Abstract
Lyme borreliosis (LB), caused by bacteria of the Borrelia burgdorferi sensu lato (Borrelia) species, is the most common tick-borne infection in the northern hemisphere. LB diagnostics is based on clinical evaluation of the patient and on laboratory testing, where the main method is the detection of Borrelia specific antibodies in patient samples. There are, however, shortcomings in the current serology based LB diagnostics, especially its inability to differentiate ongoing infection from a previously treated one. Identification of specific biomarkers of diseases is a growing application of metabolomics. One of the main methods of metabolomics is nuclear magnetic resonance (NMR) spectroscopy. In the present study, our aim was to analyze whether Borrelia growth in vitro and infection in vivo in mice causes specific metabolite differences, and whether NMR can be used to detect them. For this purpose, we performed NMR analyses of in vitro culture medium samples, and of serum and urine samples of Borrelia infected and control mice. The results show, that there were significant differences in the concentrations of several amino acids, energy metabolites and aromatic compounds between Borrelia culture and control media, and between infected and control mouse serum and urine samples. For example, the concentration of L-phenylalanine increases in the Borrelia growth medium and in serum of infected mice, whereas the concentrations of allantoin and trigonelline decrease in the urine of infected mice. Therefore, we conclude that Borrelia infection causes measurable metabolome differences in vitro and in Borrelia infected mouse serum and urine samples, and that these can be detected with NMR.
Subject terms: Infectious-disease diagnostics, Bacterial host response
Introduction
Lyme borreliosis (LB) is a multi-organ infectious disease caused by spirochetes of Borrelia burgdorferi sensu lato species (later Borrelia). Borrelia is a common tick-borne pathogen in the northern hemisphere, making LB the most important tick-borne infectious disease in this area1. In the USA, the estimated number of new LB cases is 300,000 annually. In Europe, LB incidence is the highest in Scandinavia and central Europe. For example, in Finland, the annual incidence of disseminated LB is approximately 2,300 cases, and the number of local skin infections is three to five times that of the disseminated infections2.
The clinical manifestations of LB can be classified as the local skin infection at the tick bite site (erythema migrans; EM), and as various forms of the disseminated infection, namely infection of the nervous system (Lyme neuroborreliosis), infection of the joints (Lyme arthritis), chronic skin disorders and carditis. The diagnosis of a case of disseminated LB is established based on clinical manifestations and laboratory tests1. The mainstay of LB laboratory testing is the detection of Borrelia-specific antibodies in patient serum. This method performs well in the first episode of disseminated LB in a patient. However, antibodies in patient serum remain elevated after treatment for up to several years, leading to substantial seropositivity in the population (e.g. 3.9% in Finland)3. Therefore, an obvious shortcoming in LB serodiagnostics is its inability to differentiate an ongoing case of LB from a previously treated one4,5.
Metabolomics is one approach in the field of the so-called omics sciences. It uses cutting-edge analytical chemistry techniques combined with computational methods to characterize complex mixtures of molecules. One application of metabolomics is the global analysis of low molecular mass biological molecules in order to identify biomarkers of specific disease states6,7. Using metabolomics-based approaches, biomarker panels have been identified in a variety of human conditions: for example, in coronary heart disease, diabetes, nephropathy and in various cancers8–10. Also, in infections such as tuberculosis, typhoid fever, dengue virus infection11–13, malaria and schistosomiasis14, metabolomics has been utilized for biomarker profile discovery. Importantly, there are three previous publications with promising results of LB metabolomics of human samples15–17.
The main methods of metabolomics are gas chromatography-mass spectrometry, liquid chromatography-mass spectrometry, and nuclear magnetic resonance spectroscopy (NMR). NMR is an analytical method based on the magnetic properties of atom nuclei. NMR active nuclei (e.g. 1H, 13C, 15N, and 31P) are commonly found in biological samples which makes the methodology highly usable in biomarker research6. NMR has several advantages: the sample preparation is relatively easy and not time-consuming, and during NMR measurements the samples are not destroyed, which allows multiple measurements of one sample. Probably the most important advantage of NMR is the possibility to recognize metabolites based on their chemical properties6. NMR has been shown to be an extremely useful tool in metabolomics, as different metabolites can be identified and quantified even from complex mixtures18.
The present study was initiated to evaluate whether NMR can be used to detect Borrelia- specific metabolites in vitro and in vivo in mice. Our results show that, indeed, Borrelia growth causes NMR-demonstrable metabolome changes in the in vitro culture medium samples, and importantly, also in Borrelia-infected mouse serum and urine samples.
Results
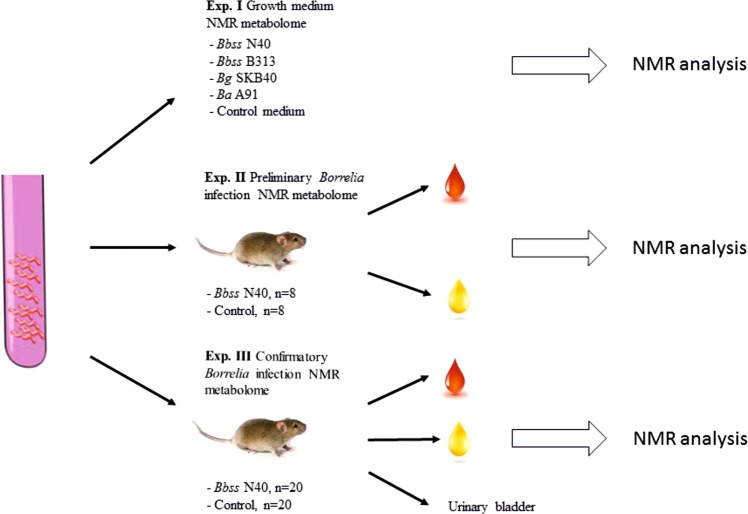
This study included three separate experiments (Fig. 1). The study was initiated with an in vitro experiment (Exp. I). Following the promising results of Experiment I, two sets of animal experiments (Exp. II and Exp. III) were performed, resulting in the in vivo NMR metabolome of mouse Borrelia infection.
Figure 1.
Overall setup of the study. The study consisted of three experiments. In Experiment I, the in vitro growth medium metabolomes of different Borrelia strains (Bbss N40, Bbss 313, Bg SKB40 and Ba A91) were analyzed using NMR. In Experiment II, in vitro grown Bbss N40 were used to infect eight mice, and eight mice were used as uninfected controls. After 4 weeks follow up, the mice were sacrificed and serum samples were collected for NMR analysis. Urine samples were collected on three consecutive days before killing on the week 4. Ear skin samples were collected for Borrelia culture. In Experiment III, in vitro grown Bbss N40 were used to infect 20 mice, and 20 mice were used as uninfected controls. After 4 weeks follow up, the mice were sacrificed, and serum samples and urinary bladders were collected for NMR analysis. Urine and ear skin samples were collected as in Experiment II.
Experiment I: In vitro NMR metabolome of Borrelia
In Experiment I, in vitro growth medium NMR metabolomes, (measured by Carr-Purcell-Meiboom-Gill pulse sequence (CPMG); see Materials and methods) of different Borrelia strains (B. burgdorferi ss N40 (Bbss N40), B. burgdorferi ss 313 (Bbss 313), B. garinii SKB40 (Bg SKB40) and B. afzelii A91 (Ba A91)) were compared. The differences between Borrelia growth- and control media were detected by PCA (principal component analysis) and PLS-DA (partial least square means discriminant analysis) and then confirmed by univariate analysis (Fig. S1). The PCA model, of which the first principal component explains 72% of the total variation, separated the samples into five distinguished groups (PCA: R2 = 0.99, Q2 = 0.95 and PLS-DA: R2 = 0.98, Q2 = 0.97). R2 represents the variation of the model whereas Q2 is an estimate of the predictive ability of the model.
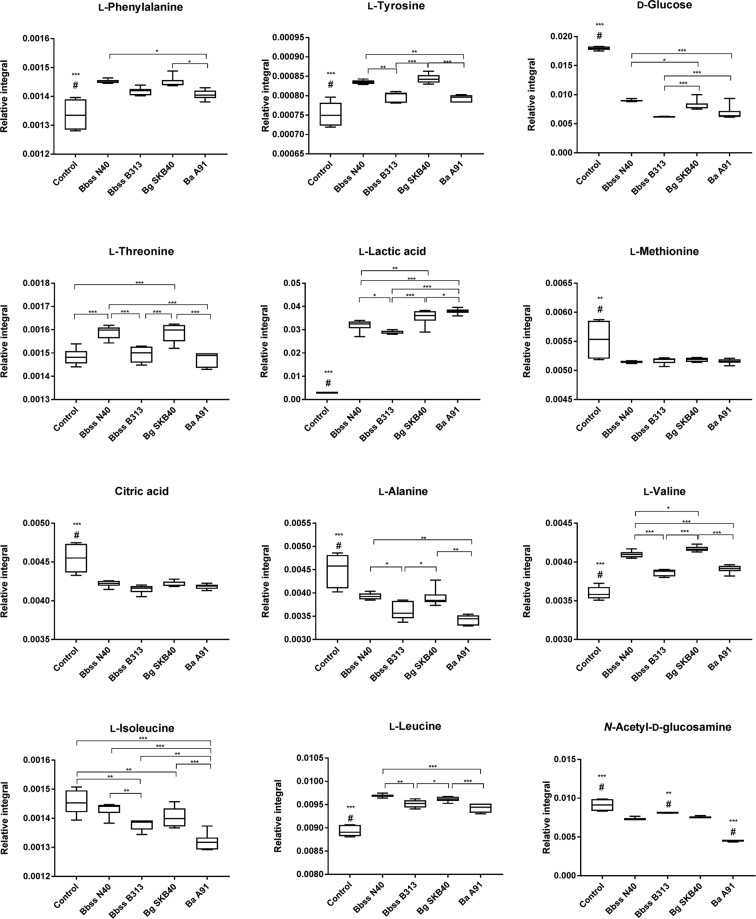
Statistically significant differences were observed in the concentrations of several amino acids and energy metabolites (Fig. 2). The results of this experiment revealed that Borrelia growth increases the concentrations of L-phenylalanine, L-tyrosine, L-threonine, L-valine, L-leucine and L-lactic, and decrease the concentrations of L-methionine, L-alanine, L-isoleucine, D-glucose, citric acid and N-acetyl-D-glucosamine in the culture media compared to the control medium. This experiment also demonstrated that there were differences between the Borrelia genotypes. Bbss N40 and Bg SKB40 caused a similar change in the concentration of the metabolites forming one pattern, whereas Bbss B313 and Ba A91 formed their own pattern (Fig. S1). All of these differences were statistically significant at least between control and Borrelia incubated medium samples, but in the most of the metabolites also between different Borrelia strains (Fig. 2). In total, 23 metabolites were identified in the growth medium samples (Table S1).
Figure 2.
Metabolites in control and Borrelia growth medium samples. Boxplot figures present the means of relative integrals of metabolites, standard deviations and 95% confidence intervals. Statistically significant difference between samples is indicated by asterisks (*<0.05, **<0.01, ***<0.001). If a group is statistically significantly different from all other groups it is annotated with #.
Experiment II: Preliminary metabolome analysis of Borrelia-infected and control mice
Because the results of Experiment I demonstrated that NMR can be used to detect Borrelia growth-induced metabolome differences in vitro, a preliminary mouse study (Exp. II) with eight Bbss N40-infected and eight control mice was performed (Fig. 1). The mice were followed up for four weeks, after which the animals were sacrificed and total blood collected. During the last week of the experiment, urine samples were also collected. The urine collection was successful in 10/16 animals. Borrelia culture of ear skin samples revealed that all infected mice were Borrelia positive, while control animals were culture negative.
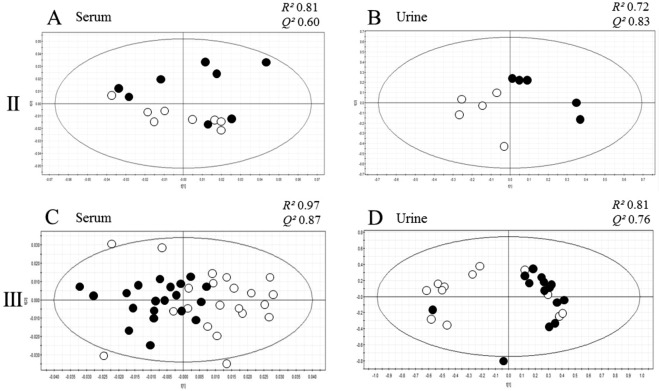
Based on CPMG spectra of the serum samples, the infected and control groups differed in the metabolite composition in the sera (Fig. 3A, Table 1, Fig. S2A). The differences between Borrelia infected and control animal sera were detected by PCA and PLS-DA and then confirmed by univariate analysis In PCA, the R2 value was 0.81 and the Q2 0.60, which means that results are not entirely reliable since the difference between R2 and Q2 is slightly over 0.2. In PLS-DA, R2 value was 0.86 and the Q2 0.75. In PCA, the first principal component explained 63% of the total variation. Statistical significance of the metabolite concentration differences observed in the PCA and PLS-DA were determined, and metabolites with statistically significant concentration difference between infected and control mice are presented in Table 1, along with the relative integral means, SD values and fold change values.
Figure 3.
Metabolome differences of Borrelia infected and control mice. Scores plot figures of multivariate principal component analyses (PCA) of Experiment II and III serum and urine samples (A: Exp. II serum, B: Exp. II urine, C: Exp. III serum, D: Exp. III urine) show the difference between infected mouse (black circles) and control mouse metabolomes (empty circles). R2 and Q2 values, representing the reliability of the model of the model, are shown above each scatter plot. The values indicate that the results of all analyses, except the PCA of urine in Experiment II, are reliable.
Table 1.
Serum metabolites differing in concentration in the infected and control mice in Experiments II and III based on PCA.
| Metabolite | Trend | Statistical significance, p-value | Control serum mean (SD) | Infected serum mean (SD) | Serum fold change | ¹H signal ppm |
|---|---|---|---|---|---|---|
| Amino acids | ||||||
| L-phenylalanine | ↑ | 0.06/0.001 | 0.0110 (0.0066)/0.0971 (0.0329) | 0.0159 (0.0094)/0.1402 (0.0066) | 1.45/1.44 | 7.435 |
| L-threonine | ↑ | 0.0004 | 0.0018 (0.0003) | 0.0022 (0.0005) | 1.27 | 4.335 |
| L-glutamine | ↓ | 0.049/0.02 | 0.0065 (0.0008)/0.0129 (0.0029) | 0.0054 (0.0012)/0.0109 (0.0020) | 1.20/1.18 | 2.025 |
| L-leucine | ↑ | 0.01 | 0.0071 (0.0008) | 0.0097 (0.0006) | 1.36 | 0.965 |
| Energy metabolites | ||||||
| D-glucose | ↑ | 0.04/0.0001 | 0.0176 (0.0026)/0.0127 (0.0016) | 0.0206 (0.0026)/0.0157 (0.0018) | 1.17/1.23 | Multiple signals |
| L-lactic acid | ↓ | 0.02/0.006 | 0.1284 (0.0149)/0.1950 (0.0215) | 0.1074 (0.0063)/0.1469 (0.0167) | 1.20/1.32 | 4.125, 1.335 |
| Citric acid | ↑ | 0.5/0.0001 | 0.0039 (0.0011)/0.0708 (0.0029) | 0.0046 (0.0021)/0.1368 (0.0041) | 1.18/1.93 | 2.695 |
| Others | ||||||
| Urea | ↓ | 0.04/0.03 | 0.0082 (0.0020/0.0114 (0.0032) | 0.0056 (0.0023)/0.0095 (0.0019) | 1.46/1.19 | 5.325 |
| 2-hydroxybutyric acid | ↓ | 0.01 | 0.0290 (0.0059) | 0.0245 (0.0044) | 1.18 | 1.735, 1.595, 0.895 |
The same differences were observed also using PLS-DA. Metabolites occurring in higher concentration in infected mice are annotated with an upward arrow (↑), whereas metabolites occurring in higher concentration in control mice are annotated with a downward arrow (↓). Metabolites differing statistically significantly in both Experiments II and III are indicated by bold whereas concentration of non-bold metabolites differed only in Experiment III. Statistical significance of metabolite differences is presented as p-values. In addition, relative means of signal integrals, SDs and fold changes are presented. If a metabolite difference is detected in both experiments, p-values, means, SDs and fold changes of both experiments are presented (Exp II/Exp III). Signal positions, by which the metabolite identification was made, are presented as ppm values. Signals based on which the statistical analyses were made are indicated by bold.
Ten urine samples were analyzed using 1D-Nuclear Overhauser Effect Spectroscopy (1D-NOESY) method. Five of the samples were from the infected and five from the control mice. Based on these measurements, the metabolite composition of the urine samples of the infected mice differed from the composition of the control mouse samples as shown in Fig. 3B. In the PCA of urine metabolite data, the R2 value was 0.72 and the Q2 value 0.83, and in PLS-DA the R2 value was 0.74 and the Q2 value 0.81. The R2 value, which describes the reliability of the multivariate analysis, is just below the border value of 0.80 in both cases, likely due to the low sample number. In PCA, the first component explained 68% of the total variation. Metabolites with statistically significant concentration difference between infected and control mice are presented in Table 2.
Table 2.
Urine metabolites differing in concentration in the infected and control mice in Experiments II and III. Metabolites occurring in higher concentration in infected mice are annotated with an upward arrow (↑), whereas metabolites occurring in higher concentration in control mice are annotated with a downward arrow (↓).
| Metabolite | Trend | Statistical significance, p-value | Control urine mean (SD) | Infected urine mean (SD) | Urine fold change | ¹H signal ppm |
|---|---|---|---|---|---|---|
| Amino acids | ||||||
| L-glutamine | ↑ | 0.08/0.002 | 0.0054 (0.0004)/0.0018 (0.0006) | 0.0059 (0.0003)/0.0025 (0.0003) | 1.09/1.39 | 2.025 |
| L-glycine | ↓ | 0.4/0.03 | 0.0382 (0.0073)/0.0284 (0.0162) | 0.0341 (0.0074)/0.0284 (0.0162) | 1.12/1.48 | 3.275 |
| Energy metabolites | ||||||
| Acetic acid | ↑ | 0.02/0.25 | 0.0051 (0.0003)/0.0074 (0.0006) | 0.0056 (0.0001)/0.0074 (0.0006) | 1.11/1.03 | 1.925 |
| Aromatic compounds | ||||||
| Allantoin | ↓ | 0.8/0.01 | 0.0282 (0.0048)/0.0119 (0.0074) | 0.0277 (0.0028)/0.0119 (0.0074) | 1.02/1.66 | 5.385 |
| Trigonelline | ↓ | 0.01/0.047 | 0.0020 (0.0014)/0.0022 (0.0002) | 0.0005 (0.0002)/0.0013 (0.0002) | 4.00/1.69 | 8.125, 8.084 |
| Hippuric acid | ↓ | 0.2/0.002 | 0.0074 (0.0021)/0.0031 (0.0016) | 0.0059 (0.0008)/0.0031 (0.0016) | 1.25/2.03 | 7.845, 7.555 |
| Indoxyl sulphate | ↓ | 0.046/0.1 | 0.0072 (0.0008)/0.0051 (0.0008) | 0.0048 (0.0015)/0.0051 (0.0008) | 1.50/1.64 | 7.195 |
| Others | ||||||
| Creatinine | ↑ | 0.02/0.5 | 0.0121 (0.0009)/0.018 (0.0027) | 0.0177 (0.0046)/0.018 (0.0027) | 1.46/1.13 | 3.935, 3.041 |
Metabolites differing statistically significantly in both Experiments II and III are indicated by bold whereas concentration of non-bold metabolites differed only in Experiment II or III. Statistical significance of metabolite differences is presented as a p-value. In addition, means of the relative signal integrals, SDs and fold changes of both experiments are presented (Exp II/Exp III). Signal positions, by which the metabolite identification was made, are presented as ppm values. Signals based on which the statistical analyses were made are indicated by bold.
All identified metabolite differences shown in Tables 1 and 2 were quantitative, meaning that no compound was totally lacking (below the detection limit of NMR) in any sample. In addition to the described major differences in Tables 1 and 2, some smaller differences occurred between the infected and control mice, especially in the spectrum area 6.5–9 ppm, but due to the low concentration variability of these molecules, we were not able to determine their identity.
Experiment III: Confirmatory metabolome analysis of Borrelia-infected and control mice
Due to the observed metabolome differences in Experiment II, we conducted a confirmatory mouse experiment as shown in Fig. 1. We also extended our study to semi-solid state NMR analysis of urinary bladders of the mice.
In this experiment, 40 mice in total were used. Half of them were infected and the other half served as control animals. Ear skin sample culture verified that all infected mice were Borrelia positive, while control animals were culture negative.
Also in this experiment, control and infected mice differed based on the metabolite composition of both serum and urine samples as shown in Fig. 3C,D (PCA) and in Fig. S2C and D (PLS-DA). In the PCA of the CPMG analysis of the sera, the R2 value was 0.97 and Q2 was 0.87, and in the PLS-DA R2 value was 0.98 and the Q2 value 0.89. Thus, the results of Experiment III are in line with the results of Experiment II. In PCA, the first principal component explained 81% of the total variation. Statistical significance of the concentration differences was determined as in Experiment II for both serum and urine metabolites. P-values, means, SDs and fold change values of statistically significantly differing metabolites are presented in Tables 1 and 2. All PCA identified metabolites are shown in Fig. 4. Differences observed in the PLS-DA analysis are presented in Fig. S3. The same metabolite differences were observed using both multivariate analysis methods, and therefore the same annotations are used in Figs 4 and S3.
Figure 4.
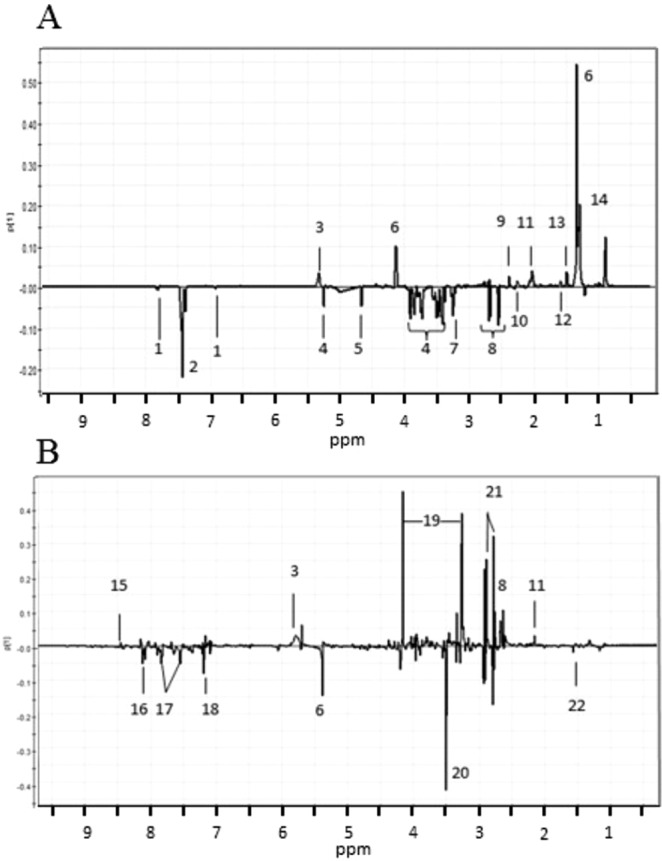
Loadings plots of the serum (A) and urine (B) analyses of Experiment III show the differentiating metabolites. Positive y-axis represents metabolites occurring in higher concentrations in the control mice, and negative y-axis represents metabolites occurring in higher concentrations in the infected mice. Identified metabolites are: 1: L-histidine, 2: L-phenylalanine, 3: urea, 4: D-glucose, 5: L-threonine, 6: L-lactic acid, 7: carnitine, 8: citric acid, 9: pyruvic acid, 10: L-valine, 11: L-glutamine, 12: 2-hydroxybutyric acid, 13: L-alanine, 14: L-leucine, 15: formic acid, 16: trigonelline, 17: hippuric acid, 18: indoxyl sulphate, 19: allantoin, 20: creatinine, 21: L-glycine, 22: acetic acid.
Major differences between the infected and control serum samples were mainly similar to the differences observed in Experiment II. However, several additional differentiating metabolites were also identified (mainly essential amino acids) due to the larger sample number and refined CPMG analysis protocol.
In this experiment, 27 urine samples were analyzed, 14 of which were from the infected mice and 13 from the control mice. 1D-NOESY measurements of the urine samples clearly showed that the metabolites in the urine of Borrelia-infected mice differed from the metabolites in the samples of the control mice (Fig. 3D). The statistically significantly differing metabolites are listed in Table 2 and all metabolites observed are shown in the loadings plots (PCA: Fig. 4B and PLS-DA: Fig. S3B). These results are mainly in line with the urine metabolite results of Experiment II, but due to the bigger number of samples, the results are more reliable. In PCA, the first principal component explained 79% of the total variation.
As a part of Experiment III, the urinary bladders of mice (n = 40) were analyzed with semi-solid state NMR. No considerable differences between the urinary bladders of the infected and the control animals could be detected with PCA (Fig. S4) or with PLS-DA (data not shown).
Discussion
In this study, we evaluated whether NMR can be used to detect Borrelia-specific metabolites in vitro and in vivo in mice. Our results show that, indeed, Borrelia infection causes metabolome differences both in BSK II growth medium incubated with and without bacteria, and in control and Borrelia-infected mouse serum and urine samples. On the other hand, no differences between Borrelia-infected and control bladder tissues samples were found.
Recently, Molins et al. and Pegalajar-Jurado et al. have elegantly shown that LB-specific metabolite panels can be identified in human LB patient serum and urine samples using a liquid chromatography-mass spectrometry (LC-MS) approach15–17. NMR, on the other hand, has not been previously used in LB metabolomics studies. However, we know that NMR has been successfully applied, for example, in the identification of biomarker profiles for malaria and schistosomiasis14,19,20. Especially concerning malaria, NMR has proven to be very useful, since an entirely new malaria-related molecule has been identified19. Based on these previous achievements, we decided to conduct our LB in vitro and mouse experiments. Importantly, samples originating from laboratory mice have the benefit over human patient samples that the housing conditions and nutrition of the animals are controlled and standardized, the time point of sample collection in regard to the duration of the infection is similar, and the infection status of mice can be confirmed by tissue Borrelia culture. There are also some disadvantages in the mouse model. Mice have slightly different metabolic routes than humans, which may cause differences in the metabolomes of Borrelia infected mice and humans. Therefore, the metabolite signatures observed in mice may not be present in human patients.
NMR was selected as the method because it provides untargeted data allowing the identification of all molecules of potential interest, and due to the non-destructive nature of the measurements, which is important considering the not-too-large number of mice (in accordance with the 3Rs policy), limited sample volume, and our setup in which the samples were first analyzed multiple times during the method validation.
In general, NMR as a method in metabolomics has its pros and cons6. One benefit of the method is the fact mentioned above that using NMR, it is possible to measure one sample multiple times. Measuring the samples is also fast, and less validation, internal standardization and instrument calibration than in LC-MS is required. In addition, different NMR instruments in different laboratories result in robust, identical results. The most important quality of NMR is the reliable recognition of metabolites based on their chemical bond properties. Unfortunately, NMR has also some major drawbacks. The sample volume required is relatively high, which leads to challenges in animal experimentation. Although NMR can recognize most organic molecules, inorganic compounds like salts can’t be identified. The most important disadvantage of NMR is the low sensitivity6. In this study we used three different NMR instruments suitable for different sample materials. Our 400 MHz instrument is designed for solid samples, 500 MHz for simple sample materials like the growth medium, and 600 MHz with cryoprobe for complex biological samples like serum and urine21.
NMR methodology allows the use of different types of pulse sequences to collect different type of information. In biological samples, like serum and urine, the most problematic signal is the water signal, which covers all other signals and makes the interpretation of the spectrum challenging. Broad signals caused by large molecules, like proteins, are also problematic. These signals are difficult to identify, and they may also hide other signals21. Therefore, different pulse sequences have been developed to eliminate the interference of these signals. For example, the Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence is developed to fade the broad signals. The 1D-Nuclear Overhauser Effect Spectroscopy (NOESY) pulse sequence is used for solvent signal suppression in the case when no wide signal suppression is required, such as in urine samples, where no or only a little proteins and lipids are present. Based on these properties of different NMR instruments and pulse sequences, we used the 500 MHz instrument and the CPMG pulse sequence in Experiment I; the 600 MHz instrument with cryoprobe and automatic sample changer for the analysis of serum (CPMG pulse sequence) and urine samples (NOESY pulse sequence) in Experiments II and III; and the 400 MHz instrument with the CPMG pulse sequence for the analysis of the urinary bladder samples in Experiments III.
In Experiment I, statistically significant differences (Fig. 2) in the metabolite profiles of the in vitro-grown bacterial strains and control samples were detected. These differences are mainly due to the conversion of glucose in glycolysis of the bacteria into lactic acid, and the lactic acid is then excreted outside of the bacterial cell. However, in addition to energy metabolites, statistically significant differences in the concentrations of several amino acids were also observed. Overall, it appears that Bbss N40 and Bg SKB40 alter amino acid and energy metabolite concentrations differently than B313 and Ba A91. All of the medium samples incubated with the different Borrelia-strains differ significantly from the control medium. Interestingly, the overall variability of the metabolites in the control medium samples is higher than the variability in Borrelia-culture samples. However, there is no clear explanation for this phenomenon. All sample and data handling steps were identical in the case of every specimen.
The above observations imply that Bbss N40 and Bg SKB40 have different metabolic rates compared to Bbss B313 and Ba A91. This finding is rather interesting since it might be expected that the two Bbss strains would cause more or less similar metabolic changes. One explanation for the difference between the two Bbss strains might be the different plasmid content of the strains, since Bbss N40 contains 16 plasmids, while the genetically modified laboratory strain Bbss B313 carries only six plasmids. Plasmid contents of Bg SKB40 and Ba A91 have not been characterized22–24.
One weakness of Experiment I is that the concentration fold change differences between the groups are small despite the statistical significance. Therefore, no far reaching conclusions were made based only on these results, thus we continued the study with mouse experiments.
In Experiments II and III, we analyzed both serum and urine samples, because these sample materials are also relevant and easy to collect for human diagnostic purposes. In both experiments, statistically significant differences in the metabolomes of the infected and the control mouse sera and urine samples were detected. In Experiments II and/or III, differences were observed in the serum concentration of L-phenylalanine, L-threonine, L-glutamine, L-leucine, D-glucose, L-lactic acid, citric acid, urea and 2-hydroxybutyric acid (Table 1). The urine metabolite differences were identical in both Experiments II and III. Metabolites with different concentrations in urine samples of the infected and control mice were L-glutamine, L-glycine, acetic acid, allantoin, trigonelline, hippuric acid, indoxyl sulphate and creatinine (Table 2).
The observed metabolites can be divided in four classes: amino acids, energy metabolites, aromatic compounds and others. According to our interpretation, the most interesting findings are the differences in amino acid and aromatic compound concentrations. These differences were mainly observed in the serum, probably because amino acids are found in much smaller concentrations in urine than in serum when the kidney function is normal.
One of the most interesting findings was the increased L-phenylalanine concentration in the infected mice compared with the control ones. Wannemacher et al. have shown that serum L-phenylalanine concentrations and phenylalanine-tyrosine ratio significantly increase during bacterial (gram positive and negative) and viral infections25 and this appears to be the case also in the Borrelia infection. Interestingly, the concentration of L-phenylalanine was significantly increased also in the Borrelia culture medium samples. Interesting is also the increased L-threonine concentration in infected mice since L-threonine is known to have immunostimulatory properties, which might explain the increased concentration26,27.
One interesting finding in this study was that L-glutamine and urea concentration differences were opposite in serum and urine samples. It is unclear why these metabolites occur in higher concentrations in control mouse sera and, on the other hand, in the urine infected mice. This phenomenon might, however, be due to the different NMR pulse sequences used to analyze the different sample materials. The CPMG sequence, used for the analysis of serum samples, is designed to reduce protein and lipid signals. However, it might also unintentionally reduce signals of small molecules locating close to protein or lipid signals. This phenomenon doesn’t appear with 1D-NOESY used for the analysis of urine samples, thus perhaps explaining the contradictory results28.
Another group of metabolites with differing concentrations is formed of the aromatic substances, allantoin (uric acid metabolite), trigonelline (a vitamin B3 metabolite), hippuric acid (metabolite of phenolic compounds) and indoxyl sulphate (L-tryptophan metabolite). All of these are common urine metabolites29, which in our study were observed in statistically significantly higher concentrations in the urine samples of control mice. Interestingly, the fold change differences of these metabolites are relatively large, although the role of the differences is difficult to explain. If the clear differences in concentrations of these metabolites appeared also in human samples, they might be potential indicators of human LB but this must be investigated in further studies.
Importantly, the results of our study are mainly in line with the results of earlier studies performed with human LB serum and urine samples15–17. Several altered metabolic routes reported in these publications (L-phenylalanine, citric acid cycle and vitamin B metabolism) were also observed in our study. In addition, several other metabolites with changed concentration were found in the present study which were not detected in the previous human studies and the other way around. This can most likely be explained by the different analytical methods used and by the different host species.
Analysis of urinary bladder was included in Experiment III to find out whether NMR is a functional tool to detect Borrelia infection in a sample of infected mouse tissue. Previous results of our group have shown that urinary bladder samples are always Borrelia culture positive, and therefore, we used this tissue type in our solid-state NMR analysis30,31. However, no differences were discovered between infected and control mouse bladders.
In conclusion, the three experiments with three different sample types resulted in a representative picture of Borrelia infection NMR metabolome. These results, however, are not a comprehensive representation of the effects of Borrelia infection on the metabolite profile of mice due to the obvious lack of sensitivity of the method. There were, however, some identified metabolites, for example L-phenylalanine and allantoin, with biomarker potential, but their applicability for biomarker use remains to be determined. In future studies, it appears to be necessary to use more sensitive methods, such as ultra-high pressure liquid chromatography-mass spectrometry (UHPLC-MS/MS), to be able to identify biomarkers with high specificity for Lyme borreliosis.
Materials and Methods
Experimental design
In our study design, the experiments were divided into three parts (Fig. 1). Experiment I was a growth medium metabolomics study in which we compared metabolite profiles of different Borrelia strains grown in Barbour-Stoenner-Kelly II (BSK-II) medium. The study was followed by two mouse experiments (Exp. II and III) in which serum and urine sample metabolomes of Borrelia-infected and control mice were analyzed by NMR and compared by PCA. Experiment II was a preliminary experiment with 16 mice, and Experiment III was a confirmatory study with 40 mice. In Experiment III, the urinary bladders of the mice were also analyzed using a semi-solid state NMR method.
Bacteria and bacterial culture
We used four different Borrelia strains: B. burgdorferi ss N40 (Bbss N40), B. burgdorferi ss 313 (Bbss 313), B.garinii SKB40 (Bg SKB40) and B. afzelii A91 (Ba A91). All of these strains were used in the growth medium experiment, while the in vivo experiments were performed using only Bbss N40.
Bacteria were grown in BSK II medium at 33 °C. Bacteria used for the infection were at the stationary growth phase. Before infection, the bacteria were washed with PBS, diluted to the 107 bacteria/ml concentration in PBS and then transferred to a syringe for mouse infection.
To verify the infection status of the mice, ear skin samples were collected in Experiments II and III at the end of the study. Collected skin samples were placed in BSK II medium at 33 °C for Borrelia cultivation. Rifampicin (5 µg/ml) and phosphomycin (10 µg/ml) were added to the culture medium to avoid contaminating bacterial growth. The cultures were observed every two weeks for six weeks by dark field microscopy.
Experiment I: In vitro NMR metabolome of borrelia
Four different Borrelia strains (Bbss N40, Bbss 313, Bg SKB40, Ba A91) were cultured in BSK-II medium at 33 °C as six parallel samples for four days until the bacteria reached a stationary growth phase. The cultures were then centrifuged at 3000 rpm for five minutes to remove the bacterial cells. Supernatant was further sterilized by passing the medium through 0.22 µm filters. As a control, two samples of BSK-II medium without bacteria were incubated and processed similarly. In total, the number of samples was 30 including six control medium samples and six medium samples after cultivation of each of the four strains. After incubation and processing, sample aliquots of 600 µl were stored at −80 °C.
Experiments II and III: In vivo metabolome of borrelia infection in mice
In the preliminary mouse Experiment II, 16 female C3H/HeNHsd mice (Envigo, Netherlands), and in the confirmatory Experiment III, 40 mice, were used as in vivo models of Borrelia infection. In both experiments, half of the mice were Bbss N40-infected, while the other half formed the control group receiving PBS injections. Mice were infected by intradermal injection, containing 106 bacteria in 100 µl of PBS, in the lower back. Infected and control mice were placed in different cages, four mice/cage, but otherwise treated equally. Water and food were given ad libitum during the whole study. Mice were observed weekly to monitor their general health. Ear markings were used for mouse identification.
Urine samples were collected on the fouth week after infection on three consecutive days before the endpoint of the experiment. Urine samples were collected by placing mice on a plastic-covered paper, where the mice were allowed to urinate freely. Urine drops were collected from the paper by pipetting to sterile Eppendorf tubes. The samples of each animal from all collection time points were pooled together.
On the week 4 after infection, the mice were sacrificed by cardiac puncture under isoflurane anesthesia followed by neck break. The total blood sample from each mouse was collected by cardiac puncture with a heparinized syringe and placed in 1.5 ml Eppendorf tubes on ice. If cardiac puncture resulted in blood volume under 0.8 ml, the mouse’s chest was opened and puncture performed directly to the heart. This procedure was successful in most cases, but resulted in hemolysis disturbing the NMR analysis of the serum samples. After mice were sacrificed, ear skin samples were collected for Borrelia culture. Urinary bladders were collected for semi-solid state NMR in Experiment III. Forceps and scissors used for sample collection were disinfected with 70% ethanol and rinsed with sterile water after every mouse during tissue sample collection.
After sample collection, all urine samples were placed in Eppendorf tubes at −80 °C. Blood samples were first centrifuged at 1000 g for 10 min and serum was carefully pipetted avoiding erythrocyte contamination. Serum was transferred to a new tube and placed also in −80 °C. Urinary bladders for semi-solid state NMR were weighted and placed in the semi-solid state NMR adapters. Approximately 10 µl of deuterated phosphate buffer (0.1 M KH2PO4 + 2 mM NaN3 + 0.1% 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TSP)) was added, and the adapter was closed before freezing it with liquid nitrogen. Samples were stored in −80 °C until NMR measurements.
Animal experiments in this project were conducted according to the recommendations of the Finnish Act of the Use of Animals for experimental purposes of the ministry of agriculture and forestry of Finland and the principle of 3 R. All efforts were made to minimize the suffering of the animals. All experimental infection studies were approved by the National Animal Experiment Board in Finland (permission ESAVI/6265/04.10.07/2017).
Sample handling/processing for NMR
Medium samples of Experiment I were first freeze-dried for 48 h to remove water. Before the NMR analyses, the samples were dissolved in 600 µl of buffer solution (0.5 M phosphate buffer, pH 7.0, 0.1 m-% TSP, 2 mM NaN3, pH adjusted with NaOD and DCl (D meaning deuterium, 2H isotope of hydrogen)) by vortexing for 15 minutes. Then, the samples (600 µl) were transferred into 5 mm NMR tubes (Cambridge Isotope Laboratories, Inc., Andover, MA).
Serum samples of Experiments II and III were thawed on ice and thoroughly mixed in a shaker. 200 µl serum was mixed with 400 µl 1 M NaCl in D2O, samples were centrifuged 1650 g 5 min 4 °C and 540 µl sample was mixed with 60 µl deuterated 1.5 M KH2PO4 + 2 mM NaN3 + 0.1% TSP buffer. Samples were transferred to 5 mm NMR tubes (VWR, Randor, Pennsylvania, USA) and stored at 4 °C until measurements. Sodium azide was used in the sample buffer to prevent bacterial growth during storage.
Urine samples of Experiments II and III were thawed on ice, thoroughly mixed in a shaker, centrifuged 1650 g 7.5 min +4 °C, and 120 µl of the supernatant was mixed with 60 µl 1.5 M KH2PO4 + 2 mM NaN3 + 0.1% TSP buffer. Urine samples were transferred to 3 mm NMR tubes (VWR) and stored at +4 °C until measurements32. Due to the low amount of urine samples, the samples of one animal from all collection time points were pooled.
NMR experiments
NMR measurements of growth medium samples were performed using a Bruker Avance 500 MHz spectrometer with BBI room temperature probe (Bruker BioSpin AG, Fällanden, Switzerland) operating at 500.13 MHz for 1H at 25 °C in batches of five samples in random order.
NMR experiments on serum and urine samples were conducted by Bruker Avance III 600 MHz instrument with TCI PRODIGY cryoprobe (Bruker BioSpin AG, Fällanden, Switzerland) operating at 600.16 MHz for 1H. SampleJet automatic sample changer (Bruker BioSpin AG, Fällanden, Switzerland) was utilized to store samples at 4 °C before and between measurements, and to change samples automatically. All samples were pre-warmed for 10 minutes before measurement in order to do the measurements at 25 °C. Sample measurement order was randomized.
NMR experiments on urinary bladders were conducted by Bruker Avance III 400 MHz (Bruker BioSpin AG, Fällanden, Switzerland) instrument with HR-MAS 4 mm solid state probe. The spinning rate was 4000 Hz and the samples were measured at 4 °C to reduce tissue degradation and metabolic changes. For the measurement, the samples were first placed in 4 mm ZrO2 rotors inside the inserts directly from the freezer, and the rotor was then placed inside the magnet. Shimming and tuning were done manually for each sample. The measurement was started within 20 minutes after taking the sample out of the freezer. Sample measurement order was randomized.
Growth medium NMR measurements were performed using a Bruker Avance 500 MHz spectrometer (Bruker BioSpin AG, Fällanden, Switzerland) operating at 500.13 MHz for 1H at 25 °C in batches of five samples in random order and CPMG, 1D-NOESY, DQF-COSY (double-quantum filtered correlation spectroscopy), CH2-modified HSQC (heteronuclear single quantum coherence), NOESY and TOCSY (total correlated spectroscopy) pulse sequences were utilized. From serum and urinary bladder samples, CPMG spectra were measured. CPMG pulse sequence with presaturation was used in order to reduce the water signal and broad signals caused by lipids and proteins which cover small molecule signals. Urine samples, naturally containing only low concentrations of proteins and lipids, were measured with 1D-NOESY pulse sequence with presaturation to reduce only the water signal.
The following parameters were used in the CPMG measurement of serum samples: acquisition time 3.41 s, echo time (d20) 0.0015 s, loop counter (l4) 64, 90° pulse length (p1) 10.65 µs and relaxation delay (d1) 5.0 s. Spectral width was 16 ppm, 64 k data points were collected, and number of scans was 256.
All 1D-NOESY measurements of urine samples were also conducted using the following parameters: acquisition time 3.41 s, mixing time (d8) 0.10 s, p1 9.0 µs and d1 5.0 s. Spectral width was 16 ppm and 64 k data points were collected, number of scans was 128.
The following parameters were used in the CPMG measurement of urine bladder samples: acquisition time 2.56 s, d20 0.0015 s, l4 128 and d1 5.0 s. Spectral width was 16 ppm, 32 k data points were collected and number of scans was 256. P1 was determined for each sample, and ranged from 4.45 to 5.80 µs.
Growth medium sample analyses were conducted with the following parameters: The pulse program used was CPMGPR with water suppression (4.71 ppm) and exclusion of the large biomolecules using the T2 filter. Acquisition time for one scan of 1.64 µs, a 90° pulse of 6.90 µs and four dummy scans were used. The total FID was a sum of 256 scans collected with 64 k data points. The dwell time between two data points in the FID was 50 µs in spectral width of 20 Hz, and line broadening was set to 0.30 Hz. TSP was used as an internal reference compound.
NMR data processing
NMR data was handled with Bruker TopSpin 3.5 pl3 software (Bruker BioSpin GmbH, Rheinstetten, Germany). All spectra were manually phase corrected, baseline corrected using polynomic function method and calibrated using α-glucose or TSP as reference signal. Data was normalized to total spectral area.
After manual spectra correction, data were bucketed for statistical analyses by Bruker Amix 3.9.15 software (Bruker BioSpin GmbH, Rheinstetten, Germany). Bucket size 0.01 ppm was used for all other spectra, but growth medium spectra were bucketed in 0.05 ppm buckets. A larger bucket size was used in growth medium dataset in order to minimize the effect of pH and varying ion content induced signal position variation, which were observed in the medium samples. Data was normalized to the whole spectrum integral in a manner that the whole spectrum integral is assumed to be 1. Bucket tables were transferred to Microsoft Excel 2013, where data was transformed from text to numeric format. Peak alignment correction of urine data was conducted by matlab based icoshift algorithm33.
All of the results presented in this study are based on a three step data analysis approach. First, PCA analysis was done to obtain unsupervised information of metabolite differences between sample types. Secondly, PLS-DA was used as a supervised model to confirm the differences observed in PCA. Thirdly, statistical significances of differences observed in PCA and PLS-DA were determined by univariate statistics.
Numeric data were analyzed using multivariate analysis by Umetrics Simca-P +12.0.1 software (Umetrics, Umeå, Sweden). In both urine and serum data analyses, Pareto scaling of data was used and analysis itself was conducted using PCA modeling (unsupervised linear mixture model which explains the variance within a dataset) as the main analysis, and PLS-DA (Partial least square discriminant analysis) as a confirmatory analysis.
In our dataset, the principal components are vectors of metabolite contributions. All principal components are decorrelated to each other. Unsupervised PCA is used as linear mixture model, in which the data matrix is divided into two parts. One is a score matrix, which contains the positions of the observations, and the other is a loading matrix, which contains the weights for the original variables. Supervised PLS-DA relates the multivariate metabolomic data to the response vector by a linear regression model and attempts to find an optimal decomposition of the predictor dataset34.
The scaling for PCA and PLS-DA was selected in order to achieve the highest possible R2 and Q2 values. R2 represents the variation of the model, whereas Q2 is an estimate of the predictive ability of the model35,36. One (1) is the highest value of both parameters35. R2 is over 0.8 when the model is considered to be reliable, and the difference between R2 and Q2 should not exceed 0.2.
The presentation of results was made using a scores plots to show differences between samples and loadings plots to show which metabolites differentiate in concentration between the samples of infected and control mice.
Based on the multivariate results, univariate analysis was performed to determine the statistical significance of the concentration differences of each metabolite observed in PCA and in PLS-DA. Univariate analysis was performed using merged bucket values corresponding to the area of analyzed metabolite’s 1H signal. The data was normalized to the total spectral area. Statistical analyses were performed using JMP Pro 13.1.0 (SAS, Cary, NC, USA) software. The normal distribution of the data was first tested using Shapiro-Wilk W –test. If the relative integral values of a metabolite were normally distributed, Student’s t-test was used in Experiment I and t-test was used in Experiments II and III to analyze statistical significance. If the data was not normally distributed, Kruskal-Wallis test (Exp. I) or Mann-Whitney U test (Exp. II and III) was used. The difference was regarded as statistically significant if p-value was <0.05.
Signal identification was performed using the Human Metabolome Database (HMDB)37–39 and literature searches29,40. Raw identification of the metabolites was made based on the literature, followed by confirmation using HMDB.
Supplementary information
Acknowledgements
Julia Cuellar and Tuula Rantasalo are acknowledged for their assistance in animal experiments and Heli Elovaara for inventing the study approach. This study was funded by Jane and Aatos Erkko foundation, Jenny and Antti Wihuri foundation and the Academy of Finland.
Author Contributions
O.G., E.P., J.J., M.K., J.S. and J.H. designed the study, O.G., E.P., J.J. and J.S. performed the NMR analyses, O.G. performed the animal work, O.G., E.P. and J.J. performed data analyses, M.K. and J.S. provided metabolomics expertise O.G., E.P., J.J. and J.H. wrote the paper. All co-authors edited the paper.
Data Availability
The data of this project will be deposited to IDA to ensure that the research community has a long-term access to the data (https://openscience.fi/ida). The service is offered to Finnish universities and polytechnics by the Ministry of Education and Culture in Finland.
All metadata of Metabo-Lyme project are made available in Etsin (https://etsin.avointiede.fi/). Etsin offers access to datasets in various fields via a joint metadata model. The descriptive metadata stored in the service includes information on the authors, subject, format and licensing of the dataset. Anyone can use Etsin to search for research datasets.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44540-5.
References
- 1.Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet. 2012;379:461–473. doi: 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- 2.Sajanti E, et al. Lyme Borreliosis in Finland, 1995-2014. Emerg Infect Dis. 2017;23:1282–1288. doi: 10.3201/eid2308.161273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Beek J, et al. Population-based Borrelia burgdorferi sensu lato seroprevalence and associated risk factors in Finland. Ticks Tick Borne Dis. 2018;9:275–280. doi: 10.1016/j.ttbdis.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Aguero-Rosenfeld ME, Wang G, Schwartz I, Wormser GP. Diagnosis of lyme borreliosis. Clin Microbiol Rev. 2005;18:484–509. doi: 10.1128/CMR.18.3.484-509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilske B, Fingerle V, Schulte-Spechtel U. Microbiological and serological diagnosis of Lyme borreliosis. FEMS Immunol Med Microbiol. 2007;49:13–21. doi: 10.1111/j.1574-695X.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- 6.Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 7.Kraus Virginia B. Biomarkers as drug development tools: discovery, validation, qualification and use. Nature Reviews Rheumatology. 2018;14(6):354–362. doi: 10.1038/s41584-018-0005-9. [DOI] [PubMed] [Google Scholar]
- 8.Choi SH, Kim CK, Park JJ, Park BK. Assessment of Early Therapeutic Changes to Concurrent Chemoradiotherapy in Uterine Cervical Cancer Using Blood Oxygenation Level-Dependent Magnetic Resonance Imaging. J Comput Assist Tomogr. 2016;40:730–734. doi: 10.1097/RCT.0000000000000424. [DOI] [PubMed] [Google Scholar]
- 9.Kalantari S, Nafar M, Samavat S, Parvin M. 1 H NMR-based metabolomics study for identifying urinary biomarkers and perturbed metabolic pathways associated with severity of IgA nephropathy: a pilot study. Magn Reson Chem. 2017;55:693–699. doi: 10.1002/mrc.4573. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, et al. Identification of metabolic biomarkers in patients with type 2 diabetic coronary heart diseases based on metabolomic approach. Sci Rep. 2016;6:30785. doi: 10.1038/srep30785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau SK, et al. Metabolomic Profiling of Plasma from Patients with Tuberculosis by Use of Untargeted Mass Spectrometry Reveals Novel Biomarkers for Diagnosis. J Clin Microbiol. 2015;53:3750–3759. doi: 10.1128/JCM.01568-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Näsström E, et al. Diagnostic metabolite biomarkers of chronic typhoid carriage. PLoS Negl Trop Dis. 2018;12:e0006215. doi: 10.1371/journal.pntd.0006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voge NV, et al. Metabolomics-Based Discovery of Small Molecule Biomarkers in Serum Associated with Dengue Virus Infections and Disease Outcomes. PLoS Negl Trop Dis. 2016;10:e0004449. doi: 10.1371/journal.pntd.0004449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tritten L, et al. Metabolic profiling framework for discovery of candidate diagnostic markers of malaria. Sci Rep. 2013;3:2769. doi: 10.1038/srep02769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molins CR, et al. Development of a metabolic biosignature for detection of early Lyme disease. Clin Infect Dis. 2015;60:1767–1775. doi: 10.1093/cid/civ185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molins Claudia R., Ashton Laura V., Wormser Gary P., Andre Barbara G., Hess Ann M., Delorey Mark J., Pilgard Mark A., Johnson Barbara J., Webb Kristofor, Islam M. Nurul, Pegalajar-Jurado Adoracion, Molla Irida, Jewett Mollie W., Belisle John T. Metabolic differentiation of early Lyme disease from southern tick–associated rash illness (STARI) Science Translational Medicine. 2017;9(403):eaal2717. doi: 10.1126/scitranslmed.aal2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegalajar-Jurado A, et al. Identification of Urine Metabolites as Biomarkers of Early Lyme Disease. Sci Rep. 2018;8:12204. doi: 10.1038/s41598-018-29713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markley JL, et al. The future of NMR-based metabolomics. Curr Opin Biotechnol. 2017;43:34–40. doi: 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh S, Sengupta A, Sharma S, Sonawat HM. Early prediction of cerebral malaria by (1)H NMR based metabolomics. Malar J. 2016;15:198. doi: 10.1186/s12936-016-1256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Junfang, Xu Wenxin, Ming Zhenping, Dong Huifen, Tang Huiru, Wang Yulan. Metabolic Changes Reveal the Development of Schistosomiasis in Mice. PLoS Neglected Tropical Diseases. 2010;4(8):e807. doi: 10.1371/journal.pntd.0000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.W, C. T. D. High-resolution NMR techniques in organic chemistry. Tetrahedron organic chemistry series27 (2009).
- 22.Zückert WR, Lloyd JE, Stewart PE, Rosa PA, Barbour AG. Cross-species surface display of functional spirochetal lipoproteins by recombinant Borrelia burgdorferi. Infect Immun. 2004;72:1463–1469. doi: 10.1128/IAI.72.3.1463-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heikkilä T, et al. Cloning of the gene encoding the decorin-binding protein B (DbpB) in Borrelia burgdorferi sensu lato and characterisation of the antibody responses to DbpB in Lyme borreliosis. J Med Microbiol. 2002;51:641–648. doi: 10.1099/0022-1317-51-8-641. [DOI] [PubMed] [Google Scholar]
- 24.Anderson JF, Barthold SW, Magnarelli LA. Infectious but nonpathogenic isolate of Borrelia burgdorferi. J Clin Microbiol. 1990;28:2693–2699. doi: 10.1128/jcm.28.12.2693-2699.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wannemacher RW, Klainer AS, Dinterman RE, Beisel WR. The significance and mechanism of an increased serum phenylalanine-tyrosine ratio during infection. Am J Clin Nutr. 1976;29:997–1006. doi: 10.1093/ajcn/29.9.997. [DOI] [PubMed] [Google Scholar]
- 26.Li P, Yin YL, Li D, Kim SW, Wu G. Amino acids and immune function. Br J Nutr. 2007;98:237–252. doi: 10.1017/S000711450769936X. [DOI] [PubMed] [Google Scholar]
- 27.Duval D, Demangel C, Munier-Jolain K, Miossec S, Geahel I. Factors controlling cell proliferation and antibody production in mouse hybridoma cells: I. Influence of the amino acid supply. Biotechnol Bioeng. 1991;38:561–570. doi: 10.1002/bit.260380602. [DOI] [PubMed] [Google Scholar]
- 28.Gowda GA, Raftery D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal Chem. 2014;86:5433–5440. doi: 10.1021/ac5005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouatra S, et al. The human urine metabolome. PLoS One. 2013;8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yrjänäinen H, Hytönen J, Hartiala P, Oksi J, Viljanen MK. Persistence of borrelial DNA in the joints of Borrelia burgdorferi-infected mice after ceftriaxone treatment. APMIS. 2010;118:665–673. doi: 10.1111/j.1600-0463.2010.02615.x. [DOI] [PubMed] [Google Scholar]
- 31.Salo J, Jaatinen A, Söderström M, Viljanen MK, Hytönen J. Decorin Binding Proteins of Borrelia burgdorferi Promote Arthritis Development and Joint Specific Post-Treatment DNA Persistence in Mice. PLoS One. 2015;10:e0121512. doi: 10.1371/journal.pone.0121512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehtonen HM, et al. 1H NMR-based metabolic fingerprinting of urine metabolites after consumption of lingonberries (Vaccinium vitis-idaea) with a high-fat meal. Food Chem. 2013;138:982–990. doi: 10.1016/j.foodchem.2012.10.081. [DOI] [PubMed] [Google Scholar]
- 33.Savorani F, Tomasi G, Engelsen S. B. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. J Magn Reson. 2010;202:190–202. doi: 10.1016/j.jmr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Bartel J, Krumsiek J, Theis FJ. Statistical methods for the analysis of high-throughput metabolomics data. Comput Struct Biotechnol J. 2013;4:e201301009. doi: 10.5936/csbj.201301009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worley B, Powers R. Multivariate Analysis in Metabolomics. Curr Metabolomics. 2013;1:92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eriksson, L., Byrne, T., Johansson, E., Trygg, J. & Vikström, C. Multi- and Megavariate Data Analysis: Basic principles and Applications. 3RD edn, 39–62 (MKS Umetrics AB, 2013).
- 37.Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wishart DS, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wishart DS, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Psychogios N, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of this project will be deposited to IDA to ensure that the research community has a long-term access to the data (https://openscience.fi/ida). The service is offered to Finnish universities and polytechnics by the Ministry of Education and Culture in Finland.
All metadata of Metabo-Lyme project are made available in Etsin (https://etsin.avointiede.fi/). Etsin offers access to datasets in various fields via a joint metadata model. The descriptive metadata stored in the service includes information on the authors, subject, format and licensing of the dataset. Anyone can use Etsin to search for research datasets.