Abstract
Positron emission tomography (PET) has been substantially used recently. To minimize the potential health risk caused by the tracer radiation inherent to PET scans, it is of great interest to synthesize the high-quality PET image from the low-dose one to reduce the radiation exposure. In this paper, we propose a 3D auto-context-based locality adaptive multi-modality generative adversarial networks model (LA-GANs) to synthesize the high-quality FDG PET image from the low-dose one with the accompanying MRI images that provide anatomical information. Our work has four contributions. First, different from the traditional methods that treat each image modality as an input channel and apply the same kernel to convolve the whole image, we argue that the contributions of different modalities could vary at different image locations, and therefore a unified kernel for a whole image is not optimal. To address this issue, we propose a locality adaptive strategy for multi-modality fusion. Second, we utilize 1×1×1 kernel to learn this locality adaptive fusion so that the number of additional parameters incurred by our method is kept minimum. Third, the proposed locality adaptive fusion mechanism is learned jointly with the PET image synthesis in a 3D conditional GANs model, which generates high-quality PET images by employing large-sized image patches and hierarchical features. Fourth, we apply the auto-context strategy to our scheme and propose an auto-context LA-GANs model to further refine the quality of synthesized images. Experimental results show that our method outperforms the traditional multi-modality fusion methods used in deep networks, as well as the state-of-the-art PET estimation approaches.
Keywords: Image synthesis, Positron emission topography (PET), Generative adversarial networks (GANs), locality adaptive fusion, multi-modality
I. Introduction
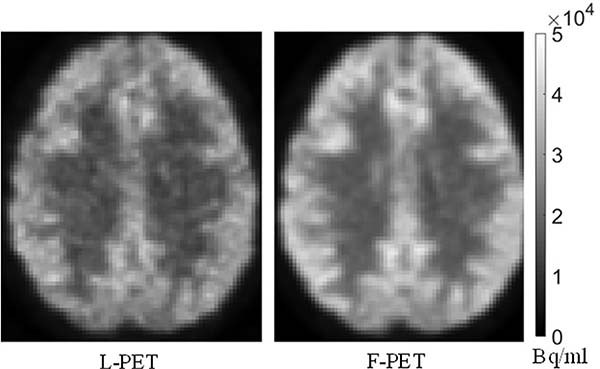
AS a nuclear imaging technology, positron emission tomography (PET) enables the visualization of metabolic processes of human body, and has been increasingly used in clinics for disease diagnosis and intervention [1]. By detecting pairs of gamma rays emitted indirectly from the radioactive tracer injected into the human body, the PET system usually uses manufacturer-provided software to do the triangulation of the source of the emissions, thus reconstructing 3D PET images of the tracer concentrations within the human body. Usually, a full-dose tracer is required to obtain PET images of diagnostic quality. However, the radioactive exposure inevitably raises concerns of potential health hazards. The risks are accumulated for patients who experience multiple PET scans as part of their treatments. To tackle the radiation problem, some researchers have tried to reduce the tracer dose (e.g., using half of the full dose) during the PET scans [2]. Since the PET imaging is a quantum accumulation process, lowering the tracer dose inevitably involves unnecessary noises and artifacts, thus degrading the PET image quality to a certain extent. This work targets on the [18F]FDG PET images. As shown in Fig. 1, the quality of the low-dose [18F]FDG PET (L-PET) image is obviously worse than that of the full-dose [18F]FDG PET image (F-PET), involving more noise and less functional details. This kind of L-PET images may not satisfy the diagnostic requirements. Therefore, it is of great interest to synthesize the high-quality F-PET image from the low-dose one to reduce the radiation exposure while maintaining the image quality.
Fig. 1.
Comparison between the low-dose PET (L-PET) image and the corresponding full-dose PET (F-PET).
Modern PET scans are usually accompanied with other modalities, such as computed tomography (CT) and magnetic resonance image (MRI). By combining functional and morphological information, PET/CT and PET/MRI systems could increase diagnostic accuracy in a variety of malignancies [3–5]. Previous research also indicates the benefit brought by multi-modality data for medical image quality enhancement [6, 7]. In this paper, to incorporate anatomical information into PET synthesis, we propose to synthesize the high-quality F-PET image from both the L-PET image and the accompanying MRI including T1-weighted MRI (T1-MRI) and diffusion tensor image (DTI).
There have been several works for F-PET image synthesis. Most of them, however, are based on voxel-wise estimation methods, e.g., random forest based regression method [8], mapping based sparse representation method [9], semi-supervised tripled dictionary learning method [10], and multi-level canonical correlation analysis framework [11]. Although these methods present good performance for PET image quality enhancement at low dose, there are two major disadvantages that limit the potential clinical usability. The first one is that they are all based on small patches, and the final estimation of each voxel is determined by averaging the overlapping patches. This strategy inevitably results in over-smoothed images that lack the texture of a typical F-PET image, thus limiting the quantification of small structures in the synthesized images. Another disadvantage is that these voxel-wise based estimation methods usually need to solve plenty of optimization problems online, and thus are quite time-consuming when tested on new subjects. That is, the estimation procedure is quite burdensome.
In recent years, deep learning has shown an explosive popularity in computer vision and medical imaging fields [12–16]. In the particular case of image synthesis, Dong et al. [17] proposed a convolutional neural networks (CNNs) method for image super-resolution. Using similar deep architecture, Li et al. [18] estimated the missing PET image from the MRI for the same subject. By integrating multiple CNN modules following the auto-context strategy, Xiang et al. [19] proposed an auto-context CNN method for F-PET image estimation. However, CNNs using small patches tend to ignore the neighborhood information in the synthesized image [20]. To preserve structural information for the entire image, Long et al. [21] first proposed a fully-convolutional-networks (FCNs) architecture for semantic segmentation; note that FCNs have been widely utilized for image segmentation and synthesis [22–24]. Particularly, Ronneberger et al. [25] proposed a special FCNs architecture, namely U-net, for neuronal structures segmentation. Using the idea of skip connection, the U-net has been successfully applied to many tasks [26, 27].
More recently, as one of the advancement of deep learning techniques, generative adversarial networks (GANs) have been extensively applied to many unsupervised and semi-supervised learning tasks [28] as well as image synthesis tasks [29]. GANs consider a two-player min-max adversarial game between two agents, i.e., a generator network and a discriminator network. The goal of the discriminator is to tell the real inputs from the generated ones by the generator, while the generator is optimized to synthesize samples that are not distinguishable by the discriminator. Subsequently, some variations of traditional GANs have been developed [30–33]. Particularly, the conditional GANs facilitate the training of a deep model to generate images conditioned on particular auxiliary information. Wang et al. [34] proposed to synthesize the F-PET image from the low-dose PET using a conditional GANs model. To address the problem of multi-modality inputs, Bi et al. [35] proposed a multi-channel GANs method to synthesize PET images from CT images and their corresponding label images. In their method, they treated each image modality as an input channel and simply applied the same kernel to convolve the whole image. However, the contributions of different modalities could vary at different image locations, and therefore a unified kernel for a whole image is not appropriate. On the other hand, Nie et al. [36] proposed a fully convolutional networks model for multi-modality infant brain segmentation. Different from the multi-channel method which stacks multi-modality data from the low-level feature maps, they proposed to train one network for each modality and then fuse their high-level information using a fusion layer. However, they did not consider the various contributions of different image locations as well. In addition, training one network for each modality would significantly increase the number of training parameters, making it quite challenging when the training sample size is relatively small, which is quite common in medical image analysis field.
In this paper, inspired by the appealing success of GANs and also motivated to tackle the limitation of the current multi-channel deep architectures for modality fusion, we propose an auto-context-based “locality adaptive” multi-modality GANs (LA-GANs) model to synthesize the F-PET image from both the L-PET and the accompanying multimodal MRI images including T1-MRI and DTI. Note that the common DTI measures include fractional anisotropy (FA), mean diffusivity (MD), radial diffusivity (RD), etc. Here, we compute FA and MD images from diffusion images for F-PET image synthesis. The contributions of our method are as follows. 1) We propose a new mechanism to fuse multi-modality information in deep neural networks. The weight of each imaging modality varies with image locations for better serving the synthesis of F-PET. 2) Using multi-modality (especially making it locality-adaptive) may induce many additional parameters to learn. We therefore propose to utilize 1×1×1 kernel to learn such locality-adaptive fusion mechanism to minimize the increase of the number of parameters. Doing so also naturally leads to a fused image that acts as a pseudo input for the subsequent learning stages. 3) We develop a 3D conditional GANs model for PET image synthesis, and jointly learn the proposed locality-adaptive fusion with the synthesis process in an end-to-end training manner. Our 3D GANs model generates high-quality PET images by employing large-sized image patches and hierarchical features. 4) Auto-context model can effectively leverage the context information which plays a vital role in interpreting image content. Therefore, we apply the auto-context strategy on our scheme and propose an auto-context LA-GANs model to further refine the quality of synthesized images. Compared with the traditional multi-modality fusion methods used in deep networks [19, 34], our method can achieve better performance while incurring less number of additional parameters.
II. Methodology
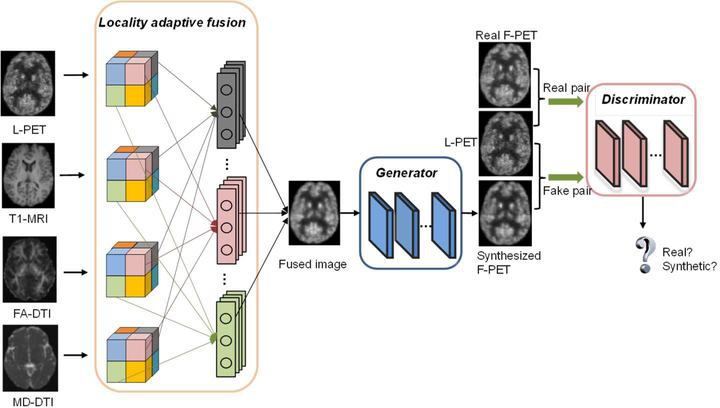
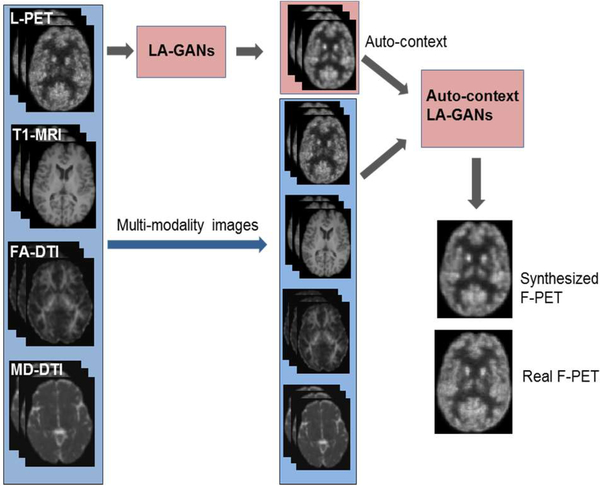
Our proposed LA-GANs model is illustrated in Fig. 2, which consists of three parts: 1) the locality-adaptive fusion network, 2) the generator network, and 3) the discriminator network. Concretely, the locality-adaptive fusion network takes an L-PET, a T1-MRI, an FA-DTI and an MD-DTI images as inputs, and generates a fused image by learning different convolutional kernels at different image locations. After that, the generator network is trained to produce a synthesized F-PET from the fused image, while the discriminator network subsequently takes a pair of images as input, i.e., the L-PET and the real or synthetic F-PET, aiming to distinguish between the real and synthetic pairs. If the discriminator can easily distinguish between them, which means the synthesized PET image has not well resembled the real one, and also that the fusion network and the generator network should be further improved to produce more realistic synthesis. Otherwise, the discriminator should be enhanced instead. Therefore, the three networks are trained simultaneously with discriminator trying to correctly distinguish between the real and synthetic data, while the fusion and generator networks trying to produce realistic images that can fool the discriminator. Please note that, we use 3D operations for all the networks to better model the 3D spatial information and thus could alleviate the discontinuity problem across slices of 2D networks. The details of the architecture as well as the objective function are described in the following sections.
Fig. 2.
Overview of our proposed pipeline for full-dose PET synthesis from low-dose counterpart and the accompanying multimodal MR images.
A. Architecture
Locality-adaptive fusion network
This is a module for multi-modality information fusion. As mentioned before, in most multi-channel based networks, image convolution is performed in a global manner, i.e., for each modality the same filter is applied to all image locations for generating the feature maps that will be combined in higher layers. This could not effectively handle the location-varying contributions from different imaging modalities. To tackle this problem, locality-adaptive convolution should be enforced. However, if the locality-adaptive convolution is simply conducted in the multi-channel framework, many additional parameters will have to be learned due to inclusion of new imaging modalities. This is not favorable for medical applications where the number of the training samples is often limited. Therefore, we propose to add a module that produces a fused image from multi-modality images and use the fused image as the pseudo input to the generator network. In this way, the increase of the number of modalities will not cause any increase on the number of parameters in the generator. Moreover, we propose to utilize 1×1×1 kernel for locality-adaptive convolution to minimize the number of necessary parameters to learn in this fusion module. The fusion network will be jointly learned with the generator and the discriminator to ensure that they can effectively negotiate with each other to achieve the best possible performance on image synthesis. Specifically, the entire L-PET and multimodal MR images are partitioned, respectively, into N non-overlapping small regions, i.e., , , and (i = 1, …, N) as indicated by the regions in different colors in Fig. 2. Then, the regions at the same location (indicated by the same color) from the four modalities, i.e., , , and , are convolved, respectively, using four different 1×1×1 filters with parameters , , and . For instance, in the fusion block in Fig. 2, the four gray filters are respectively operated on the four gray regions of the L-PET, T1-MRI, FA-DTI and MD-DTI images to generate their corresponding combined region. Formally, the combined region is obtained as follows:
| (1) |
In this way, we will learn N groups of different convolution kernels for N local regions. The outputs of the fusion are further assembled to form an entire fused image as the input of the following generator network.
Generator network
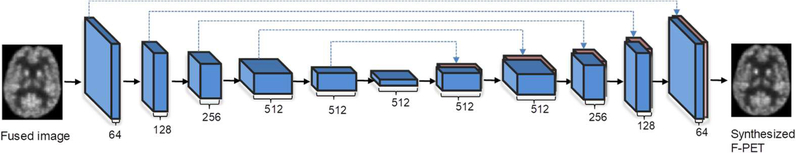
In our generator network, we adopt both the convolutional layers and de-convolutional layers to ensure the same size of the input and output images. Since the low-dose and full-dose PET images belong to the same modality, there are lots of low-level information shared between them. As such, we follow the U-net and add skip connections between the convolutional and de-convolutional layers, thus combining hierarchical features for better synthesis [25]. Also, the skip connection strategy mitigates the vanishing gradient problem, allowing the network architecture to be possibly much deeper. The advantage of using skip connections over traditional CNN has been well demonstrated in the U-net related literature [36]. Fig. 3 illustrates the architecture of our 3D U-net-like generator network, including a contracting encoder part to analyze the input fused image and an expansive decoder part to generate an output of synthetic F-PET image. Since the pooling layers could reduce the spatial resolution of feature maps, we do not employ any pooling layers in our generator architecture.
Fig. 3.
Architecture of the U-net-like generator network. Blue boxes represent feature maps while the brown boxes represent the copied feature maps. The number of feature maps is denoted under each feature map. The black solid arrows denote convolutional operations while the blue dotted arrows mean the copy and concatenate operations.
We follow the basic network architecture in [37] to build layers with multiple Convolution-BatchNormalizaion-Leaky Relu components. Specifically, the entire network constitutes 12 convolutional layers. In the encoder part which includes the first 6 convolutional layers, we use 4×4×4 filters and a stride of 2 for convolution, and 0.2 negative slope for the leaky ReLu. The number of feature maps increases from 64 in the 1st layer to 512 in the 6th layer. The number of feature maps in each convolutional layer is denoted in Fig. 3 (see the number under each blue block). In addition, since we apply zero padding with 1×1×1 kernel, the output of each convolutional layer of the encoder part halves the size of the feature maps. In the decoder part, we perform up-sampling with a factor of 2. Using the skip connections, the feature maps from the encoder part are copied and concatenated with the feature maps of the decoder part, as indicated by the dotted arrows in Fig. 3. The batch normalization is also introduced in each convolutional layer to ease the training of deep neural networks. Finally, the output of the generator network is considered as the synthetic F-PET image.
Discriminator network
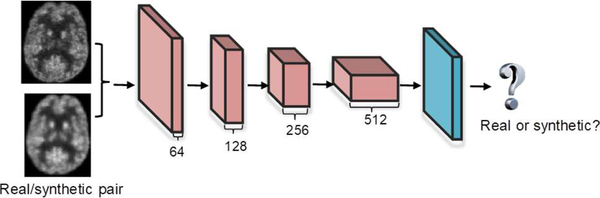
The same Convolutional Batch Normalization Leaky Relu blocks are used in our discriminator network. As shown in Fig. 4, the discriminator network is a typical CNN architecture consisting of 4 convolutional layers, and each of them uses 4×4×4 filters with a stride of 2, similar to the encoder structure of the generator. The first convolution layer produces 64 feature maps, and this number is doubled at each of the following convolutional layers. On top of the convolutional layers, a fully connected layer is further applied and followed by a sigmoid activation to determine whether the input is the real pair or the synthetic one.
Fig. 4.
Architecture of the discriminator network. The number of feature maps is denoted under each feature map. The green block means sigmoid activation.
B. Objective functions
Let us denote xL an L-PET image, xT1, xFA and xMD the accompanying multimodal MR images, and yF the corresponding real F-PET image (i.e., the ground-truth annotation). In this study, we learn three function mappings. The first mapping is for the locality-adaptive fusion network, which produces a fused image from xL, xT1, xFA and xMD. The second mapping is for the generator network, which maps the fused image to a synthetic F-PET image . The third mapping corresponds to the discriminator network function , whose task is to distinguish the synthetic pair (ideally d → 0) from the real pair YF := (xL, yF) (ideally d → 1). The symbols α, β and γ denote the parameter sets of the three networks, respectively, and are automatically learned from a training set . Formally, we solve the following optimization problem:
| (2) |
where λ > 0 is a trade-off parameter. The last term is an L1 loss, used to ensure that the synthetic F-PET image stays close to its real counterpart. The L1 loss is defined as:
| (3) |
Please note that, the fusion network F and the generator network G, in a sense, can be regarded as a whole network whose goal is to synthesize realistic-looking F-PET images that can fool the discriminator network D. Following the approximation scheme in [38], the minimization of term can be replaced by minimizing a simpler form . Therefore, training the fusion network F and the generator network G equals to minimizing the following problem:
| (4) |
On the other hand, the discriminator network D tries to tell the real pair (xL, yF) from the synthetic pair by maximizing Equation (2). Therefore, training the discriminator network corresponds to maximizing:
| (5) |
C. Training the LA-GANs
The fusion network F together with the generator network G and the discriminator network D are trained in an alternating manner, which is similar to the standard approach of [38]. Specifically, we first fix F and G to train D for one step using the gradients computed from the loss function, and then fix D to train F and G. As shown in Equation (2), the training of F, G and D is just like playing a min-max game: F and G try to minimize the loss function while D tries to maximize it. With continuation of the training, the three networks become more and more powerful. Finally, the generator will be able to generate the synthetic F-PET image that is extremely close to the real one. In the testing stage, only the fusion and generator networks are needed for synthesis. The only difference from the usual protocol is that we apply batch normalization using the statistics of the testing batch, instead of aggregated statistics of the training batch. To balance the contribution of each modality to train our LA-GANs model, we initialize the learning parameters of 0.25 for all patches in L-PET and MR images. All networks are trained by Adam solver with mini-batch stochastic gradient descent (SGD), and the mini-batch size is set to 128. The training process runs for 200 epochs, and the learning rate is set to 0.0002 for the first 100 epochs and then linearly decays to 0 in the second 100 epochs. The weight of the estimation error term λ is empirically set as 200 in all our experiments.
D. Auto-context LA-GANs
It has been known that the context information plays a crucial role in interpreting image content and the auto-context model can effectively leverage the context information [20]. To harness the integration of the high-level auto-context information and the low-level image appearance, we further propose an auto-context LA-GANs model to improve the quality of the synthesized F-PET image generated by the LA-GANs model, as illustrated in Fig. 5. Specifically, given the multi-modality training images, we first train a LA-GANs
Algorithm 1.
| 1: Input: A set of training low-dose PET images , a set of training multimodal MRI images including , , , and a set of training full-dose PET images . N is the total number of training samples. |
| 2. Perform the 3D LA-GANs between IL, IT1, IFA, IMD and IS to obtain the fusion network F, the generator network G, and the discriminator network D. |
| 3. For each training sample i (i=1,2,...,N), use the above trained fusion network F and generator network G to generate the synthetic full-dose PET image . Finally, get the estimations for all training subjects . |
| 4. The synthesis of the training subjects , along with the original low-dose PET IL and multimodal MRI images IT1, IFA and IMD, are all input to the subsequent LA-GANs network, namely, auto-context LA-GANs, to obtain the updated fusion network F′, the generator network G′ and the discriminator network D′. |
| 5: Output: The trained fusion networks for LA-GANs and auto-context LA-GANs F and F’, as well as the trained generator networks G and G’. |
model using the original training modalities including L-PET, T1-MRI, FA-DTI and MD-DTI. Then, for each training subject, we generate a corresponding synthetic F-PET image by using the trained model. After that, the synthetic F-PET images for all training samples generated from the LA-GANs are used as the context information, together with the original modalities (i.e., appearance information), to train a new auto-context LA-GANs model, which further refines the synthesized F-PET image. The detailed procedure is summarized in Algorithm 1. In the testing stage, given a new L-PET image , together with its corresponding multimodal MRI images , , , we can use the trained fusion networks F and F’ as well as the trained generator networks G and G’ to obtain the final results.
Fig. 5.
Illustration of the auto-context LA-GANs architecture.
III. Experiments and results
A. Simulated data
We first test our method on the simulated phantom brain dataset to evaluate the effectiveness of our proposed method. For data acquisition, twenty simulated subjects were generated from the BrainWeb database of twenty normal brains [39, 40]. Specifically, anatomical distribution functions were downloaded from BrainWeb to simulate T1-MRI of each subject and also used as input for generating PET data. The original brain maps were defined at 1 mm voxel size with fuzzy segmentation into 11 tissue classes. PET uptake maps were created for each of the fuzzy segmentations by assigning typical uptake ratios to each tissue class. Models were made for 18F-FDG distributions. PET attenuation maps were also determined from typical values of tissue linear attenuation coefficients at 511 keV.
PET acquisition data were simulated to model the acquisition geometry of the Siemens Biograph mMR system. The PET uptake maps were blurred with a 4mm FWHM Gaussian kernel to model positron range and limited spatial resolution. Line-of-response (LOR) data were created by ray-tracing through the blurred PET uptake map with the native geometry (3.5×108 LORs) of the scanner system. Similarly, attenuation effects for each LOR were applied by ray-tracing through the attenuation maps. The attenuated LORs were collapsed into the smaller set typically used by the scanner manufacturer prior to reconstruction (7.3×107 LORs). Nonuniformity effects were applied to the collapsed projection data using a uniformity correction obtained from scanner data. After all effects were applied, Poisson noise was simulated to achieve count levels associated with full dose, based on measurement from patient image raw data, and low dose, by simulating at 25% of the normal count level. PET reconstructions were created using the Ordered Subsets-Expectation Maximization algorithm (OSEM) with 21 subsets and 3 iterations followed by a 3D Gaussian 4mm FWHM post-reconstruction filter. The PET reconstruction space was 344×344×127 with voxels of size 2.08×2.08×2.03 mm3. Attenuation correction was applied from a uniform attenuation map modeled on the original anatomy, emulating the MR-based map that would be obtained from a Dixon sequence. Uniformity correction was also applied in reconstruction. The resulting image sets included a T1 anatomical image, an F-PET reconstruction, and an L-PET reconstruction for each of the twenty simulated subjects.
Considering the small number of the training samples, we cropped and expanded the image to size of 128×128×128, and then extracted 125 large 3D image patches of size 64×64×64 from each image, rather than directly using the entire 3D image, to train the deep model. In this way, we can significantly increase the number of samples (i.e., from 20 to 2500 in total). In addition, to make full use of available samples, we followed the widely used “Leave-One-Subject-Out” strategy, i.e., we repeated the training and test for 20 times, and at each time reserved one subject in turn for test and trained our model on the patches from the rest subjects. To train the proposed locality-adaptive fusion network, we further partitioned each large image patch into non-overlapping 8×8×8 regions for fusion. In this manner, we can get 512 regions for each modality per large image patch. Since the phantom data just include T1-MRI and PET images (i.e., without DTI), we initialized the learning parameters of 0.5 for the fusion network in L-PET and T1-MR images. Our method was implemented by PyTorch, and all the experiments were carried out on an NVIDIA GeForce GTX 1080 Ti with 11GB memory.
To quantitatively characterize the synthesis accuracy, we use two popular metrics1: 1) peak signal-to-noise (PSNR) and 2) structural similarity index measurement (SSIM) [41]:
| (6) |
| (7) |
where in Equation (6), IF is the ground-truth F-PET image, the synthesized F-PET image, R the maximal intensity value of IF and , and U the number of voxels in each image; and in Equation (7), and represent the average of IF and , and the variance of IF and , and the covariance of IF and . The positive constants c2 and c2 are used to avoid a null denominator. SSIM is a ratio between 0 and 1 and it has no unit. Theoretically, synthesis result with higher PSNR and higher SSIM means better image quality.
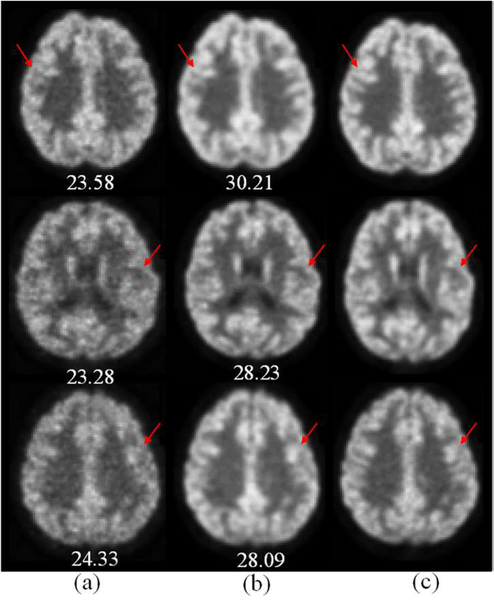
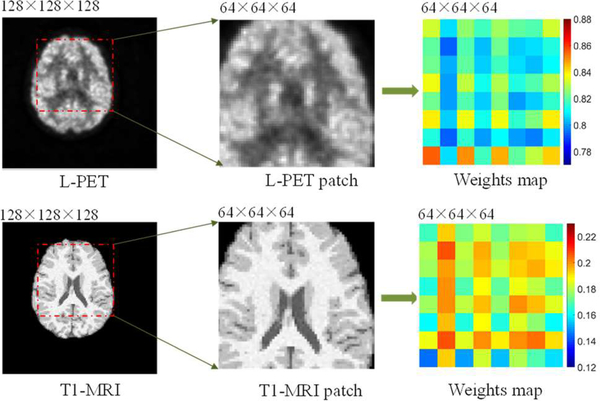
Fig. 6 shows three examples of the synthesized F-PET images by our method (the middle column). We also give the corresponding L-PET images (the left-most column) and the real F-PET images (the right-most column) for comparison. As observed, the image quality of the L-PET is obviously worse than that of the F-PET image. The synthetic results by our method significantly improve the details over the L-PET image (as indicated by the red arrows), and are quite similar to the real F-PET images. The PSNR for each image is also given under the corresponding image in Fig. 6, where we can see a significant improvement over the L-PET image. To show how the contributions of different input modalities vary at different regions, we visualize the weights with color coding for different regions in different modalities, with an example given in Fig. 7. As can be clearly seen, the contributions of different modalities vary at different image locations.
Fig. 6.
Examples of the synthetic F-PET image by our auto-context LA-GANs method from three subjects. (a) L-PET (b) Synthesized F-PET (c) real F-PET. The values under the images denote the PSNR of the corresponding image.
Fig. 7.
Locality adaptive weights of different regions in different modalities.
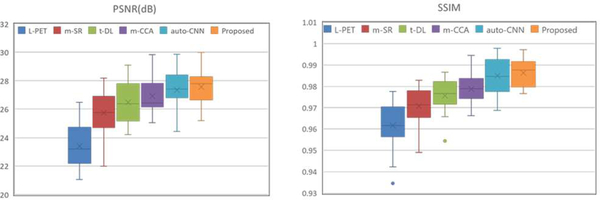
For quantitative evaluation, we further compare our method with four state-of-the-art methods: (1) mapping based sparse representation method (m-SR) [9], (2) tripled dictionary learning method (t-DL) [10], (3) multi-level CCA method (m-CCA) [11], and (4) auto-context CNN method (auto-CNN) [19]. The PSNR and SSIM over the whole brain image are showed in Fig. 8. The results show that the synthesis by our auto-context LA-GANs model is more accurate than the existing F-PET estimation methods, with the highest PSNR and SSIM. To study if our improvement is statistically significant, we perform paired t-tests to compare the existing methods against ours. Throughout the t-tests, the p-values of all the competing methods are consistently less than 0.05, indicating that the improvement by our method is statistically significant.
Fig. 8.
Quantitative comparison with the state-of-the-art PET estimation methods on the phantom dataset.
Both qualitative and quantitative experimental results conducted on the simulated phantom data demonstrate that our proposed method can achieve high quality PET synthesis at low dose. In the next section, the proposed method will be evaluated on real human brain dataset.
B. Clinical data
We further evaluate our method on the clinical real human brain dataset. The real human brain dataset consists of 8 normal control (NC) subjects and 8 mild cognitive impairment (MCI) subjects, each with an L-PET image, an F-PET image, a T1-MRI image, an FA-DTI image and an MD-DTI image. Specifically, the PET scans were acquired by a Siemens Biograph mMR PET-MR scanner, accompanying with the T1-MRI sequences and the DTI images. For each subject, the PET images and the DTI image were respectively aligned to their T1-MRI to build the voxel-level correspondence via affine transformation [42]. Finally, the FA-DTI and MD-DTI images can be computed from the resulting registered DTI image. The reconstruction was performed iteratively with the OSEM algorithm (3 iterations, 21 subsets, and post-reconstruction filtered with a 3D Gaussian with FWHM of 2 mm). Each aligned image has the resolution of 2.09×2.09×2.03 mm3 and the image size of 128×128×128. The clinical data used in this work is the same as our previous work in [34]. To train the deep network, we used the similar strategies as for the phantom data: 1) extracted 125 large 3D image patches of size 64×64×64 from each image, rather than directly using the entire 3D image, 2) used “Leave-One-Subject-Out” strategy to make full use of available samples, and 3) used the NC and MCI data together in training to maximally utilize the available samples.
Compared with the simulated phantom data, the clinical data could more realistically reflect clinical imaging applications. Therefore, we perform abundant experiments to investigate the contributions of our proposed method, including 1) the locality-adaptive fusion network, 2) the multimodal MRI images, 3) the adversarial learning, and 4) the auto-context model. We also compare our method with the state-of-the-art multi-modality PET synthesis approaches.
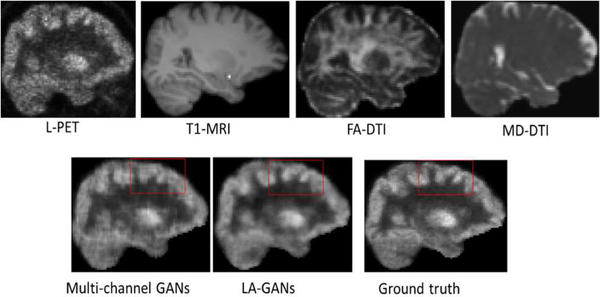
1). Comparison between the multi-channel based method and our proposed LA-GANs method
To study the contribution of the locality-adaptive fusion network of our proposed model, we conduct comparison experiments between the traditional multi-channel GANs model and our proposed LA-GANs model that does not employ the auto-context (AC) strategy (denoted as LA-GANs w/o AC). We visualize the comparison results in Fig. 9, where the first row images are the input L-PET, T1-MRI, FA-DTI and MD-DTI images, and the last image in the second row is the ground-truth F-PET. Note that the PET images displayed below share the same grayscale coding as in Fig. 1. We can clearly see that the synthesized F-PET image of our proposed model has less artifacts than that of the multi-channel model, as indicated by the red rectangles. The averaged quantitative comparison in terms of PSNR and SSIM are also provided in Table 1 and Table 2, with Table 1 showing the performances on the NC subjects and Table 2 showing the performances on the MCI subjects, respectively. We can see that, compared with multi-channel GANs method, the averaged PSNR of our method increases approximately 0.25 and 0.2 for NC and MCI groups, respectively. The standard deviation of our method is also smaller than that of the multi-channel GANs, while the median is higher. Also, the paired t-test shows that our improvement against the multi-channel one is statistically significant with p<0.05 (p = 0.0482 for NC subjects and p = 0.0161 for MCI subjects). The SSIM values in the two tables indicate the same conclusion. Moreover, the number of additional learning parameters incurred by adding multimodal MRI is 2048 for our method and 16384 for the multi-channel GANs, suggesting that our model produces better performance with less increase on the number of parameters.
Fig. 9.
Visual comparison with multi-channel GANs method.
Table 1.
Quantitative comparison with the multi-channel GANs method on NC subjects. Med. means median.
| Method | PSNR(dB) | SSIM | ||
|---|---|---|---|---|
| Mean (std.) | Med. | Mean (std.) | Med. | |
| L-PET | 19.88(2.34) | 20.68 | 0.979(0.0076) | 0.980 |
| Multi-channel | 24.36(1.93) | 24.78 | 0.981(0.0065) | 0.983 |
| LA-GANs (w/o AC) | 24.61(1.79) | 25.32 | 0.986(0.0053) | 0.987 |
Table 2.
Quantitative comparison with the multi-channel GANs method on MCI subjects. Med. means median.
| Method | PSNR(dB) | SSIM | ||
|---|---|---|---|---|
| Mean (std.) | Med. | Mean (std.) | Med. | |
| L-PET | 21.33(2.53) | 21.62 | 0.976(0.0102) | 0.979 |
| Multi-channel | 24.99(2.03) | 25.36 | 0.9795(0.0098) | 0.982 |
| LA-GANs (w/o AC) | 25.19(1.98) | 25.54 | 0.9843(0.0097) | 0.988 |
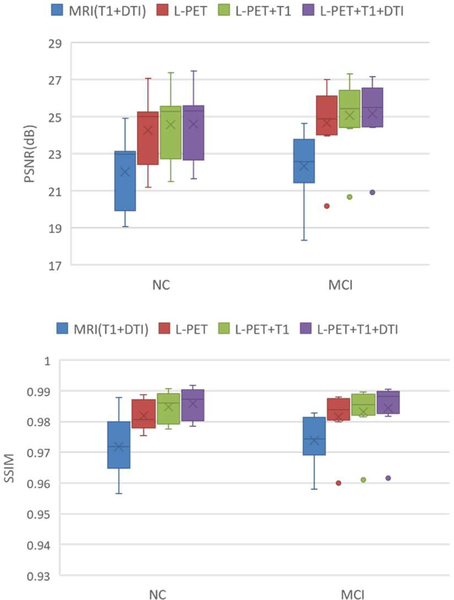
2). Contribution of the multimodal MRI images
To study the contributions of the multimodal MRI images for F-PET synthesis, we respectively use 1) MRI images (T1+DTI), 2) L-PET image, 3) L-PET+T1 images, and 4) L-PET+T1+DTI images, for synthesis. Note that, the model just using L-PET for F-PET synthesis exactly follows the single modality GANs model used in [34]. The detailed quantitative comparison in terms of PSNR and SSIM is given in Fig. 10.
Fig.10.
Comparison results of our LA-GANs model using different modalities in terms of PSNR and SSIM.
From Fig. 10, we can see that, just using the MRI images for PET synthesis obtains the lowest PSNR and lowest SSIM. The main reason is that the imaging mechanisms between PET and MRI are different. Therefore, just using MRI to estimate F-PET images leads to unsatisfactory results. Compared with using single modality of L-PET, employing two modalities (L-PET and T1) achieves a better result, with the PSNR improved from 24.29 to 24.58 and the SSIM increased from 0.982 to 0.985, respectively, for NC dataset. This is because the T1-MRI contains abundant anatomical structural information which can help synthesize the F-PET image from the L-PET image. When we try to also incorporate the DTI information (FA-DTI and MD-DTI) for synthesis, the model further produces an improved estimation, though not as significantly as it does by adding T1-MRI. This indicates that the fiber path reflected in DTI may probably be less helpful for PET image synthesis, compared with the anatomical information from T1-MRI. Moreover, as demonstrated by paired t-test, our locality adaptive multi-modality GANs statistically significantly outperforms the single modality GANs in [34], with p-value 0.0089 for NC subjects and 0.0035 for MCI subjects, respectively. In addition, by analyzing the relevance between the distribution of weights and the underlying anatomy of different modalities, we found that, compared with the DTI image, the T1-weighted image contributes more in both grey matter and white matter for PET synthesis, i.e., with larger weights. This is mainly because the T1-weighted image can show both white matter and grey matter reasonably well. For the DTI image, the weights in the white matter regions are larger than those in the grey matter regions, suggesting that the DTI image contributes more in white matter than in grey matter for PET synthesis.
3). Contribution of the adversarial network
To investigate the contribution of the adversarial network in our proposed model, we conduct comparison experiments between the proposed LA-GANs model and the model that removes the discriminator network (i.e., just the locality fusion network and the generator network shown in Fig. 2). The PSNR values are 24.35(1.84) / 24.61(1.79) for NC subjects, and 24.76(2.12) / 25.19(1.98) for MCI subjects, by the models without / with adversarial network, respectively. Note that these results provided here do not include the auto-context strategy. From these quantitative results, we can clearly see that the generated images using the adversarial training approach have better synthesis quality, indicating the essentials of the adversarial training in our 3D LA-GANs model.
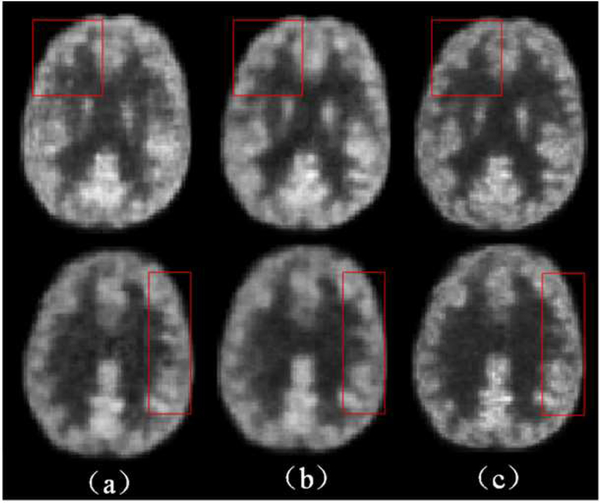
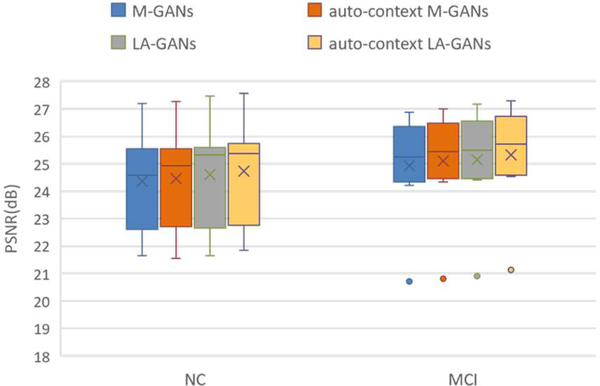
4). Contribution of the Auto-context model
We now show the contribution of the auto-context model. As observed in Fig. 11, the estimation quality of the auto-context LA-GANs model improves notably compared with the original LA-GANs model, as indicated by the red rectangles. This can also be seen from the quantitative comparison results given in Fig. 12, where the values of PSNR are improved with the use of context information for both multi-channel GANs (M-GANs) model and our LA-GANs model, and the best performance is achieved by our auto-context LA-GANs. Actually, we can further iteratively refine the generated results as the original auto-context algorithm, however, we found that the following iterative refinements give marginal improvement (PSNR<0.08), but bring much computational cost. Therefore, to balance the computational time and the synthesis performance, we choose the output of auto-context LA-GANs as the final result.
Fig. 11.
Visual comparison between the LA-GANs model and auto-context LA-GANs model. (a) results produced by the proposed LA-GANs, (b) results produced by the proposed auto-context LA-GANs model, (c) real full-dose PET images.
Fig. 12.
Qualitative comparison between the LA-GANs model and auto-context LA-GANs model.
5). Comparison with the state-of-the-art methods
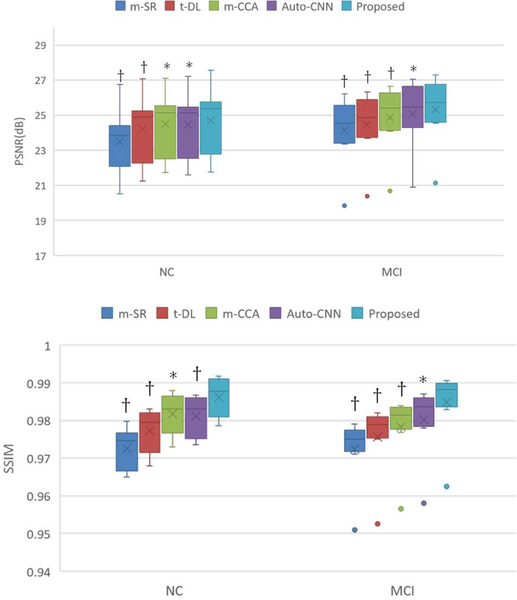
Similar to the experiments on the phantom data, we also compare our method with the state-of-the-art multi-modality based PET estimation methods, including (1) m-SR[9], (2) t-DL[10], (3) m-CCA[11], and (4) auto-CNN[19]. The PSNR and SSIM results are given in Fig. 13, from which we can see that our proposed method outperforms all the other competing methods for both image quality and structural information preservation, demonstrating its effectiveness and advantage. In addition, the small p-values from the t-test further demonstrate the statistical significance of the achieved improvement.
Fig. 13.
Qualitative comparison with the state-of-the-art PET estimation methods in terms of PSNR and SSIM. † indicates p<0.01 in the t-test while * means p<0.05.
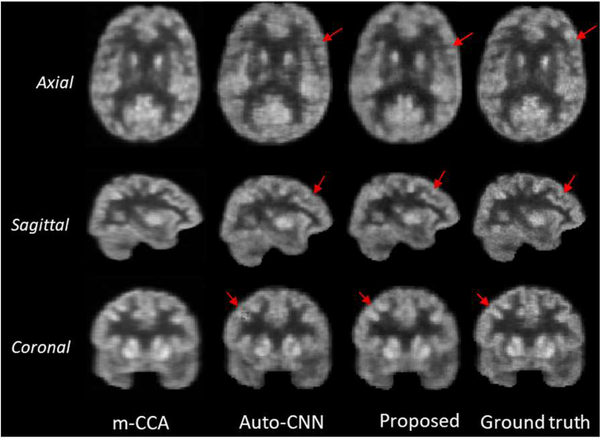
In Fig. 14, we visualize an example result of our method and compared it with those from the two methods (m-CCA and auto-CNN) which produce the top two results in the literature. As observed, the estimated images by the m-CCA method are over-smoothed compared with the real F-PET images due to the averaging of the overlapping patches to construct the final output images. Compared with the auto-CNN network, our model tends to better preserve the detailed information in the estimated F-PET images, as indicated by the red arrows. We argue that this is because the auto-context CNN method does not consider the varying contributions across image locations. Also, the adversarial training network used in our model constrains the synthesized images to be similar to the real ones.
Fig. 14.
Qualitative comparison with the state-of-the-art PET estimation methods.
6). Clinical evaluation on lesions of MCI subjects
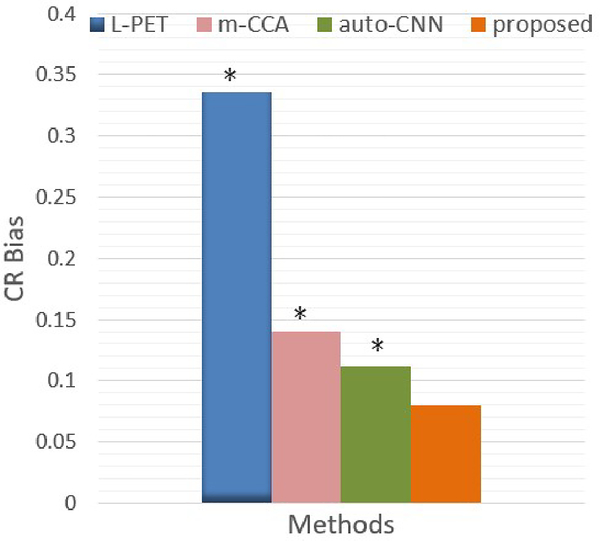
In addition to the image quality metrics, it is also important that the lesions in the synthetic F-PET image could be well preserved in terms of clinical quantification, as compared with the real F-PET image, since lesions could suffer reduced contrast. Clinically, the hippocampus is among the first brain structures to be affected by MCI pathology. To investigate this aspect, we evaluate the contrast recovery (CR) [43] in the hippocampal regions of MCI subjects. Specifically, the hippocampi are selected as the ROIs and the cerebellum is chosen as the background. The averaged CR bias for MCI subjects are shown in Fig. 15. For comparison, we also provide the CR results of both m-CCA and auto-context CNN methods that produce the top two performances in the literature.
Fig. 15.
CR bias for MCI subjects. * indicates p<0.05 in the t-test.
As can be seen, the CR bias in the original L-PET images is significant. This value is reduced in the synthetic F-PET image from m-CCA and auto-context CNN methods. Compared to these two state-of-the-art methods, the proposed LA-GANs model further mitigates this bias. Through paired t-tests, the small p-values demonstrate the statistically significant improvement achieved by our method.
IV. Discussion
Our work aims to reduce the tracer dose in PET scans while maintaining image quality. Although the idea of using multiple imaging modalities for F-PET image synthesis has been presented in previous work [8–11], however, these methods are based on voxel-wise, which have some drawbacks that limit the potential clinical usability (e.g., resulting in over-smoothed images and the synthesis is very time-consuming).
In this paper, we propose a totally different synthesis method using locality adaptive GANs model. Our locality adaptive fusion, learned in an end-to-end trained deep network, has never been proposed in the literature (to the best of our knowledge). Different from the standard convolutional approach which applies the same kernel to convolve the whole image, we argue that the contributions of different modalities could vary at different image locations. In our method, the weights at different image locations of each modality are automatically learned in the deep neural network to better serve the objective of PET synthesis. From the experimental results in Section III-A, we can see that the contributions of different modalities vary at different image locations. Also, as shown in Section III-B(1), we can see that the our locality adaptive GANs model presents better results than the multi-channel based GANs model with also much less number of additional parameters to learn. These two experiments fully demonstrate the superiority of our locality adaptive approach, which cannot be achieved by a standard convolutional approach. The adversarial training network used in our model constrains the synthesized images to be similar to the real ones. Such an objective is evaluated via a deep CNN classification network (the discriminator) rather than a single cost function used in the conventional approaches without adversarial training. Since the generator and discriminator in GANs are competing against each other, the improvement on one indicates a higher loss on the other. With the increase of training epochs, both the discriminator and the generator losses are converged to certain constant numbers, indicating that the GANs model finally finds a Nash equilibrium between the generator and discriminator networks. The experimental results in Section III-B(3) demonstrate the essentials of the adversarial training in our model.
The idea of using 3D conditional GANs has been presented in our previous work [34]. However, there is distinct difference between the two works. Specifically, [34] focuses on single-modality GANs. In contrast, this work innovatively explores the fusion of multiple imaging modalities to synthesize full-dose PET images via GANs. With tactically fusing multi-modalities, our work achieves better performance than that work. Also, our locality adaptive fusion provides a general methodology, which can be applied to a variety of deep neural networks beyond GANs model.
In addition, the fusion strategy for image synthesis has also been studied in [44, 45], by using the mean & variance fusion or the max fusion. However, the fusion strategies used in these two papers and in our work are significantly different. First and most importantly, either the mean & variance fusion or the max-fusion does not include learnable fusion weights. In contrast, in our locality adaptive fusion, the fusion weights are automatically and jointly learned with the image synthesis task, therefore being able to better serve the ultimate goal of the applications. Second, our locality adaptive fusion network directly takes multi-modality images as input, and outputs a fused image as the pseudo input to the following generator network. In contrast, the two fusion strategies in [44, 45] are implemented on the features obtained from the previous encoder network, without considering locality adaption as well.
To train reliable deep model with small size of training samples, we employ large 3D image patches rather than directly using the entire 3D image as input. In this way, we significantly increase the number of training samples. Experimental results show that this strategy significantly mitigates the over-smoothed problem of small patches used in previous work. Although we can increase the number of training samples by extracting even smaller patches from the original images, however, it is found that the results are inferior to our current settings. In contrast, using small patches could possibly bring the problems of over-smoothing as well as additional computational cost.
Moreover, in addition to the image quality metrics, we also explore whether the lesions in the synthetic F-PET image could be well preserved in terms of clinical quantification, as compared to the real F-PET image. Experimental results indicate an improved clinical usability, as compared to the L-PET images and the results by the-state-of-the-art methods.
Our current work has the following limitations. First, in the current study, only a limited number of training images are available to evaluate the proposed method. In our future work, we will involve more subjects into the study to further increase the generalization capacity of the proposed method. Second, for the phantom data, only healthy brains were simulated. It would be interesting to simulate AD-related hypometabolism or tumors since lesions could suffer reduced contrast. In our future work, we plan to simulate lesions in some phantom cases [40] and use more quantitative metrics (e.g. root mean squared analysis and overlap quantification) [46–48] to comprehensively evaluate the algorithm. Third, our current model does not deal with missing modalities, e.g., some subjects may not have a complete set of image modalities, which will make them excluded from the study and thus reduce the number of applicable cases. Therefore, making use of all available data to achieve enhanced synthesis performance is one of our research focuses in future. Fourth, the proposed method is evaluated on the [18F]FDG PET images. We expect our method, as a general methodology, could be applied to other PET tracers, but its performance has to be consolidated by acquiring different training sets for different types of tracers. Also, it is not yet clear how the performance of the method relates to the distributions of tracers. For example, there are likely differences for tracers that distribute widely and uniformly across regions (such as amyloid tracers) versus those with very specific focal targets (dopamine, serotonin). In the future, it would be beneficial to train and evaluate our method for each specific tracer and tailor it according to specific applications in PET. In addition, the affine transformation of different modalities, which is a common step in multi-modality fusion, may change image quality due to the use of image intensity interpolation during the transformation. Although multi-modality images could complement to each other to some extent, this cannot completely remedy the image quality loss caused by the transformation. Our future work will target at integrating image transformation and multi-modality fusion procedures into deep neural networks, in order to further alleviate the influence of transformation/alignment of different image modalities.
V. Conclusion
In this work, we proposed a 3D auto-context locality-adaptive GANs model for synthesizing high-quality PET images from the low-dose PET and multimodal MRI images. Different from traditional multi-channel networks which often directly use multiple channels of low-dose PET and MRI as the inputs for the deep model, we proposed a locality adaptive fusion network to identify local patches that are useful for PET synthesis. Also, the auto-context strategy is adopted to make our LA-GANs model context-aware. Experiments conducted on both phantom and real human brain datasets show that our method can effectively synthesize F-PET images. Both qualitative and quantitative results also demonstrate that our method significantly outperforms the traditional multi-modality fusion methods used in deep networks, as well as the state-of-the-art PET estimation approaches. In addition, please note that, our proposed model can be used in wider applications where a mapping from one or multiple modalities to another modality is needed. Our model can also potentially boost the training data for deep learning algorithms that depend on large PET data collections. In the future, we will further investigate the potential of our model for general image synthesis tasks.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC61701324), Australian Research Council (ARC DE160100241), and NIH grant EB006733.
Footnotes
We also use another metric Mean squared error (MSE), which is close to PSNR, and the results are available in the supplementary files.
Contributor Information
Yan Wang, School of Computer Science, Sichuan University, China.
Luping Zhou, School of Electrical and Information Engineering, University of Sydney, Australia.
Biting Yu, School of Computing and Information Technology, University of Wollongong, Australia.
Lei Wang, School of Computing and Information Technology, University of Wollongong, Australia.
Chen Zu, School of Computing and Information Technology, University of Wollongong, Australia.
David S. Lalush, Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, USA.
Weili Lin, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, USA..
Xi Wu, School of Computer Science, Chengdu University of Information Technology, China..
Jiliu Zhou, School of Computer Science, Chengdu University of Information Technology, China..
Dinggang Shen, IDEA Lab, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, USA; Department of Brain and Cognitive Engineering, Korea University, Republic of Korea.
References
- [1].Chen W, “Clinical applications of PET in brain tumors,” Journal of nuclear medicine, vol. 48, no. 9, pp. 1468–1481, 2007. [DOI] [PubMed] [Google Scholar]
- [2].Alessio A, Vesselle H, Lewis D, Matesan M, Behnia F, Suhy J, de Boer B, Maniawski P, and Minoshima S, “Feasibility of low-dose FDG for whole-body TOF PET/CT oncologic workup,” Journal of Nuclear Medicine, vol. 53, no. supplement 1, pp. 476–476, 2012. [Google Scholar]
- [3].Lee BJ, Grant AM, Chang C-M, Watkins RD, Glover GH, and Levin CS, “MR performance in the presence of a radio frequency-penetrable positron emission tomography (PET) insert for simultaneous PET/MRI,” IEEE Transactions on Medical Imaging, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Delbeke D, Coleman RE, Guiberteau MJ, Brown ML, Royal HD, Siegel BA, Townsend DW, Berland LL, Parker JA, and Hubner K, “Procedure guideline for tumor imaging with 18F-FDG PET/CT 1.0,” Journal of nuclear Medicine, vol. 47, no. 5, pp. 885–895, 2006. [PubMed] [Google Scholar]
- [5].Song Y, Cai W, Huang H, Wang X, Zhou Y, Fulham MJ, and Feng DD, “Lesion detection and characterization with context driven approximation in thoracic FDG PET-CT images of NSCLC studies,” IEEE transactions on medical imaging, vol. 33, no. 2, pp. 408–421, 2014. [DOI] [PubMed] [Google Scholar]
- [6].Huang Y, Shao L, and Frangi AF, “Cross-Modality Image Synthesis via Weakly-Coupled and Geometry Co-Regularized Joint Dictionary Learning,” IEEE Transactions on Medical Imaging, 2017. [DOI] [PubMed] [Google Scholar]
- [7].Cao X, Gao Y, Yang J, Wu G, and Shen D, “Learning-based multimodal image registration for prostate cancer radiation therapy,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2016, pp. 1–9: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kang J, Gao Y, Shi F, Lalush DS, Lin W, and Shen D, “Prediction of standard-dose brain PET image by using MRI and low-dose brain [18F] FDG PET images,” Medical physics, vol. 42, no. 9, pp. 5301–5309, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang Y, Zhang P, An L, Ma G, Kang J, Shi F, Wu X, Zhou J, Lalush DS, and Lin W, “Predicting standard-dose PET image from low-dose PET and multimodal MR images using mapping-based sparse representation,” Physics in Medicine & Biology, vol. 61, no. 2, p. 791, 2016. [DOI] [PubMed] [Google Scholar]
- [10].Wang Y, Ma G, An L, Shi F, Zhang P, Lalush DS, Wu X, Pu Y, Zhou J, and Shen D, “Semisupervised tripled dictionary learning for standard-dose PET image prediction using low-dose PET and multimodal MRI,” IEEE Transactions on Biomedical Engineering, vol. 64, no. 3, pp. 569–579, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].An L, Zhang P, Adeli E, Wang Y, Ma G, Shi F, Lalush DS, Lin W, and Shen D, “Multi-level canonical correlation analysis for standard-dose PET image estimation,” IEEE Transactions on Image Processing, vol. 25, no. 7, pp. 3303–3315, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu M, Cheng D, Wang K, Wang Y, and A. s. D. N. Initiative, “Multi-Modality Cascaded Convolutional Neural Networks for Alzheimer’s Disease Diagnosis,” Neuroinformatics, pp. 1–14, 2018. [DOI] [PubMed] [Google Scholar]
- [13].Schlemper J, Caballero J, Hajnal JV, Price A, and Rueckert D, “A deep cascade of convolutional neural networks for dynamic MR image reconstruction,” arXiv preprint arXiv:170402422, 2017. [DOI] [PubMed] [Google Scholar]
- [14].Bahrami K, Shi F, Rekik I, and Shen D, “Convolutional neural network for reconstruction of 7T-like images from 3T MRI using appearance and anatomical features,” in Deep Learning and Data Labeling for Medical Applications: Springer, 2016, pp. 39–47. [Google Scholar]
- [15].Yuan Y, Chao M, and Lo Y-C, “Automatic skin lesion segmentation using deep fully convolutional networks with Jaccard distance,” IEEE transactions on medical imaging, vol. 36, no. 9, pp. 1876–1886, 2017. [DOI] [PubMed] [Google Scholar]
- [16].de Vos BD, Wolterink JM, de Jong PA, Leiner T, Viergever MA, and Išgum I, “ConvNet-Based Localization of Anatomical Structures in 3-D Medical Images,” IEEE transactions on medical imaging, vol. 36, no. 7, pp. 1470–1481, 2017. [DOI] [PubMed] [Google Scholar]
- [17].Dong C, Loy CC, He K, and Tang X, “Image super-resolution using deep convolutional networks,” IEEE transactions on pattern analysis and machine intelligence, vol. 38, no. 2, pp. 295–307, 2016. [DOI] [PubMed] [Google Scholar]
- [18].Li R, Zhang W, Suk H-I, Wang L, Li J, Shen D, and Ji S, “Deep learning based imaging data completion for improved brain disease diagnosis,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2014, pp. 305–312: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xiang L, Qiao Y, Nie D, An L, Lin W, Wang Q, and Shen D, “Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI,” Neurocomputing, vol. 267, pp. 406–416, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, Wang Q, and Shen D, “Medical Image Synthesis with Deep Convolutional Adversarial Networks,” IEEE Transactions on Biomedical Engineering, 2018, vol. 62, pp. 2720–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Long J, Shelhamer E, and Darrell T, “Fully convolutional networks for semantic segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [22].Li Y, Shen L, and Yu S, “HEp-2 specimen image segmentation and classification using very deep fully convolutional network,” IEEE transactions on medical imaging, vol. 36, no. 7, pp. 1561–1572, 2017. [DOI] [PubMed] [Google Scholar]
- [23].Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, and Shen D, “Medical image synthesis with context-aware generative adversarial networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2017, pp. 417–425: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xiang L, Wang Q, Nie D, Qiao Y, and Shen D, “Deep Embedding Convolutional Neural Network for Synthesizing CT Image from T1-Weighted MR Image,” arXiv preprint arXiv:170902073, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, 2015, pp. 234–241: Springer. [Google Scholar]
- [26].Salehi SSM, Erdogmus D, and Gholipour A, “Auto-context convolutional neural network (auto-net) for brain extraction in magnetic resonance imaging,” IEEE transactions on medical imaging, vol. 36, no. 11, pp. 2319–2330, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, and Ronneberger O, “3D U-Net: learning dense volumetric segmentation from sparse annotation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2016, pp. 424–432: Springer. [Google Scholar]
- [28].Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, and Chen X, “Improved techniques for training gans,” in Advances in Neural Information Processing Systems, 2016, pp. 2234–2242. [Google Scholar]
- [29].Zhang H, Xu T, Li H, Zhang S, Huang X, Wang X, and Metaxas D, “Stackgan: Text to photo-realistic image synthesis with stacked generative adversarial networks,” in IEEE Int. Conf. Comput. Vision (ICCV), 2017, pp. 5907–5915. [DOI] [PubMed] [Google Scholar]
- [30].Mirza M and Osindero S, “Conditional generative adversarial nets,” arXiv preprint arXiv: 14111784, 2014. [Google Scholar]
- [31].Denton EL, Chintala S, and Fergus R, “Deep generative image models using a Laplacian pyramid of adversarial networks,” in Advances in neural information processing systems, 2015, pp. 1486–1494. [Google Scholar]
- [32].Chen X, Duan Y, Houthooft R, Schulman J, Sutskever I, and Abbeel P, “Infogan: Interpretable representation learning by information maximizing generative adversarial nets,” in Advances in Neural Information Processing Systems, 2016, pp. 2172–2180. [Google Scholar]
- [33].Arjovsky M, Chintala S, and Bottou L, “Wasserstein gan,” arXiv preprint arXiv:170107875, 2017. [Google Scholar]
- [34].Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, Wu X, Zhou J, Shen D, and Zhou L, “3D conditional generative adversarial networks for high-quality PET image estimation at low dose,” NeuroImage, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bi L, Kim J, Kumar A, Feng D, and Fulham M, “Synthesis of Positron Emission Tomography (PET) Images via Multi-channel Generative Adversarial Networks (GANs),” in Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment: Springer, 2017, pp. 43–51. [Google Scholar]
- [36].Nie D, Wang L, Gao Y, and Shen D, “Fully convolutional networks for multi-modality isointense infant brain image segmentation,” in Biomedical Imaging (ISBI), 2016 IEEE 13th International Symposium on, 2016, pp. 1342–1345: IEEE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Radford A, Metz L, and Chintala S, “Unsupervised representation learning with deep convolutional generative adversarial networks,” arXiv preprint arXiv:151106434, 2015. [Google Scholar]
- [38].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, “Generative adversarial nets,” in Advances in neural information processing systems, 2014, pp. 2672–2680. [Google Scholar]
- [39].Aubert-Broche B, Griffin M, Pike GB, Evans AC, and Collins DL, “Twenty new digital brain phantoms for creation of validation image data bases,” IEEE transactions on medical imaging, vol. 25, no. 11, pp. 1410–1416, 2006. [DOI] [PubMed] [Google Scholar]
- [40].Aubert-Broche B, Evans AC, and Collins L, “A new improved version of the realistic digital brain phantom,” NeuroImage, vol. 32, no. 1,pp. 138–145, 2006. [DOI] [PubMed] [Google Scholar]
- [41].Wang Z, Bovik AC, Sheikh HR, and Simoncelli EP, “Image quality assessment: From error visibility to structural similarity,” IEEE Trans. Image Process, vol. 13, no. 4, pp.600–612, 2004. [DOI] [PubMed] [Google Scholar]
- [42].Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, and Flitney DE, “Advances in functional and structural MR image analysis and implementation as FSL,” Neuroimage, vol. 23, pp. S208–S219, 2004. [DOI] [PubMed] [Google Scholar]
- [43].Oehmigen M, Susanne Z, Bjoern WJ, Georgi J, Paulus DH, and Harald HQ, “Radiotracer Dose Reduction in Integrated PET/MR: Implications from National Electrical Manufacturers Association Phantom Studies,” Journal of Nuclear Medicine, vol. 55, no. 8, pp. 1361–1367, 2014. [DOI] [PubMed] [Google Scholar]
- [44].Havaei M, Guizard N, Chapados N, and Bengio Y, “HeMIS: Hetero-Modal Image Segmentation” International Conference on Medical Image Computing and Computer-Assisted Intervention Springer, Cham, vol. 3896, pp. 469–477, 2016. [Google Scholar]
- [45].Chartsias A, Joyce T, Giuffrida MV, and Tsaftaris SA, “Multimodal MR Synthesis via Modality-Invariant Latent Representation”. IEEE Trans Med Imaging, vol. 37, no. 3, pp. 803–814, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Baete K, Nuyts J, Van PW, Suetens P, and Dupont P, “Anatomical-based FDG-PET reconstruction for the detection of hypo-metabolic regions in epilepsy”. IEEE Trans Med Imaging, vol. 23, no. 4, pp. 510–519., 2004. [DOI] [PubMed] [Google Scholar]
- [47].Vunckx K, Atre A, Baete K, Reilhac A, Deroose CM, Laere KV, and Nuyts J, “Evaluation of Three MRI-Based Anatomical Priors for Quantitative PET Brain Imaging”. IEEE Transactions on Medical Imaging, vol. 31, no. 3, pp. 599–612, 2012. [DOI] [PubMed] [Google Scholar]
- [48].Vunckx K, Dupont P, Goffin K, Van PW, Van LK, and Nuyts J, “Voxel-based comparison of state-of-the-art reconstruction algorithms for 18F-FDG PET brain imaging using simulated and clinical data”. Neuroimage, vol.102, pp. 875–884, 2014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.