Abstract
Accurate modeling of structural dynamics of proteins and their differentiation across different species can help us understand generic mechanisms of function shared by family members and the molecular basis of the specificity of individual members. We focused here on the family of lipoxygenases, enzymes that catalyze lipid oxidation, the mammalian and bacterial structures of which have been elucidated. We present a systematic method of approach for characterizing the sequence, structure, dynamics, and allosteric signaling properties of these enzymes using a combination of structure-based models and methods and bioinformatics tools applied to a data set of 88 structures. The analysis elucidates the signature dynamics of the lipoxygenase family and its differentiation among members, as well as key sites that enable its adaptation to specific substrate binding and allosteric activity.
Introduction
Understanding the structural dynamics and allosteric mechanisms of proteins and their differentiation among family members is still a challenge despite its significance for accurate design of proteins with finely tuned activities and for understanding their response to intermolecular or environmental effects. The accumulation of structural data on well-studied proteins now permits us to learn from the evolution of sequence and structure toward gaining insights into key sites and interactions that underlie the stability and function.1−6 Equally important is to assess the molecular mechanisms/dynamics that underlie the adaptability of the same protein to evolving functions. Recent advances in both molecular modeling and bioinformatics tools now offer the possibility of quantitatively characterizing the shared properties of family members as well as member-specific features. The present study aims at introducing such a computational approach and providing insights into the biologically significant family of lipoxygenases (LOXs) – enzymes crucial for catalyzing lipid oxidation, thus regulating a broad range of cellular activities.
LOXs are found in both prokaryotes (e.g., bacteria) and eukaryotes (plants, fungi, and animals). LOXs are involved in formation of lipid mediators - signaling molecules involved in inflammatory cascades in animals, including a variety of eicosanoids (e.g., leukotrienes, hydroxyeicosatetraenoic acid [HETE], and 15-hydroperoxyeicosatetraenoic acid [15-HPETE],7,8 to name a few). In plants, they play a role in the defense system against pests, synthesis of oxylipins, germination, and senescence.9 LOXs are also present in some prokaryotes, although only a few have been biochemically characterized.8 The most common substrates of LOXs are polyunsaturated fatty acids (PUFAs).7,10−12 The specificity of LOX catalytic activity (the position of the oxygenation site in the PUFA) has been an intriguing question for biologists.13 There exist LOXs specific to most of the available oxidizable positions on linoleic acid (LA) and arachidonic acid (AA) - two common substrates of LOXs. LOX family members are named after the PUFA carbon they oxygenate; for example, 12LOX oxygenates AA at carbon 12 (C12), 15LOX at C15, etc. The human genome contains six functional arachidonate LOX (ALOX) genes.14 Two of these encode 15LOX forms, which have been extensively studied due to their involvement in ferroptosis15,16 and aberrant metabolic reactions associated with asthma, brain, kidney, and intestinal injuries.17 ALOX15 encodes 15LO1, which is expressed at high levels in eosinophils, interleukin-4 treated airway epithelial cells, and monocytes;18−20 ALOX15B encodes 15LO2, which is highly expressed in a variety of epithelial cells.18,21
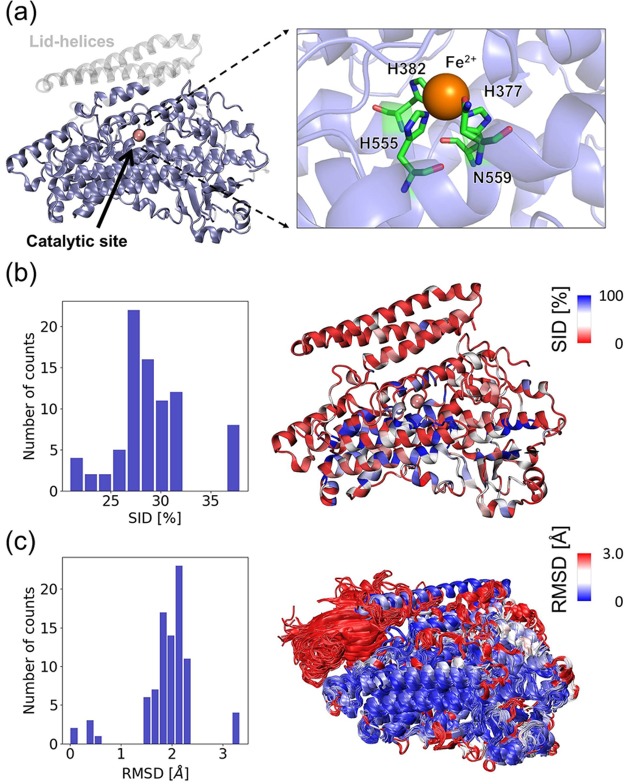
The members of the LOX superfamily share a common structural core irrespective of their origin–bacterial, plant, fungal, invertebrate, or vertebrate. These are single polypeptide chains with a molecular mass of ∼75–80 kDa in animals and ∼94–104 kDa in plants and a highly conserved catalytic center.22−26Figure 1a illustrates the shared structural core and catalytic site in the LOX from Pseudomonas aeruginosa (also called pLoxA),27 which we use as our reference. LOXs have an N-terminal β-barrel domain, also called a PLAT domain that assists in association with the lipid bilayer (except in prokaryotes where it is replaced by the lid helices) and a larger catalytic domain. The catalytic site contains a nonheme iron liganded to at least three conserved histidines and a conserved isoleucine at the C-terminus. The active LOX is in the ferric (Fe3+) form, but the enzymes isolated experimentally tend to be in the inactive, ferrous (Fe2+) form.
Figure 1.
Sequence and structure properties of the lipoxygenase family members. (a) Structural core of LOXs shared by 88 family members colored in ice blue, generated using the pLoxA structure (PDB id: 5ir5) as reference. Transparent gray regions display the lid helices that are not present in all LOXs. The catalytic site is enlarged on the right. (b) Distributions of percent sequence identity (SID) with respect to pLoxA. Six structures (pLoxA mutants) with 99–100% SID are excluded for better visualization. The ribbon diagram on the right is color-coded by the average % SID of each residue in the 88 PDB structures with respect to pLoxA sequence. Residues with high levels of SID (i.e., evolutionarily conserved residues) are in blue, residues with intermediate levels of SID are in white, and residues with low levels of SID are in red. The mean value of SID percentage and its standard deviation is 28.6 ± 3.7. The pink sphere is the iron (Fe2+) ion at the catalytic site. (c) Distribution of RMSDs among LOX structures with respect to pLoxA (left). The mean value is 2.1 ± 0.3 Å. A superposition of the 88 structures (right) is colored as above to highlight regions with structural similarity and variation. Note the largest structural variation at the β-barrel domain on the upper-left part, which is shared by mammalian LOXs but is absent in bacterial LOXs.
We recently reported that phosphatidylethanolamine (PE)-binding protein 1 (PEBP1), a small promiscuous scaffolding protein, allosterically modulates the oxygenase activity of 15LOX by changing its substrate specificity from free PUFA to PUFA esterified in phosphatidylethanolamine PUFA-PE, thus regulating ferroptotic cell death.15,16 Computational modeling revealed that the PEBP1-binding site on 15LO1 includes residues K156, F166, D173, and A179 and the highly conserved R402 (which correspond to P161, F171, I179, V185, and G419 in pLoxA). The interface closely neighbors the catalytic site of 15LOX. Anisotropic Network Model (ANM)28 analysis of the 15LOX-PEBP1 complex revealed the role of PEBP1 in altering the conformational dynamics of 15LOX.15 We also showed that the bacterial lipoxygenase, pLoxA, from Pseudomonas aeruginosa, hijacks this cell-death mechanism to infect human host epithelial cells.29
The biological significance of the above-described interactions and the versatility of LOX family members to bind a range of substrates and perform a diversity of catalytic actions, while sharing the same structure, call for a thorough evaluation of their sequence-structure-dynamics-function relations. While the catalytic regio- and stereospecificities of LOXs are known,13 the structural basis of their mechanochemical activity and the critical sites that control their conformational dynamics and allosteric interactions remain to be elucidated. Here we propose to shed light on the functional dynamics of LOXs as well as the identity of key residues that mediate their mechanics and allostery, by an ensemble analysis of family members combined with sequence analysis. An integrative method of approach that takes advantage of the wealth of sequences and structural data available for LOXs (Table S1) is adopted. As explained below, the method of approach integrates modules existing in the ProDy(30) interface and DynOmics(31) server with newly implemented modules, to enable an automated evaluation of sequence-structure-dynamics features. Specifically, we determine here the shared structural dynamics of the LOX family of enzymes, as well as unique features of selected members; the type and strength of couplings between catalytic sites and functional motifs; and the identities of the residues that act as sensors and effectors of allosteric signals. Our findings help gain a deeper understanding of the molecular basis of recent experimental observations, including the change in the PUFA-oxygenation specificity of LOX elicited by the complexation of LOX with PEBP1.
Figure 1b displays the distributions of percent sequence identity (SID) of the 88 members that will be examined here with respect to the reference (see also the matrix in Figure S1a). We note that the percent SID varies in the range 21 to 38% (excluding a few cases corresponding to mutants), with a mean SID percent of 28.6 ± 3.7 (Figure 1b). Despite this relatively low SID, family members share the same fold, as evidenced by their root-mean-square deviations (RMSDs) that are less than ∼2.3 Å (Figure 1c and Figure S1b), except for manganese LOXs which deviate from others by ∼3.2 Å. Note that the largest structural variation occurs at the β-barrel domain (see the color-coded diagram in Figure 1c, right panel) and at the lid helices (not displayed in Figure 1c). The method of approach proposed here, which will characterize the signature dynamics of the family as well as distinctive features of family members required to achieve their specific catalytic and allosteric activities, is broadly applicable to any protein with sufficient structural and sequence data.
Materials and Methods
Ensemble Analysis of Structurally Characterized LOXs
We analyzed a data set of 88 structures (Table S1) retrieved from the Protein Data Bank (PDB32,33) with the Dali server34 using the bacterial pLoxA crystal structure determined at 1.48 Å resolution (PDB code: 5ir5)27 as query. The sequence and structure distributions of this data set of LOXs are presented in Figure 1 and Figures S1 and S2, and a detailed list is provided in Table S1. The methods utilized here allow for a comparison and classification of the LOXs in this data set based on their (i) sequence, (ii) structure, (iii) dynamics, and (iv) allostery.
An ensemble of superposed structures was created with the Python advanced programming interface ProDy30 using the alignments provided by Dali. This ensemble included only the conserved core residues that could be aligned to the pLoxA structure used as a reference. The ensemble was subjected to principal component analysis (PCA) to determine the principal changes in structure and used for the comparative analyses described below.
The dynamics of individual members was analyzed using a simple elastic network model (ENM), the Gaussian Network Model (GNM),35 complemented by molecular simulations for selected members (see below). We focused on the softest (lowest frequency) modes that are usually relevant to function.36−39 The system-environment framework40−42 was adopted for analyzing collective dynamics of the conserved core (system) in the context of its environment. Here the environment refers to the remaining (noncore) portions of each structure such as extra domains or loops belonging to selected members that are different from the reference. The analysis characterizes the “signature dynamics” of the family, i.e. the average fluctuation profile of residues and their cross-correlations under physiological conditions, as well as the departures of the individual members from the signature behavior, thus permitting us to identify both generic (shared) and specific (divergent) features. To ensure comparison of equivalent modes, we reordered the modes from each structure to match those in the reference structure using the framework of a linear assignment problem (Kuhn 1955). The correlation cosine, ρkl(A, B) = νkA·νl, between each pair of modes k and l belonging to proteins A and B, was used to evaluate the cost of matching them as [1 – ρkl(A, B)], and the set of pairs that minimizes the total cost was selected.
Allosteric properties were deduced from perturbation response scanning (PRS) analysis,43−46 which is an extension of linear response theory.47 PRS allows for the calculation of the response of residue i to perturbation at residue j. The N-dimensional vector ΔRd of node displacements in response to the application of a perturbation (an N-dimensional force vector F) obeys Hooke’s law F = ΓΔRd, where Γ is the N × N Kirchhoff matrix in the GNM theory. The ith element of ΔRd designates displacement of the ith residue away from its equilibrium position in response to the exerted force F. We evaluate the response ΔRd = Γ–1F of all residues to a unit force applied to the j site using the operation, Γ–1Fj, where Fj is composed of all zeros, except for the jth element that is equal to one. Repeating (scanning) this procedure for all sites yields a response matrix, P, the ijth element of which provides a measure of the sensitivity of i to perturbation at j. Elements in each row are normalized with respect to the diagonal term. The averages over the rows and columns of the normalized P yield the sensitivity and effectiveness profiles, with peaks therein designated as sensors and effectors, respectively. We also applied mechanical stiffness (MechStiff) analysis, which calculates the resistance of all residue pairs to uniaxial tension.48,49
The ProDy(30) application programming interface (API) and the DynOmics server(31) were used for ENM-based studies together with in-house codes. The structures and properties were visualized using the molecular graphics software VMD.50
Sequence Analysis of Pfam Data
We also analyzed a larger set of LOX sequences based on a multiple sequence alignment (MSA) of LOXs retrieved from the Pfam database.51 The MSA was refined by removing the outliers (poorly aligned sequences) as well as highly similar sequences that provide redundant data. These filtering criteria led to a final alignment of 218 sequences. We used MUSCLE52 for sequence alignment and sorting, and manual curation, where applicable. We analyzed the sequence conservation properties of the final MSA using the Evol module of ProDy(30) as previously described.29
Molecular Dynamics (MD) Simulations
We performed full-atomic MD simulations of human 15LO1 (PDB code: 1lox) alone and in the presence of PEBP1 (PDB: 1beh) using the NAMD53 MD simulation software with CHARMM2754 force fields. Docking simulations with the Gramm-X55 were used to generate structural models for the PEBP1-15LO1 complex, as reported previously.15 In preparatory simulations with explicit water models (TIP3P), we adopted the following protocol: 0.2 ns of water equilibration, 10,000 steps of minimization, 0.35 ns of heating from 0 to 300 K, and 0.15 ns equilibration of the whole system before initiating the production MD run. Two MD trajectories containing 15LO1 and the 15LO1-PEBP1 complex were generated, each 200 ns duration with 2 fs time step. A cutoff of 12 Å for nonbonded interactions was applied. Langevin dynamics and the Langevin piston algorithm were used to maintain the temperature at 300 K and the pressure at 1 atm. We used VMD50 for visualization and the ProDy30 API for trajectory analysis. PRS analysis was performed as described above with the covariance matrix from the MD simulation substituted for Γ–1.
Generation of Sequence- and Structure-Based Cladograms
We present sequence and structure classifications using as metrics the pairwise Hamming distances for sequences and RMSDs for structures (see Figure S1). Dendrograms are visualized using in-house code to interface ProDy(30) with the iTOL server.56
Results and Discussion
Gaussian Network Model Analysis Reveals the Signature Dynamics of LOX Family Members
Elastic network models (ENMs) serve as efficient tools for characterizing the collective dynamics of proteins, albeit at low resolution.37 Low frequency modes (also called soft or global modes) evaluated using ENMs provide robust information on large-scale (often allosteric) motions uniquely encoded by the architecture, which would otherwise require extensive MD simulations to be visualized.39,57−59 As such, ENMs found wide applications in modeling global dynamics and comparative analyses of ensembles or families of proteins.60−64
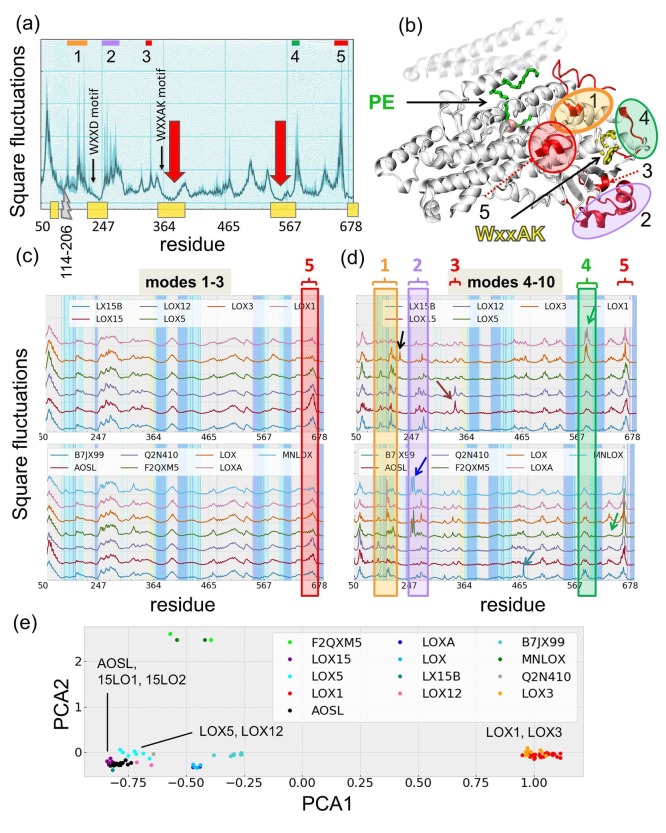
Figure 2 presents the signature dynamics of the LOX family evaluated using the GNM. Panel a shows the mean-square fluctuations (MSFs) of residues based on the first 10 global modes, averaged over the 88 structures in our data set (blue curve) and its standard deviation (light blue band). The curve will be shortly called the signature profile. The catalytic sites (indicated by the red vertical arrows) occupy minima, i.e. they undergo minimal movements if any, in accord with the precise and tight positioning of catalytic residues - a requirement for mechanochemical activity of enzymes.65 Closer examination indeed shows that the three deepest minima in the signature profile are located at (i) the region enclosing the catalytic residues H555 and N559; (ii) the segment W357-A391 that includes both the strictly conserved motif, W357 xxAK361, characteristic of the LOX family,8 and the catalytic H377; and (iii) F228-R248 centered at P242-N243, which includes the conserved motif WXXD (replaced by F228 xxD231 in pLoxA). Finally, we also note the minimum at the C-terminus P676-I685 that includes I685 which coordinates iron at the catalytic site and another at both sides of the helical lid (residues V114-D206) delimiting the core domain. These regions are indicated by the yellow bars along the lower abscissa. Residue numbers refer to those of the reference structure, pLoxA, unless specified otherwise. Corresponding residues in 15LO1 and 15LO2 can be found in Figure S2 and Table S2.
Figure 2.
ENM results for LOX family members.(a) Mean square fluctuation profile (dark line) and standard deviation (lighter bands) for residues as predicted by the GNM softest 10 modes computed for the data set of 88 PDB structures. Residues along the abscissa refer to the pLoxA catalytic domain (residues I50–I685, excluding V114-D206 indicated by the gray breakpoint). Regions labeled 1–5 display the highest differences among family members. Yellow bars along the abscissa highlight functional sites with minimal fluctuations. (b) Regions 1–5 shown on the representative crystal structure (pLoxA, PDB code: 5ir5). Sites 1–5 are centered around D207-R211, I250-E276, A338, S585-S588, and A666-R668. This structure includes a PE lipid (green sticks) bound at the catalytic site and the catalytic iron ion (pink sphere). Yellow sticks represent the WxxAK motif comprising residues W357–K361 in pLoxA. The transparent region represents the lid helices (V114-D206) of pLoxA, which are excluded from the plots as they are not present in all LOXs. (c-d) Soft modes (modes 1–3; c) and low-to-intermediate frequency (LTIF) modes (modes 4–10; d) for different types of LOX family members. Regions labeled 1–5 correspond to those indicated in panels a and b. Results are presented for 13 representative LOXs, which are indicated by their UniProt IDs: two manganese lipoxygenases (F2QXM5, MNLOX), arachidonate 15-lipoxygenase (LOX15, also designated as 15LO1), arachidonate 12-lipoxygenase (LOX12), arachidonate 5-lipoxygenase (LOX5), arachidonate 15-lipoxygenase B (LX15B also designated as 15LO2), allene oxide synthase-lipoxygenase protein (AOSL), 11R-lipoxygenase (Q2N410), seed linoleate 13S-lipoxygenase-1 (LOX1), seed linoleate 9S-lipoxygenase-3 (LOX3), linoleate 9/13-lipoxygenase (LOX), arachidonate 15-lipoxygenase (LOXA), and arachidonate 15-lipoxygenase (B7JX99). (e) Distribution of LOXs in the subspace spanned by the two principal components obtained by PCA of the 88 PDB structures.
Minima in the global modes indicate key sites whose perturbation would impact the function. All these regions are therefore suggested to be implicated in LOX catalytic activity and/or the communication between catalytic residues and others in close proximity, which support the activity, as will be detailed below. We have highlighted in Figure 2c the residues located within 14 Å of the catalytic site by cyan vertical lines and those interacting with PUFA in the pLoxA crystal structure (PDB code: 5ir5)27 by blue shades. Strikingly, both regions coincide with the minima in the signature profile, in support of their significance as finely tuned and/or structurally conserved regions (with minimal fluctuations) that enable binding and catalysis.
The signature profile is closely shared among all LOXs as evidenced by the narrow band, except for a few peak regions (labeled 1–5, indicated by colored bars along the upper abscissa and shown in panel b) where large variations occur. Sites 1, 2, and 4 are highlighted by the respective orange, violet, and green ovals on the pLoxA crystal structure27 in Figure 2b. The peaks in fluctuation profiles usually indicate substrate/ligand recognition sites, and the variations among members in those regions may reflect their substrate specificity.65 Sites 3–5 are located around the WxxAK motif (Figure 2b, yellow sticks). This motif, also noted above, may assist in accommodating different substrates at the catalytic site, assisted by these adaptable sites 3–5 in its close vicinity. Moreover, site 5 is also within 7 Å from the catalytic site (indicated by the nonheme iron shown as a pink sphere). The member-dependent spatial fluctuations at site 5 (highlighted by the red box in Figure 2b) may help customize the precise positions of specific PUFA carbons next to the iron at the catalytic site.
Low-to-Intermediate Frequency Motions Highlight the Differences between Specific Members
We dissected the GNM mobility profiles in two frequency regimes: the low frequency regime represented by modes 1–3 (Figure 2c) and the low-to-intermediate frequency (LTIF) regime represented by modes 4–10 (Figure 2d). Representative members were selected based on the sequence identity (<20%) with respect to the reference (pLoxA) and sequence coverage (>60%) to ensure a diverse set with no significant gaps. As mentioned above, low-frequency modes typically relate to functional (or allosteric) changes in structure, robustly shared among members. The low frequency profile indeed maintains the same generic shape for all members, except for a few regions (e.g., site 5 enclosed in the red box) that exhibit some variations among members.
Differences in mobility profiles, potentially reflecting the specificity of LOXs at their substrate binding sites, can be detected in the low-to-intermediate frequency range (Figure 2d). The definition of these two regimes is indeed based on the observations that the first three modes of LOXs were closely conserved among members, while those in the range 4–10 exhibited member-specific features. We focus in particular on sites 1–4 depicted in Figure 2a and b, also enclosed in color-coded boxes in Figure 2d. Site 1 residues are noted to neighbor conserved residues located within 14 Å of the catalytic site (cyan bars), suggesting that their member-specific fluctuation behavior directly affects the sequentially (or spatially) neighboring sites exhibited in the generic dynamics of the family. In contrast, site 2 (I247-F264 in pLoxA) is not close to the active site. In pLoxA as well as the mammalian LOXs (e.g., LOX15 and LOX15B), this region is composed of two short helices linked by a coiled segment, and it is connected to a three-strand β-sheet. Its solvent exposure, spatial decoupling from the catalytic site, and coiled conformation indicate high flexibility to adapt to substrates without interfering with enzymatic reactivity. Site 3, which also exhibits distinctive peaks at specific LOXs (LOX15B and LOX12), is spatially adjacent to site 2, and their fluctuations are coupled (also confirmed by inter-residue cross-correlation analysis below). Finally, member-specific fluctuations at site 4, indicated by the green box in Figure 2d and green oval in Figure 2b, invite attention to two plant lipoxygenases, LOX1 and LOX3, which share unique features characterized by high fluctuations near A590.
Principal Component Analysis Clusters LOX Structures with Similar GNM Dynamics
The above analysis also shows which LOXs share similar dynamics (Figure 2d), e.g. two strains of pLoxA, linoleate 9/13-lipoxygenase (LOX) and arachidonate 15-lipoxygenase (LOXA). Likewise, the coral LOXs 11R-lipoxygenase from Gersemia Fruticosa (Q2N410) and allene oxide synthase-lipoxygenase (AOSL) from Plexaura homomalla exhibit highly similar GNM profiles. The two manganese lipoxygenases (F2QXM5 and MNLOX) exhibit a distinctive behavior (higher mobility) compared to all other LOXs at two regions, around site 2 (I247-F264) and around H610 (green arrow). The preferred substrate for manganese LOXs from Ascomycete fungi is LA,66 while for the mammalian LOXs and pLoxA it is AA. Overall, evolutionarily close LOXs seem to also share similar structural dynamics. Our analysis includes four types of human AA LOXs: LOX5, LOX12, LOX15 (or 15LO1), and LOX15B (or 15LO2). While their overall dynamics is similar, a slightly different pattern is observed around D207, and seed linoleate LOX3 is distinguished by a peak at this region too (Figure 2d, top panel; black arrow). This region is exposed to the environment, therefore making it more amenable to substrate recognition.
In order to assess to what extent these similarities in the LTIF regime could be traced back to similarities in structures, we performed a PCA of the data set of LOX structures. Interestingly, PCA identified clearly separated clusters (Figure 2e) formed by (i) the two plant LOXs, LOX1 and LOX3; (ii) a cluster of bacterial LOXs, including LOXA from Pseudomonas aeruginosa and B7JX99 from Cyanothece sp. (strain PCC 8801); (iii) human LOX5 and LOX12; (iv) LOX15, LOX15B, and AOSL nearby; and (v) MNLOX and F2QXM5. These clusters are in accord with the similarities/differences observed in Figure 2d, indicating that the dynamic features elucidated by the GNM for the individual members can be traced back to their structural differences.
Shared Family Properties and Their Differentiation Evidenced by Cross-Correlations between Residue Fluctuations
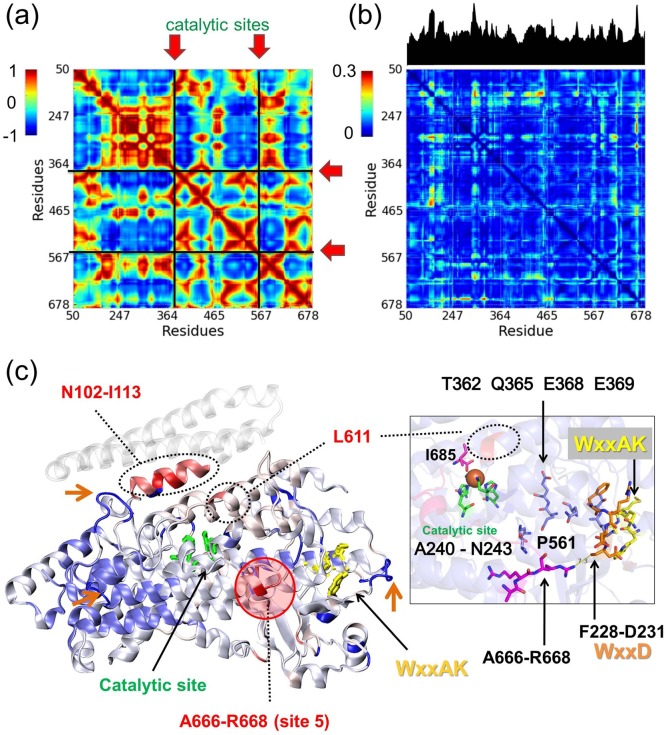
We analyzed the cross-correlations between the spatial fluctuations of residue pairs, i.e. examined whether they tend to move in the same (correlated) or in opposite (anticorrelated) directions with respect to each other in the global modes, or whether they are simply uncorrelated. Figure 3a displays the results averaged over all family members, and Figure 3b shows the standard deviation from the mean behavior at each entry, thus permitting us to assess the generic and specific correlations, respectively. The correlations are normalized with respect to MSFs such that they vary from −1 (fully anticorrelated; dark blue) to 1 (fully correlated; dark red). The dark red blocks refer to groups of residues that undergo highly correlated (same direction) motions, and dark blue blocks represent groups undergoing highly anticorrelated motions. We note that the catalytic sites lie at the interface between anticorrelated regions (indicated by black lines and red arrows in panel a), again a feature typical of the catalytic site of enzymes (e.g., the cleft region between two domains). Also, the highly conserved WxxAK (357–361) and WxxD (228–231) motifs move in concert with each other and are anticorrelated with respect to regions that have been proposed to be on oxygen access pathways, near L367 in 12/15 lipoxygenase67 (L383 in pLoxA, denoted by the red arrow in Figure S3).
Figure 3.
Cross-correlation analysis of LOXs. (a) Mean values and (b) standard deviations for the cross-correlations between residue fluctuations evaluated for the data set of structurally resolved LOXs. Note that catalytic residues are at the crossover regions between anticorrelated blocks of residues, indicated by the black lines and red arrows in panel a. Black bars above the matrix in panel b show the column averages, which indicate the overall differentiation of residues from the generic cross-correlation pattern in a. (c) Location of specific residues of interest in the pLoxA structure (PDB code: 5ir5) based on the black bars in panel b. A high level of differentiation in cross-correlation among LOXs is shown in red, an intermediate level is shown in white, and a low level is shown in blue. Residues in the former group (N102-I113, L611, and site 5 residues A666-R667-R668) are labeled. Orange arrows point to regions that exhibit highly conserved cross-correlations. The inset illustrates the communication path between the catalytic site and the WxxAK motif (yellow sticks) involving the WXXD motif (F228-D231 in pLoxA) and polar/charged residues (T362, Q365, E368, and E36) subject to highly conserved (minimal standard deviation) fluctuations and cross-correlations. Note the close proximity of iron-coordinating I685 to L611.
To visualize the residues that deviate from the generic behavior in Figure 3a, we calculated the average of each column in Figure 3b (black bars above the matrix). The strongest deviations are observed in three regions (circled red regions in Figure 3c): (i) N102-I113, (ii) L611, and (iii) A666-R668. The first region is the PEBP1-LOX binding site region in human LOXs (15LO1 and 15LO2).15 It has been shown that the substrate specificity of 15LO1 (or 15LO2) is changed from AA to esterified AA-PE in the presence of PEBP1,15 while in bacterial pLoxA a function similar to PEBP1 is performed by the two lid helices.29 The second region (L611) interacts directly with the bound substrate in the pLoxA crystal structure and is localized close to the catalytic site. The third region comprising two arginines (R667-R668) was already noted above to exhibit divergent dynamics (site 5 in Figure 2a, b, and d) and suggested to control a gating motion that regulates the access to the catalytic site to accommodate different substrates among LOXs. This region (A666RR668) is in direct interaction with the moderately conserved WxxD motif, which in turn interacts with the highly conserved WxxAK motif (see inset in Figure 3c). We also note the conserved I685 at the catalytic site, which coordinates the nonheme iron. Thus, a dynamically divergent region is juxtaposed next to a dynamically conserved region, suggesting these two regions play a significant functional role, perhaps via an allosteric modulation of the shape or size of the catalytic cavity.
In contrast, three regions exhibit almost invariant cross-correlations (orange arrows in Figure 3c): (i) A240-N243, (ii) P561, and (iii) W357-E369 that also includes the WxxAK motif. All three regions are tightly packed and buried inside the LOX helical domain and are in direct contact with the catalytic site histidines (H377, H382, H555; see Figures 1a and 3c insets). These “conserved” cross-correlations emerge as generic properties of all LOX family members.
Residues Acting As Sensors and Effectors of Allosteric Communication Are Identified by Perturbation-Response Scanning
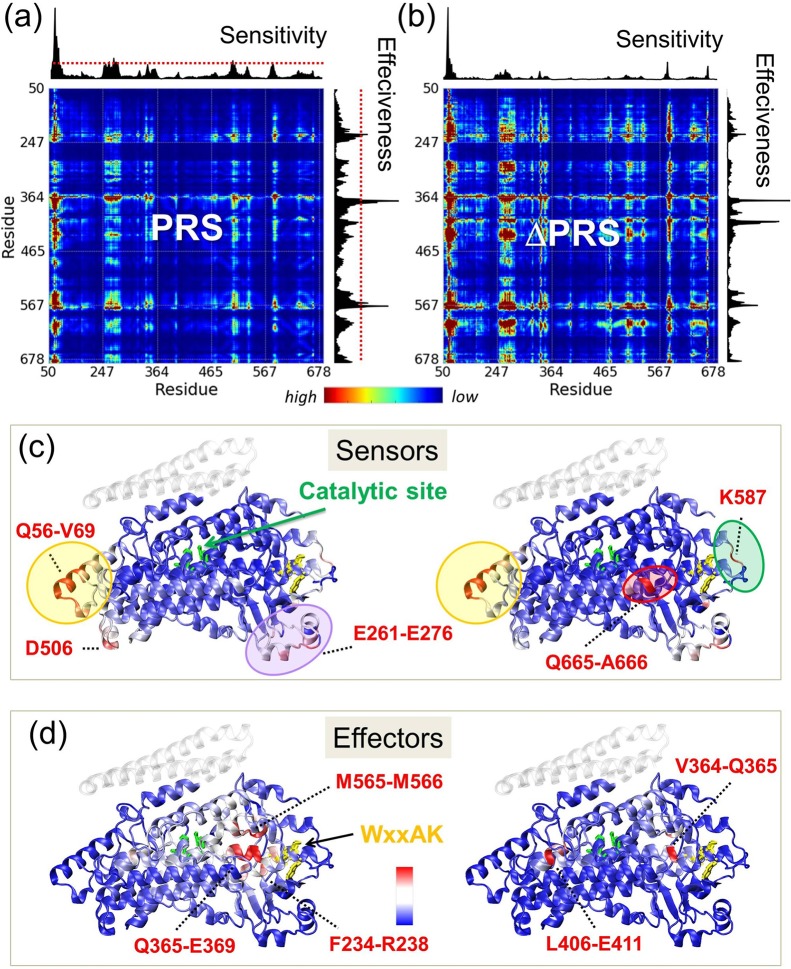
We examined the potential mechanisms of allosteric signaling shared by LOX family members by perturbation response scanning (PRS) analysis. The resulting average PRS heat map and its standard deviation (ΔPRS) are presented in Figure 4a and b. The ijth entry in the PRS map describes the effect of perturbing residue i on the dynamics of residue j. Each row corresponds to the perturbation (random displacement) of a given residue i, and the elements of that row show the response of all residues. Likewise, the jth column represents the response of residue j to the perturbations of every single residue. Large/small effects are shown in red/blue.
Figure 4.
Evaluation of the role of LOX residues as sensors and effectors of allosteric signals and the variations among family members. (a) Average PRS heat map and (b) its standard deviation among LOX family members. The bar plots on the upper abscissa (sensitivity) and right ordinate (effectiveness) describe the propensity of residues to serve as sensors and effectors, respectively. The highest values represent the strongest sensors and effectors, shown in red on the 3D structure of pLoxA in the left parts of panels c and d. Dotted red lines mark the cutoff for defining the strongest sensors and effectors. (c-d) Structures colored by sensitivity (c) and effectiveness (d) propensities. Catalytic residues are shown in green sticks, and the WxxAK motif is shown in yellow sticks. The diagrams on the right in the respective panels c and d highlight the regions where maximal differentiation in the respective sensor and effector properties are observed among LOXs family members.
PRS maps describe the propensity of residues to sense and transmit perturbations and thus elicit cooperative responses, such as an allosteric conformational change induced at site j upon ligand binding to a highly “sensitive” site i.43,44 Residues distinguished in PRS analysis by their ability to sense and to transmit signals have been proposed to serve as sensors and effectors of signals, respectively.44 Along the axes of the heat maps in Figure 4a and b, we display the sensitivity and effectiveness of each residue, averaged over the elements in the corresponding column and row, respectively. Many residues that show high signals in panel a also exhibit peaks in panel b, suggesting that sites distinguished by their strong role in allosteric communication also have member-specific roles.
The locations of the strongest sensors (peaks in the sensitivity plot along the upper abscissa of Figure 4a) on the structure are displayed on the left side of panel c. These include the exposed residues Q56-V69 and D506 and five charged residues E261, E262, D271, D272, and E276 at the above identified site 2 (see Figure 2). Some of these residues are also characterized by the highest variances (peaks in Figure 4b and right ribbon diagram in Figure 4c).
The high propensity to serve as sensor observed at the N-terminal region Q56-V69 prevails equally for both the mean PRS sensitivity profile and its variance profile (Figure 4a-b, highlighted yellow circle). Notably, this particular region, which also appeared as the highest peak in Figure 2a around D58-G62, is involved in the association of the N-terminal β-barrel domain that is present in mammalian LOXs to the catalytic domain (see Figure 1c). In human LOXs, the β-barrel serves as an anchor to the membrane, whereas in bacterial (pathogenic) LOXs such as pLoxA, this β-barrel is absent.29 It is instead replaced by a helical hairpin (called the lid helices and shown in semitransparent gray in Figure 1a). Its ability to serve as sensors and therefore bind substrates is consistent with its role in stabilizing the β-barrel domain in mammalian LOXs. Finally, we also note that site 5 residues Q665-A666 noted earlier (Figure 2c) to be implicated in specific substrate recognition are distinguished by their significant variation in sensitivity (Figure 4c, right, red oval), also consistent with a member-specific substrate-recognition role.
Effectors of allosteric signals typically occupy positions near the active site and potentially form a pathway to the response site. Often times, because of their crucial role in mediating activity, they belong to the protein core which is buried and thus protected from external factors.68 LOX effectors, Q365-E369, M565-M566, and F234-R238, are also localized close to the catalytic site, notably lining the path between catalytic residues and the highly conserved WxxAK motif (Figure 4d, left; see also Figure 3c inset), and some exhibit strong dependencies on the member type (Figure 4d, right). Note that all three groups of residues lie in minima in the signature profile (Figure 2a), i.e. their spatial fluctuations are severely restricted. The latter property presumably underlies their effectiveness as signal propagators/effectors as they themselves resist movement and cause other residues to move instead. The high variance observed for Q365 suggests that the effector role at this site is specific to selected LOXs only.
Complexation with PEBP1 Changes the Sensitivity of LOX without Altering the Signal Propagation Properties of Effector Residues
We recently reported that PEBP1 binding allosterically modulates the activity of 15LO1 by changing its substrate specificity from PUFA to PUFA-PE. In order to examine the effect of PEBP1 binding on 15LO1 allosteric properties, we analyzed the sensor and effector residues in a 15LO1 structure using the PRS method. The results are presented in Figure S4 panels a and b for 15LO1 alone and for its complex with PEBP1, respectively. We note that the residues in the β-barrel domain of this human LOX exhibit a high propensity to serve as sensors (colored red in the middle diagram in Figure S4a). This domain apparently serves as a sensor for membrane binding/localization. The same property is maintained in the complex (red arrow, middle diagram in Figure S4a). Notably, this region has been associated with a potent activator for LOX5.69 Another region, located on the back side of 15LO1 (corresponding to Q431-F435), has been pointed out as a sensor region. Interestingly, this region is located on the protein surface and leads to a buried site that has been linked to allosteric activation in 15LOX.70,71
The heat maps on top of the two panels show little difference, except for the increased sensitivity of 15LO1 residues toward the C-terminus gained upon binding PEBP1 (which can be seen clearly from the comparison of the bar plots along the upper abscissa). In particular, the residues G638-K643 are distinguished by their newly gained sensor role, shown in the middle diagram in Figure S4b. The bottom diagrams show that the enzyme signal propagation properties (mediated by effector residues) remain almost unchanged - apart from a small uniform enhancement in the complex.
This analysis thus reveals that the change in substrate specificity gained upon PEBP1 binding, experimentally detected in earlier studies,15,16 is essentially conferred by a gain of function at the segment G638-K643 near the C-terminus of 15LO1. The sequence analysis presented next will further show the evolutionary significance (strong conservation) of several residues in this segment.
Sequence Analysis Reveals the Conservation or Variation of Amino Acids Consistent with Their Generic or Specific Roles
We next examined to what extent residues identified here to play a key unifying role in LOX dynamics and allostery are sequentially conserved and, conversely, to what extent those responsible for differentiation among members are sequentially variable. The residues identified to be highly constrained in the GNM modes have been shown in previous application to a diverse data set of proteins to be also conserved evolutionarily.72
Figure 5a displays the conservation profile of LOX family members, using the sequence numbering of pLoxA. The highest peaks therein indicate the most conserved residues (colored in Figure S2). Among the peaks we note the residues that coordinate the iron atom (H377, H382, H555, I685, and N559; indicated by red stars), which are absolutely essential for the enzymatic function.7 All those conserved sites exhibit minimal fluctuations in the global modes (indicated by red arrows in Figure 2a), and they were pointed out above to lie in the two deepest minima of the signature profile. In contrast, site 2 residues (E261-E276) distinguished by their large-amplitude member-specific motions (Figure 2a, b and d; violet circle/box) and by their role to serve as sensors for binding substrates (Figure 4c) exhibit low conservation (indicated by the violet box in Figure 5a). The low mobility of conserved residues and high mobility of variable residues are consistent with the concept of coupling between sequence- and structure-variations validated in an earlier study.72
Figure 5.
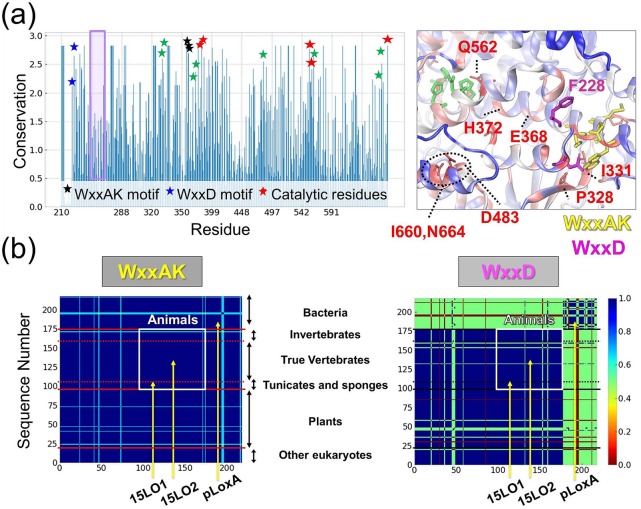
Conservation of characteristic motifs present in LOXs. (a) Conservation propensity of LOX residues. Shannon entropy subtracted from maximum entropy is shown for the LOX catalytic domain (residues G217-Y671 in pLoxA) based on Pfam data. The highest values correspond to the most conserved residues. Stars show the WxxAK motif (black), the WxxD motif (blue), the catalytic residues (red), and the residues I331, P328, I660, N664, E368, H372, and D483 (green) with a relatively high level of conservation, displayed in the inset. The inset in the right panel displays conservation mapped on the 3D structure of pLoxA. Residues with the highest conservation are displayed in red, residues with a moderate level of conservation are displayed in white, and residues with a weak level of conservation are displayed in blue with the exception of the highly conserved WxxAK (yellow sticks) and WxxD (magenta) motifs. (b) Sequence identity matrices corresponding to the WxxAK (left) and WxxD (right) motifs obtained from 218 LOX sequences from all domains of life. High sequence similarity is in blue, intermediate sequence similarity is in yellow/green, and low sequence similarity is in red. The matrix generated for the entire LOX sequence in our previous study29 shows blocks evident for plants, animals, and bacteria that are indicated here. Residues from the WxxAK motif (left panel) are highly conserved in all species, whereas residues from the WxxD motif (right panel) exhibit a moderate level of conservation with species-specific differentiation with bacteria having dissimilar sequences to eukaryotes.
We now turn our attention to the WxxAK motif (W357-K361; black stars in Figure 5a), seen to be conserved across different species (Figure 5b, left). Likewise, the WxxD motif (F228-D231 in pLoxA) localized next to WxxAK in the structure shows a relatively high level of conservation (blue stars), while also exhibiting some species-specific evolutionary trends (Figure 5b, right). Detailed analysis on the 3D structure (Figure 5a, right panel) provides additional insights. We see that the WxxAK motif interacts with I331 and P328, which are also highly conserved (indicated by green stars on the left panel), thus forming a tight network of interactions. It is interesting to note that the residues indicated by the PRS analysis to serve as effectors are adjacent to this conserved network. Among them, E368 and H372 (Figure 5a, green stars) are conserved at a moderate level. We also noticed a very high conservation level of Q562, which is in close vicinity to the catalytic site.
Interestingly, we can see three highly conserved residues, I660, R663 and N664, that are sequentially neighboring site 5 (Figure 2a) and spatially neighboring the conserved D483 (Figure 5a). Likewise, MechStiff analysis demonstrates that the region T659-N664 is distinguished by its high stiffness (or resistance to conformational change; see Figure S5). In contrast, the site 5 residues themselves (A665, R666, and R667) are highly variable consistent with their high fluctuations and variation among family members discussed in Figure 2. Significance of site 5 has been evidenced in the LTIF regime distinctive behavior (Figure 2d), as well as the cross-correlation (Figure 3c) and PRS (Figure 4c, right) analyses. Such juxtaposition of sequentially conserved (and dynamically constrained) and sequentially variable (and dynamically flexible) residues appears to be a design feature to mutually support the respective generic and specific properties of the enzyme. Presumably site 5 serves as a regulator of conformational motions that trigger the opening and closing of the active site to accommodate different substrates among LOXs.
Conclusion
The present study introduces an integrated approach for automated analysis of sequence-structure-dynamics properties of protein family members toward assessing the shared mechanisms of function across family members (termed generic features), as well as the differences between family members (specific features). The approach takes advantage of the wealth of structural data accumulated for well-studied proteins, on the one hand, and recent advances in structure- and sequence-based computational models and methods, on the other. The conservation of internal dynamics among enzyme family members is not a new concept and has been demonstrated in numerous case studies.1−5,62,63,73−75 The approach adopted here allows for utilizing sequence- and structure-based modules in our Python advanced programming interface ProDy,30 together with other well-established software, such as the Dali structural alignment tool34 and the interactive tree of life (iTOL) visualization software,56 in a unified platform to facilitate fully automated data retrieval, output generation, analysis, and visualization.
Application to the family of lipoxygenases, enzymes crucially important for regulating cellular responses upon oxidation or peroxidation of phospholipids and/or PUFAs esterified into lipids, highlighted several key sites and interactions, some consistent with our recent experiments,15,16 others yet to be tested/validated. A list of such residues distinguished by their dynamic and allosteric properties is presented in Table S2 for the bacterial pLoxA, along with their counterparts in the human lipoxygenases 15LO1, 15LO2, and LOX12. Conserved residues among them refer to those supporting family properties, and others are proposed to enable specificity.
First, our analysis of the signature dynamics of the LOX family indicated a series of regions highly constrained in the global motions of the enzyme (minima in Figure 2a) all of which coincide with the active and/or AA-binding sites of the LOXs (indicated by the respective cyan and blue bars in Figure 2c and d). The same regions also include the signature motifs WxxD and WxxAK. The signature profile thus reveals the critical loci, whose perturbations would potentially impact the function, which include evolutionarily conserved residues (Figures S2 and 5). Note that this profile is uniquely defined by the shared architecture of family members, without knowledge/input on substrate-binding or active sites.
Second, the signature profile, especially the modes in the low-to-intermediate frequency (LTIF) regime, reveals certain sites (labeled 1–5) whose conformational dynamics deviate from the average behavior (Figure 2d). These presumably underlie the specificity of family members, also supported by their sequence variations. Among them we note site 5 (A666-R668 in pLoxA) distinguished by its member-specific cross-correlations (Figure 3) and site 2 (E261-E276) exhibiting a high propensity to serve as a sensor of allosteric signaling–typical of substrate recognition sites, confirmed by MD simulations (Figure S4). Our analysis also reveals the sites that are likely to play a key role in transmitting allosteric signals (effectors), such as Q367-E369 (see Figures 4d and 5b).
While the WxxAK motif has been known to be a highly conserved motif in LOXs,7 its function has not been characterized. This motif is connected to at least two catalytic histidines that coordinate the nonheme iron and to the conserved WxxD motif. The latter interacts with site 5 which is likely to mediate specificity. The two conserved motifs (WxxAK and WxxD) along with site 5 and near-neighboring conserved residues (Figures 3c inset and 5a right) may be important in modifying the cavity shape/size allosterically, to facilitate precise positioning of the respective carbon atom of the AA/PUFA whose (per)oxidation is catalyzed by a particular LOX. Another interesting observation is anticorrelated motion observed around the putative oxygen access channel with the conserved blocks including the WxxAK and WxxD motifs. This anticorrelated motion may be facilitating access of oxygen to the catalytic site. The fact that this region shows a conserved mobility profile in all the LOXs, which are all oxygen-dependent for their catalytic function, supports this putative role, which is yet to be experimentally validated.
Notably, the method of approach is applicable to any protein provided that a sufficiently large number of structures, preferably from different domains of life, are available. Sequence data are also needed for evaluating sequence conservation. For example, multiple sequence alignments of 218 bacterial sequences were used in the present study to examine the sequence features of LOXs. Typically, shared biological functions would be associated with conserved sequence, structure, and dynamics features, whereas specificity would be manifested as nonconserved aspects of both structural and dynamics properties. We noted that the differences are mostly observed in the regions surrounding the catalytic site (but not in the immediate vicinity), which are exposed to the environment, or are in the spatial neighborhood of highly conserved motifs. Another design principle emerges from the comparison of the fluctuation behavior of sensors and effectors (Figures 4 and S4). Sensors are usually subject to large fluctuations (being usually on the enzyme surface), and their exposure and high mobility enable their adaptability to binding substrates. In contrast, effectors lie at the minima of the fluctuation profile; their tight interactions on a local scale and/or key mechanical role (as hinge centers) help effectively mediate between anticorrelated regions. We anticipate the current methodology to be of utility for rigorous evaluation of family based mechanisms of motions and their differentiation in general and assist in the design, evaluation, and/or alterations of the specific functionalities of structural homologues.
Acknowledgments
Support from National Institutes of Health awards P41 GM103712, P30 DA035778, and U19 AI068021 is gratefully acknowledged. K.M.R. would like to thank Dr. Lukasz Peplowski from NCU in Poland for his helpful advice on CHARMM force field parameters.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jcim.9b00006.
Figure S1, matrices and dendrograms; Figure S2, sequence alignments; Figure S3, details of generic cross-correlations; Figure S4, PRS results for 15LO1 alone and in complex with PEBP1; Figure S5, generic mechanical stiffness properties; Table S1, 88 lipoxygenases used in this study; and Table S2, key residues dominating dynamics and allostery of LOX family members (PDF)
Author Contributions
¶ K.M.R. and I.S. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Zou T.; Risso V. A.; Gavira J. A.; Sanchez-Ruiz J. M.; Ozkan S. B. Evolution of Conformational Dynamics Determines the Conversion of a Promiscuous Generalist into a Specialist Enzyme. Mol. Biol. Evol. 2015, 32, 132–143. 10.1093/molbev/msu281. [DOI] [PubMed] [Google Scholar]
- Nevin Gerek Z.; Kumar S.; Banu Ozkan S. Structural Dynamics Flexibility Informs Function and Evolution at a Proteome Scale. Evol. Appl. 2013, 6, 423–433. 10.1111/eva.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haliloglu T.; Bahar I. Adaptability of Protein Structures to Enable Functional Interactions and Evolutionary Implications. Curr. Opin. Struct. Biol. 2015, 35, 17–23. 10.1016/j.sbi.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglebakk E.; Tiwari S. P.; Reuter N. Comparing the Intrinsic Dynamics of Multiple Protein Structures Using Elastic Network Models. Biochim. Biophys. Acta, Gen. Subj. 2015, 1850, 911–922. 10.1016/j.bbagen.2014.09.021. [DOI] [PubMed] [Google Scholar]
- Tiwari S. P.; Reuter N. Conservation of Intrinsic Dynamics in Proteins—What Have Computational Models Taught Us?. Curr. Opin. Struct. Biol. 2018, 50, 75–81. 10.1016/j.sbi.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Echave J. Beyond Stability Constraints: A Biophysical Model of Enzyme Evolution with Selection for Stability and Activity. Mol. Biol. Evol. 2019, 399154. 10.1093/molbev/msy244. [DOI] [PubMed] [Google Scholar]
- Brash A. R. Lipoxygenases: Occurrence, Functions, Catalysis, and Acquisition of Substrate. J. Biol. Chem. 1999, 274, 23679–23682. 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- Hansen J.; Garreta A.; Benincasa M.; Fusté M. C.; Busquets M.; Manresa A. Bacterial Lipoxygenases, a New Subfamily of Enzymes? A Phylogenetic Approach. Appl. Microbiol. Biotechnol. 2013, 97, 4737–4747. 10.1007/s00253-013-4887-9. [DOI] [PubMed] [Google Scholar]
- Kusaka T.; Ikeda M. Liquid Chromatography-Mass Spectrometry of Fatty Acids Including Hydroxy and Hydroperoxy Acids as Their 3-Methyl-7-Methoxy-1, 4-Benzoxazin-2-One Derivatives. J. Chrom. 1993, 639, 165–173. 10.1016/0021-9673(93)80251-3. [DOI] [Google Scholar]
- Grechkin A. Recent Developments in Biochemistry of the Plant Lipoxygenase Pathway. Prog. Lipid Res. 1998, 37, 317–352. 10.1016/S0163-7827(98)00014-9. [DOI] [PubMed] [Google Scholar]
- Gerwick W. H. Structure and Biosynthesis of Marine Algal Oxylipins. Biochim. Biophys. Acta, Lipids Lipid Metab. 1994, 1211, 243–255. 10.1016/0005-2760(94)90147-3. [DOI] [PubMed] [Google Scholar]
- Funk C. D. The Molecular Biology of Mammalian Lipoxygenases and the Quest for Eicosanoid Functions Using Lipoxygenase-Deficient Mice. Biochim. Biophys. Acta, Lipids Lipid Metab. 1996, 1304, 65–84. 10.1016/S0005-2760(96)00107-5. [DOI] [PubMed] [Google Scholar]
- Newcomer M. E.; Brash A. R. The Structural Basis for Specificity in Lipoxygenase Catalysis. Protein Sci. 2015, 24, 298–309. 10.1002/pro.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C. D.; Chen X.-S.; Johnson E. N.; Zhao L. Lipoxygenase Genes and Their Targeted Disruption. Prostaglandins Other Lipid Mediators 2002, 68, 303–312. 10.1016/S0090-6980(02)00036-9. [DOI] [PubMed] [Google Scholar]
- Wenzel S. E.; Tyurina Y. Y.; Zhao J.; Croix C. M. S.; Dar H. H.; Mao G.; Tyurin V. A.; Anthonymuthu T. S.; Kapralov A. A.; Amoscato A. A. Pebp1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell 2017, 171, 628–641.e26. 10.1016/j.cell.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthonymuthu T. S.; Kenny E. M.; Shrivastava I.; Tyurina Y. Y.; Hier Z. E.; Ting H.-C.; Dar H. H.; Tyurin V. A.; Nesterova A.; Amoscato A. A.; Mikulska-Ruminska K.; Rosenbaum J. C.; Mao G.; Zhao J.; Conrad M.; Kellum J. A.; Wenzel S. E.; VanDemark A. P.; Bahar I.; Kagan V. E.; Bayir H. l. Empowerment of 15-Lipoxygenase Catalytic Competence in Selective Oxidation of Membrane Ete-Pe to Ferroptotic Death Signals, Hpete-Pe. J. Am. Chem. Soc. 2018, 140, 17835–17839. 10.1021/jacs.8b09913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; O’Donnell V. B.; Balzar S.; Croix C. M. S.; Trudeau J. B.; Wenzel S. E. 15-Lipoxygenase 1 Interacts with Phosphatidylethanolamine-Binding Protein to Regulate Mapk Signaling in Human Airway Epithelial Cells. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 14246–14251. 10.1073/pnas.1018075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. J.; Kuhn H.; Mulkins M.; Highland E.; Sigal E. Specific Inflammatory Cytokines Regulate the Expression of Human Monocyte 15-Lipoxygenase. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 217–221. 10.1073/pnas.89.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal E.; Dicharry S.; Highland E.; Finkbeiner W. E. Cloning of Human Airway 15-Lipoxygenase: Identity to the Reticulocyte Enzyme and Expression in Epithelium. Am. J. Physiol. 1992, 262, L392–L398. 10.1152/ajplung.1992.262.4.L392. [DOI] [PubMed] [Google Scholar]
- Sigal E.; Craik C. S.; Highland E.; Grunberger D.; Costello L. L.; Dixon R. A.; Nadel J. A. Molecular Cloning and Primary Structure of Human 15-Lipoxygenase. Biochem. Biophys. Res. Commun. 1988, 157, 457–464. 10.1016/S0006-291X(88)80271-7. [DOI] [PubMed] [Google Scholar]
- Jisaka M.; Kim R. B.; Boeglin W. E.; Nanney L. B.; Brash A. R. Molecular Cloning and Functional Expression of a Phorbol Ester-Inducible 8s-Lipoxygenase from Mouse Skin. J. Biol. Chem. 1997, 272, 24410–24416. 10.1074/jbc.272.39.24410. [DOI] [PubMed] [Google Scholar]
- Boyington J. C.; Gaffney B. J.; Amzel L. M. The Three-Dimensional Structure of an Arachidonic Acid 15-Lipoxygenase. Science 1993, 260, 1482–1486. 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- Garreta A.; Val-Moraes S. P.; García-Fernández Q.; Busquets M.; Juan C.; Oliver A.; Ortiz A.; Gaffney B. J.; Fita I.; Manresa À. Structure and Interaction with Phospholipids of a Prokaryotic Lipoxygenase from Pseudomonas Aeruginosa. FASEB J. 2013, 27, 4811–4821. 10.1096/fj.13-235952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N. C.; Bartlett S. G.; Waight M. T.; Neau D. B.; Boeglin W. E.; Brash A. R.; Newcomer M. E. The Structure of Human 5-Lipoxygenase. Science 2011, 331, 217–219. 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor S. A.; Villaseñor A.; Fletterick R.; Sigal E.; Browner M. F. The Structure of Mammalian 15-Lipoxygenase Reveals Similarity to the Lipases and the Determinants of Substrate Specificity. Nat. Struct. Biol. 1997, 4, 1003–1009. 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- Neau D. B.; Bender G.; Boeglin W. E.; Bartlett S. G.; Brash A. R.; Newcomer M. E. Crystal Structure of a Lipoxygenase in Complex with Substrate: The Arachidonic Acid Binding Site of 8r-Lipoxygenase. J. Biol. Chem. 2014, 289, 31905–31913. 10.1074/jbc.M114.599662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banthiya S.; Kalms J.; Yoga E. G.; Ivanov I.; Carpena X.; Hamberg M.; Kuhn H.; Scheerer P. Structural and Functional Basis of Phospholipid Oxygenase Activity of Bacterial Lipoxygenase from Pseudomonas Aeruginosa. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2016, 1861, 1681–1692. 10.1016/j.bbalip.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Eyal E.; Lum G.; Bahar I. The Anisotropic Network Model Web Server at 2015 (Anm 2.0). Bioinformatics 2015, 31, 1487–1489. 10.1093/bioinformatics/btu847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar H. H.; Tyurina Y. Y.; Mikulska-Ruminska K.; Shrivastava I.; Ting H.-C.; Tyurin V. A.; Krieger J.; St. Croix C. M.; Watkins S.; Bayir E. Pseudomonas Aeruginosa Utilizes Host Polyunsaturated Phosphatidylethanolamines to Trigger Theft-Ferroptosis in Bronchial Epithelium. J. Clin. Invest. 2018, 128, 4639–4653. 10.1172/JCI99490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan A.; Dutta A.; Mao W.; Liu Y.; Chennubhotla C.; Lezon T. R.; Bahar I. Evol and Prody for Bridging Protein Sequence Evolution and Structural Dynamics. Bioinformatics 2014, 30, 2681–2683. 10.1093/bioinformatics/btu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Chang Y.-Y.; Lee J. Y.; Bahar I.; Yang L.-W. Dynomics: Dynamics of Structural Proteome and Beyond. Nucl. Aci. Res. 2017, 45, W374–W380. 10.1093/nar/gkx385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucl. Aci. Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P. W.; Prlić A.; Altunkaya A.; Bi C.; Bradley A. R.; Christie C. H.; Costanzo L. D.; Duarte J. M.; Dutta S.; Feng Z. The Rcsb Protein Data Bank: Integrative View of Protein, Gene and 3d Structural Information. Nucl. Aci. Res. 2017, 45, D271–D281. 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L.; Rosenström P. Dali Server: Conservation Mapping in 3d. Nucleic Acids Res. 2010, 38, W545–W549. 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Chang Y.-Y.; Yang L.-W.; Bahar I. Ignm 2.0: The Gaussian Network Model Database for Biomolecular Structural Dynamics. Nucleic Acids Res. 2016, 44, D415–D422. 10.1093/nar/gkv1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I. On the Functional Significance of Soft Modes Predicted by Coarse-Grained Models for Membrane Proteins. J. Gen. Physiol. 2010, 135, 563–573. 10.1085/jgp.200910368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I.; Lezon T. R.; Yang L.-W.; Eyal E. Global Dynamics of Proteins: Bridging between Structure and Function. Annu. Rev. Biophys. 2010, 39, 23. 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Blanco J. R.; Chacón P. New Generation of Elastic Network Models. Curr. Opin. Struct. Biol. 2016, 37, 46–53. 10.1016/j.sbi.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Leioatts N.; Romo T. D.; Grossfield A. Elastic Network Models Are Robust to Variations in Formalism. J. Chem. Theory Comput. 2012, 8, 2424–2434. 10.1021/ct3000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsen K.; Petrescu A.-J.; Dellerue S.; Bellissent-Funel M.-C.; Kneller G. R. Harmonicity in Slow Protein Dynamics. Chem. Phys. 2000, 261, 25–37. 10.1016/S0301-0104(00)00222-6. [DOI] [Google Scholar]
- Ming D.; Wall M. E. Allostery in a Coarse-Grained Model of Protein Dynamics. Phys. Rev. Lett. 2005, 95, 198103. 10.1103/PhysRevLett.95.198103. [DOI] [PubMed] [Google Scholar]
- Zheng W.; Brooks B. R. Probing the Local Dynamics of Nucleotide-Binding Pocket Coupled to the Global Dynamics: Myosin Versus Kinesin. Biophys. J. 2005, 89, 167–178. 10.1529/biophysj.105.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilgan C.; Atilgan A. R. Perturbation-Response Scanning Reveals Ligand Entry-Exit Mechanisms of Ferric Binding Protein. PLoS Comput. Biol. 2009, 5, e1000544 10.1371/journal.pcbi.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- General I. J.; Liu Y.; Blackburn M. E.; Mao W.; Gierasch L. M.; Bahar I. Atpase Subdomain Ia Is a Mediator of Interdomain Allostery in Hsp70 Molecular Chaperones. PLoS Comput. Biol. 2014, 10, e1003624 10.1371/journal.pcbi.1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerek Z. N.; Ozkan S. B. Change in Allosteric Network Affects Binding Affinities of Pdz Domains: Analysis through Perturbation Response Scanning. PLoS Comput. Biol. 2011, 7, e1002154 10.1371/journal.pcbi.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilgan C.; Gerek Z.; Ozkan S.; Atilgan A. Manipulation of Conformational Change in Proteins by Single-Residue Perturbations. Biophys. J. 2010, 99, 933–943. 10.1016/j.bpj.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeguchi M.; Ueno J.; Sato M.; Kidera A. Protein Structural Change Upon Ligand Binding: Linear Response Theory. Phys. Rev. Lett. 2005, 94, 078102. 10.1103/PhysRevLett.94.078102. [DOI] [PubMed] [Google Scholar]
- Eyal E.; Bahar I. Toward a Molecular Understanding of the Anisotropic Response of Proteins to External Forces: Insights from Elastic Network Models. Biophys. J. 2008, 94, 3424–3435. 10.1529/biophysj.107.120733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulska-Ruminska K.; Kulik A. J.; Benadiba C.; Bahar I.; Dietler G.; Nowak W. Nanomechanics of Multidomain Neuronal Cell Adhesion Protein Contactin Revealed by Single Molecule Afm and Smd. Sci. Rep. 2017, 7, 8852. 10.1038/s41598-017-09482-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. Vmd: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Finn R. D.; Coggill P.; Eberhardt R. Y.; Eddy S. R.; Mistry J.; Mitchell A. L.; Potter S. C.; Punta M.; Qureshi M.; Sangrador-Vegas A. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016, 44, D279–D285. 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Muscle: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucl. Aci. Res. 2004, 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R.; Kale L.; Schulten K. Scalable Molecular Dynamics with Namd. J. Comput. Chem. 2005, 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell A. D. Jr; Feig M.; Brooks C. L. III Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas Phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J. Comput. Chem. 2004, 25, 1400–1415. 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A.; Vakser I. A. Gramm-X Public Web Server for Protein–Protein Docking. Nucleic Acids Res. 2006, 34, W310–W314. 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I.; Bork P. Interactive Tree of Life (Itol) V3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekpinar M.; Yildirim A. Only a Subset of Normal Modes Is Sufficient to Identify Linear Correlations in Proteins. J. Chem. Inf. Model. 2018, 58, 1947–1961. 10.1021/acs.jcim.8b00486. [DOI] [PubMed] [Google Scholar]
- Mahajan S.; Sanejouand Y. H. Jumping between Protein Conformers Using Normal Modes. J. Comput. Chem. 2017, 38, 1622–1630. 10.1002/jcc.24803. [DOI] [PubMed] [Google Scholar]
- Zheng W.; Brooks B. R.; Thirumalai D. Low-Frequency Normal Modes That Describe Allosteric Transitions in Biological Nanomachines Are Robust to Sequence Variations. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 7664–7669. 10.1073/pnas.0510426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakan A.; Bahar I. The Intrinsic Dynamics of Enzymes Plays a Dominant Role in Determining the Structural Changes Induced Upon Inhibitor Binding. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 14349–14354. 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi F.; Orozco M.; Fanelli F. Deciphering the Deformation Modes Associated with Function Retention and Specialization in Members of the Ras Superfamily. Structure 2010, 18, 402–414. 10.1016/j.str.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Tiwari S. P.; Reuter N. Similarity in Shape Dictates Signature Intrinsic Dynamics Despite No Functional Conservation in Tim Barrel Enzymes. PLoS Comput. Biol. 2016, 12, e1004834 10.1371/journal.pcbi.1004834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zen A.; Carnevale V.; Lesk A. M.; Micheletti C. Correspondences between Low Energy Modes in Enzymes: Dynamics Based Alignment of Enzymatic Functional Families. Protein Sci. 2008, 17, 918–929. 10.1110/ps.073390208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.-T.; del Valle Marcos M. L.; Hwang J.-K.; Echave J. A Mechanistic Stress Model of Protein Evolution Accounts for Site-Specific Evolutionary Rates and Their Relationship with Packing Density and Flexibility. BMC Evol. Biol. 2014, 14, 78. 10.1186/1471-2148-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.-W.; Bahar I. Coupling between Catalytic Site and Collective Dynamics: A Requirement for Mechanochemical Activity of Enzymes. Structure 2005, 13, 893–904. 10.1016/j.str.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Wennman A.; Karkehabadi S.; Engstrom A.; Oliw E. H. Crystal Structure of Linoleate 13r-Manganese Lipoxygenase in Complex with an Adhesion Protein. J. Lipid Res. 2016, 57, 1574–1588. 10.1194/jlr.M069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saam J.; Ivanov I.; Walther M.; Holzhütter H.-G.; Kuhn H. Molecular Dioxygen Enters the Active Site of 12/15-Lipoxygenase Via Dynamic Oxygen Access Channels. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 13319–13324. 10.1073/pnas.0702401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A.; Krieger J.; Lee J. Y.; Garcia-Nafria J.; Greger I. H.; Bahar I. Cooperative Dynamics of Intact Ampa and Nmda Glutamate Receptors: Similarities and Subfamily-Specific Differences. Structure 2015, 23, 1692–1704. 10.1016/j.str.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisastra R.; Kok P. A.; Eleftheriadis N.; Baumgartner M. P.; Camacho C. J.; Haisma H. J.; Dekker F. J. Discovery of a Novel Activator of 5-Lipoxygenase from an Anacardic Acid Derived Compound Collection. Bioorg. Med. Chem. 2013, 21, 7763–7778. 10.1016/j.bmc.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H.; McClendon C. L.; Dai Z.; Li K.; Zhang X.; He S.; Shang E.; Liu Y.; Lai L. Discovery of Novel 15-Lipoxygenase Activators to Shift the Human Arachidonic Acid Metabolic Network toward Inflammation Resolution. J. Med. Chem. 2016, 59, 4202–4209. 10.1021/acs.jmedchem.5b01011. [DOI] [PubMed] [Google Scholar]
- Meng H.; Dai Z.; Zhang W.; Liu Y.; Lai L. Molecular Mechanism of 15-Lipoxygenase Allosteric Activation and Inhibition. Phys. Chem. Chem. Phys. 2018, 20, 14785–14795. 10.1039/C7CP08586A. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Bahar I. Sequence Evolution Correlates with Structural Dynamics. Mol. Biol. Evol. 2012, 29, 2253–2263. 10.1093/molbev/mss097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguid S.; Fernandez-Alberti S.; Echave J. Evolutionary Conservation of Protein Vibrational Dynamics. Gene 2008, 422, 7–13. 10.1016/j.gene.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Micheletti C. Comparing Proteins by Their Internal Dynamics: Exploring Structure–Function Relationships Beyond Static Structural Alignments. Phys. Life Rev. 2013, 10, 1–26. 10.1016/j.plrev.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Marcos E.; Crehuet R.; Bahar I. On the Conservation of the Slow Conformational Dynamics within the Amino Acid Kinase Family: Nagk the Paradigm. PLoS Comput. Biol. 2010, 6, e1000738 10.1371/journal.pcbi.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.