Abstract
Disease incidence is intimately associated with an animal’s commensal bacteria populations (microbiome), as microbes that are involved with morbidity and mortality are commonly found in animals with no sign of disease. An understanding of the animal’s resident respiratory pathogens, in the upper nasal cavity prior to weaning, may help us to understand the impact of these pathogens on incidence of respiratory disease. For this research, the overall goal was to characterize bacterial populations associated with calves at an early age and through time periods prior to weaning in 3 herds at the U.S. Meat Animal Research Center. Nasal swabs from the upper nasal cavity were collected at initial vaccination (approximately 40 d of age), preconditioning (approximately 130 d of age), and weaning (approximately 150 d of age) in 2015 and 2016. DNA was extracted from nasal swabs and combined into 2 pools of 10 animals for each sampling time point, in each herd, for a total of 6 pools at each sampling time point and 18 pools for all sampling time points within each year. To evaluate and compare the microbiome of each pooled sample, hypervariable regions 1 through 3 along the 16S ribosomal RNA (rRNA) gene were amplified by PCR and sequenced using next-generation sequencing (Illumina MiSeq) for identification of the bacterial taxa present. Alpha and beta diversity were also measured. Overall, microbial communities were different between combinations of sampling year, herd location, and sampling time prior to weaning as shown by beta diversity. Analysis of these specific respiratory pathogens prior to weaning will present a clearer picture of the distribution of microbial populations in animals prior to weaning and not exhibiting clinical signs of respiratory disease. Therefore, evaluation of the animal’s resident bacterial populations in the upper nasal cavity during different phases of the beef production system may help us to understand the impact of the microbiome on incidence of respiratory disease in cattle.
Keywords: cattle; microbiome; nasal; respiratory disease, 16S
INTRODUCTION
There is growing recognition that microbial populations (microbiome) associated with animals have profound effects on their physiology, phenotype, and disease incidence. More specifically, bacterial pathogens appear to play an integral role in the overall incidence of bovine respiratory disease (BRD; Holman et al., 2015, 2017; Timsit et al., 2016; Johnston et al., 2017). Bovine respiratory disease is a multifactor disease that commonly occurs in the feedlot when stress levels increase in the animals due to weaning, subsequent transport to the feedlot, and commingling. As a result, BRD is the most expensive animal disease afflicting herds in U.S. beef cattle industry, costing the industry over US$1 billion annually (Griffin et al., 2010), so an understanding of the interaction of the respiratory tract microbiome with potential pathogens at time points prior to development of BRD is crucial.
Animals may be predisposed to develop BRD by a variety of bacterial agents that compose the microbiome of the upper nasal cavity prior to weaning. Recent microbiome research in cattle has primarily focused on the rumen and nonrumen compartments of the digestive tract (McCann et al., 2014; Myer et al., 2015a, 2015b, 2015c; Myer et al., 2016a, 2016b, 2016c; Martinez-Fernandez et al., 2016). Studies focusing on regions of the nasal cavity have also been performed; however, these studies have focused on sampling time points after cattle are weaned and diagnosed with BRD in the feedlot (Holman et al., 2015, 2017; Timsit et al., 2016; Johnston et al., 2017). Therefore, evaluation of the animal’s resident bacterial populations in the upper nasal cavity from early age through weaning in multiple herds will enable study of the variation of the microbial populations over times prior to entry into the feedlot. This study aims to characterize the microbiome of animals from 3 herds born in different locations at U.S. Meat Animal Research Center and sampled at multiple time points prior to entry into the feedlot.
MATERIALS AND METHODS
Animal Populations
Data were collected in 2015 and 2016 in advanced generations of the U.S. Meat Animal Research Center (USMARC) GPE (Germplasm Evaluation Program; Schiermiester et al., 2015) herd, Clay Center, NE. This particular GPE subset of approximately 800 animals each year was a product of multiple-sire matings of crossbred cows to F1 and purebred bulls from various breeds. The resulting animals used within this study consisted of variable fractions of 18 breeds: Angus, Hereford, Red Angus, Brahman, Charolais, Gelbvieh, Limousin, Simmental, Brangus, Beefmaster, Shorthorn, Maine Anjou, Santa Gertrudis, Chiangus, Salers, Braunvieh, South Devon, and Tarentaise. For each year, 3 research herds (approximately 800 animals total) were evaluated that originated and were managed in separate locations (locations 1, 2, and 3) at USMARC. Each location is separated by at least 3 miles and does not intersect with the other locations. Calves were raised under similar management, receiving standardized vaccinations and diets. Calves at any one location never had direct contact with calves at other locations until weaning. Calves were evaluated at the same 3 locations at USMARC in 2015 and 2016. Locations 1, 2, and 3 included 507, 153, and 160 calves, respectively, for 2015 and 376, 256, and 162 calves, respectively, for 2016. All animal use was approved by the U.S. Meat Animal Research Center Animal Care and Use Committee.
Nasal Swabs and DNA Samples
Nasal swabs were collected from the upper nasal cavity of all calves (820 in 2015 and 794 in 2016) using 6-inch nasal swabs at initial vaccination (approximately 40 d of age), preconditioning (approximately 130 d of age), and weaning (approximately 150 d of age; Table 1). For sampling, the nose of the animal was wiped cleaned with a single-use towel if fecal material was present. The unguarded 6-inch nasal swab was then gently inserted into the nasal cavity at an approximate depth of 6 inches. The nasal swab was then rotated and removed. After collection of the sample, all swabs were placed in buffered peptone water with 12% glycerol and stored at −80 °C. Total DNA was extracted from each swab using a commercial kit (PowerSoil DNA kit; Qiagen, Germantown, MD) and initial DNA quantity was evaluated with a DNA spectrophotometer (DeNovix DS-11 FX Series; Wilmington, DE). Equal amounts of DNA from each swab were then pooled within location (1, 2, or 3) and sampling time point after weaning (initial vaccination, preconditioning, or weaning) resulting in 2 pools of 10 different randomly selected animals for each location at each sampling time point. Hence, 180 animals were placed in pools of 10 each year for sequencing over 2 yr for a total of 360 animals contributing nasal swab samples for sequencing. Amplification of the 16S rRNA gene V1 to V3 hypervariable region was then completed for each pool of DNA using standard PCR (AccuPrime, Invitrogen, Carlsbad, CA) and primers with index sequences as previously described that amplify hypervariable regions 1 through 3 of the 16S rRNA gene (Myer et al., 2015a). Quality and quantity of the resulting 16S rRNA gene amplification was checked on the Fragment Analyzer (Advanced Analytical, Ankeny, IA) and then sequenced utilizing the MiSeq Illumina Sequencer (Illumina, San Diego, CA) with a MiSeq Reagent Kit v3 to generate 2 × 300 paired end reads. By using indexed primers to amplify the 16S rRNA gene, pools were combined into a single sequencing run. As samples were collected and processed across 2 yr (2015 and 2016), initially one sequencing run was completed for each year evaluated. Pools that did not pass the initial quality score cutoff of Q20 > 75% for sequence reads were run in a second sequencing run. Approximately 1–1.5 million reads were further evaluated for each pool.
Table 1.
Animal number and age (d) of calves sampled for each year
| Year | Animal number | Sampling time point | ||
|---|---|---|---|---|
| Initial vaccination, d | Preconditioning, d | Weaning, d | ||
| 2015 | 820 | 42.9 | 137.9 | 157.5 |
| 2016 | 794 | 36.6 | 124.2 | 145.3 |
The upper nasal cavity of calves was sampled at initial vaccination, preconditioning, and weaning in 2015 and 2016.
d, days of age.
Data Analysis
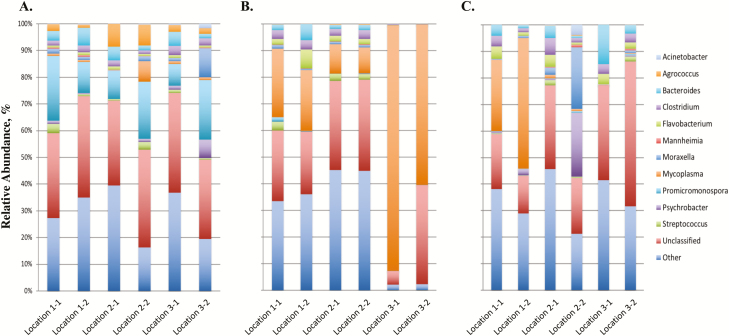
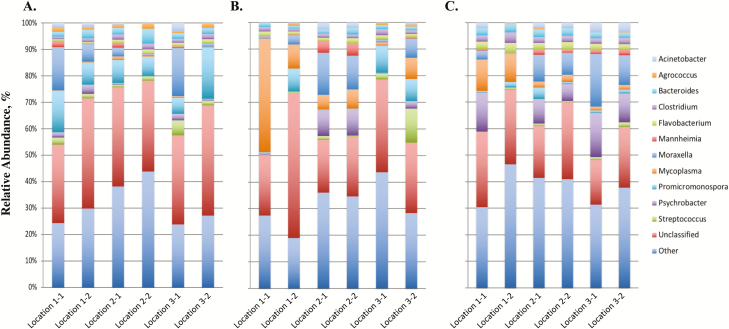
The paired-end data files for each DNA pool were downloaded from the MiSeq Illumina Sequencer and initially processed through Geneious (10.2.3; www.geneious.com). Briefly the paired-end files were transferred to Geneious where paired-end reads were identified, merged, and low-quality (Q20 < 75%) sequence reads were removed. Reads that did not merge were also removed from the data set. Resulting sequence reads were then submitted to the 16S App in Basespace (Illumina, San Diego, CA), which is the cloud computing environment for sequence data. The 16S App pipeline performs taxonomic classification against the GreenGenes database (13.5, May 2013) and uses Naïve Bayesian assignment based on composition similarity (Garcia-Etxebarria et al., 2014). Data were then evaluated for common contaminants (Salter et al., 2014) that may have originated from contaminated reagents or consumables during the DNA extraction. If bacterial genera of common contaminants were identified in the data set, the second swab collected from the animal was extracted for DNA and subsequent 16S rRNA gene amplification. Data are presented as a relative abundance (%) of each bacterial genus in the sample. Overall, approximately 30% of the sequence reads could not be classified to the genus level and are identified as unclassified (Figures 1 and 2). Top 11 bacterial genera in abundance are reported and remaining genera of low abundance are grouped and identified as other (Figures 1 and 2). Information of top 6 phylum and 10 family is also reported (Supplementary Figures 1–4).
Figure 1.
16S profiles of bacterial genera in 2015. 16S profiles (relative abundance %) were evaluated in calves at 3 locations (1, 2, and 3) at USMARC at initial vaccination, preconditioning, and weaning for 2015 (Figure 1A–C, respectively). Two pools of 10 different randomly selected animals were evaluated for each location at each sampling time point (e.g., 1-1 and 1-2 for location 1). Data present the 11 most relatively abundant genera in the upper respiratory tract microbiota.
Figure 2.
16S profiles of bacterial genera in 2016. 16S profiles (relative abundance %) were evaluated in calves at 3 locations (1, 2, and 3) at USMARC at initial vaccination, preconditioning, and weaning for 2016 (Figure 2A–C, respectively). Two pools of 10 different randomly selected animals were evaluated for each location at each sampling time point (e.g., 1-1 and 1-2 for location 1). Data present the 11 most relatively abundant genera in the upper respiratory tract microbiota.
Statistical Methods
Comparison of samples for bacterial pathogens requires a multivariate approach because there are many microbial genera and species and a characterization of the microbiome involves evaluation of multiple genera. Effects tested included location (L; 1, 2 or 3), sampling time (ST; initial vaccination, preconditioning, or weaning), year (Y; 2015 or 2016), and interactions (L × ST, L × Y, ST × Y, and L × ST × Y) using PERMANOVA (Table 2; Tang et al., 2016). Euclidean distance among relative taxa abundance of 36 pooled samples with the PERMANOVA-F test was used, which would be heavily influenced by the most abundant taxa at the genus level. All statistical tests were based on 500,000 permutations. Eight hundred two total bacterial taxa were identified in our nasal swab samples. To evaluate individual bacterial genera for each source and all 2-way and 3-way interactions, correction for multiple testing was done using false discovery rate (FDR; Benjamini and Hochberg, 1995; Supplementary Table 1). Additionally, alpha diversity was estimated using the diversity() function of the Vegan package of R with the index set at “shannon,” “simpson,” and “invsimpson” (Supplementary Table 2) and beta diversity was estimated as the mean of vegdist() of Vegan (Oksanen et al., 2018). Beta diversity using Bray–Curtis was further characterized by year, sampling time, location, and all interactions (2 and 3 way) using Adonis(), which is a package in Vegan (Supplementary Table 3).
Table 2.
Analysis of variance based on PERMANOVA with 500,000 permutations1
| Source | PERMANOVA-F | Subclasses | Distribution under null hypothesis by permutation | P | |
|---|---|---|---|---|---|
| Mean | 95% Quantile | ||||
| Location (L) | 0.111 | 3 | 0.069 | 0.158 | 1.53E-01 |
| Sampling Time (ST) | 0.494 | 3 | 0.069 | 0.158 | 1.80E-05* |
| Year (Y) | 0.144 | 2 | 0.034 | 0.101 | 1.35E-02* |
| L × ST | 0.446 | 9 | 0.189 | 0.376 | 2.34E-02* |
| L × Y | 0.143 | 6 | 0.095 | 0.224 | 1.84E-01 |
| ST × Y | 0.176 | 6 | 0.095 | 0.224 | 1.07E-01 |
| L × ST × Y | 1.296 | 18 | 0.235 | 0.483 | 2.24E-04* |
1The effect of location (L), sampling time (ST), and year (Y) on bacterial profiles in calves from 3 locations at USMARC.
*P value < 0.05.
RESULTS AND DISCUSSION
A comparison of 3 sampling time points (ST; initial vaccination, preconditioning, and weaning) from early age through weaning was completed in calves at 3 locations (L; locations 1, 2, and 3) at the U.S. Meat Animal Research Center in 2015 and 2016 to evaluate the bacterial populations present in the upper nasal cavity at multiple sampling time points (and calf age) prior to entry into the feedlot. Differences among bacterial profiles (expressed as a relative abundance of the bacterial genus in the sample; Figures 1 and 2) due to location (L), year (Y), sampling time (ST), and all 2- and 3-way interactions (L × ST, L × Y, ST × Y, and L × ST × Y) were evaluated by computing the PERMANOVA-F statistic and comparing it with the expected distribution under the null hypothesis of no effect (Table 2). Overall, the 3-way interaction of location, sampling time, and year (L × ST × Y) was significant (P = 2.24 × 10−4), which implies that bacterial profiles differed among L × ST × Y subclasses (L = location 1 vs. location 2 vs. location 3; ST = initial vaccination vs. preconditioning vs. weaning; Y = 2015 vs. 2016). This significance of the 3-way interaction indicates that differences among any one of the main effects (L, ST, or Y) depend on the subclass defined by the other 2 main effects.
Upon evaluation of the main effects, a significant difference of microbial profiles was identified for sampling time (ST) and year (Y) when averaged over L × Y and L × ST subclasses, respectively (Table 2). Conversely, microbial profiles were not different among locations (L) when averaged over ST × Y subclasses. When evaluating specific genera, there were differences in classification to 24 genera for sampling time (ST), 10 genera for year (Y), 8 genera for L × ST, and 98 genera for L × ST × Y (FDR < 0.05; Supplementary Table 1). With a significant effect of sampling time (ST), the predominant bacterial genera at initial vaccination, preconditioning, and weaning were evaluated. This sampling of the population through time allows one to evaluate temporal changes of the upper nasal cavity that have not previously been reported in the literature. Initial vaccination was the first sampling time point evaluated when the calves averaged approximately 40 d of age. For this sampling time point, the Promicromonospora genus was identified as the predominant genus in the core microbiome (Figures 1A and 2A). Furthermore, this predominance of the Promicromonospora genus was present in the upper nasal cavity of calves located at all 3 locations at USMARC and across the 2 yr evaluated, indicating that Promicromonospora is a common bacterial genus of the commensal microbiome at this sampling time point and age of calf. This bacterial genus has been reported previously (Zeineldin et al., 2017) as a pathogen present in nasal samples collected from cattle after weaning in the feedlot. However, the data presented herein demonstrate that this pathogen is also present in calves prior to weaning and as early as 40 d of age. Further evaluation of the Promicromonospora genus in the upper nasal cavity may help elucidate the association of Promicromonospora with respiratory disease of feedlot cattle. Previous research by McMullen et al. (2018) also evaluated the microbial profiles of the upper nasal cavity of calves prior to weaning. For the McMullen et al. (2018) study, nasopharyngeal samples were initially collected from calves at 3 to 7 wk (21–49 d) of age. This is a similar time point to our sample at initial vaccination (approximately 40 d of age). Comparison of McMullen et al. (2018) findings to the data herein shows a similar high abundance of the phylum Actinobacteria (Supplemental Figures 1A and 2A). Additionally, common phyla at the approximately 40-d time point for both studies included Proteobacteria, Tenericutes, and Firmacutes (Supplementary Figures 1A and 2A).
Evaluation of the subsequent sampling time points (preconditioning and weaning) at approximately 130 and 150 d of age identified changes in the microbiome profiles from the initial vaccination sampling time point. This change in the microbiome profiles included an increase in abundance of bacterial genera including Mycoplasma, Psychrobacter, and Moraxella (Figures 1B, C and 2B, C) that have been previously reported to be associated with the upper nasal cavity in cattle diagnosed with BRD in the feedlot after weaning (Holman et al., 2015, 2017; Timsit et al., 2016; Johnston et al., 2017). McMullen et al. (2018) also identified changes in the microbiome profiles of calves from nasopharyngeal samples collected at spring processing (21–49 d of age) to arrival at the feedlot (average 123 d of age). Together, these data suggest that although these bacterial genera are in the microbiome of cattle that are diagnosed with BRD, they are also detected in the commensal microbiome of healthy calves at sampling time points (preconditioning and weaning) prior to entry into the feedlot. However, it is of interest to note that these bacterial genera are not present in high abundance in the microbiome of the upper nasal cavity of calves at the earliest sampling time point evaluated at 40 d of age (initial vaccination). These changes in the microbiome profiles across sampling time points, in addition to the differences between locations, suggest that there is not a common pattern of bacterial pathogens in the upper nasal cavity throughout the time points evaluated. This adds to the complexity of BRD and of identifying the bacterial pathogens associated with cattle that develop BRD. Further research to include more intensive sampling time points and cattle populations from multiple locations and sources is needed.
Contamination throughout sample collection and processing is of concern and has been reported in the literature (Salter et al., 2014). To identify contamination of our samples, sequence data were initially evaluated for known contaminants. We have previously identified known contaminants including the genera Delftia and Ralstonia (Salter et al., 2014) in our sequence data (data not presented) from extracted DNA samples. Upon identification of these contaminants, DNA from a second nasal sample was extracted with new reagents. This typically resulted in the removal of the bacterial contaminants. We have also identified the family Chitninophagaceae in the data presented herein (Supplementary Figures 3 and 4) and similar to Delftia and Ralstonia, this is also a well-known contaminant (Salter et al., 2014). However, we have been unable to remove this contaminant through re-extraction of a second sample; unlike the Delftia and Ralstonia contamination, we do see traditional bacterial pathogens that are associated with nasal sampling such as the genera Mycoplasma, Moraxella, and Pasteurella at a relative abundance greater than 5% with Chitinophagaceae contamination. This suggests that Chitinophagaceae contamination may be occurring prior to the DNA extraction and may occur during the sampling even though we do not observe the contamination in all samples that are collected at a single sampling time point.
When evaluating alpha and beta diversity of the pooled samples, alpha diversity of the taxa within a sample pool of nasal swabs of 10 individuals was much lower for both replicates of location 3 at preconditioning in 2015 compared with all the other 34 pools (Supplementary Table 2). In both pooled samples, Mycoplasma was by far the predominant genus. Coincidently, Chitinophagaceae (classified only to the family level) was the second most prevalent family in these 2 pooled samples though at a much lower level of abundance. Differences in microbial communities across pooled samples (beta diversity) associated with sampling time were significant by the PERMANOVA analysis (Euclidian distance; Supplementary Table 3). Additionally, Adonis analysis using Bray–Curtis demonstrated that all factors (Y, ST, and L) in this study affected beta diversity among the pools (Supplementary Table 3), indicating that microbial communities are different due to those factors and combination of those factors. As a result, microbial communities in a random sample of 10 individuals within a location, sampling time, and year are more similar to a similar sample of 10 individuals in the same situation (combination of location, sampling time, and year) than the communities in pooled samples of 10 animals that are from a different location, sampling time, or year. The spatial and temporal factors the we considered in our study contribute to beta diversity.
For the data presented herein, DNA was extracted from nasal swabs of individual animals randomly selected and 36 pools were constructed with 10 animals per pool with equal contributions of DNA from each animal sample by pipetting volumes inversely proportional to DNA concentrations. Previous sequencing work has shown (data not shown) that at least 99% of the DNA in a nasal swab is of bovine origin. As a result, the 16S rRNA gene is targeted by PCR and amplified to focus on the sequencing of microbes; however, it cannot normalize the sequencing for samples in a pool with differing proportions of microbial and bovine DNA. Hence, pools with low microbial alpha diversity (microbiome dominated by a few species) might indicate that the pool was overwhelmed by microbial DNA from 1 or 2 animals. Conversely, pools with high-microbial alpha diversity might represent equal representation of animals with low alpha diversity or unequal representation of animals with high alpha diversity. Alpha diversity of individual animals is not estimable because of the pooling design. As a result, one might conclude that we cannot evaluate beta diversity by spatial and temporal factors. However, the design of the experiment makes estimation of beta diversity possible. Our experiment included 3 locations x 3 sampling times x 2 yr for a total of 18 cells. Each of the cells includes 2 pools of 10 animals each with no common animals between the 2 pools. The number of microbes contributed by nasal swabs from individual animals is likely unequal between pools within cell (location x sampling time x year subclass); hence, differences between pools would reflect these differences in animal representation. Two analyses were run to compare beta diversity among sampling times, location and year, and interactions with beta diversity between pools within cell. These 2 analyses based on different distance matrices (Euclidean for PERMANOVA and Bray–Curtis for Adonis) consistently demonstrate more beta diversity among these spatial and temporal classifications than within cell. Although interactions (Table 2) indicate that we are currently unable to characterize how microbiome distributions change with location and time, sampling times (initial vaccination, preconditioning, and weaning) are consistently defined from year to year. Therefore, both sampling time and location can be replicated over multiple years, whereas year effects cannot be replicated.
For the calves evaluated in 2015 and 2016, an outbreak of BRD did not occur in the feedlot after weaning. Therefore, we were unable to evaluate the effect of respiratory disease incidence on the microbiome of the calves in our study. Additional populations of calves will be sampled at the time points prior to entry into the feedlot and also sampled after incidence of BRD in the feedlot to provide further data to characterize the microbiome of the upper nasal cavity and if the microbiome is significantly altered upon diagnosis of BRD.
When classifying 16S rRNA gene sequence by taxonomy, approximately 30% of the sequence reads could not be classified from family to the genus level and were subsequently identified as unclassified at genus (Figures 1 and 2). Further evaluation of these unclassified sequences at the family level revealed that 21.4% to 94% of the unclassified reads at the genus level were from the family Chitinophagaceae. Although analysis was able to classify 0.07% to 2.4% of Chitinophagaceae family members to the five genera, which included Flavisolibacter, Niastella, Segetibacter, Chitinophaga, and Niabella, additional analysis will be completed to determine further classification of the Chitinophagaceae family members that did not classify to the genus level. Furthermore, as a high proportion of the unclassified sequences at the genus level were from a single family, this indicates that failure to classify these sequences was not a result of random error in classification but instead suggests that these unclassified sequences are a result of genera that are not yet well classified in the 16S database.
Although previous research has documented the change in bacterial profiles after weaning in beef cattle diagnosed with BRD (Holman et al., 2015, 2017; Timsit et al., 2016; Johnston et al., 2017), the data presented herein document changes in bacterial profiles across multiple sampling time points prior to weaning, in addition to the comparison of bacterial profiles of a large population of calves born and raised in 3 different locations and 2 consecutive years.
Conclusions
Overall, we were able to demonstrate through next-generation sequencing of the 16S rRNA gene that the composition of the core microbiome varies among subclasses for location, sampling time, and year. With the microbial profile in the upper nasal cavity differing with sampling time and location, this indicates that calves from the same sampling time and location appear to have a unique microbiome signature as random pools of calves sampled without replacement at the same sampling time and location were more similar to one another than calves from a different sampling time or location. Through this study, the microbiome of the upper nasal cavity in calves changed from early age through time periods prior to weaning and bacterial genera of the core microbiome for these sampling time points have also been reported in cattle diagnosed with BRD. The data reported herein evaluate sampling a large group of calves through time at multiple locations and across multiple years. Furthermore, evaluation of these changes in the animal’s bacterial populations in the upper nasal cavity prior to weaning will improve our understanding of the impact of the microbiome on incidence of BRD in cattle.
Supplementary Material
ACKNOWLEDGMENTS
We would like to recognize Tammy Sorensen and Sam Nejezchleb for technical assistance.
Footnotes
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA).
USDA is an equal opportunity provider and employer.
LITERATURE CITED
- Benjamini Y., and Hochberg Y.. . 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. of the Royal Stat. Soc. Series B (Methodological). 57(1):289–300. [Google Scholar]
- Garcia-Etxebarria K., Garcia-Garcerà M., and Calafell F.. . 2014. Consistency of metagenomic assignment programs in simulated and real data. BMC Bioinformatics. 1:90. doi:10.1186/1471-2105-15-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D., Chengappa M. M., Kuszak J., and McVey D. S.. . 2010. Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. North Am. Food Anim. Pract. 26:381–394. doi:10.1016/j.cvfa.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Holman D. B., Timsit E., Amat S., Abbott D. W., Buret A. G., and Alexander T. W.. . 2017. The nasopharyngeal microbiota of beef cattle before and after transport to a feedlot. BMC Microbiol. 17:70. doi: 10.1186/s12866-017-0978-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman D. B., McAllister T. A., Topp E., Wright A. D., and Alexander T. W.. . 2015. The nasopharyngeal microbiota of feedlot cattle that develop bovine respiratory disease. Vet. Microbiol. 180(1–2):90–95. doi: 10.1016/j.vetmic.2015.07.031 [DOI] [PubMed] [Google Scholar]
- Johnston D., Earley B., Cormican P., Murray G., Kenny D. A., Waters S. M., McGee M., Kelly A. K., and McCabe M. S.. . 2017. Illumina MiSeq 16S amplicon sequence analysis of bovine respiratory disease associated bacteria in lung and mediastinal lymph node tissue. BMC Vet. Res. 13(1):118. doi: 10.1186/s12917-017-1035-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Fernandez G., Denman S. E., Yang C., Cheung J., Mitsumori M., and McSweeney C. S.. . 2016. Methane inhibition alters the microbial community, hydrogen flow, and fermentation response in the rumen of cattle. Front. Microbiol. 7:1122. doi: 10.3389/fmicb.2016.01122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J. C., Wiley L. M., Forbes T. D., Rouquette F. M. Jr, and Tedeschi L. O.. . 2014. relationship between the rumen microbiome and residual feed intake-efficiency of brahman bulls stocked on bermudagrass pastures. PLoS ONE. 9(3):e91864. doi: 10.1371/journal.pone.0091864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen C., Orsel K., Alexander T. W., van der Meer F., Plastow G., and Timsit E.. . 2018. Evolution of the nasopharyngeal bacterial microbiota of beef calves from spring processing to 40 days after feedlot arrival. Vet. Microbiol. 225:139–148. doi: 10.1016/j.vetmic.2018.09.019 [DOI] [PubMed] [Google Scholar]
- Myer P. R., Wells J. E., Smith T. P., Kuehn L. A., and Freetly H. C.. . 2015b. Microbial community profiles of the colon from steers differing in feed efficiency. Springerplus. 4:454. doi:10.1186/s40064-015-1201-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer P. R., Wells J. E., Smith T. P., Kuehn L. A., and Freetly H. C.. . 2015c. Cecum microbial communities from steers differing in feed efficiency. J. Anim. Sci. 93(11):5327–5340. doi:10.2527/jas.2015-9415 [DOI] [PubMed] [Google Scholar]
- Myer P. R., Wells J. E., Smith T. P., Kuehn L. A., and Freetly H. C.. . 2016c. Microbial community profiles of the jejunum from steers differing in feed efficiency. J. Anim. Sci. 94(1):327–338. doi:10.2527/jas.2015-9839 [DOI] [PubMed] [Google Scholar]
- Myer P. R., Kim M., Freetly H. C., and Smith T. P.. . 2016a. Evaluation of 16S rRNA amplicon sequencing using two next-generation sequencing technologies for phylogenetic analysis of the rumen bacterial community in steers. J. Microbiol. Methods. 127:132–140. doi:10.1016/j.mimet.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Myer P. R., Kim M., Freetly H. C., and Smith T. P.. . 2016b. Metagenomic and near full-length 16S rRNA sequence data in support of the phylogenetic analysis of the rumen bacterial community in steers. Data Brief. 8:1048–1053. doi:10.1016/j.dib.2016.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer P. R., Smith T. P., Wells J. E., Kuehn L. A., and Freetly H. C.. . 2015a. Rumen microbiome from steers differing in feed efficiency. PLoS One.10(6):e0129174. doi:10.1371/journal.pone.0129174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H., Szoecs E. and Wagner H.. . 2018. Vegan: Community Ecology Package; R package version 2.5-2. https://CRAN.R-project.org/package=vegan. [Google Scholar]
- Salter S. J., Cox M. J., Turek E. M., Calus S. T., Cookson W. O., Moffatt M. F., Turner P., Parkhill J., Loman N. J., and Walke A. W.. . 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology. 12:87. doi:10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiermiester L. N., Thallman R. M., Kuehn L. A., Kachman S. D., and Spangler M. L.. . 2015. Estimation of breed-specific heterosis effects for birth, weaning, and yearling weight in cattle. J. Anim. Sci. 93:46–52. doi:10.2527/jas.2014-8493 [DOI] [PubMed] [Google Scholar]
- Tang Z. Z., Chen G., and Alekseyenko A. V.. . 2016. PERMANOVA-S: association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics. 32(17):2618–2625. doi: 10.1093/bioinformatics/btw311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit E., Workentine M., Schryvers A. B., Holman D. B., van der Meer F., and Alexander T. W.. . 2016. Evolution of the nasopharyngeal microbiota of beef cattle from weaning to 40 days after arrival at a feedlot. Vet. Microbiol. 187:75–81. doi:10.1016/j.vetmic.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Zeineldin M. M., Lowe J. F., Grimmer E. D., de Godoy M. R. C., Ghanem M. M., Abd El-Raof Y. M., and Aldridge B. M.. . 2017. Relationship between nasopharyngeal and bronchoalveolar microbial communities in clinically healthy feedlot cattle. BMC Microbiology. 17:138. doi: 10.1186/s12866-017-1042-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.