagakos WS, Keshwani M, Olefsky JM. Liver-specific p70 S6 kinase depletion protects against hepatic steatosis and systemic insulin resistance. J Biol Chem 2012;287:18769–18780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A 2008;105:17414–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Divakaruni AS, Brand MD. The Regulation and Physiology of Mitochondrial Proton Leak. Physiology 2011;26:192–205. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Liu X, Nie J, Zhang J, Kimball SR, Zhang H, Zhang WJ, et al. ALCAT1 controls mitochondrial etiology of fatty liver diseases, linking defective mitophagy to steatosis. Hepatology 2015;61:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Begriche K, Massart J, Robin M-A, Bonnet F, Fromenty B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology 2013;58:1497–1507. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Carreras M, Del Hoyo P, Martín MA, Rubio JC, Martín A, Castellano G, Colina F, et al. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology 2003;38:999–1007. [DOI] [PubMed] [Google Scholar]
- 40.Satapati S, Kucejova B, Duarte JA, Fletcher JA, Reynolds L, Sunny NE, He T, et al. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J Clin Invest 2015;125:4447–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gariani K, Menzies KJ, Ryu D, Wegner CJ, Wang X, Ropelle ER, Moullan N, et al. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology 2016;63:1190–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arruda AP, Pers BM, Parlakgul G, Guney E, Inouye K, Hotamisligil GS. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat Med 2014;20:1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.