Abstract
Despite significant advances in the treatment of human immunodeficiency virus type-1 (HIV) infection, antiretroviral therapy only suppresses viral replication but is unable to eliminate infection. Thus, discontinuation of antiretrovirals results in viral reactivation and disease progression. A major reservoir of HIV latent infection resides in resting central memory CD4+ T cells (TCM) that escape clearance by current therapeutic regimens and will require novel strategies for elimination. Here, we evaluated the therapeutic potential of autophagy-inducing peptides, Tat-Beclin 1 and Tat-vFLIP-α2, which can induce a novel Na+/K+-ATPase dependent form of cell death (autosis), to kill latently HIV-infected TCM while preventing virologic rebound. In this study, we encapsulated autophagy inducing peptides into biodegradable lipid-coated hybrid PLGA (poly lactic-co-glycolic acid) nanoparticles for controlled intracellular delivery. A single dose of nanopeptides was found to eliminate latent HIV infection in an in vitro primary model of HIV latency and ex vivo using resting CD4+ T cells obtained from peripheral blood mononuclear cells of HIV-infected patients on antiretroviral with fully suppressed virus for greater than 12 months. Notably, increased LC3B lipidation, SQSTM1/p62 degradation and Na+/K+-ATPase activity characteristic of autosis, were detected in nanopeptide treated latently HIV-infected cells compared to untreated uninfected or infected cells. Nanopeptide-induced cell death could be reversed by knockdown of autophagy proteins, ATG5 and ATG7, and inhibition or knockdown of Na+/K+-ATPase. Importantly, viral rebound was not detected following the induction of the Na+/K+-ATPase dependent form of cell death induced by the Tat-Beclin 1 and Tat-vFLIP-α2 nanopeptides. These findings provide a novel strategy to eradicate HIV latently infected resting memory CD4+ T cells, the major reservoir of HIV latency, through the induction of Na+/K+-ATPase dependent autophagy, while preventing reactivation of virus and new infection of uninfected bystander cells.
Subject terms: Macroautophagy, HIV infections
Introduction
At present, an estimated 37 million people live with human immunodeficiency virus type-1 (HIV) infection worldwide1. Despite the tremendous success of antiretroviral therapy (ART) in suppressing the virus and changing HIV from an invariably fatal disease to a chronic illness, current treatment strategies have failed to eradicate the virus and achieve a virologic cure. Additionally, despite prolonged virologic suppression, the discontinuation of ART in most cases leads to the rapid return of viremia2–4. The difficulty in eradicating HIV from latent reservoirs has changed the focus of much research to be directed towards achieving a “functional cure”. As part of a functional cure strategy, numerous investigators are attempting strategies to reactivate HIV from latent reservoirs (shock) followed by killing the virus. However, the “shock and kill” approach has, to date, been unsuccessful because methods attempted have failed to reactivate latent virus5.
The primary reservoirs of HIV latently infected cells are thought to be long-lived, resting central memory CD4+ T cells (TCM), which are established early in infection, harbor integrated proviral DNA, and fail to produce replication-competent virus6–8. These latently infected cells are not targeted by the immune system and ART is ineffective in eradicating the virus. Although the precise mechanisms that promote the long-term survival of HIV latently infected TCM are largely unknown, it is likely that cell death pathways are critical to cell survival, and that anti-apoptotic proteins and modulation of autophagy play an important role in prolonged cell survival.
Macroautophagy (referred to here as autophagy) is a critical cyto-adaptive response to environmental stresses including starvation, ischemia, cancer and infection9,10. The hallmark of autophagy is a double-membraned autophagosome that engulfs bulk cytoplasm and cytoplasmic organelles, such as mitochondria and endoplasmic reticulum11. The accumulation of autophagosomes and autophagic proteins within a cell represents the failure of autophagy to rescue the cell from toxic stress12. Although the major role of autophagy is cell survival and maintenance of cellular homeostasis, the over induction of autophagy can lead to autophagic cell death13,14. During initial HIV infection of host cells, there is an induction of autophagy and HIV uses autophagy-related proteins (ATG) to promote its own replication15. During permissive infection, HIV down-regulates autophagy to prolong cell survival. In the case of CD4+ T cells, autophagy is dysfunctional and most infected cells die16. However, infection of macrophages and microglial cells leads to a balance between the levels of autophagy required for cell survival and low-level viral replication without killing the cell.
In addition to extending cell survival, autophagy plays a central role in the degradation and elimination of intracellular pathogens10,17–19. Our laboratory has had a particular interest in examining the role of autophagy in HIV pathogenesis20–23, and identified that the induction of autophagy inhibits HIV replication and promotes the degradation of viral proteins23–26. Once HIV establishes a productive infection, HIV Nef binds Beclin 1 resulting in mTOR activation, TFEB phosphorylation and cytosolic sequestration, and the inhibition of autophagy23. These findings help to explain how HIV modulates autophagy to promote cell survival and viral persistence, and further establishes Beclin 1 as an important target to eliminate latent reservoirs of HIV. The region of Beclin 1 binding to Nef has been mapped to an 18 aa conserved region27. In conjunction with the laboratory of Dr. Beth Levine, we showed that a Tat-Beclin 1 fusion peptide consisting of the Tat transduction domain and the identified region of Beclin 1 that binds Nef is a potent inducer of autophagy27. Additionally, we showed that pre-treatment of macrophages with this Tat-Beclin 1 peptide inhibits HIV infection. Similar proteins that inhibit autophagy have been found in other viruses. One of these, viral FLIP (vFLIP) present in Kaposi’s sarcoma-associated herpesvirus, herpesvirus saimiri and molluscum contagiosum virus, was found to inhibit autophagy and cell death by preventing Atg3 from binding and processing LC3, a critical protein in autophagosome biogenesis28.
Liu et al. have described a novel form of autophagic cell death that they have termed “autosis” which has unique morphologic features, depends on cellular Na+/K+-ATPase, and occurs during treatment with autophagy inducing peptides, starvation and hypoxia29,30. In a previous study, we found that autophagy inducing peptides, Tat-Beclin 1 and Tat-vFLIP-α2, exhibit robust anti-HIV activity through induction of autophagy31. Both autophagy-inducing peptides induce cell death through autosis that is dependent on alteration of Na+/K+-ATPase. Of interest, we found that intracellular delivery of Tat-Beclin 1 and Tat-vFLIP-α2 peptides by lipid-coated hybrid PLGA (poly lactic-co-glycolic acid) nanoparticles can preferentially induce autosis and selectively kill chronically infected macrophages. In this study, we further evaluated the potential of these nanoformulated autophagy-inducing peptides (nanopeptides) to kill latently HIV-infected central memory CD4+ T cells (HIV-TCM). Our findings demonstrate that nanopeptides can induce a unique Na+/K+-ATPase dependent form of cell death that has the potential to eliminate latently HIV-infected TCM without reactivation of HIV replication, and protect bystander cells.
Results
Nanopeptides preferentially kill CD4+ T memory cells with latent infection
We have previously shown the ability of lipid-coated hybrid PLGA nanoparticles loaded with Tat-vFLIP-α2 peptides to kill selectively HIV-infected macrophages31. However, the most common reservoir of HIV is thought to reside in resting central memory CD4+ T cells (TCM)32,33. Thus, our initial experiments were designed to demonstrate the potential of these nanopeptides to preferentially kill HIV-TCM. Because the frequency of latently HIV-infected CD4+ T cells in infected persons on fully suppressive ART is extremely low, our laboratory has adapted a primary in vitro latency model of HIV-infected resting CD4+ central memory T cells for screening novel anti-HIV latency altering agents34. In this model, HIV-TCM lack both cell surface activation and cell cycle markers, do not synthesize DNA, do not proliferate, and contain an average of 1 copy per cell of integrated HIV DNA. Importantly, HIV can be reactivated from these cells using αCD3/αCD28-conjugated beads, phytohemagglutinin, M form (PHA-M) or interleukin 7. In the studies presented here, we further optimized this latency model through co-incubation with the HIV protease inhibitor, atazanavir, and the nucleoside reverse transcriptase inhibitor, tenofovir (Supplementary Fig. 1).
We formulated and characterized nanopeptides following our previous established protocol31. Using single step nanoprecipitation, the formulated PLGA nanoparticles loaded Tat-Beclin 1 (NP-Beclin 1) and Tat-vFLIP-α2 (NP-vFLIP-α2) had an average 127 nm and 147 nm in diameter, respectively (Supplementary Fig. 2). All of the formulated nanopeptides had more than 15% (wt:wt) loading yield, were stable in both PBS and water over the 96 h evaluation period, and did not exhibit any burst release of Tat-Beclin 1 or Tat-vFLIP-α2 in peptide release kinetics studies (Supplementary Fig. 2).
To assess the ability of our nanopeptides to induce preferential killing, NP-Beclin 1 and NP-vFLIP-α2 at increasing concentrations were incubated with HIV-TCM and uninfected cells. After 24 h, both nanopeptides demonstrated a dose dependent killing of HIV-TCM. NP-Beclin 1 treatment resulted in 43.6, 85.7, and 92.3% cell death of HIV-TCM at 1, 5, and 10 µM, respectively. Similarly, following NP-vFLIP-α2 treatment at the same concentrations, we observed 36.6, 87.3, and 94.6% killing of HIV-TCM (Fig. 1a). In contrast, uninfected TCM when treated under the same conditions at the highest concentration of 10 µM of nanopeptide resulted in 11.2 and 12.9% cell killing for NP-Beclin 1 or NP-vFLIP-α2, respectively.
Fig. 1. Nanopeptides preferentially kill latent primary HIV-TCM cells.
a At day 32 post infection (p.i.), latent HIV-TCM cells were treated with increasing doses of NP-Beclin 1 or NP-vFLIP-α2 for 24 h. The cytotoxicity of NP-Beclin 1 and NP-vFLIP-α2 were measured by trypan blue staining. b The collected cell culture supernatants were incubated with TZM-bl cells. After 48 h, TZM-bl cells were measured for ß-galactosidase activity. c The collected cell culture supernatants were also tested for RT activity. Data are plotted from four different donors with means. M = Mock infection, H = 200 TCID50 HIVNL-43 virus, L = latent HIV-TCM cells, NP-S1 = 10 μM nanoformulated Tat-Beclin-1 scrambled peptides, NP-S2 = 10 μM nanoformulated Tat-vFLIP-α2 scrambled peptides. RLU = relative luminescence units. F.S. = fluorescent signaling. *P < 0.05, ***P < 0.001
Nanopeptides induced selective killing does not reactivate HIV replication
We next evaluated whether nanopeptides would reactivate infectious virus following treatment of HIV-TCM. HIV infectious virus was assessed using the TZM-bl assay system35. After co-culture with TZM-bl for 48 h, we did not detect the presence of any infectious virus activated from the HIV-TCM (Fig. 1b). These findings were further confirmed by quantification of viral reverse transcriptase (RT) activity in the collected supernatants from nanopeptide-treated HIV-TCM. No increase in RT activity was observed following administration of the nanopeptides (Fig. 1c), further confirming that our nanopeptides kill latently infected TCM without reactivating HIV replication.
Killing of latently infected CD4+ T cells is autophagy dependent
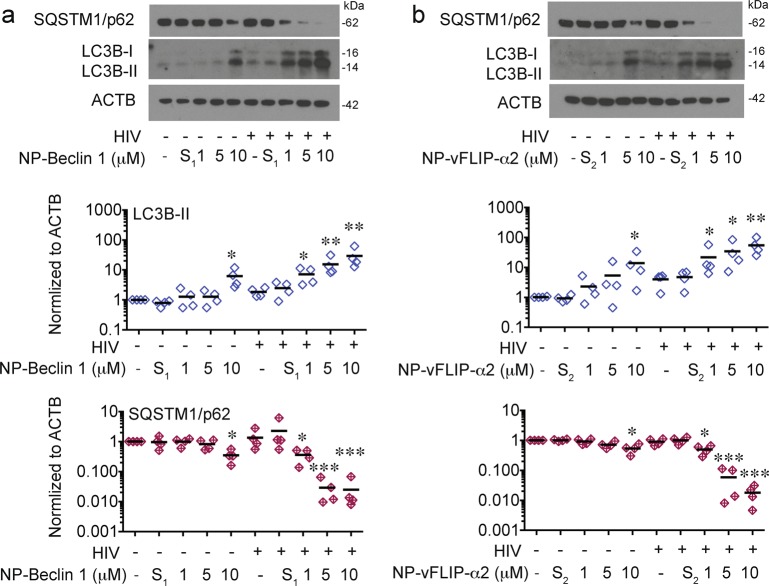
Based on our previous research, we suspected that the mechanism for the selective killing observed of HIV-TCM would be dependent on an autophagy dependent form of cell death31. In these experiments, HIV-TCM and uninfected TCM were treated with increasing concentrations of nanopeptides. In HIV-TCM for both NP-Beclin 1 and NP-vFLIP-α2, we observed a significant dose response increase in LC3B-II while sequestosome 1 (SQSTM1/p62) was significantly decreased indicating that autophagy was being induced and going to completion (P < 0.001; Fig. 2). In contrast for the HIV-uninfected TCM, except at the highest concentration of the nanopeptides (10 μM), there was little change in LC3B-II or SQSTM1/p62 (Fig. 2). However, even at the highest concentration, the decrease in SQSTM1/p62 in uninfected TCM was significantly less than that observed in HIV-TCM (NP-Beclin 1, P < 0.01; NP-vFLIP-α2, P < 0.01).
Fig. 2. Nanopeptides induce enhanced autophagy in latent HIV-TCM cells.
a, b At day 32 p.i., HIV-TCM cells were treated with NP-Beclin 1 and NP-vFLIP-α2, respectively for 24 h. Cell lysates were harvested for analysis of LC3B-II lipidation and SQSTM1/p62 degradation. Representative western blots are shown. Densitometric analysis are summarized from 4 different donors and normalized to loading control ACTB. NP-S1 = 10 μM nanoformulated Tat-Beclin-1 scrambled peptides, NP-S2 = 10 μM nanoformulated Tat-vFLIP-α2 scrambled peptides. *P < 0.05, **P < 0.01, ***P < 0.001
To further confirm the importance of autophagy in NP-Beclin 1 and NP-vFLIP-α2 mediated cell death, we assessed the effect of ATG5 and ATG7 silencing. Knockdown of ATG5 and ATG7 reversed nanopeptide-induced cell death (Fig. 3), and inhibited LC3B-II lipidation and SQSTM1/p62 degradation further confirming that NP-Beclin 1 and NP-vFLIP-α2 induced preferential cell death is through an autophagy dependent mechanism.
Fig. 3. RNA interference of ATG5 and ATG7 inhibits nanopeptide-induced autophagy dependent cell death in latent HIV-TCM cells.
a, d Lentiviral shATG5 and shATG7 transduced latently infected resting CD4+ T cells were tested for knockdown efficiency by western blot. b, e shATG5 and shATG7 transduced latent CD4+ TCM cells were challenged with 10 μM NP-Beclin 1 or 10 μM NP-vFLIP-α2 for 24 h. Autophagy was evaluated in cell lysates by western blot. c, f Cytotoxicity of NP-Beclin 1 and NP-vFLIP-α2 was measured in cell culture supernatants. Densitometric analyses are summarized from four different donors and normalized to loading control ACTB with means. NP-S1 = 10 μM nanoformulated Tat-Beclin-1 scrambled peptides, NP-S2 = 10 μM nanoformulated Tat-vFLIP-α2 scrambled peptides. *P < 0.05, **P < 0.01, ***P < 0.001
Having demonstrated an important role of autophagy in our nanopeptides induced killing of HIV-TCM, it was important to exclude other potential causes of cell death including apoptosis and necroptosis. In these experiments, HIV-TCM were treated with the specific pharmacologic inhibitors of interest followed by exposure to either NP-Beclin 1 or NP-vFLIP-α2 nanopeptides. We used the pan-caspase inhibitor z-VAD-FMK for assessing apoptosis and the RIPK1 inhibitor necrostatin-1 for testing necroptosis. Treatment with either inhibitor, however, had no effect on nanopeptide induced cell death (Fig. 4). To further verify that the observed cell death is not due to apoptosis or necroptosis, we assessed HIV-TCM and TCM cultures for the presence of cleaved caspase (CASP)3. Cleaved CASP3, the active form of CASP336,37, is induced by CASP8 and CASP9, and is considered a key initiator of apoptosis, pyroptosis and necroptosis. Consistent with our previous findings, the expression of cleaved CASP3 in HIV-TCM or uninfected TCM was unchanged following treatment with NP-Beclin 1 and NP-vFLIP-α2, further supporting that the observed cell death was not related to apoptosis and necroptosis.
Fig. 4. Nanopeptides induce caspase-independent cell death.
a HIV-TCM cells were pretreated with 20 μM Z-VAD-FMK, 50 μM necrostatin-1, and 200 nM bafilomycin A1 for 2 h, and further challenged with 10 μM NP-Beclin 1 or 10 μM NP-vFLIP-α2 for an additional 24 h. The cell culture supernatants were collected for LDH cytotoxicity assay. b Cleaved CASP3 was analyzed in the harvested cell lysates by western blot. All densitometric analyses are summarized from four different donors and normalized to loading control ACTB with means. NP-S1 = 10 μM nanoformulated Tat-Beclin 1 scrambled peptides, NP-S2 = 10 μM nanoformulated Tat-vFLIP-α2 scrambled peptides. *P < 0.05, **P < 0.01, ***P < 0.001
Tat-Beclin 1 and Tat-vFLIP-α2 induce a Na+/K+-ATPase dependent form of cell death, autosis
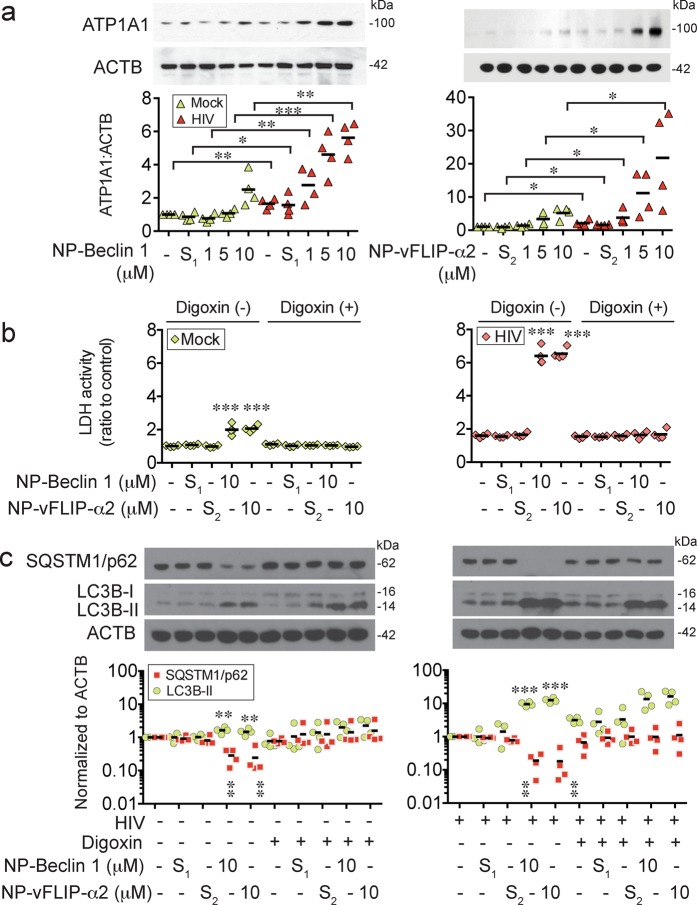
In previous studies performed by our lab and others, Tat-Beclin 1 and Tat-vFLIP-α2 have been shown to induce a Na+/K+-ATPase dependent form of cell death that has been designated as autosis29–31,38. To determine if autosis is responsible for the selective killing of HIV-TCM, we evaluated the expression of Na+/K+-ATPase in nanopeptide-treated HIV-TCM (Fig. 5a). At increasing concentrations of both nanopeptides, we observed an increasing expression of the alpha 1 subunit of Na+/K+-ATPase (ATP1A1), which is the critical catalytic component for activating Na+/K+-ATPase. We also found that both nanopeptides induced a dose dependent increase of ATP1A1 in the mock-infected TCM. However, comparing with the same dose of nanopeptide induced ATP1A1 in the HIV-TCM cells, NP-Beclin 1 induced a mean of 72.1, 76.7, and 55.4% less expression of ATP1A1 in mock-infected cells; similarly, NP-vFLIP-α2 induced 64.6, 69.1, and 75% less expression of ATP1A1 in the mock infected cells.
Fig. 5. NP-Beclin 1 and NP-vFLIP-α2 kills latent HIV-TCM cells through a Na+/K+-ATPase dependent mechanism.
a HIV-TCM were incubated with increasing concentrations of NP-Beclin 1 or NP-vFLIP-α2 for 24 h. The collected cell lysates were tested for Na+/K+-ATPase-α1 subunit (ATP1A1) expression by western blot. b After pretreating with 50 nM digoxin for 2 h, latent HIV-TCM were incubated with NP-Beclin 1 or NP-vFLIP-α2 for 24 h. The cytotoxicity was monitored by LDH assay. c Autophagy activity was analyzed in collected cell lysates by western blot. All densitometric analyses are summarized from four different donors and normalized to loading control ACTB with means. NP-S1 = 10 μM nanoformulated Tat-Beclin-1 scrambled peptides, NP-S2 = 10 μM nanoformulated Tat-vFLIP-α2 scrambled peptides. *P < 0.05, **P < 0.01, ***P < 0.001
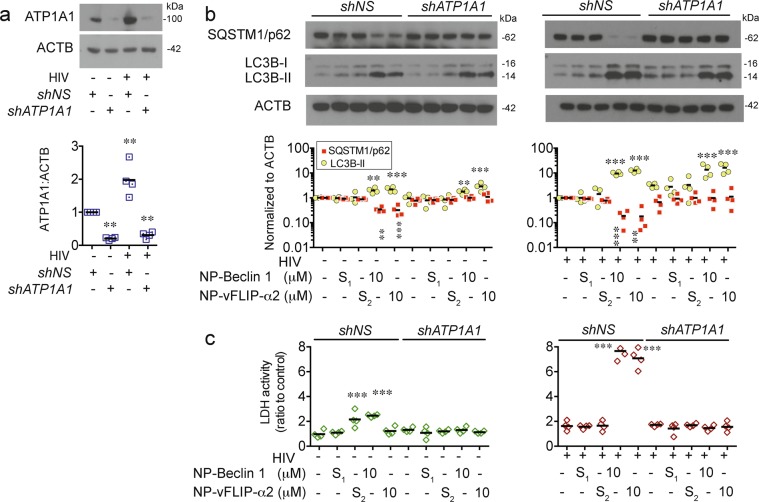
To further establish that induction of the Na+/K+-ATPase is the mechanism of cell death, we pre-treated TCM cultures with digoxin, a known inhibitor of Na+/K+-ATPase, 2 h prior to exposure to either NP-Beclin 1 or NP-vFLIP-α2. After 24 h, digoxin treated cultures demonstrated a marked reduction in cell death (Fig. 5b, c). Digoxin treated cultures also showed a marked reduction in autophagic flux as demonstrated by the absence of SQSTM1/p62 degradation. A similar effect was observed in experiments where ATP1A1 was knocked down using shRNA (Fig. 6). In total, these findings confirm that Na+/K+-ATPase is essential to Tat-Beclin 1 and Tat-vFLIP-α2 induced TCM cell death.
Fig. 6. Knockdown of Na+/K+-ATPase inhibits NP-Beclin 1 and NP-vFLIP-α2 induced killing of latent HIV-TCM cells.
a HIV HIV-TCM cells were transduced with shATP1A1 for knockdown of Na+/K+-ATPase. The knockdown efficiency was evaluated by western blot in cell lysates. b shATP1A1 transduced latent HIV-TCM cells were treated with 10 μM NP-Beclin 1 or 10 μM NP-vFLIP-α2 for an additional 24 h. The effect of shATP1A1 transduction was tested by western blot in cell lysates. c Cytotoxicity was measured by LDH assay. All densitometric analyses are summarized from four different donors and normalized to loading control ACTB with means. NP-S1 = 10 μM nanoformulated Tat-Beclin-1 scrambled peptides, NP-S2 = 10 μM nanoformulated Tat-vFLIP-α2 scrambled peptides. **P < 0.01, ***P < 0.001
HIV infection increases Na+/K+-ATPase in CD4+ T memory cells with acute and latent infection
Recently, cardiac glycosides/aglycones which inhibit Na+/K+-ATPase were identified to inhibit HIV replication39–41. Combined with our findings presented here, we hypothesized that cells latently infected with HIV might have increased Na+/K+-ATPase activity that contributes to the generation of latency. For these experiments, HIV-TCM were established and evaluated for the presence of ATP1A1. Of interest, as the replication of virus in TCM progressed to latency, the amount of ATP1A1 detected steadily increased over 30 days (Fig. 7a). To explore the potential function of Na+/K+-ATPase in generating HIV-TCM, we incubated Na+/K+-ATPase inhibitor digoxin with HIV-infected CD4+ T cells, and maintained digoxin treatment throughout the 30 days as the infection progressed to latency. As the infection progressed, the production of HIV p24 was significantly lower in the digoxin treated cells (Fig. 7b). When treated and untreated cultures reached latency, those cells treated with digoxin had a mean of 0.2 copies of proviral DNA per cells compared to 1.3 of HIV-TCM controls (Fig. 7c).
Fig. 7. HIV infection increases Na+/K+-ATPase in CD4+ resting memory T cells.
a ATP1A1 expression level was measured by western blot of cell lysates of CD4+ T memory cells. b CD4+ T cells were pretreated with 50 nM digoxin for 2 h followed by infection with HIVNL-43 (M.O.I = 0.1). The infected CD4+ T cells were incubated with 50 nM digoxin for 12 days. The cell culture supernatants were tested for HIVp24 by ELISA. c At day 12 p.i, digoxin treated CD4+ T cells under went magnetic negative selection to enrich for central memory CD4+ T cells. The purified CD4+ T memory cells were treated with 50 nM digoxin, 100 nM atazanavir, and 200 nM tenofovir for another 20 days. The harvested cells were measured for integrated HIV DNA using Alu-gag QPCR. All analyses are summarized from four different donors and normalized to loading control ACTB with mean. *P < 0.05, ***P < 0.001
Nanopeptides preferentially kill ex vivo latent CD4+ T cells in patient blood
To confirm that nanopeptides can preferentially kill HIV latently infected cells, we performed ex vivo studies on patient samples. PBMC were obtained from the blood of HIV-infected patients with viral suppression on ART, had undetectable viral loads defined as <20 copies HIV RNA/ml for at least 12 months and a CD4+ count of >400/mm3. Resting CD4+ T cells were isolated from PBMC and treated with nanopeptides for 24 h following the methods previously described by our laboratory34. Following treatment, there was no discernable increase in the number of dead cells in treated versus control cultures. This is not surprising given that latently infected cells comprise <10 per 106 cells in vivo (Fig. 8a, b).
Fig. 8. Nanopeptides reduce HIV latent infection in resting CD4+ T cells from HIV patients ex vivo.
a HIV-infected patients who were receiving suppressive antiretroviral treatment, were virologically suppressed for >12 months (<20 copies HIV RNA/μL) and had >400 CD4+ cell/mL were recruited for blood donation. The purified resting CD4+ T cells were treated with 10 μM NP-Beclin 1 or 10 μM NP-vFLIP-α2 for 24 h, and then activated by PHA and γ-irradiation. Replication competent virus reactivated from CD4+ T cells was measured using a quantitative viral outgrowth assay (QVOA). b The cytotoxicity of NP-Beclin 1 and NP-vFLIP-α2 were tested with trypan blue staining in the purified patient resting CD4+ T cells after 24 h treatment. c The QVOA results were determined by HIVp24 ELISA and analyzed by maximum likelihood statistics. Data are summarized from seven different donors and plotted with means. IUPM = infectious units per million resting CD4+ T cells, NP-S1 = 10 μM nanoformulated Tat-Beclin-1 scrambled peptides, NP-S2 = 10 μM nanoformulated Tat-vFLIP-α2 scrambled peptides. **P < 0.01
To identify the elimination of HIV latently infected cells that produce replication competent virus, the quantitative viral outgrowth assay (QVOA) was used34. Resting CD4+ T cells were isolated and treated with nanopeptides for 24 h, and CD4+ T cells were serially diluted and subjected to a limiting dilution quantitative outgrowth assay. Following a single treatment of NP-Beclin 1 and NP-vFLIP-α2, the amount of replication competent virus was reduced 70.8 and 71.8%, respectively (Fig. 8c), whereas the scrambled control nanopeptides had no effect. Thus, these findings provide further evidence that Tat-Beclin 1 and Tat-vFLIP-α2 have the potential to preferentially eliminate HIV latently infected cells with minimal cytotoxicity to uninfected cells in vivo.
Inhibition of Na+/K+-ATPase prevents nanopeptide induced killing of ex vivo patient HIV latent CD4+ T cells
To determine the mechanism of nanopeptide mediated preferential killing of patient latently infected CD4+ T cells, we tested whether the decline in replication competent virus was dependent on the induction of Na+/K+-ATPase leading to autosis. For these studies, patient resting CD4+ T cells were incubated with digoxin for 2 h followed by treatment with nanopeptides for an additional 24 h at which time replication competent virus was assessed by QVOA. Following digoxin treatment, the anti-HIV and cell killing of latently infected cells of NP-Beclin 1 and NP-vFLIP-α2 was reversed and little difference was found in the quantity of infectious virus between the untreated and digoxin plus nanopeptide treated patient (Supplementary Fig. 3). Thus, these results confirm that our nanopeptides induce a Na+/K+-ATPase dependent cell death that can be reversed by inhibiting the Na+/K+-ATPase.
Discussion
Considerable data support that HIV infection induces apoptosis in activated CD4+ T cells, impairing host immune function42–46. However, in some cells, HIV alters the transcriptional profile to promote the upregulation of anti-apoptotic proteins, leading to prolonged cell survival47–51. Additionally, during permissive infection, HIV Nef binds to Beclin 1, a key protein in autophagy, resulting in the down-regulation of autophagy that can lead to the prevention of autolysosomal degradation of the virus23,52. We have previously demonstrated that treatment of macrophages with Tat-Beclin 1 and Tat-vFLIP-α2 triggers the selective killing of HIV-infected macrophages while sparing uninfected macrophages31. However, a primary site of the HIV reservoir in persons on ART is believed to reside in long-lived, resting memory CD4+ T cells. Thus, in the current work, we have focused our research on this cell population. Our findings demonstrate both in an in vitro model and ex vivo studies that NP-Beclin 1 and NP-vFLIP-α2 can induce the selective killing of latent HIV-infected resting memory CD4+ T cells, while sparing uninfected cells and preventing new infection of bystander cells. Our research has further shown that the preferential killing of cells latently infected with HIV is due at least in part to the increased sensitivity of infected cells to alterations in the Na+/K+-ATPase, driving cells to a specific form of autophagy mediated cell death, autosis.
Numerous HIV cure strategies are currently under investigation. Although transplantation with CCR5Δ32 homozygous hematopoietic stem cells appears to have resulted in an HIV cure for one or two patients53–55, this strategy is impractical and has only been applied to patients with malignancies. Gene therapy approaches, on the other hand, are attempting to mimic this strategy using zinc finger nucleases to target CCR5 and more recently clustered regularly interspaced short palindromic repeats (CRISPR)/Cas-956-61. Other gene editing approaches are attempting to use host restriction factors to engineer cells to resist HIV infection. Additional strategies aimed at reactivating HIV from latently infected cells using latency reversing agents in what has been termed the “shock and kill” approach whereby virus is reactivated and subsequently killed by antiretrovirals, and in some cases, combined with broadly neutralizing antibodies5,62. Numerous other approaches are attempting to modulate the immune response to HIV to control infection without the need for antiretroviral treatment in order to achieve what has been term a “functional cure”. Here, we have developed a novel approach that is capable of preferential killing HIV latently infected cells while sparing uninfected cells. This approach is based on several important principles: (1) Autophagy is essential for the maintenance of cellular homeostasis and persistence of HIV latent reservoirs; (2) Despite being an essential survival mechanism, excessive levels of autophagy are able to induce autophagy dependent cell death; (3) Autosis is an autophagy-dependent non-apoptotic form of cell death and can be triggered by autophagy-inducing peptides through the induction of Na+/K+-ATPase; and (4) The induction of autosis in addition to killing cells with replicating HIV, can kill HIV latently infected cells that are not undergoing viral replication without reactivation of virus.
Resting CD4+ TCM cells, carrying replication-competent latent proviral DNA, are widely considered to remain at a quiescent status over years only to produce HIV upon activation. Although the role of autophagy in the establishment of HIV latent infection in resting CD4+ TCM cells is unknown, many studies have shown that autophagy is essential for generating T memory cells and maintaining its immune function63–65. In memory CD8+ T cells, depletion of ATG5 and ATG7 directly induces cell death and impairs the memory phenotype66. Our previous studies have identified the antiviral effect of autophagy in controlling HIV infection in human primary macrophages and memory CD4+ T cells, through the formation of autophagosome and autolysosome mediated capture and degradation of viral proteins24–26,31,34. In this study, we further identify that autophagy-inducing peptides have the potential to eliminate HIV latent infection in TCM cells using an autophagy-dependent mechanism.
To improve the translational potential of our approach, we have loaded the autophagy-inducing peptides into lipid-coated hybrid PLGA nanoparticles using biocompatible and biodegradable materials. Combining the unique features of liposomes and polymeric nanoparticles, our lipid-coated hybrid PLGA nanoparticles are capable of effective delivery of encapsulated cargos67. In this study, we successfully loaded Tat-Beclin 1 or Tat-vFLIP-α2 into the hybrid nanoparticles resulting in sustained release of autophagy-inducing peptides over 96 h. This lipid hybrid PLGA nanoformulation has been shown to extend the bioavailability of autophagy-inducing peptides in human macrophages for almost one week in our previous study31. Indeed, the hybrid nanoparticles have improved the intracellular bioavailability of our peptides to HIV-infected cells, and transformed the encapsulated peptide drug into biocompatible and translational drug candidate68. Recently, we developed a novel targeted version of the PLGA nanoparticles with surface coating of CD4+ T cell plasma membrane, which broadly recognizes HIV envelope protein, glycosylated protein 120 (gp120)69. This CD4+ T cell plasma membrane coated polymeric nanoparticle has high binding affinity to HIV gp120 leading to robust antiretroviral activity through neutralizing cell free HIV entering into CD4+ T cells and macrophages. This new formulation has the potential to improve the targeted killing of all HIVgp120 positive cells including chronically infected and reactivated latently infected cells.
In summary, we have identified that two nanopeptides, Tat-Beclin 1 and Tat-vFLIP-α2, targeting proteins critical to autophagy induce Na+/K+-ATPase and selectively kill HIV latently infected resting memory CD4+ T cells with little effect on uninfected cells. The preferential killing of latently infected cells is dependent on increased Na+/K+-ATPase as the induction of cell death can be reversed by digoxin, an inhibitor of Na+/K+-ATPase. These peptides when loaded into PLGA nanoparticles can be delivered and incorporated into HIV-infected cells, and are synthesized from FDA-approved material that will facilitate their ability to be used in humans. Thus, we believe that these peptides have great potential to be used as part of an overall strategy designed to eliminate HIV from infected persons.
Materials and methods
Ethics statement
Venous blood was obtained from HIV seronegative and HIV seropositive donors. The protocol was reviewed and approved by the Human Research Protections Program of the University of California, San Diego (Project 09-0660). Written informed consent was obtained from all blood donors.
Preparation of NP-Beclin 1 and NP-vFLIP-α2
Autophagy-inducing peptides including Tat-Beclin 1 (RRRQRRKKRGY-GG-TGFEGDHWIEFTANFVNT), scrambled Tat-Beclin 1 (RRRQRRKKRGY-GG-WETAFGTTEHNIFFDNGV), Tat-vFLIP-α2 (RRRQRRKKRGY-GFVNLLFLVVE) and scrambled Tat-vFLIP-α2 (RRRQRRKKRGY-GFVNLAAAVVE), were synthesized in D-isomer sequence and obtained from New England Peptide, Inc. Autophagy-inducing peptides loaded lipid-coated PLGA nanoparticles were synthesized using single step nanoprecipitation. Poly (D,L-lactic-co-glycolic acid) PLGA (50:50, 0.67 dl/g, Pelham, AL) with ester-terminated and autophagy-inducing peptides were dissolved in organic solvent acetonitrile at 10 mg/ml. Soybean lecithin and DSPE-PEG2000-COOH (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-carboxy (polyenylene glycol)2000) (Alabaster, AL) was mixed in chloroform at 20% of the PLGA polymer weight and air-dried as a lipid film. The dissolved PLGA/autophagy-inducing peptides organic solution was added into the lipid film under gentle stirring and further vortexed for 3 min. PLGA nanoparticles were then washed with 10 mM tris-HCl pH 8 buffer and loaded with autophagy-inducing peptides. The remaining acetonitrile and free polymers were removed by washing using an Amicon Ultra-4 centrifugal filter (Millipore, Billerica, MA) with a molecular weight cut-off of 10 kDa.
Characterization of NP-Beclin 1 and NP-vFLIP-α2
The morphological characteristics of NP-Beclin 1 and NP-vFLIP-α2 were evaluated by JEOL Gatan transmission electron microscopy (TEM) using negative staining. The size, polydispersity index and surface zeta potential were measured by dynamic light scattering Malvern Zetasizer Nano ZS for each of nanoformulations. The peptide loading capacity was evaluated by Slide-A-Yzer MINI dialysis microtube with a molecular weight cutoff of 3.5 kDa (Thermofisher, Rockford, IL), and determined by the weight ratio of the peptide payload to the PLGA nanoparticles.
Generation of HIV latently infected primary resting memory CD4+ T cells
HIV latently infected resting memory T cells were generated following our previously published protocol34. Briefly, resting memory CD4+ T cells from HIV-uninfected donors were suspended with RPMI 1640 medium with 10% fetal bovine serum supplemented with 29 nM CCL19 (PeproTech, Rocky Hill, NJ) for 48 h. Next, the CD4+ T cells were infected with HIVNL4-3 at a multiplicity of infection (M.O.I.) of 0.1, and incubated with 250 ng/mL staphylococcal enterotoxin B (Sigma, St.Louis, MO) and 25 U/mL IL-2 (Roche, Basel, Switzerland) for an additional 3 days. After removal of staphylococcal enterotoxin, cells were cultured with 25 U/mL IL-2 for another 9 days. Then, the central memory CD4+ T cells were purified from HIV-infected CD4+ T cells using negative magnetic isolation, and further cultured with 1 ng/mL IL-7 (PeproTech, Rocky Hill, NJ), 100 nM atazanavir and 200 nM tenofovir (Selleckchem, Houston, TX) for another 20 days.
Quantitative viral out growth assay of HIV latent ex vivo CD4+ T in the isolated patient blood
Our laboratory has adapted the Quantitative viral out growth assay (QVOA) assay previously established by the Siliciano laboratory34,70. HIV-infected patients who were virologically suppressed on ART, had a viral load of <20 copies/mL for at least 12 months and had a CD4+ count of >400 cells/mm3 were recruited and obtained informed consent at University of California San Diego Mother-Child-Adolescent HIV Program clinic, and Peripheral blood mononuclear cells (PBMC) were isolated from collected venous blood using Ficoo-PaqueTM density centrifugation (GE Healthcare, Pittsburgh, PA). Resting CD4+ T cells were purified through negative magnetic cell isolation of human CD25, CD69 and anti-HLA-DR following the manufacturer’s protocols, and reactivated with gamma irradiation (5000R in Cs-source) and phytohemagglutinin (PHA-M, 1 μg/mL). The CD8+ T cell depleted PBMC were reactivated with PHA-M and used as feeder cells to co-culture with limiting dilution of resting CD4+ T cells for 3 weeks. The collected cell cultured supernatants were tested with HIV p24 antigen using by ELISA.
Western blotting
The collected cell lysates were mixed with pierce lane marker reducing sample buffer (ThermoFisher Scientific, Waltham, MA) and boiled for 5 min to achieve protein denaturation. The protein samples were separated by ExpressPlus PAGE Gels and electrophoretic transferred to PVDF or nitrocellulose membrane. The targeted proteins were detected by primary anti-LC3B (Novus Biologicals, Littleton, CO), anti-ATP1A1, anti-ATG5, anti-ATG7 (Cell Signaling Technology, Danvers, MA) and anti-SQSTM1 antibodies (Abcam, Cambridge, MA). The horseradish peroxidase conjugated goat anti-mouse and anti-rabbit secondary antibodies were used to amplify the detected antigen.
Cytotoxicity assay
The collected cell culture supernatants were co-incubated with LDH cytotoxicity assay reaction buffer following the manufacture’s protocol (Clontech, Mountain View, CA). The colorimetric results were quantified using a BioTek microplate reader at 490 nm wavelength. The dead cells were counterstained with 0.4% trypan blue solution and quantified by Nexcelom cell counter.
TZM-bl HIV infectivity assay
TZM-bl HIV infectivity was tested following an established protocol35. Briefly, the collected cell culture supernatants were loaded into 96-well plates containing 50,000 TZM-bl cells per well in RPMI 1640 medium with 10% FBS, and incubated at 37 °C for 48 h. After washing, the incubated TZM-bl cells were measured by Beta-Glo assay kit (Promega, Madison, WI).
Measurement of HIV infection
Genomic DNA from TCM was isolated through the PureLink® Genomic DNA Kits (Invitrogen, Carlsbad, CA). Following the established protocol71–73, integrated viral DNA was measured with Alu-gag QPCR. The isolated genomic DNA from ACH-2 and 8E5 cell lines was used as HIV standards. The collected cell culture supernaturants were tested for HIVp24 ELISA (PerkinElmer, Waltham, MA) and RT activity (ThermoFisher, Carlsbad, CA) following the manufacture protocol.
RNA interference
Short hairpin RNA lentiviral transduction particles kits were purchased from Sigma-Aldrich for silencing ATG5, ATG7 and ATP1A1 in primary central CD4+ T memory cells (ATG5-TRCN0000151963, ATG7-TRCN0000435480 and ATP1A1-TRCN0000424769). The lentiviral shRNA were transduced following the manufacture’s protocols. shRNA control vector was also obtained from Sigma-Aldrich used as non-specific targeting control (SHC002).
Statistics
All results were assessed in GraphPad Prism 6.0 (GraphPad, La Jolla, CA). The normalized data including fold and ratio changes were transformed into log2 value to normalize the data. Two-tailed Student t test, ANOVA, Pearson correlation and Wilcoxon rank test were applied for statistical analysis. P values < 0.05 two-tailed were considered statistically significant.
Supplementary information
Acknowledgements
We thank Erin Maule, Jonathan Hana and Morcel Hamidy for experimental assistance, and Siyu Zhu and Zhe Zhong for assistance with illustration and statistical analysis. This work was supported, in whole or in part, by the National Institute of Neurological Disorders and Stroke of the NIH under Grant R01 NS084912 and R01 NS104015; International Maternal Pediatric Adolescent AIDS Clinical Trials Network. Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials (IMPAACT) Network was provided by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under Grant UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH), National Institute of Allergy and Infectious Diseases (NIAID) [UM1AI068632] and National Institute of Allergy and Infectious Diseases (NIAID) [UM1AI106716].
Authors contributions
G.Z., L.Z., and S.A.S designed and conceived the research. G.Z., B.T.L, X.W., G.R.C., R.H.F. performed the experiments. G.Z., L.Z., and SAS analyzed the data. G.Z., L.Z., and S.A.S. wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by T. Kaufmann
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1661-7).
References
- 1.Yoshimura K. Current status of HIV/AIDS in the ART era. J. Infect. Chemother. 2017;23:12–16. doi: 10.1016/j.jiac.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Rainwater-Lovett K, Luzuriaga K, Persaud D. Very early combination antiretroviral therapy in infants: prospects for cure. Curr. Opin. Hiv Aids. 2015;10:4–11. doi: 10.1097/COH.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun TW, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. Aids. 2010;24:2803–2808. doi: 10.1097/QAD.0b013e328340a239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun TW, Moir S, Kovacs C, Fauci AS. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication Reply. Aids. 2011;25:872–873. doi: 10.1097/QAD.0b013e328344c25a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deeks SG. HIV Shock and kill. Nature. 2012;487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 6.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 7.Murray AJ, Kwon KJ, Farber DL, Siliciano RF. The latent reservoir for HIV-1: how immunologic memory and clonal expansion contribute to HIV-1 persistence. J. Immunol. 2016;197:407–417. doi: 10.4049/jimmunol.1600343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad. Sci. USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi Y, Bowman JW, Jung JU. Autophagy during viral infection—a double-edged sword. Nat. Rev. Microbiol. 2018;16:340–353. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell. Bio. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 12.Button RW, Roberts SL, Willis TL, Hanemann CO, Luo SQ. Accumulation of autophagosomes confers cytotoxicity. J. Biol. Chem. 2017;292:13599–13614. doi: 10.1074/jbc.M117.782276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulda S, Kogel D. Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene. 2015;34:5105–5113. doi: 10.1038/onc.2014.458. [DOI] [PubMed] [Google Scholar]
- 14.Doherty J, Baehrecke EH. Life, death and autophagy. Nat. Cell. Biol. 2018;20:1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nardacci R, et al. Role of autophagy in HIV infection and pathogenesis. J. Intern. Med. 2017;281:422–432. doi: 10.1111/joim.12596. [DOI] [PubMed] [Google Scholar]
- 16.Gomez-Mora E, et al. Brief Report: Impaired CD4 T-Cell Response to Autophagy in Treated HIV-1-Infected Individuals. J. Acquir. Immune Defic. Syndr. 2017;74:201–205. doi: 10.1097/QAI.0000000000001201. [DOI] [PubMed] [Google Scholar]
- 17.Mao K, Klionsky DJ. Xenophagy: a battlefield between host and microbe, and a possible avenue for cancer treatment. Autophagy. 2017;13:223–224. doi: 10.1080/15548627.2016.1267075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wileman T. Autophagy as a defence against intracellular pathogens. Essays Biochem. 2013;55:153–163. doi: 10.1042/bse0550153. [DOI] [PubMed] [Google Scholar]
- 19.Nardacci R, et al. Autophagy plays an important role in the containment of HIV-1 in nonprogressor-infected patients. Autophagy. 2014;10:1167–1178. doi: 10.4161/auto.28678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou DJ, Masliah E, Spector SA. Autophagy is increased in postmortem brains of persons with HIV-1-associated Encephalitis. J. Infect. Dis. 2011;203:1647–1657. doi: 10.1093/infdis/jir163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou DJ, Kang KH, Spector SA. Production of interferon alpha by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J. Infect. Dis. 2012;205:1258–1267. doi: 10.1093/infdis/jis187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou DJ, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. Aids. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell GR, Rawat P, Bruckman RS, Spector SA. Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription Factor EB sequestration. Plos Pathog. 2015;11:e1005018. doi: 10.1371/journal.ppat.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell GR, Spector SA. Vitamin D Inhibits human immunodeficiency virus type 1 and Mycobacterium Tuberculosis infection in macrophages through the induction of autophagy. Plos Pathog. 2012;8:e1005018. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell GR, Spector SA. Hormonally active vitamin D3 (1 alpha,25-Dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection. J. Biol. Chem. 2011;286:18890–18902. doi: 10.1074/jbc.M110.206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell GR, et al. Induction of autophagy by PI3K/MTOR and PI3K/MTOR/BRD4 inhibitors suppresses HIV-1 replication. J. Biol. Chem. 2018;293:5808–5820. doi: 10.1074/jbc.RA118.002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoji-Kawata S, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JS, et al. FLIP-mediated autophagy regulation in cell death control. Nat. Cell. Biol. 2009;11:1355–U1225. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl Acad. Sci. USA. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015;22:367–376. doi: 10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, Luk BT, Hamidy M, Zhang LF, Spector SA. Induction of a Na+/K+-ATPase-dependent form of autophagy triggers preferential cell death of human immunodeficiency virus type-1-infected macrophages. Autophagy. 2018;14:1359–1375. doi: 10.1080/15548627.2018.1476014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soriano-Sarabia N, et al. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J. Virol. 2014;88:14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung JM, Margolis DM. HIV persistence on antiretroviral therapy and barriers to a cure. Adv. Exp. Med. Biol. 2018;1075:165–185. doi: 10.1007/978-981-13-0484-2_7. [DOI] [PubMed] [Google Scholar]
- 34.Campbell GR, Bruckman RS, Chu YL, Trout RN, Spector SA. SMAC mimetics induce autophagy-dependent apoptosis of HIV-1-infected resting memory CD4+T Cells. Cell Host Microbe. 2018;24:689–702.e7. doi: 10.1016/j.chom.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanyal A, et al. Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4(+) T cells. Nat. Med. 2017;23:885–889. doi: 10.1038/nm.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf BB, Schuler M, Echeverri F, Green DR. Caspase-3 is the primary activator of apoptotic DNA fragmentation via DNA fragmentation factor-45/inhibitor of caspase-activated DNase inactivation. J. Biol. Chem. 1999;274:30651–30656. doi: 10.1074/jbc.274.43.30651. [DOI] [PubMed] [Google Scholar]
- 37.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Csh Perspect. Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kheloufi M, Boulanger CM, Codogno P, Rautou PE. Autosis occurs in the liver of patients with severe anorexia nervosa. Hepatology. 2015;62:657–658. doi: 10.1002/hep.27597. [DOI] [PubMed] [Google Scholar]
- 39.Wong RW, Lingwood CA, Ostrowski MA, Cabral T, Cochrane A. Cardiac glycoside/aglycones inhibit HIV-1 gene expression by a mechanism requiring MEK1/2-ERK1/2 signaling. Sci. Rep-Uk. 2018;8:850. doi: 10.1038/s41598-018-19298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laird GM, Eisele EE, Rabi SA, Nikolaeva D, Siliciano RF. A novel cell-based high-throughput screen for inhibitors of HIV-1 gene expression and budding identifies the cardiac glycosides. J. Antimicrob. Chemoth. 2014;69:988–994. doi: 10.1093/jac/dkt471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong RW, Balachandran A, Ostrowski MA, Cochrane A. Digoxin suppresses HIV-1 replication by altering viral RNA processing. PLoS Pathogens. 2013;9:e1003241. doi: 10.1371/journal.ppat.1003241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badley AD, et al. Upregulation of Fas ligand expression by human immunodeficiency virus in human macrophages mediates apoptosis of uninfected T lymphocytes. J. Virol. 1996;70:199–206. doi: 10.1128/jvi.70.1.199-206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyrhol-Riise AM, et al. The Fas/FasL system and T cell apoptosis in HIV-1-infected lymphoid tissue during highly active antiretroviral therapy. Clin. Immunol. 2001;101:169–179. doi: 10.1006/clim.2001.5101. [DOI] [PubMed] [Google Scholar]
- 44.Fevrier M, Dorgham K, Rebollo A. CD4+ T cell depletion in human immunodeficiency virus (HIV) infection: role of apoptosis. Viruses. 2011;3:586–612. doi: 10.3390/v3050586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano A, et al. Dysregulation of apoptosis and autophagy gene expression in peripheral blood mononuclear cells of efficiently treated HIV-infected patients. Aids. 2018;32:1579–1587. doi: 10.1097/QAD.0000000000001851. [DOI] [PubMed] [Google Scholar]
- 46.Gougeon ML, Piacentini M. New insights on the role of apoptosis and autophagy in HIV pathogenesis. Apoptosis. 2009;14:501–508. doi: 10.1007/s10495-009-0314-1. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Huertas MR, et al. The presence of HIV-1 Tat protein second exon delays fas protein-mediated apoptosis in CD4+ T lymphocytes: a potential mechanism for persistent viral production. J. Biol. Chem. 2013;288:7626–7644. doi: 10.1074/jbc.M112.408294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf D, et al. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat. Med. 2001;7:1217–1224. doi: 10.1038/nm1101-1217. [DOI] [PubMed] [Google Scholar]
- 49.Kim Y, Anderson JL, Lewin SR. Getting the “Kill” into “Shock and Kill”: strategies to eliminate latent HIV. Cell Host Microbe. 2018;23:14–26. doi: 10.1016/j.chom.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cummins NW, Badley AD. Anti-apoptotic mechanisms of HIV: lessons and novel approaches to curing HIV. Cell Mol. Life Sci. 2013;70:3355–3363. doi: 10.1007/s00018-012-1239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo HH, et al. Anti-apoptotic protein BIRC5 maintains survival of HIV-1-infected CD4(+) T Cells. Immunity. 2018;48:1183–1194 e1185. doi: 10.1016/j.immuni.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kyei GB, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hutter G. Stem cell transplantation in strategies for curing HIV/AIDS. AIDS Res. The.r. 2016;13:31. doi: 10.1186/s12981-016-0114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiem HP, Jerome KR, Deeks SG, McCune JM. Hematopoietic-stem-cell-based gene therapy for HIV disease. Cell Stem Cell. 2012;10:137–147. doi: 10.1016/j.stem.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta RK, et al. HIV-1 remission following CCR5Delta32/Delta32 haematopoietic stem-cell transplantation. Nature. 2019;568:244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas based antiviral strategies against HIV-1. Virus Res. 2018;244:321–332. doi: 10.1016/j.virusres.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, et al. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 58.Owens B. Zinc-finger nucleases make the cut in HIV. Nat. Rev. Drug Discov. 2014;13:321–322. doi: 10.1038/nrd4316. [DOI] [PubMed] [Google Scholar]
- 59.Manjunath N, Yi G, Dang Y, Shankar P. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5:2748–2766. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu W, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drake MJ, Bates P. Application of gene-editing technologies to HIV-1. Curr. Opin. Hiv Aids. 2015;10:123–127. doi: 10.1097/COH.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Halper-Stromberg A, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murera D, et al. CD4 T cell autophagy is integral to memory maintenance. Sci. Rep.-Uk. 2018;8:5951. doi: 10.1038/s41598-018-23993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Botbol Y, Guerrero-Ros I, Macian F. Key roles of autophagy in regulating T-cell function. Eur. J. Immunol. 2016;46:1326–1334. doi: 10.1002/eji.201545955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weil J, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 2016;17:277–285. doi: 10.1038/ni.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu XJ, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, et al. Self-assembled lipid–polymer hybrid nanoparticles: a robust drug delivery platform. ACS Nano. 2008;2:1696–1702. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu CM, et al. Half-antibody functionalized lipid-polymer hybrid nanoparticles for targeted drug delivery to carcinoembryonic antigen presenting pancreatic cancer cells. Mol. Pharm. 2010;7:914–920. doi: 10.1021/mp900316a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei X, et al. T-Cell-mimicking nanoparticles can neutralize HIV infectivity. Adv. Mater. 2018;30:e1802233. doi: 10.1002/adma.201802233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laird GM, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. Plos Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang G, et al. The mixed lineage kinase-3 inhibitor URMC-099 improves therapeutic outcomes for long-acting antiretroviral therapy. Nanomedicine. 2016;12:109–122. doi: 10.1016/j.nano.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo D, et al. Endosomal trafficking of nanoformulated antiretroviral therapy facilitates drug particle carriage and HIV clearance. J. Virol. 2014;88:9504–9513. doi: 10.1128/JVI.01557-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puligujja P, et al. Pharmacodynamics of long-acting folic acid-receptor targeted ritonavir-boosted atazanavir nanoformulations. Biomaterials. 2015;41:141–150. doi: 10.1016/j.biomaterials.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.