Abstract
Dendrites and axons are delicate neuronal membrane extensions that undergo degeneration after physical injuries. In neurodegenerative diseases, these processes often degenerate prior to neuronal death. Understanding the mechanisms of neurite degeneration has been an intense focus of neurobiology research in the last two decades. As a result, many discoveries have been made in the molecular pathways that lead to neurite degeneration and the cell-cell interactions responsible for the subsequent clearance of neuronal debris. Drosophila melanogaster has served as a prime in vivo model system for identifying and characterizing the key molecular players in neurite degeneration, thanks to its genetic tractability and easy access to its nervous system. The knowledge learned in the fly provided targets and fuel for studies in other model systems that have further enhanced our understanding of neurodegeneration. In this review, we will introduce the experimental systems developed in Drosophila to investigate injury-induced neurite degeneration, and then discuss the biological pathways that drive degeneration. We will also cover what is known about the mechanisms of how phagocytes recognize and clear degenerating neurites, and how recent findings in this area enhance our understanding of neurodegenerative disease pathology.
Keywords: Drosophila, neurite degeneration, axon, dendrite, Wallerian degeneration, phagocyte, phagocytosis, PS exposure, injury assay
1. Introduction
Drosophila melanogaster has been a boon to neuroscience for more than a century. In the early 20th century, scientists learned critical genetic concepts and developed important genetic tools using this model organism. This foundation later led to many critical discoveries in neuroscience, for example, the cell biology of neurogenesis and the molecular basis of circadian rhythm, learning, and memory (reviewed by Bellen et al., 2010). In the last several decades, Drosophila melanogaster has also become a powerful model for studying many facets of neurodegeneration, such as neuronal pruning and remodeling, injury-induced and pathological degeneration, and drug-induced peripheral neuropathies. While the Drosophila nervous system is simpler than its vertebrate counterparts, it is analogous to the mammalian nervous system in many key ways, and can be utilized to address complex questions more rapidly than in vertebrate model organisms. In particular, the fly can be used as a versatile model for studying the cell biology of neurodegeneration and for dissecting the intricate relationships between neurons and their neighboring cells.
In this review, we will primarily focus on neurite degeneration that occurs after neuronal injury. First, we will discuss models of injury-induced neurodegeneration developed in Drosophila. We will then review key intrinsic molecular pathways that drive injury-induced neurodegeneration and address the consequences of extrinsic forces on neurodegeneration, specifically the role of phagocytes. Phagocytes are important for recognizing and engulfing degenerating neurites. Do they also contribute to the process of neurodegeneration? We will review recent findings which support the idea that phagocytes play a more active role during neurite degeneration. Although the focus is on how the Drosophila model system has contributed to the many important discoveries in these research areas, we will also touch upon related discoveries made in other model organisms. For other topics of neurodegeneration, such as axon and dendrite pruning, and neurological disease models in Drosophila, please refer to these excellent reviews (Luo and O’Leary, 2005; Charng et al., 2014; Yu and Schuldiner, 2014; McGurk et al., 2015; Riccomagno and Kolodkin, 2015; Schuldiner and Yaron, 2015; Xiong and Yu, 2018; Yuva-Aydemir et al., 2018).
2. Injury-induced neurite degeneration
2.1. Injury-induced neurodegeneration
Injury-induced neurite degeneration was first described by Augustus Waller in 1850 (Waller, 1850), who found that the distal portions of severed frog glossopharyngeal and hypoglossal nerves broke into separate particles of various sizes after the surgery. This type of local disintegration of axons after nerve severing or crushing was later called “Wallerian degeneration”. Axon degeneration typically follows a latent phase, then manifests as a rapid breakdown of the axon cytoskeleton and internal organelles, and ends with complete fragmentation of the axon distal to the injury site (Lee, 1963; Thomas, 1964; Thomas and Sheldon, 1964; reviewed by Coleman and Freeman, 2010). While Wallerian degeneration was initially thought to be a passive process, a dominant mutation dubbed Wallerian Degeneration Slow (Wlds) was identified in mice that caused axons to persist for weeks post axotomy, demonstrating that Wallerian degeneration is a highly regulated and active process (Lunn et al., 1989). Although Wallerian degeneration typically only refers to injury-induced axon degeneration, injured dendrites follow a similar process of fragmentation that can be suppressed by Wlds (Tao and Rolls, 2011), arguing that the molecular pathways that lead to degeneration in these two cellular compartments are likely similar. Morphologically, both injured axons and injured dendrites undergo local enlargement and thinning, generating a beads-on-a-string appearance, before or during fragmentation. However, dendrites reportedly degenerate faster than axons and show features such as protrusion and membrane vesicle shedding (Han et al., 2011; Sapar et al., 2018), which have not been reported for axon degeneration (Kerschensteiner et al., 2005; Martin et al., 2010; Tao and Rolls, 2011). The neuronal debris resulting from the degeneration is subsequently removed. In both the central nervous system (CNS) and the peripheral nervous system (PNS), resident phagocytes play an important role in clearing the neuronal debris.
Local degeneration is not unique to injured neurites. During nervous system development, excessive or aberrant neurites are pruned to ensure the establishment of functional neural circuits (Liu et al. 2005, Low et al. 2008). Metabolous insects also remodel their nervous systems during metamorphosis by trimming existing dendrites or axons (Watts et al., 2003; Williams and Truman, 2005). These pruning events employ local degeneration that bears morphological similarities to injury-induced neurite degeneration. In this review, we primarily focus on the mechanisms of injury-induced degeneration but will touch upon findings derived from studies of neurite pruning when we discuss the clearance of degenerating neurites.
2.2. Models of injury-induced neurite degeneration in Drosophila
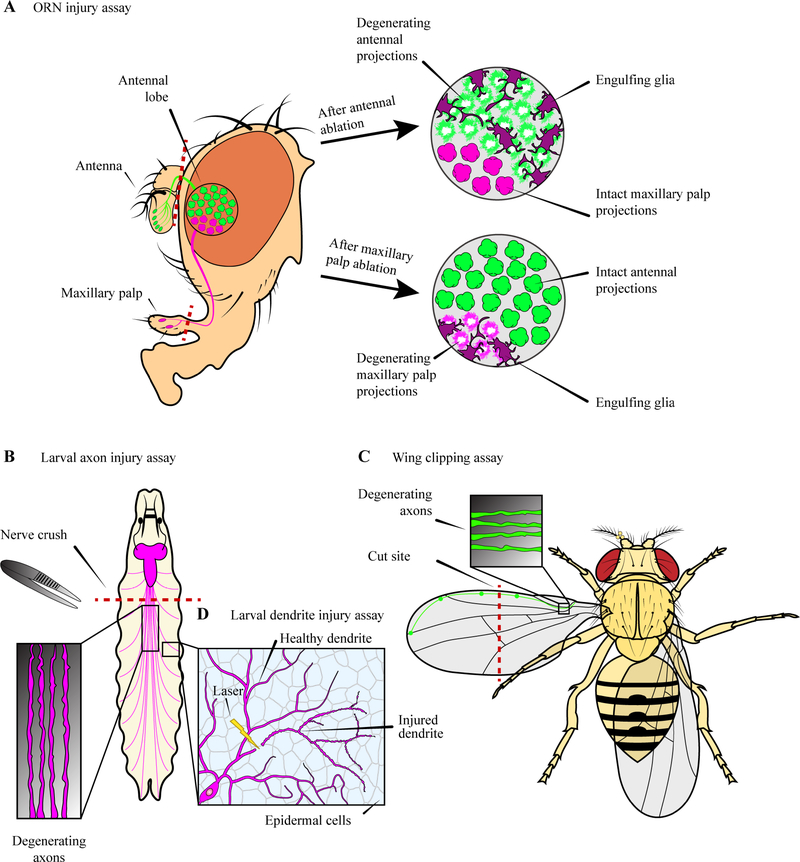
The fly’s genetic tractability and relatively simple nervous system led to the development of several nerve injury assays. After molecular tools were developed for fluorescent labeling of specific subsets of neurons in the fly olfactory circuit, the olfactory receptor neurons (ORNs) became a critical nerve injury model for studying Wallerian degeneration (MacDonald et al., 2006) (Fig. 1A). Drosophila ORN cell bodies are located in the antenna and maxillary palp and project their axons to the antennal lobe in the central brain. Physical removal of either the antenna and maxillary palps causes distal ORN axons to degenerate, and the axons can be imaged in the brain using confocal microscopy. The maxillary palp ORNs project significantly fewer axons into the central brain than the antenna ORNs and therefore are useful for studying the degeneration of a subset of ORNs. Another injury model called the nerve crush assay was developed in the larva for studying axon degeneration (Fig. 1B). The nerve bundles containing axons of larval motor neurons and sensory neurons are damaged by pinching the larva with forceps (Xiong et al., 2010). The state of the degenerating neurons can be easily imaged after minimal dissection.
Fig. 1: Neurite injury assays in Drosophila Melanogaster.
A: The olfactory receptor neurons (ORN) injury assay showing a sagittal view of the fly olfactory system. ORNs are damaged by removal of peripheral organs, antenna and maxillary palps, where neuron cell bodies are located. Maxillary palp ORNs (magenta) project fewer axons into the antennal lobe than antennal ORNs (green). Injured axons begin to show signs of degeneration by day 1 post axotomy and are mostly cleared away by day 3. Glial membrane infiltration can be detected by day 1 post axotomy. B: The larval axon injury assay. Motor neurons are damaged by pinching the larva with forceps. The axons distal to the injury site degenerate. C: The wing clipping assay. Glutamatergic sensory neuron cell bodies are located along the anterior margin of the wing and their axons run in parallel in a bundle called L1 vein towards the fly body. Clipping the wing results in axon degeneration distal from the cut site. Axons can be imaged where L1 vein ends in the proximal part of the wing. D: The larval dendrite injury assay. Sensory da neuron dendrites innervate the basal side of the larval epidermis. Using a high energy laser, injury can be induced on a single dendrite branch. Injured dendrites degenerate distal to the cut site and their debris is engulfed by epidermal cells.
While both ORN removal and nerve crush assays are extremely useful, they still require invasive techniques to access the axons, which limits the direct and real-time visualization of injury-induced axon degeneration and makes it challenging to perform large-scale genetic screens for regulators of degeneration. A simpler in vivo assay was later developed in the Drosophila wing to overcome these limitations (Fig. 1C). The wing is innervated by glutamatergic sensory neurons whose cell bodies are located along the anterior margin. The axons run in parallel in a bundle called L1 vein back to the fly body. Because the wing is translucent and nonessential for survival, it is easily accessible for microscopy and manipulation. The sensory axons can be injured simply by clipping one of the wings while the intact wing serves as an internal control (Fang et al., 2013). This assay was used to develop a toolkit for performing genome-wide forward genetic screens with single axon resolution (Neukomm et al., 2014).
Lastly, an in vivo injury assay was developed for dendrite degeneration using the larval dendritic arborization (da) neurons (Fig. 1D). Da neurons are peripheral somatosensory neurons that elaborate dendritic arbors along the body wall epidermis (Grueber et al., 2002; Han et al., 2012; Kim et al., 2012). Individual da dendrites in intact animals can be severed to induce degeneration by using a high energy laser (Tao and Rolls, 2011; Han et al., 2014). Furthermore, a technique of long-term time-lapse imaging developed for da neurons proved to be especially useful for studying dendrite dynamics and dendrite degeneration for up to 10 hours at a time (Poe et al., 2017; Sapar et al., 2018).
2.3. Signaling pathways in injury-induced neurite degeneration
The discovery of Wlds suggested that there is an axon-death pathway that actively promotes axon degeneration. This revelation prompted enthusiastic investigations on the mechanisms of Wlds-mediated axon protection and stimulated searches in Drosophila for genetic mutations that can produce similar effects. As a result, several critical components in this axon-death pathway have been identified. Their characterization demonstrates that Wallerian degeneration is a complex interplay of evolutionarily conserved pro-survival and pro-degenerative factors. Below, we summarize the core components of the axon-death pathway that are shared between flies and vertebrates.
2.3.1. NAD+ and NMNATs
Wlds encodes a chimeric protein containing a functional nicotinamide mononucleotide adenyltransferase (NMNAT1), an enzyme that synthesizes nicotinamide adenine dinucleotide (NAD+) (Mack et al., 2001). Uncovering how NAD+ metabolism promotes axonal survival became a central focus for delineating the signaling pathways behind axonal degeneration. NAD+ and its reduced form, NADH, serve as cofactors in many important cellular metabolic pathways such as glycolysis, TCA cycle, protein modifications, and redox reactions. Following injury, the axon distal to the damage site undergoes a rapid local depletion of NAD+ (Wang et al., 2005). Because of the nature of the Wlds fusion protein, NMNAT1, which is normally localized in the nucleus, is thought to mislocalize to the axon where it functions autonomously to maintain NAD+ levels. Driving wild-type NMNAT1 localization to axons leads to a Wlds-like axon protection (Sasaki et al., 2009; Beirowski et al., 2010) and the NMNAT enzymatic activity is required to confer the axonal protection (Araki et al., 2004; Gerdts et al., 2016). Further research in mammalian model systems showed that although the NMNAT1 function is vital to Wlds axon protection, endogenous nuclear NMNAT1 may not have a role in axon destruction or maintenance. Instead, the mislocalized NMNAT1 protects axons by replacing the function of its paralog NMNAT2, which is a labile axon maintenance factor and whose degradation results in axon degeneration (Gilley and Coleman, 2010; Hicks et al., 2012). Drosophila has only one Nmnat gene, whose loss of function (LOF) causes severe neurodegeneration of eye photoreceptors and glutamatergic sensory neurons in the wing (Zhai et al., 2006; Neukomm et al., 2017). Similarly, da neurons and motor neurons mutant for Nmnat showed severe axon degeneration (Wen et al., 2011). These studies provided evidence of a critical link between NAD+ metabolism and axon degeneration.
The axon protective features of NMNAT turned out to be evolutionary conserved. Overexpression of mouse Wlds or Drosophila Nmnat protein significantly delayed axon degeneration, by up to 20 days or 5 days, respectively, following ORN axotomy (Hoopfer et al., 2006; MacDonald et al., 2006). In the larval PNS, Wlds overexpression also blocks da sensory neuron dendrite degeneration after laser-induced injury (Tao and Rolls, 2011; Sapar et al., 2018).
2.3.2. dSarm
Using the ORN axotomy assay, the Freeman group performed an F2 forward genetic screen in conjunction with mosaic analysis with a repressible cell marker (MARCM) and identified mutations that mirrored the Wlds overexpression phenotype (Osterloh et al., 2012). These mutations were mapped to Drosophila Toll receptor adaptor dSarm (sterile alpha/Armadillo/Toll-Interleukin receptor homology domain protein), the loss of which suppressed axon degeneration for weeks after injury, demonstrating that dSarm is a major pro-degenerative factor. In mice, the loss of homologous SARM1 also suppresses axon degeneration after severing for up to 18 h, further demonstrating that the degeneration pathway is highly conserved. The shorter period of protection in mammalian neurons, however, indicates that additional pathways could drive axon degeneration in vertebrates (Osterloh et al., 2012).
The identification of dSarm/SARM1 led to a better understanding of how NAD+ pathways regulate axon degeneration. Further studies of SARM1 conducted in cultured mouse dorsal root ganglia (DRG) neurons revealed a link between SARM1 and NAD+ metabolism. SARM1 degrades NAD+ via its intrinsic NADase activity in the TIR domain, which may be regulated by NMNAT2 in live neurons (Gerdts et al., 2015; Gilley et al., 2015; Essuman et al., 2017). Either increasing cytoplasmic NMNAT1 level or blocking the SARM1 pathway reduced the rate of axon degeneration in several in vivo neurodegenerative disease and peripheral neuropathy models (Zhu et al., 2013; Geisler et al., 2016; Henninger et al., 2016; Yin et al., 2016; Turkiew et al., 2017; Williams et al., 2017; Ziogas and Koliatsos, 2018). These observations are consistent with a model that NAD+ depletion triggers axon degeneration (Wang et al., 2005; Gerdts et al., 2015). However, further studies examining other components in NAD+ metabolic pathways suggest that this model may be too simple to explain the trigger for axon degeneration, at least in vertebrates.
Two lines of evidence suggest that accumulation of nicotinamide mononucleotide (NMN), a substrate of NMNAT2 and a precursor of NAD+, is required for axon degeneration: Pharmacological inhibition of nicotinamide phosphoribosyltransferase (NAMPT), the enzyme synthesizing NMN, leads to a modest delay in axon degeneration. Expression of a bacterial NMN deamidase that converts NMN to nicotinic acid mononucleotide (NaMN) causes a delay in degeneration comparable to Wlds (Di Stefano et al., 2015). Consistent with this idea, Sasaki et al. further found that NMN deamidase protects axons despite also reducing NAD+ levels (Sasaki et al., 2016). But by measuring NMN and NAD+ levels in healthy and injured DRG axons, they also found that neither NAD+ reduction nor NMN accumulation alone is sufficient to trigger axon degeneration, a conclusion that is corroborated by a more recent study that tried to decouple NMN and NAD+ levels in DRG neurons (Liu et al., 2018). Therefore, it appears that both NMN accumulation and NAD+ reduction are necessary for the activation of SARM1. However, ultimately, the further catastrophic depletion of NAD+ induced by SARM1 activity seems still responsible for triggering axon degeneration (Sasaki et al., 2016), a conclusion supported by the fact that NMN is not part of the NAD+ biosynthetic pathways in Drosophila (Gossmann et al., 2012).
2.3.3. Axed and Pebbled
Genetic screens conducted using the wing clipping assay identified two more factors involved in neurite degeneration. The first one is Axundead (Axed), a BTB (bric-à-brac, tramtrack, broad complex) and a BACK (BTB and C-terminal Kelch) containing protein (Neukomm et al., 2017). Neukomm et al. demonstrated that the loss of axed protected ORN axons up to 50 days post axotomy. Like dSarm, Axed is not involved in programmed cell death or developmental neurite pruning of mushroom body (MB) γ neurons, suggesting that it may be unique to injury-induced pro-degeneration program. Moreover, severed axed mutant axons remained functional and elicited responses to optogenetic stimulation post axotomy. Remarkably, axed mutations completely suppressed neuronal degeneration caused by the expression of a constitutively active dSarm (dSarmGOF), suggesting that Axed functions downstream of dSarm.
A further comparison of the effects of dSarm and axed mutations by Neukomm et al. (2017) revealed interesting details of the axon-death pathway. While dSarm is required for the degeneration of injured axons, dSarm loss did not rescue the degeneration of Nmnat mutant sensory neurons. In contrast, axed/Nmnat double mutant neurons showed no signs of degeneration for up to 3 weeks. These findings seem to support a hypothesis that both Nmnat loss and dSarm activation can dominantly cause NAD+ depletion, which triggers axon degeneration by activating Axed. Characterizing the biochemical activity of Axed would be an important next step for understanding the role of Axed in axon degeneration.
Another factor identified using the wing clipping assay was Pebbled (Peb), a C2H2 zinc finger transcription factor (Farley et al., 2018). The loss of peb resulted in either fully preserved axons or large axon fragments that lingered for weeks. Expression of the C-terminal 10–14 DNA-binding zinc finger domains of Peb rescued the axon death defect in peb mutant neurons, suggesting that Peb establishes the potential of an axon to undergo degeneration after injury through its transcription factor activity. The human homolog of Peb, RREB1, can also rescue the peb mutant axon death defect, demonstrating that the Peb function is conserved. Interestingly, the loss of peb suppressed degeneration of the glutamatergic but not the cholinergic sensory neurons innervating the wing, suggesting the existence of distinct signaling in the axon death pathway among different neuron subtypes. It is unclear whether Peb functions upstream of or in parallel to dSarm, as it does not bind to dsarm locus or to axed or highwire loci. The peb mutation did not suppress dSarmGOF, but the loss of a single dsarm allele enhanced peb mutant phenotype, suggesting that Peb and dSarm could act in the same pathway. Intriguingly, removing a copy of axed did not enhance the peb phenotype. How precisely Peb modulates the axon death pathway and why it is important for axon degeneration of only some types of neurons remain to be answered.
2.3.4. Highwire
The nerve crush assay was instrumental for the characterization of Highwire (Hiw), an E3 ubiquitin ligase that promotes Wallerian degeneration. The loss of hiw is axonal protective to a comparable level as Wlds overexpression (Xiong et al., 2012). hiw mutant neurons have higher Nmnat levels prior to injury which remain elevated even after injury, most likely contributing to axonal protection (Xiong et al., 2012). Conversely, overexpressing de-ubiquitinating enzyme UBP2 delayed the degeneration of Drosophila motoneuron axons, likely by counteracting the effect of Hiw on Nmnat. These results led to the conclusion that Hiw ubiquitinates and promotes the degradation of endogenous NMNAT protein. The role of Hiw in axon degeneration is also conserved, as the deletion of Phr1, the hiw ortholog in mice, also increased NMNAT2 levels in uninjured axons and protected severed sciatic axons (Babetto et al., 2013). However, NMNAT2 levels still fell in injured Phr1 LOF axons, suggesting that additional mechanisms can degrade NMNAT2 in mice. Highwire’s RING domain, which is involved in the direct transfer of ubiquitin (Ub) from the E2-Ub complex to the target substrate, is important for its function, as a hiw dominant negative allele lacking this domain blocks degeneration (Xiong et al., 2012). This was also confirmed when Neukomm and colleagues identified dominant and recessive alleles of hiw using the wing clipping assay for ethyl methane sulphonate (EMS) mutagenesis screens. The hiw alleles that suppressed degeneration had mutations in the protein’s RING domain (Neukomm et al., 2014).
2.3.5. Wallenda
Besides Nmnat, Hiw also downregulates Wallenda (Wnd), a MAP kinase kinase kinase (MAPKKK) and dual leucine kinase (DLK) involved in axon degeneration. wnd is one of the first genes identified to promote axon degeneration, demonstrated with the Drosophila ORN axotomy assay (Miller et al., 2009). Loss of wnd delayed the degeneration of injured axons for up to 2 days in Drosophila and loss of mouse DLK protected the axons of cultured mouse DRG neurons for up to one day. Xiong and colleagues also found that fly motoneuron axons are more resilient to degeneration if they were injured previously and this requires the function of Wnd/DLK (Xiong and Collins, 2012). Wnd/DLK is thought to promote degeneration through its target, the MAPK JNK (c-Jun N-terminal kinase). In the mammalian system, additional members of the MAPKKK family, MEKK4 and MLK2, promote degeneration and the combined knockdown of each with DLK led to longer lasting axon protection than DLK alone (Yang et al., 2015b). The three MAPKKKs most likely activate the JNK1–3, whose simultaneous knockdown protected axons after injury as well.
DLK proteins are downregulated by Phr1-family ubiquitin ligases from worms to mammals (Nakata et al., 2005; Collins et al., 2006; Babetto et al., 2013). However, this regulation does not explain the protective role of Phr1 LOF in axon degeneration, because it is the loss, not the increase, of DLK that is protective. How is the DLK/MAPK pathway activated in injured axons then? Yang et al. (2015b) later showed that MAPK pathway activation in injured mouse optical nerves depends on SARM1 and is inhibited by Wlds expression, suggesting that DLK acts downstream of SARM1. However, mutant alleles of MKK4, MKK7 and JNK do not suppress dSarmGOF-induced degeneration in the fly (Neukomm et al., 2017). Considering that the axon protection due to DLK inactivity is modest compared to Wlds overexpression, a plausible explanation is that the MAPK pathway acts together with other parallel pathways downstream of SARM1 activation during axon death.
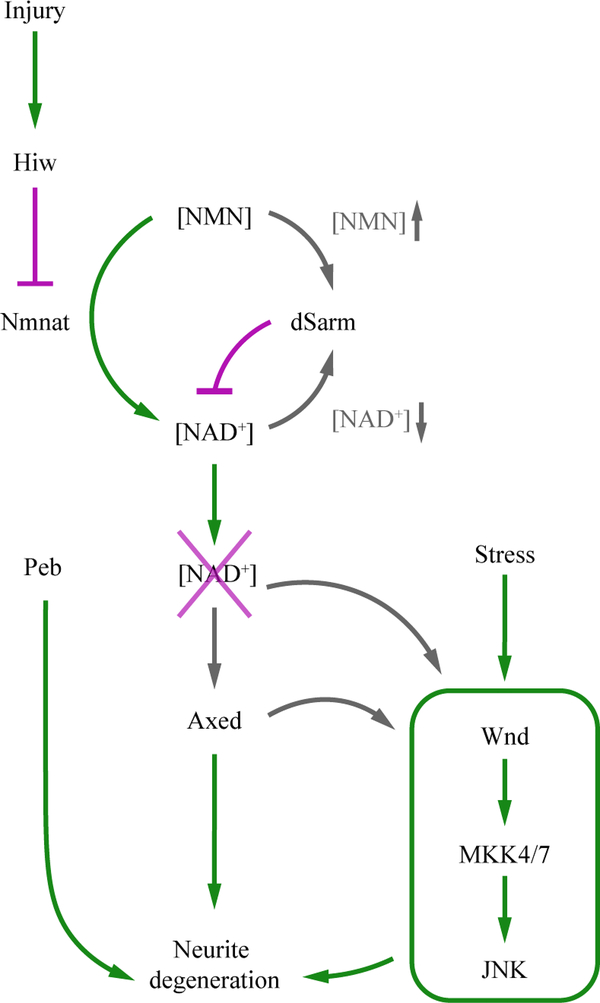
Together, the studies conducted in Drosophila, which were further corroborated by analyses in mammalian systems, delineate evolutionarily conserved and highly regulated signaling pathways of axon degeneration (Fig. 2). In the core degeneration pathway, Highwire/Phr1 mediates the degradation of Nmnat after axon injury, resulting in the fall of NAD+ levels below a threshold needed to maintain neuronal integrity. dSarm/SARM1 is activated, likely by a combination of NAD+ reduction and NMN accumulation, to further deplete NAD+. Axed is subsequently activated, perhaps by NAD+ depletion, to commit the axon to self-destruction. The Wnd/DLK and the JNK pathway may serve to integrate inputs from diverse stress stimuli and from the core pathway to promote degeneration. Additional factors such as Peb may modulate the susceptibility of certain types of neurons to axon degeneration. Based on the similar role of Wlds in dendrite protection, it is reasonable to suspect that these pathways may similarly regulate injury-induced dendrite degeneration.
Fig. 2: Injury-induced neurodegeneration pathway.
In the core pathway, injury induces accumulation of Hiw/Phr1, which in turn destabilizes Nmnat. The loss of Nmnat results in accumulation of NMN and reduction of NAD+. High levels of NMN and low levels of NAD+ likely contribute to the activation of dSarm/SARM1, which further depletes NAD+ and commits the axon to degeneration possibly by activating Axed. Wnd/DLK and the JNK pathway promote axon death downstream of dSarm/SARM1, likely also downstream of Axed. Peb promotes degeneration but its position in the pathway is unclear. The green arrows represent activation; magenta lines represent blocking. Gray arrows represent uncertain activation.
3. Clearance of degenerating neurites
While the degeneration pathways orchestrate the autonomous cellular processes to break neurites apart, the clearance of the resultant neurite fragments has to rely on other phagocytic cells that specifically recognize degenerating neural tissues. In Drosophila, neuronal debris is engulfed by different cell types in the CNS and the PNS, but the cellular processes and the molecular mechanisms by which the phagocytes recognize and engulf neuronal fragments are nevertheless very similar. Here, we first discuss the phagocytes in the CNS and the PNS and then summarize what we know about the recognition of neuronal debris by phagocytes.
3.1. Phagocytes that engulf degenerating neurites
3.1.1. Glia: phagocytes in the CNS
During nervous system development, neurite pruning and synapse elimination are important steps in circuit refinement (reviewed by Riccomagno and Kolodkin, 2015). Resident phagocytes of the CNS, specifically glia, are closely involved in the process of axon pruning. The Drosophila nervous system contains several glial subtypes that share morphological and functional features with cells in the mammalian nervous system. Perineural and subperineural glia are found along the surface of the fly brain and the peripheral nerves. Subperineural glia provide chemical and physical barriers to neurons analogous to the blood-brain barrier in mammals. Within the CNS, there are three neuron-associated glial cell types, cortex glia, astrocytes, and ensheathing glia, which share structural and functional similarities with mammalian astrocytes. In the PNS, besides perineural and subperineural glia, an additional glial type, wrapping glia, ensheathes peripheral axon projections similarly to how Schwann cells form Remak bundles in the mammalian PNS (reviewed by Freeman, 2015). Key studies done in the Drosophila CNS have shown definitively that astrocytes play an active role in the developmental axon pruning of the MB γ axons: they not only engulf axonal debris but also promote fragmentation of MB axons (Watts et al., 2003; Awasaki and Ito, 2004; Hakim et al., 2014). Recent studies showed that microglia and astrocytes are involved in synaptic elimination and axon pruning in the mammalian CNS (reviewed by Riccomagno and Kolodkin, 2015; Presumey et al., 2017).
The clearance of injured axons in the Drosophila CNS was first investigated using the adult ORN injury model (MacDonald et al., 2006), in which the response of neuronal support cells such as glia can be scored and dissected during axon degeneration. Ensheathing glia were found to play the primary role in neuronal debris clearance after axon injury. MacDonald et al. demonstrated that glia membranes infiltrate the olfactory lobe and clear axons after axotomy and are also capable of discriminating healthy axons from damaged axons. They accomplished this by removing only the subset of ORNs located on the maxillary palps. The glia infiltrated and cleared the injured axon bundles projected from the maxillary palps and ignored the axons projected from the antenna, supporting the idea that glia can differentiate intact axons from injured ones. Interestingly, astrocytes do not seem to play any role in clearing injured axons of ORNs despite being associated with the synaptic regions of ORNs normally (Doherty et al., 2009). In the mammalian CNS, microglia are the first responders to brain injury and other pathological insults when they invade areas of injury and inflammation (reviewed by Gadani et al., 2015).
3.1.2. Phagocytes in the PNS
In the fly PNS, other resident cells have been implicated as the phagocytes required for clearance of neuronal debris after developmental pruning and injury-induced degeneration. Epidermal cells, which make up the tissue that the da sensory dendrites directly innervate, function as the primary phagocytes that break down and engulf injured and pruned dendrites (Han et al., 2014). Han and colleagues demonstrated that when the phagocytic ability of epidermal cells is disrupted by overexpressing a temperature-sensitive dominant allele of shibire (a gene homolog of dynamin), which is required for the formation of the phagocytic cup, da dendrite debris clearance was blocked. This epidermal cell-mediated clearance of sensory neurites is conserved in vertebrates ―in zebrafish, epidermal cells clear injured sensory axons post-axotomy as well (Rasmussen et al., 2015). Besides epidermal cells, both peripheral glia and muscle cells can also function as phagocytes to engulf immature synapses at the Drosophila neuromuscular junction (Fuentes-Medel et al., 2009). In the mammalian PNS, satellite glial cell (SGC) precursors clear neuron corpses during peripheral ganglia development (Wu et al., 2009). Importantly, efficient clearance of neuronal debris, whether it is due to developmental pruning or injury/disease, is critical for maintaining tissue homeostasis and minimizing inflammation (reviewed by Salter and Stevens, 2017). Efficient debris clearance is also important for axon regeneration as lingering axon debris can repel regenerating axons (Martin et al., 2010).
3.2. Recognition of degenerating neurites by phagocytes
Phagocytes remove neuronal debris without engulfing uninjured neurites, suggesting that specific signals allow phagocytes to recognize neuronal debris. Interestingly, in both CNS and PNS, phagocytes also actively participate in breaking down neurites destined to be removed, as blocking phagocytosis not only suppresses the clearance of neuronal debris but also delays neurite fragmentation (Awasaki and Ito, 2004; Hakim et al., 2014; Han et al., 2014; Tasdemir-Yilmaz and Freeman, 2014). How do phagocytes recognize degenerating neurites and neuronal debris? Specifically, what are the signal(s) and receptor(s) that mediate the recognition? How are the signals regulated in degenerating neurites? Lastly, what are the consequences when the recognition pathway is perturbed? We discuss our current understanding of these questions below.
3.2.1. Engulfment receptors involved in neurodegeneration
Phagocytes express transmembrane engulfment receptors that allow them to recognize dying cells. These engulfment receptors interact with cell surface ligands, dubbed “eat-me” signals, found on apoptotic cells directly or indirectly through bridging molecules (reviewed by Ravichandran, 2010). Eat-me signals activate the engulfment receptors on the phagocytes, resulting in a signaling cascade that promotes cytoskeletal rearrangements required for cell corpse engulfment (reviewed by Ravichandran and Lorenz, 2007). An engulfment receptor involved in clearing neuronal debris in Drosophila is Draper (Drpr), a homolog of Caenorhabditis elegans CED-1. Loss of drpr strongly suppresses debris clearance during axon pruning in MB (Awasaki et al., 2006) and blocks glial activation and subsequent axon clearance after axotomy in the olfactory bulb (MacDonald et al., 2006). Drpr is also required for the clearance of injured da sensory dendrites in the PNS by the larval epidermal cells, and loss of drpr strongly delayed clearance of pruned da sensory dendrites in the pupal stage (Williams et al., 2006; Han et al., 2014). Interestingly, these studies conducted in both the CNS and the PNS consistently demonstrated that Drpr is necessary for engulfment during injury-induced degeneration but is only partially required for clearance of degenerating neurites during developmental pruning.
The engulfment receptor redundant to Drpr during neurite pruning is currently unknown. A transmembrane protein implicated in engulfment in Drosophila is Six-microns-under (SIMU), which is important for cell corpse clearance during embryonic CNS development (Kurant et al., 2008). However, SIMU is unlikely to be an engulfment receptor because expressing a secreted version of its extracellular domain rescued the clearance defects of simu embryos. Phenotypically, simu is epistatic to drpr, leading to the conclusion that SIMU functions as a bridging molecule between apoptotic neurons and phagocytes (Kurant et al., 2008). SIMU is not required for the clearance of dendrite debris by epidermal cells after injury and during pruning in the PNS (Han et al., 2014), suggesting that its role is stage or context dependent. Croquemort (Crq) is another membrane protein identified as an engulfment receptor in Drosophila and it shares around 20% identity with human CD36, a scavenger receptor that facilitates engulfment of apoptotic cells (Franc et al., 1996). However, during PNS dendrite degeneration, loss of crq led to an excessive phagosome fusion and inefficient debris degradation in phagocytes rather than an engulfment defect (Han et al., 2014).
Drpr’s mammalian homologs, Jedi-1 and MEGF10, also function as engulfment receptors in the developing peripheral ganglia where they clear apoptotic neurons (Wu et al., 2009). All these three receptors have single-pass transmembrane domains that link extracellular EGF repeats to intracellular tyrosine (Tyr) phosphorylation sites. Drpr signals through its immunoreceptor tyrosine-based activation motif (ITAM) domain to recruit downstream effectors, the Src family kinase Src42a, SH2 domain containing kinase Shark, and the adaptor protein Crk (Ziegenfuss et al., 2008; Ziegenfuss et al., 2012). Mammalian Shark homolog, Syk, interacts with Jedi-1 and MEGF10 through their ITAM domains as well, suggesting a high conservation of this ancient engulfment pathway (Scheib et al., 2012; Williamson and Vale, 2018).
Another signaling component downstream of Drpr is tissue necrosis factor receptor associated factor 4 (TRAF4). TRAF4 binds to Drpr and is important for Drpr-mediated gene expression changes in glia in response to axon injury by activating the JNK pathway (Lu et al., 2017). Knockdown of TRAF4 in glia resulted in diminished glial responses and delay of ORN axon debris clearance after axotomy. While it is unclear whether MEGF10 also interacts with TRAF4, activation of JNK/c-Jun signaling has been implicated in astrocyte proliferation and reactive gliosis in mammals (Gadea et al., 2008).
Additional engulfment receptors have been identified in mammals that lack homologs in Drosophila, which is to be expected as mammals have a more sophisticated innate immune system. The mammalian TAM receptors (Tyro3, Axl, and Mer) are a family of receptor tyrosine kinases that have an important role in engulfment and phagocytic clearance of apoptotic cells (reviewed by Lemke, 2013). Microglia express Axl and Mer (Gautier et al., 2012) and loss of these genes in adult mouse CNS resulted in impaired cell clearance after viral infection (Tufail et al., 2017). Axl and Mer were also shown to be involved in clearance of apoptotic cells in neurogenic regions of adult CNS, specifically the subgranular and subventricular zones of the mouse hippocampus (Fourgeaud et al., 2016). Also, Axl expression is upregulated in glia during inflammatory insults. Another mammalian engulfment receptor, αvβ3 integrin, is involved in the diurnal phagocytosis of photoreceptor outer segments (POS) in the mouse retina (Nandrot et al., 2007). Its Drosophila homolog, αPS3, together with Drpr, has been implicated in hemocyte priming for efficient engulfment in the embryo (Nonaka et al., 2017). Whether αPS3 is involved in neuron debris clearance is unclear, but αPS3 is expressed in glia and is important for glia wrapping of Drosophila peripheral nerves and development of retinal glia (Xie and Auld, 2011; Tavares et al., 2015).
The mammalian innate immune system has also been implicated in activity-dependent synapse elimination in the developing mouse visual system, where engulfment is carried out by resident microglia via the activation of the classical complement cascade (Stevens et al., 2007; Schafer et al., 2012; Bialas and Stevens, 2013; Hong et al., 2016). There is also evidence showing that overactivation of the complement system is the cause of excess synaptic pruning pathology found in schizophrenia disease (Sekar et al., 2016). The roles of microglia in synapse elimination and related pathology are covered in detail in other excellent reviews (Neniskyte and Gross, 2017; Presumey et al., 2017; Salter and Stevens, 2017).
3.2.2. PS exposure during neurite degeneration
What are the ligands that allow these engulfment receptors to recognize neuronal debris? A highly conserved eat-me signal for apoptotic cells is phosphatidylserine (PS). PS is a phospholipid that resides exclusively in the cytoplasmic leaflet of the plasma membrane of most healthy cells (reviewed by Ravichandran, 2010). The establishment and maintenance of PS asymmetry is an active and highly regulated cellular process mediated by transmembrane aminophospholipid flippases encoded by the P4-ATPase family (reviewed by Leventis and Grinstein, 2010). During apoptosis, PS is externalized by lipid scramblases to the outer leaflet and tags the cell for engulfment (reviewed by Ravichandran, 2010; Segawa and Nagata, 2015). One of the scramblases, Xk-related protein 8 (XKR8), is activated by caspase-mediated cleavage and is responsible for PS exposure in apoptotic cells (Suzuki et al., 2013a). Another scramblase, TMEM16F, is activated by intracellular Ca2+ and is required for PS exposure on platelets during blood clotting (Suzuki et al., 2010; Suzuki et al., 2013b; Fujii et al., 2015). Both TMEM16F and XKR8 can cause elevated PS exposure when overexpressed (Segawa et al., 2011; Suzuki et al., 2013a).
Do degenerating neurites expose PS? Visualizing PS exposure on degenerating neurons in vivo has been difficult because tissue fixation inevitably disrupts the lipid asymmetry of the cell membrane and results in false positive labeling. Using the secreted versions of two PS-binding proteins Annexin V and lactadherin (Koopman et al., 1994; Andersen et al., 2000; Hanayama et al., 2002; Mapes et al., 2012) to label PS exposure in live animals, we recently demonstrated that Drosophila sensory dendrites expose PS after injury in the larval stage and during pruning in the pupal stage (Sapar et al., 2018). We took advantage of the fly’s open circulatory system and expressed the two PS sensors in the fat body so that the sensors were secreted into the hemolymph and diffused to the sensory dendrites located at the basal membrane of the epidermal cells. The PS sensors only bind to the PS on the outer surface of cell structures, thus are capable of labeling specific dendritic branches that expose PS. Since the system does not require dissection or immunostaining, we were able to take long-term time-lapse videos of live animals and demonstrate that PS exposure always occurred prior to dendrite fragmentation and phagocytic engulfment of debris, supporting the role of PS as an eat-me signal. Mammalian neurons expose PS during degeneration as well. Rat sciatic nerves expose PS after injury in vivo and cultured mouse DRG neurons expose PS after nerve growth factor (NGF) withdrawal (Kim et al., 2010; Shacham-Silverberg et al., 2018).
3.2.3. PS exposure recognition during neurodegeneration
Does PS exposure mediate recognition of neurites destined for phagocytosis? Both in vitro and in vivo evidence suggests that PS sensing plays a role in Drpr-mediated engulfment in Drosophila. In vitro, Drpr has been shown to bind to PS (Tung et al., 2013). Furthermore, Drosophila S2 cells transfected to express Drpr become proficient phagocytes and can engulf PS-exposing cell corpses and PS-containing liposomes (Williamson and Vale, 2018). In this system, PS is sufficient to locally trigger Drpr phosphorylation at the ITAM to activate downstream engulfment signaling. In vivo, high levels of PS exposure on injured and pruned dendrites precedes Drpr-mediated engulfment. Importantly, da neuron dendrites forced to expose PS at a low level are engulfed by larval epidermal cells in a Drpr-dependent manner (Sapar et al., 2018), arguing that epidermal cells can recognize degenerating dendrites through externalized PS. However, in these experiments whether Drpr binds PS directly or interacts with PS indirectly via bridging molecules is difficult to determine. Notably, a bridging molecule TTR-52 mediates the interaction between PS and the Drpr homolog CED-1 in C. elegans (Wang et al., 2010).
Besides PS, Drpr possibly recognizes other eat-me signals on apoptotic cells. Drpr reportedly binds to protein ligands Pretaporter (Prtp) and calcium-binding protein 1 (CaBP1), both of which are implicated in cell corpse clearance in developing embryos of Drosophila but are not involved in larval axon pruning (Kuraishi et al., 2009; Okada et al., 2012). Whether Prtp and CaBP1 play any role in the clearance of injured neurites is unknown.
Several lines of evidence suggest that PS recognition is also important for clearance in the mammalian CNS. Members of TAM receptor family, Axl and Mer, depend on interactions with PS binding proteins, growth arrest-specific protein 6 (Gas6) and protein S (Pros1), for cell corpse clearance in the CNS (Fourgeaud et al., 2016). Moreover, the αvβ3 integrin-mediated phagocytosis of POS in the mouse retina is stimulated by lactadherin (MFG-E8), which has been shown to bind to PS and to link apoptotic cells to the integrin receptor on macrophages for engulfment (Andersen et al., 2000; Hanayama et al., 2002; Nandrot et al., 2007). Interestingly, lactadherin lacking its integrin binding domain can potentiate Drosophila epidermal cells to phagocytose and damage PS-exposing dendrites of da neurons in a Drpr-dependent manner (Sapar et al., 2018), suggesting that lactadherin may be able to interact with non-integrin engulfment receptors. Lastly, there is also growing evidence that C1q of the complement cascade may bind to PS (Paidassi et al., 2008; Gyorffy et al., 2018).
3.2.4. Regulation of PS exposure during neurite degeneration
Relatively little is known about how PS exposure is regulated during neurite degeneration at this point. However, existing evidence points to a connection with NAD+ metabolism, perhaps not surprisingly as NAD+ depletion is a key factor required for triggering intrinsic injury-induced axon and dendrite degeneration. In vivo, Wlds overexpression in da sensory neurons blocked PS exposure on injured dendrites for at least 8–10 h (possibly longer, as at this time point dendrites showed minimal morphological signs of degeneration) (Sapar et al., 2018). Similarly, in cultured DRG neurons, Wlds overexpression significantly delayed PS exposure after axon transection (Almasieh et al., 2017). Supplementing transected neurons in culture with NAD+ also inhibits PS exposure on degenerating axons (Shacham-Silverberg et al., 2018). As NAD+ reduction does not seem to be required for developmental neurite pruning, it remains to be determined whether PS exposure during neurite pruning is related to NAD+ metabolism.
Interestingly, Shacham-Silverberg and colleagues showed that PS exposure could be decoupled from degeneration process itself (Shacham-Silverberg et al., 2018). ATP depletion by inhibition of mitochondrial activity, which is also an expected consequence of NAD+ depletion in injured axons (Yang et al., 2015b), was sufficient to expose PS on the outer membrane of cultured mouse DRG neurons, while the neurons remained intact and showed minimal signs of degeneration for up to 24 h post-treatment. This finding suggests that PS exposure may be regulated separately from other intrinsic processes of axon degeneration such as cytoskeletal disruption and membrane severing.
What other pathways may also be involved in PS exposure in neurons? Mammalian studies have provided some important clues. First, autophagy seems to play a positive role in PS exposure. When the components of the autophagy pathway, Atg5 and Atg7, were knocked down in cultured DRG neurons, local ATP production was disrupted in the degenerating axons, and PS exposure was significantly reduced (Wakatsuki et al., 2017). Consistent with these observations, inhibition of autophagy in the optic nerve axons led to lower recruitment of phagocytes to the axonal debris, presumably because neurons exposed a lower level of PS. Second, knockdown of the scramblase XKR8 or a lipid translocase ABC1 reduced PS exposure during degeneration as well, although it is unclear if this is related to the induction of autophagy or a separate pathway (Wakatsuki and Araki, 2017). Third, neurons can expose PS through necroptosis, which is a nonapoptotic form of death initiated by mixed lineage kinase domain-like (MLKL) protein (Zargarian et al., 2017). Zargarian and colleagues demonstrated that necroptotic cells expose PS after phosphorylated MLKL is translocated to the cell membrane. When the translocation of phosphorylated MLKL was blocked, PS exposure was reversed, and cell viability was restored. Interestingly, MLKL and other necroptotic machinery have been implicated in a mouse model of amyotrophic lateral sclerosis (ALS) (Ito et al., 2016), indicating a possible link with abnormal PS regulation.
3.2.5. Association of PS asymmetry dysregulation and neurodegeneration
If PS acts as an eat-me signal for degenerating neurites, is PS exposure alone sufficient to induce surrounding phagocytes to attack live neurons and kill them? This type of cell death has been described as primary phagocytosis or phagoptosis, a term referring to cell death resulting from engulfment of live cells by phagocytes. Phagoptosis is distinct from regular apoptosis in that the engulfed cell is stressed but viable, exposes an eat-me signal but in a reversible manner, and is executed by the activity of surrounding phagocytes, which can be prevented by blocking recognition or disrupting the engulfment machinery of the attacking phagocytes (reviewed by Brown and Neher, 2014). Below we discuss the consequences of ectopic PS exposure in otherwise healthy neurons and briefly cover how improper PS exposure and aberrant phagocyte-neuron interactions could contribute to neurodegenerative disease pathology.
The first evidence supporting a dominant effect of PS exposure in degeneration came from a study of a mouse mutant called wabbler-lethal (wl), which shows neurodegeneration in both the central and peripheral nervous systems (Zhu et al., 2012). wl carries a mutation in ATP8A2, a P4-ATPase family member that specifically translocates PS to the inner leaflet of membranes. Although spontaneous PS exposure was not demonstrated in wl neurons, this study suggests that dysregulation of PS asymmetry may be behind the degeneration phenotype. A more direct demonstration for the association of PS exposure and neurite degeneration was shown in our recent characterization of Drosophila PS flippases. The Drosophila genome encodes six P4-ATPases, five of which require a putative chaperone, CDC50, for proper subcellular localization and function (Saito et al., 2004; Paulusma et al., 2008; Tanaka et al., 2011). By utilizing a tissue-specific CRISPR/Cas9 system (Poe et al., 2019), we characterized the Drosophila genes CG9947 and CG42321, which encode fly homologs of CDC50 and ATP8A P4-ATPase, respectively. The loss of either gene in class IV da neurons resulted in low levels of ectopic PS exposure at distal terminal dendrites (Sapar et al., 2018). Such moderate PS exposure due to flippase loss strongly suggests the existence of additional PS externalization mechanisms that promote rapid and high levels of PS exposure during neurite degeneration. These results also suggest that neurons are compartmentalized in such a way that their distinct processes are differentially sensitive to PS asymmetry disruption.
Consistent with the idea of phagoptosis, CDC50 knockout (KO) neurons lost membranes at dendrite tips in a phagocyte-dependent manner. The interpretation is that nearby epidermal cells detected the low level of PS exposure on dendrites, and subsequently broke and engulfed the PS-exposing dendrite tips. This phenomenon can be further enhanced by combining CDC50 KO with the overexpression of a hypersensitive mutant of the scramblase TMEM16F (Segawa et al., 2011). In the CNS, knocking out CDC50 in postmitotic OR22a ORNs led to a progressive age-dependent axon degeneration, suggesting that even a low amount of PS externalization can have a cumulative effect in causing axon loss over time. Overexpression of TMEM16F and subsequent activation of OR22a ORNs by their odorant, ethyl butyrate (Dobritsa et al., 2003), to promote TMEM16F scramblase activity by elevating intracellular Ca2+, led to a striking axon loss in flies within 5 days (Sapar et al., 2018). These findings suggest that loss of flippase function and/or activation of scramblases can induce enough ectopic PS exposure on live neurons to cause potentially devastating dendrite and axon loss due to attacks of phagocytes. This study establishes a genetic basis for phagocytosis-related disease pathology of neurodegenerative disorders due to PS asymmetry dysregulation.
It has been shown in mammalian systems that proteins associated with Alzheimer’s disease can lead to PS exposure on live neurons. Cultured neurons expose significantly more PS when treated with a nanomolar amount of Amyloid β peptide, which is involved in plaque formation in Alzheimer’s disease (Neniskyte et al., 2011). If co-cultured with microglia, Amyloid β leads to PS exposure and microglia-induced phagoptosis of neurons. The neuronal cell death can be blocked by masking PS with PS-binding proteins or by directly blocking phagocytosis. Similarly, live DRG neurons containing tau filaments expose PS in a reactive oxygen species-dependent manner and can be killed via phagoptosis when co-cultured with microglia (Brelstaff et al., 2018). Reversing PS exposure or blocking lactadherin-dependent recognition of neurons by microglia was neuroprotective. Whether neurodegeneration seen in other diseases is also associated with inappropriate PS exposure on viable neurons remains to be determined.
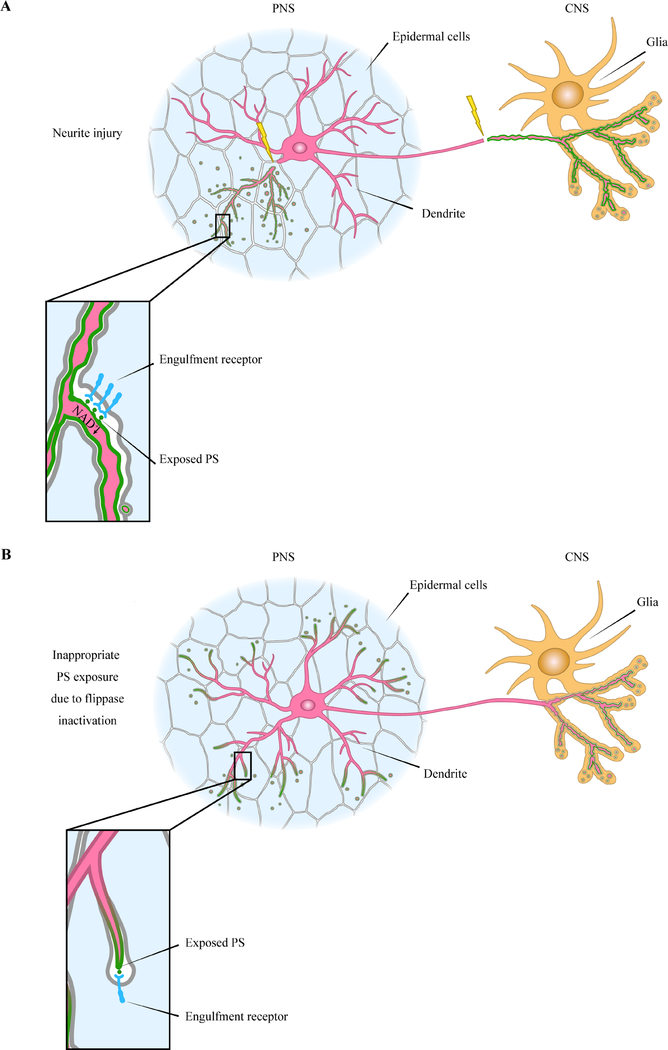
Together, the evidence summarized above pinpoints a close association between PS exposure and neurite degeneration. During both developmental pruning and injury-induced degeneration, neurites expose PS on their surface to engage engulfment receptors on nearby resident phagocytes directly or indirectly through PS-binding bridging molecules. This PS exposure, at least in injury-induced degeneration, depends on NAD+ depletion (Fig. 3A). Under pathological conditions that lead to disruptions of PS flippase function or activation of scramblases, neurons ectopically expose PS, which in turn induces attack and damage by phagocytes on otherwise healthy neurites (Fig. 3B).
Fig. 3: The association of PS exposure and neurite degeneration.
A: Phosphatidylserine (PS) exposure resulting from neurite degeneration. In the PNS, injured dendrites (magenta) expose high levels of PS (green) which is recognized via engulfment receptor (blue) on phagocytic epidermal cells. Epidermal cells clear away the injured dendrites but spare the rest of the dendritic arbor. In the CNS, glia engulf degenerating axons by recognizing exposed PS. B: Ectopic PS exposure as a cause of neurite degeneration. When PS asymmetry is perturbed, such as by the loss of PS flippase function or by scramblase activation, neurons expose low levels of PS at the distal dendritic arbor. This is sufficient to drive epidermal cells to attack and engulf the PS exposing dendrites. In the CNS, ectopic PS exposure also results in loss of axon membranes. PNS, peripheral nervous system; CNS, central nervous system.
4. Perspectives
Elucidating the mechanisms of neurite degeneration has obvious clinical relevance, and is also instrumental for understanding the development, maintenance, and dynamics of the nervous system. Over the last couple of decades, significant progress has been made in understanding the intrinsic molecular pathways responsible for breaking down injured neurites and the neurite-phagocyte interactions that lead to prompt clearance of neuronal debris. Drosophila models and assays have played an integral role in providing insights into the basic mechanisms underlying nervous system injuries and neurological diseases.
Looking forward, many important questions in neurite degeneration and clearance remain to be answered. Drosophila will undoubtedly continue to play an important role in answering these questions. Regarding the axon-death pathway, more details are needed to fully understand how dSarm is activated. How Axed activity is related to NAD+ depletion and what target proteins Axed regulates also need to be determined. In addition, the MAPK pathway appears to be turned on in a different temporal window from that of SARM1 (Yang et al., 2015a). Therefore, how the MAPK pathway contributes to neurite degeneration and how it is related to dSarm/SARM1 and Axed are still mysteries. To address these questions, sophisticated genetic experiments will likely be necessary to elucidate how these components interact in vivo, in conjunction with biochemical characterization of less understood components like Axed. Some key links may still be missing in our understanding of the pathways, even though classical LOF genetic screens have been conducted in Drosophila to saturation. Thus, modifier screens may be useful for uncovering additional or redundant components involved in neurite degeneration.
Related to neurite clearance, it is important to determine if other eat-me signals are also involved in the recognition of degenerating neurites and debris by phagocytes. Dysregulation of these signals could cause or contribute to neurodegenerative diseases as exemplified by the consequences of mis-regulated neuronal PS exposure in vivo. Although the association between neuronal PS exposure and neurite degeneration is well supported by experimental evidence in multiple animal model systems, many aspects of neuronal PS exposure are still poorly known. First, do different levels of neuronal PS exposure influence phagocyte activity under physiological and pathological conditions? Second, how is PS exposure regulated by the core axon-death pathway and does this include direct inactivation of PS asymmetry regulatory machinery (flippases/floppases) and/or activation of lipid scramblases? Third, what is the role of PS exposure in synaptic remodeling and maturation? Lastly, how does PS exposure contribute to various neurodegenerative diseases and whether targeting PS regulation and sensing can intervene in disease progression? With increasingly powerful and sophisticated genetic and cellular tools, Drosophila holds great promise for exploring these exciting new frontiers.
Acknowledgments
We apologize for not being able to include all relevant studies in the topics of this review due to the space limit. We thank Hui Ji and Ankita Sarkar for helpful discussions. This work was supported by a Cornell start-up fund and NIH grants (R01NS099125 and R21OD023824) awarded to C.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almasieh M, Catrinescu MM, Binan L, Costantino S, and Levin LA, 2017. Axonal degeneration in retinal ganglion cells is associated with a membrane polarity-sensitive Redox process. J. Neurosci 37, 3824–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MH, Graversen H, Fedosov SN, Petersen TE, and Rasmussen JT, 2000. Functional analyses of two cellular binding domains of bovine lactadherin. Biochemistry 39, 6200–6206. [DOI] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, and Milbrandt J, 2004. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013. [DOI] [PubMed] [Google Scholar]
- Awasaki T, and Ito K, 2004. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr. Biol 14, 668–677. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, and Ito K, 2006. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855–867. [DOI] [PubMed] [Google Scholar]
- Babetto E, Beirowski B, Russler EV, Milbrandt J, and DiAntonio A, 2013. The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 3, 1422–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirowski B, Morreale G, Conforti L, Mazzola F, Di Stefano M, Wilbrey A, Babetto E, Janeckova L, Magni G, and Coleman MP, 2010. WldS can delay Wallerian degeneration in mice when interaction with valosin-containing protein is weakened. Neuroscience 166, 201–211. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Tong C, and Tsuda H, 2010. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci 11, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas AR, and Stevens B, 2013. TGF-beta signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat. Neurosci 16, 1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brelstaff J, Tolkovsky AM, Ghetti B, Goedert M, and Spillantini MG, 2018. Living neurons with tau filaments aberrantly expose phosphatidylserine and are phagocytosed by microglia. Cell Rep. 24, 1939–1948 e1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC, and Neher JJ, 2014. Microglial phagocytosis of live neurons. Nat. Rev. Neurosci 15, 209–216. [DOI] [PubMed] [Google Scholar]
- Charng WL, Yamamoto S, and Bellen HJ, 2014. Shared mechanisms between Drosophila peripheral nervous system development and human neurodegenerative diseases. Curr. Opin. Neurobiol 27, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MP, and Freeman MR, 2010. Wallerian degeneration, wld(s), and nmnat. Annu. Rev. Neurosci 33, 245–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, and DiAntonio A, 2006. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron 51, 57–69. [DOI] [PubMed] [Google Scholar]
- Di Stefano M, Nascimento-Ferreira I, Orsomando G, Mori V, Gilley J, Brown R, Janeckova L, Vargas ME, Worrell LA, Loreto A, Tickle J, Patrick J, Webster JR, Marangoni M, Carpi FM, Pucciarelli S, Rossi F, Meng W, Sagasti A, Ribchester RR, Magni G, Coleman MP and Conforti L, 2015. A rise in NAD precursor nicotinamide mononucleotide (NMN) after injury promotes axon degeneration. Cell Death Differ. 22, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, and Carlson JR, 2003. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, and Freeman MR, 2009. Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci 29, 4768–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, and Milbrandt J, 2017. The SARM1 Toll/Interleukin-1 receptor domain possesses intrinsic NAD(+) cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334–1343 e1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Soares L, and Bonini NM, 2013. Design and implementation of in vivo imaging of neural injury responses in the adult Drosophila wing. Nat. Protoc 8, 810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley JE, Burdett TC, Barria R, Neukomm LJ, Kenna KP, Landers JE, and Freeman MR, 2018. Transcription factor Pebbled/RREB1 regulates injury-induced axon degeneration. Proc. Natl. Acad. Sci. U. S. A 115, 1358–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourgeaud L, Traves PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagorska A, Rothlin CV, Nimmerjahn A, and Lemke G, 2016. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, and Ezekowitz RA, 1996. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4, 431–443. [DOI] [PubMed] [Google Scholar]
- Freeman MR, 2015. Drosophila Central Nervous System Glia. Cold Spring Harb. Perspect. Biol 7 10.1101/cshperspect.a020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Medel Y, Logan MA, Ashley J, Ataman B, Budnik V, and Freeman MR, 2009. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol. 7, e1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Sakata A, Nishimura S, Eto K, and Nagata S, 2015. TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc. Natl. Acad. Sci. U. S. A 112, 12800–12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadani SP, Walsh JT, Lukens JR, and Kipnis J, 2015. Dealing with danger in the CNS: the response of the immune system to injury. Neuron 87, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea A, Schinelli S, and Gallo V, 2008. Endothelin-1 regulates astrocyte proliferation and reactive gliosis via a JNK/c-Jun signaling pathway. J. Neurosci 28, 2394–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ and Immunological Genome C, 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol 13, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Doan RA, Strickland A, Huang X, Milbrandt J, and DiAntonio A, 2016. Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain 139, 3092–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, and Milbrandt J, 2015. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science 348, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Summers DW, Milbrandt J, and DiAntonio A, 2016. axon self-destruction: new links among SARM1, MAPKs, and NAD+ metabolism. Neuron 89, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, and Coleman MP, 2010. Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 8, e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Orsomando G, Nascimento-Ferreira I, and Coleman MP, 2015. Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell. Rep 10, 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann TI, Ziegler M, Puntervoll P, de Figueiredo LF, Schuster S, and Heiland I, 2012. NAD(+) biosynthesis and salvage--a phylogenetic perspective. FEBS J. 279, 3355–3363. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, and Jan YN, 2002. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129, 2867–2878. [DOI] [PubMed] [Google Scholar]
- Gyorffy BA, Kun J, Torok G, Bulyaki E, Borhegyi Z, Gulyassy P, Kis V, Szocsics P, Micsonai A, Matko J, Drahos L, Juhasz G, Kekesi KA and Kardos J, 2018. Local apoptotic-like mechanisms underlie complement-mediated synaptic pruning. Proc. Natl. Acad. Sci. U. S. A 115, 6303–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim Y, Yaniv SP, and Schuldiner O, 2014. Astrocytes play a key role in Drosophila mushroom body axon pruning. PLoS One 9, e86178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Jan LY, and Jan YN, 2011. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. U. S. A 108, 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Song Y, Xiao H, Wang D, Franc NC, Jan LY, and Jan YN, 2014. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron 81, 544–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Wang D, Soba P, Zhu S, Lin X, Jan LY, and Jan YN, 2012. Integrins regulate repulsion-mediated dendritic patterning of Drosophila sensory neurons by restricting dendrites in a 2D space. Neuron 73, 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, and Nagata S, 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187. [DOI] [PubMed] [Google Scholar]
- Henninger N, Bouley J, Sikoglu EM, An J, Moore CM, King JA, Bowser R, Freeman MR, and Brown RH Jr., 2016. Attenuated traumatic axonal injury and improved functional outcome after traumatic brain injury in mice lacking Sarm1. Brain 139, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks AN, Lorenzetti D, Gilley J, Lu B, Andersson KE, Miligan C, Overbeek PA, Oppenheim R, and Bishop CE, 2012. Nicotinamide mononucleotide adenylyltransferase 2 (Nmnat2) regulates axon integrity in the mouse embryo. PLoS One 7, e47869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, Lemere CA, Selkoe DJ and Stevens B, 2016. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O’Leary DD, and Luo L, 2006. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron 50, 883–895. [DOI] [PubMed] [Google Scholar]
- Ito Y, Ofengeim D, Najafov A, Das S, Saberi S, Li Y, Hitomi J, Zhu H, Chen H, Mayo L, Geng J, Amin P, Dewitt JP, Mookhtiar AK, Florez M, Ouchida AT, Fan JB, Pasparakis M, Kelliher MA, Ravits J and Yuan J, 2016. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 353, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner M, Schwab ME, Lichtman JW, and Misgeld T, 2005. In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat. Med 11, 572–577. [DOI] [PubMed] [Google Scholar]
- Kim ME, Shrestha BR, Blazeski R, Mason CA, and Grueber WB, 2012. Integrins establish dendrite-substrate relationships that promote dendritic self-avoidance and patterning in Drosophila sensory neurons. Neuron 73, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Chen J, Chan JR, and Langen R, 2010. Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration. Nat. Methods 7, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, and van Oers MH, 1994. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84, 1415–1420. [PubMed] [Google Scholar]
- Kuraishi T, Nakagawa Y, Nagaosa K, Hashimoto Y, Ishimoto T, Moki T, Fujita Y, Nakayama H, Dohmae N, Shiratsuchi A, and Nakanishi Y, 2009. Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells. EMBO J.28, 3868–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant E, Axelrod S, Leaman D, and Gaul U, 2008. Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell 133, 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, 1963. Electron microscopy of Wallerian degeneration. J. Comp. Neurol 120, 65–79. [DOI] [PubMed] [Google Scholar]
- Lemke G, 2013. Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol 5, a009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis PA, and Grinstein S, 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys 39, 407–427. [DOI] [PubMed] [Google Scholar]
- Liu HW, Smith CB, Schmidt MS, Cambronne XA, Cohen MS, Migaud ME, Brenner C, and Goodman RH, 2018. Pharmacological bypass of NAD(+) salvage pathway protects neurons from chemotherapy-induced degeneration. Proc. Natl. Acad. Sci. U. S. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TY, MacDonald JM, Neukomm LJ, Sheehan AE, Bradshaw R, Logan MA, and Freeman MR, 2017. Axon degeneration induces glial responses through Draper-TRAF4-JNK signalling. Nat. Commun 8, 14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn ER, Perry VH, Brown MC, Rosen H, and Gordon S, 1989. Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve. Eur. J. Neurosci 1, 27–33. [DOI] [PubMed] [Google Scholar]
- Luo L, and O’Leary DD, 2005. Axon retraction and degeneration in development and disease. Annu. Rev. Neurosci 28, 127–156. [DOI] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, and Freeman MR, 2006. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 50, 869–881. [DOI] [PubMed] [Google Scholar]
- Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, Fernando FS, Tarlton A, Andressen C, Addicks K, Magni G, Ribchester RR, Perry VH and Coleman MP, 2001. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci 4, 1199–1206. [DOI] [PubMed] [Google Scholar]
- Mapes J, Chen YZ, Kim A, Mitani S, Kang BH, and Xue D, 2012. CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr. Biol 22, 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SM, O’Brien GS, Portera-Cailliau C, and Sagasti A, 2010. Wallerian degeneration of zebrafish trigeminal axons in the skin is required for regeneration and developmental pruning. Development 137, 3985–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk L, Berson A, and Bonini NM, 2015. Drosophila as an in vivo model for human neurodegenerative disease. Genetics 201, 377–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, and DiAntonio A, 2009. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci 12, 387–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, and Jin Y, 2005. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120, 407–420. [DOI] [PubMed] [Google Scholar]
- Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, and Finnemann SC, 2007. Essential role for MFG-E8 as ligand for alphavbeta5 integrin in diurnal retinal phagocytosis. Proc. Natl. Acad. Sci. U. S. A 104, 12005–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neniskyte U, and Gross CT, 2017. Errant gardeners: glial-cell-dependent synaptic pruning and neurodevelopmental disorders. Nat. Rev. Neurosci 18, 658–670. [DOI] [PubMed] [Google Scholar]
- Neniskyte U, Neher JJ, and Brown GC, 2011. Neuronal death induced by nanomolar amyloid beta is mediated by primary phagocytosis of neurons by microglia. J. Biol. Chem 286, 39904–39913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm LJ, Burdett TC, Gonzalez MA, Zuchner S, and Freeman MR, 2014. Rapid in vivo forward genetic approach for identifying axon death genes in Drosophila. Proc. Natl. Acad. Sci. U. S. A 111, 9965–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm LJ, Burdett TC, Seeds AM, Hampel S, Coutinho-Budd JC, Farley JE, Wong J, Karadeniz YB, Osterloh JM, Sheehan AE, and Freeman MR, 2017. Axon death pathways converge on Axundead to promote functional and structural axon disassembly. Neuron 95, 78–91 e75. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Ando Y, Kanetani T, Hoshi C, Nakai Y, Nainu F, Nagaosa K, Shiratsuchi A, and Nakanishi Y, 2017. Signaling pathway for phagocyte priming upon encounter with apoptotic cells. J. Biol. Chem 292, 8059–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada R, Nagaosa K, Kuraishi T, Nakayama H, Yamamoto N, Nakagawa Y, Dohmae N, Shiratsuchi A, and Nakanishi Y, 2012. Apoptosis-dependent externalization and involvement in apoptotic cell clearance of DmCaBP1, an endoplasmic reticulum protein of Drosophila. J. Biol. Chem 287, 3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, Macdonald JM, Ziegenfuss JS, Milde S, Hou YJ, Nathan C, Ding A, Brown RH Jr., Conforti L, Coleman M, Tessier-Lavigne M, Zuchner S & Freeman MR, 2012. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidassi H, Tacnet-Delorme P, Garlatti V, Darnault C, Ghebrehiwet B, Gaboriaud C, Arlaud GJ, and Frachet P, 2008. C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol 180, 2329–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulusma CC, Folmer DE, Ho-Mok KS, de Waart DR, Hilarius PM, Verhoeven AJ, and Oude Elferink RP, 2008. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 47, 268–278. [DOI] [PubMed] [Google Scholar]
- Poe AR, Tang L, Wang B, Li Y, Sapar ML, and Han C, 2017. Dendritic space-filling requires a neuronal type-specific extracellular permissive signal in Drosophila. Proc. Natl. Acad. Sci. U. S. A 114, E8062–E8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe AR, Wang B, Sapar ML, Ji H, Li K, Onabajo T, Fazliyeva R, Gibbs M, Qiu Y, Hu Y, and Han C, 2019. Robust CRISPR/Cas9-mediated tissue-specific mutagenesis reveals gene redundancy and perdurance in Drosophila. Genetics 211, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presumey J, Bialas AR, and Carroll MC, 2017. Complement system in neural synapse elimination in development and disease. Adv. Immunol 135, 53–79. [DOI] [PubMed] [Google Scholar]
- Rasmussen JP, Sack GS, Martin SM, and Sagasti A, 2015. Vertebrate epidermal cells are broad-specificity phagocytes that clear sensory axon debris. J. Neurosci 35, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran KS, 2010. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J. Exp. Med 207, 1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran KS, and Lorenz U, 2007. Engulfment of apoptotic cells: signals for a good meal. Nat. Rev. Immunol 7, 964–974. [DOI] [PubMed] [Google Scholar]
- Riccomagno MM, and Kolodkin AL, 2015. Sculpting neural circuits by axon and dendrite pruning. Annu. Rev. Cell Dev. Biol 31, 779–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, and Tanaka K, 2004. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 3418–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MW, and Stevens B, 2017. Microglia emerge as central players in brain disease. Nat. Med 23, 1018–1027. [DOI] [PubMed] [Google Scholar]
- Sapar ML, Ji H, Wang B, Poe AR, Dubey K, Ren X, Ni JQ, and Han C, 2018. Phosphatidylserine externalization results from and causes neurite degeneration in Drosophila. Cell Rep. 24, 2273–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Nakagawa T, Mao X, DiAntonio A, and Milbrandt J, 2016. NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. eLife 5, e19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BP, Baloh RH, and Milbrandt J, 2009. Transgenic mice expressing the Nmnat1 protein manifest robust delay in axonal degeneration in vivo. J. Neurosci 29, 6526–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B, 2012. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheib JL, Sullivan CS, and Carter BD, 2012. Jedi-1 and MEGF10 signal engulfment of apoptotic neurons through the tyrosine kinase Syk. J. Neurosci 32, 13022–13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, and Yaron A, 2015. Mechanisms of developmental neurite pruning. Cell. Mol. Life Sci 72, 101–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, and Nagata S, 2015. An Apoptotic ‘Eat Me’ Signal: Phosphatidylserine Exposure. Trends Cell. Biol 25, 639–650. [DOI] [PubMed] [Google Scholar]
- Segawa K, Suzuki J, and Nagata S, 2011. Constitutive exposure of phosphatidylserine on viable cells. Proc. Natl. Acad. Sci. U S A 108, 19246–19251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Schizophrenia Working Group Of The Psychiatric Genomics, C., Daly MJ, Carroll MC, Stevens B and Mccarroll SA, 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacham-Silverberg V, Sar Shalom H, Goldner R, Golan-Vaishenker Y, Gurwicz N, Gokhman I, and Yaron A, 2018. Phosphatidylserine is a marker for axonal debris engulfment but its exposure can be decoupled from degeneration. Cell. Death. Dis 9, 1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW and Barres BA, 2007. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, and Nagata S, 2013a. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Fujii T, Imao T, Ishihara K, Kuba H, and Nagata S, 2013b. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J. Biol. Chem 288, 13305–13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Umeda M, Sims PJ, and Nagata S, 2010. Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Fujimura-Kamada K, and Yamamoto T, 2011. Functions of phospholipid flippases. J. Biochem 149, 131–143. [DOI] [PubMed] [Google Scholar]
- Tao J, and Rolls MM, 2011. Dendrites have a rapid program of injury-induced degeneration that is molecularly distinct from developmental pruning. J. Neurosci 31, 5398–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir-Yilmaz OE, and Freeman MR, 2014. Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 28, 20- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L, Pereira E, Correia A, Santos MA, Amaral N, Martins T, Relvas JB, and Pereira PS, 2015. Drosophila PS2 and PS3 integrins play distinct roles in retinal photoreceptors-glia interactions. Glia 63, 1155–1165. [DOI] [PubMed] [Google Scholar]
- Thomas PK, 1964. Changes in the endoneurial sheaths of peripheral myelinated nerve fibres during Wallerian degeneration. J. Anat 98, 175–182. [PMC free article] [PubMed] [Google Scholar]
- Thomas PK, and Sheldon H, 1964. Tubular arrays derived from myelin breakdown during Wallerian degeneration of peripheral nerve. J. Cell Biol 22, 715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufail Y, Cook D, Fourgeaud L, Powers CJ, Merten K, Clark CL, Hoffman E, Ngo A, Sekiguchi KJ, O’Shea CC, Nimmerjahn A, 2017. Phosphatidylserine exposure controls viral innate immune responses by microglia. Neuron 93, 574–586 e578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung TT, Nagaosa K, Fujita Y, Kita A, Mori H, Okada R, Nonaka S, and Nakanishi Y, 2013. Phosphatidylserine recognition and induction of apoptotic cell clearance by Drosophila engulfment receptor Draper. J. Biochem 153, 483–491. [DOI] [PubMed] [Google Scholar]
- Turkiew E, Falconer D, Reed N, and Hoke A, 2017. Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. J. Peripher. Nerv. Syst 22, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki S, and Araki T, 2017. Specific phospholipid scramblases are involved in exposure of phosphatidylserine, an “eat-me” signal for phagocytes, on degenerating axons. Commun. Integr. Biol 10, e1296615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki S, Tokunaga S, Shibata M, and Araki T, 2017. GSK3B-mediated phosphorylation of MCL1 regulates axonal autophagy to promote Wallerian degeneration. J. Cell. Biol 216, 477–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller A, 1850. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Philosophical Transactions of the Royal Society of London 140, 423–429. [Google Scholar]
- Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, and He Z, 2005. A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol 170, 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Zhao D, Liu B, Shi Y, Chen B, Yang H, Guo P, Geng X, Shang Z, Peden E, Kage-Nakadai E, Mitani S and Xue D, 2010. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat. Cell Biol 12, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, and Luo L, 2003. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38, 871–885. [DOI] [PubMed] [Google Scholar]
- Wen Y, Parrish JZ, He R, Zhai RG, and Kim MD, 2011. Nmnat exerts neuroprotective effects in dendrites and axons. Mol. Cell Neurosci 48, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, and Truman JW, 2006. Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci 9, 1234–1236. [DOI] [PubMed] [Google Scholar]
- Williams DW, and Truman JW, 2005. Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development 132, 3631–3642. [DOI] [PubMed] [Google Scholar]
- Williams PA, Harder JM, Foxworth NE, Cochran KE, Philip VM, Porciatti V, Smithies O, and John SW, 2017. Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science 355, 756–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson AP, and Vale RD, 2018. Spatial control of Draper receptor signaling initiates apoptotic cell engulfment. J. Cell Biol 217, 3977–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Bellmunt E, Scheib JL, Venegas V, Burkert C, Reichardt LF, Zhou Z, Farinas I, and Carter BD, 2009. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat. Neurosci 12, 1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, and Auld VJ, 2011. Integrins are necessary for the development and maintenance of the glial layers in the Drosophila peripheral nerve. Development 138, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, and Collins CA, 2012. A conditioning lesion protects axons from degeneration via the Wallenda/DLK MAP kinase signaling cascade. J. Neurosci 32, 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Hao Y, Sun K, Li J, Li X, Mishra B, Soppina P, Wu C, Hume RI, and Collins CA, 2012. The Highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol 10, e1001440. 10.1371/journal.pbio.1001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, Diantonio A, and Collins CA, 2010. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J. Cell Biol 191, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, and Yu J, 2018. Modeling Parkinson’s disease in Drosophila: what have we learned for dominant traits? Front. Neurol 9, 228 10.3389/fneur.2018.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen YZ, Zhang Y, Wang X, Zhao X, Godfroy JI 3rd, Liang Q, Zhang M, Zhang T, Yuan Q, Ann Royal M, Driscoll M, Xia NS, Yin H and Xue D, 2015a. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat. Commun 6, 5717 10.1038/ncomms6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wu Z, Renier N, Simon DJ, Uryu K, Park DS, Greer PA, Tournier C, Davis RJ, and Tessier-Lavigne M, 2015b. Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell 160, 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin TC, Voorhees JR, Genova RM, Davis KC, Madison AM, Britt JK, Cintron-Perez CJ, McDaniel L, Harper MM, and Pieper AA, 2016. Acute axonal degeneration drives development of cognitive, motor, and visual deficits after blast-mediated traumatic brain injury in mice. eNeuro 3, ENEURO.0220–16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, and Schuldiner O, 2014. Axon and dendrite pruning in Drosophila. Curr. Opin. Neurobiol 27, 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Almeida S, and Gao FB, 2018. Insights into C9ORF72-Related ALS/FTD from Drosophila and iPSC Models. Trends Neurosci. 41, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zargarian S, Shlomovitz I, Erlich Z, Hourizadeh A, Ofir-Birin Y, Croker BA, Regev-Rudzki N, Edry-Botzer L, and Gerlic M, 2017. Phosphatidylserine externalization, “necroptotic bodies” release, and phagocytosis during necroptosis. PLoS Biol. 15, e2002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Cao Y, Hiesinger PR, Zhou Y, Mehta SQ, Schulze KL, Verstreken P, and Bellen HJ, 2006. Drosophila NMNAT maintains neural integrity independent of its NAD synthesis activity. PLoS Biol. 4, e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Libby RT, de Vries WN, Smith RS, Wright DL, Bronson RT, Seburn KL, and John SW, 2012. Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8, e1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Zhang L, Sasaki Y, Milbrandt J, and Gidday JM, 2013. Protection of mouse retinal ganglion cell axons and soma from glaucomatous and ischemic injury by cytoplasmic overexpression of Nmnat1. Invest. Ophthalmol. Vis. Sci 54, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenfuss JS, Biswas R, Avery MA, Hong K, Sheehan AE, Yeung YG, Stanley ER, and Freeman MR, 2008. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature 453, 935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenfuss JS, Doherty J, and Freeman MR, 2012. Distinct molecular pathways mediate glial activation and engulfment of axonal debris after axotomy. Nat. Neurosci 15, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziogas NK, and Koliatsos VE, 2018. Primary traumatic axonopathy in mice subjected to impact acceleration: a reappraisal of pathology and mechanisms with high-resolution anatomical methods. J. Neurosci 38, 4031–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]