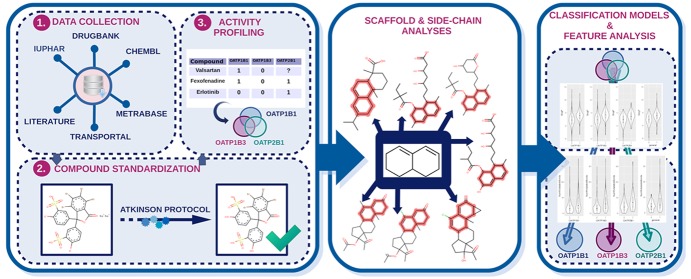
Abstract
Hepatocellular organic anion transporting polypeptides (OATP1B1, OATP1B3, and OATP2B1) are important for proper liver function and the regulation of the drug elimination process. Understanding their roles in different conditions of liver toxicity and cancer requires an in-depth investigation of hepatic OATP–ligand interactions and selectivity. However, such studies are impeded by the lack of crystal structures, the promiscuous nature of these transporters, and the limited availability of reliable bioactivity data, which are spread over different data sources in the open domain. To this end, we integrated ligand bioactivity data for hepatic OATPs from five open data sources (ChEMBL, the UCSF–FDA TransPortal database, DrugBank, Metrabase, and IUPHAR) in a semiautomatic KNIME workflow. Highly curated data sets were analyzed with respect to enriched scaffolds, and their activity profiles and interesting scaffold series providing indication for selective, dual-, or pan-inhibitory activity toward hepatic OATPs could be extracted. In addition, a sequential binary modeling approach revealed common and distinctive ligand features for inhibitory activity toward the individual transporters. The workflows designed for integrating data from open sources, data curation, and subsequent substructure analyses are freely available and fully adaptable. The new data sets for inhibitors and substrates of hepatic OATPs as well as the insights provided by the feature and substructure analyses will guide future structure-based studies on hepatic OATP–ligand interactions and selectivity.
Introduction
Organic anion transporting polypeptides (OATPs) belong to the SLCO (SLC21) superfamily of the solute carrier (SLC) group of membrane transport proteins, which mediate the transport of natural substrates as well as nutrients, clinically relevant drugs, and other xenobiotics across cellular membranes.1 Here we focus on OATP1B1, OATP1B3, and OATP2B1 (encoded by the genes SLCO1B1, SLCO1B3, and SLCO2B1, respectively), all of which are expressed at the basolateral membrane of hepatocytes mediating the uptake of endogenous compounds like bile salts and bilirubin into liver cells. Therefore, hepatocellular OATPs are important for proper liver function and physiological processes like the enterohepatic circulation of bile salts2 and bilirubin metabolism.3
Apart from the endogenous substrates (bile acids, steroid conjugates, hormones, and linear and cyclic peptides), hepatic OATPs accept a broad spectrum of structurally unrelated pharmaceuticals, including antibiotics (e.g., rifampicin, benzylpenicillin, azithromycin, clarithromycin, and erythromycin4), antivirals (e.g., telaprevir5), anticancer drugs (e.g., rapamycin, SN-38, paclitaxel, docetaxel, and imatinib6), antifungals (e.g., caspofungin7), statins (e.g., pravastatin, rosuvastatin, and cerivastatin8), antihistamines (e.g., fexofenadine9), antidiabetics (e.g., repaglinide and rosiglitazone10), cardiac glycosides (e.g., digoxin11), and anti-inflammatory drugs (e.g., diclofenac, ibuprofen, and lumiracoxib12). Importantly, impairment of the hepatic OATPs has been found to alter the pharmacokinetic profiles of various compounds and drugs, which can lead to drug–drug interactions and consequently adverse drug reactions and liver toxicity.13
The substrate and inhibitor profiles of the three hepatic OATPs are partly overlapping, and some selective substrates and inhibitors are known (e.g., pravastatin for OATP1B1 and erlotinib for OATP2B1). Whereas hepatocytes are the exclusive location for the expression of OATP1B1 and OATP1B3, OATP2B1 is additionally expressed, e.g., in the intestine, the mammary gland, and the placenta and at the blood–brain barrier.14 Also, by sequence OATP2B1 is less related to the hepatic members of the OATP1 family (approximately 30%), and knowledge about this transporter is the least among the three in terms of available ligand data and biochemical studies. As our knowledge about all three hepatic OATPs is increasing, we will learn more about their interplay with respect to the delivery and disposition of endogenous substances and drugs. These efforts are impeded by the lack of crystal or NMR structures of any member of the OATP family to be used as templates for structure-based modeling as well as the limited availability of high-quality bioactivity data, which are spread over different data sources in the public domain. Furthermore, the promiscuous nature of hepatic OATPs turns modeling efforts into even more challenging tasks.
Several ligand-based computational studies have been performed to predict hepatocellular OATP–ligand interactions, with a predominance of studies focusing on inhibitors of the structurally more closely related transporters OATP1B1 and OATP1B3 (approximately 80% sequence identity). For example, de Bruyn et al.15 carried out in vitro high-throughput screening of almost 2000 potential molecules against OATP1B1 and OATP1B3, which identified 212 inhibitors for OATP1B1 and 139 inhibitors for OATP1B3. Subsequently, proteochemometric modeling for predicting OATP1B1/1B3 inhibitors was applied. In other studies, Bayesian models for OATP1B1 and its mutated form OATP1B1*15 were employed for inhibitor prediction,16 and Kotsampasakou et al.17 used six in silico consensus classification models to predict OATP1B1 and OATP1B3 inhibition. With respect to OATP2B1, only very few computational studies are available to date, likely because of the shortage of available data for this member of the hepatic OATPs. Just recently, Giacomini and co-workers addressed this shortcoming by combining biochemical studies with in silico ligand-based and structure-based approaches for the identification of novel OATP2B1 inhibitors.18
To the best of our knowledge, only one study is available comparing the inhibitory activity profiles of 225 compounds on these three hepatocellular OATPs. In that study 27, 9, and 3 specific inhibitors of OATP1B1 (e.g., amprenavir, indomethacin, rosiglitazone, and spironolactone), OATP2B1 (e.g., erlotinib, astemizole, piroxicam, and valproic acid), and OATP1B3 (Hoechst 33342, mitoxantrone, and vincristine), respectively, were identified.19
In the present work, we expanded on the investigations by Karlgren et al.,19 including in our study different aspects related to the chemical structures of the ligands contributing to hepatic OATP–ligand interactions or selectivity. Since the major aim of this study was to perform an in-depth investigation of ligand availability, ligand profiles, and ligand properties across the three related transporters, we started our analysis with an extensive data curation exercise by integrating ligand data from various open data sources via semiautomatic KNIME20 workflows. By fusing ligand bioactivity data from five different databases (ChEMBL,21 the UCSF–FDA TransPortal database,22 DrugBank,23 Metrabase,24 and IUPHAR25), we could increase the size of the data sets, their coverage of chemical space, and the confidence in the data quality by considering data from multiple independent bioactivity measurements. In order to retrieve reliable annotations for activity and selectivity, we filtered out ambiguous compounds from multiple independent measurements. In order to be able to systematically annotate a compound as either an inhibitor or noninhibitor or as a substrate or nonsubstrate, we considered the different bioactivity end points as well as different activity annotations or activity comments available in the respective databases. As a result, a total of six high-quality data sets including selective, dual-selective, and pan-interacting ligands for OATP1B1, OATP1B3, and OATP2B1 were retrieved, treating inhibitors and substrates separately.
As we were interested in the structural determinants of ligand selectivity, scaffold decomposition was applied, and frequently occurring scaffolds per transporter were inspected further. Here the focus was on the extraction of frameworks with a higher prevalence for just one or two of the three transporters. Scaffold series of this kind will be important candidates for future detailed structure–activity relationship (SAR) studies (including, e.g., molecular docking). We also looked for pan-interacting scaffolds (e.g., the steroidal scaffold and its conjugates derived from natural substrates). These interesting cases can provide information on the influence of side chains in conferring selectivity switches.
Finally, binary classification modeling by using hierarchical levels for compound classification (sequential binary classification models) revealed important descriptors that might trigger ligand activity or selectivity.
Here, we present an integrative, semiautomatic data mining approach that combines data from various open data sources, preprocesses and curates the data, and analyzes the chemical compounds with respect to chemical features related to transporter selectivity.
The novel high-quality data sets for OATP1B1, OATP1B3, and OATP2B1 for (non)inhibitors and (non)substrates are provided in the Supporting Information, and the data mining workflows (which can be reused for ligand profiling on other related targets of interest) are described. Insights provided by the scaffold and substructure analyses as well as the binary classification modeling will be helpful for subsequent ligand- and structure-based in silico and in vitro studies investigating novel tool compounds for hepatic OATPs.
Materials and Methods
Fetching Data from Different Sources
KNIME Analytics Platform20 (version 3.4) is an open-source solution for the automatization of data integration and analysis that is extensively used in the field of chemoinformatics. Here we created (semi)automatic KNIME workflows for integrative data mining from the open domain.
Bioactivity measurements and/or annotations (substrate, nonsubstrate, inhibitor, noninhibitor) were fetched from five different sources: ChEMBL,21 the UCSF–FDA TransPortal database,22 DrugBank,23 Metrabase,24 and IUPHAR.25 In addition, three novel OATP2B1 (non)inhibitors from Khuri et al.18 as well as ten novel OATP1B1 and OATP1B3 (non)inhibitors from Kotsampasakou et al.17 were manually added to the data set.
Ligands from ChEMBL23 were collected via RESTful web services by providing UniProt protein accession numbers for OATP1B1 [Q9Y6L6], OATP1B3 [Q9NPD5], and OATP2B1 [O94956] to the “ChEMBLdb Connector” node. Data sets retrieved from the UCSF–FDA TransPortal do not contain any type of structural format. Therefore, an automated “name-to-structure” mapping workflow was created to retrieve InChIKeys according to generic names using PubChem’s (https://pubchem.ncbi.nlm.nih.gov) PUG REST services. URL links for retrieving compound identifiers (CIDs) from PubChem were created by inserting the compound names as variables. Records with CIDs were downloaded in XML file format by the “GET Request” node, and the CIDs were extracted (“XPath”). In the case of multiple CIDs for a single entity, only the first one was retained. Unmapped compounds were curated manually. Furthermore, InChIKeys for the respective CIDs were retrieved (“GET Request” node) in XML format and further extracted via an “XPath” query. The quality of the bioactivity measurements from ChEMBL was also assessed by the confidence score. This parameter is included in all CHEMBL entries and evaluates the assay-to-target relationships, ranging from 0 (i.e., so-far uncurated entries) to 9 (i.e., high confidence level of the data). The curated CHEMBL data in our data set have high confidence scores of 9 (898 bioactivities) or 8 (2487 bioactivities), which is a positive indicator of the quality of our curated data sets.
Data from DrugBank and IUPHAR were fetched from the UniProt webpage by downloading the respective XML (DrugBank) and JSON (IUPHAR) files for human OATP1B1, OATP1B3, and OATP2B1. Compound identifiers, compound names, and standard InChIKeys were further extracted via the “XPath” or “JSON Path” node. Metrabase data were fetched from its website using the “HttpRetriever” and “HtmlParser” nodes. The HTML document was processed via an “XPath” query to retrieve the compound names and the associated activity values. InChIKeys for Metrabase compounds were retrieved from PubChem using the same procedure as for UCSF–FDA TransPortal data.
Data Preprocessing and Curation and Assignment of Binary Activity Labels
For each data source, the ligand data were split into two different tables to treat the substrates and inhibitors separately. First, assignment was done on the basis of the “Activity annotation” (substrate, nonsubstrate, inhibitor, or noninhibitor), if available. If the manual activity annotation was not available, the “bioactivity_type” was used as a criterion for classification as either a substrate or inhibitor. For substrates, data entries with either Km or EC50 end points were considered. For inhibitors, data entries with Ki, IC50, and/or percentage inhibition were considered. Potential data errors (activity values greater than 108) were removed, as were data points with missing activity values.
For all end points except percentage inhibition, activity units other than nanomolar (e.g., micromolar) were converted into nanomolar units and further into their negative logarithmic molar values (−logActivity [molar]). The distribution of bioactivity measurements for each transporter was analyzed systematically in order to be able to rationally select a good cutoff for the separation of actives from inactives. A compound was defined as active if the bioactivity was <10 μM and inactive if the bioactivity was greater than or equal to 10 μM. Data with percentage inhibition values were inspected further since we noted that some of them were rather measurements of uptake stimulation. Data with such inverse expression of the inhibitory effect (i.e., “% of control”) were converted into direct inhibition values (100 – [% of control]). Values greater than 100% were interpreted as 100%.
Classification of percentage inhibition data into actives and inactives was done on the basis of recommended thresholds that were manually extracted from primary literature sources (detailed information is available in Tables S1 and S2). If no threshold was recommended but in one of the other sources the same compound concentration was used, the threshold was adopted accordingly. If such information was not available, the data point was removed from the data set.
Percentage inhibition data with negative values (interpreted as “stimulators of uptake”) were filtered out of the data set. Retrieved chemical compounds were further standardized via the Atkinson standardization protocol (available at https://wwwdev.ebi.ac.uk/chembl/extra/francis/standardiser/). This procedure includes breakage of covalent bonds between oxygen/nitrogen atoms and metal atoms, charge neutralization, application of structure normalization rules (e.g., proton shift between heteroatoms, protonation of bicyclic heterocycles, or correction of charge conjugation), and removal of salt/solvent. All of the incorrectly standardized compounds were filtered out (24 compounds). Compounds from various data sets were subsequently grouped by their standardized InChIKeys. If multiple measurements for a single compound/target pair were available, the median activity label was retained. Compounds with conflicting activity labels [median activity label (mean of middle values) = 0.5] were sorted out. All of the compounds with contradictory activity labels are listed in the Supporting Information [Tables S3–S5 for (non)substrates and Tables S6–S8 for (non)inhibitors]. A pivot table was generated by grouping the data by compounds (standardized InChIKeys) and targets. The applied data mining procedure is visually depicted in Figure S1.
Scaffold Generation and Clustering
The three hepatic OATPs were analyzed with respect to privileged scaffolds. Murcko scaffolds26 were extracted via the “RDKit Find Murcko Scaffolds” node in a targetwise manner. The obtained scaffolds were used as queries for substructure mining against the sparse data set for the respective target for the sake of enrichment of existing clusters by additional molecules with analogous scaffolds (since the addition of (a) ring(s) leads to a novel Murcko scaffold). The relative occurrences of scaffolds in the “active” and “inactive” activity classes were subsequently calculated, and only scaffolds with higher prevalence in the “active” class were kept. Generic scaffolds (i.e., those composed of only one aromatic ring with zero or one heteroatom) were filtered out. The Fisher exact test was applied to keep only statistically significant scaffolds (p < 0.05, unless otherwise stated). Hierarchical scaffold clustering [“Hierarchical Clustering (DistMatrix)” node] was applied for scaffolds that appeared in multiple data sets (for different OATPs) by calculation of their maximum common substructure as a measure of similarity. Scaffolds were assigned to discrete clusters on the basis of their distance threshold (set to 0.7). Retrieved compounds belonging to a particular cluster were selected in cases where they exerted the same pharmacological profile as the parent scaffold. All inadequate compounds were reassigned to a corresponding scaffold cluster.
The same analysis was repeated with the dense data set (compounds with measurements for all three hepatic OATPs) in order to retrieve enriched scaffolds with a full pharmacological profile. We also repeated the analysis with full dose–response curve data only (excluding percentage inhibition data) in order to be able to see whether major trends in enriched scaffolds persist with data of higher confidence.
Side-Chain Analysis
The SMARTS pattern for steroidal scaffolds was generated as a query for substructure mining with the aim of detecting all steroid-associated compounds in the sparse data set. The “A” ring (according to IUPAC nomenclature) was defined to be less structurally restricted in order to search for both sp3- and sp2-hybridized carbocycles (estrone-like and cholate-like).
The “RDKit R Group Decomposition” node was used to identify all distinct side chains across the given steroidal scaffold of retrieved compounds. The frequencies of side-chain attachment to different positions of steroidal scaffolds for the different hepatic OATPs were subsequently calculated.
Semiautomatic KNIME Workflows
Workflows for fetching data from different sources, scaffold clustering and analysis, and side-chain analysis are available from myExperiment (https://www.myexperiment.org/workflows/5097.html; https://www.myexperiment.org/workflows/5098.html).
Data Sets for Binary Classification Models: Training and Test Set Selection
Predictive binary classification models were generated in KNIME in order to identify driving factors for inhibitory activity (and eventually selectivity) in terms of molecular features. Only data on transport inhibition were considered, representing data sets more comprehensive than that for substrates/nonsubstrates. Seventy percent of each class was randomly selected to be used as the training set; the remaining compounds were considered as the test set. The compositions of the resulting data sets are shown in Table 1.
Table 1. Compositions of the Data Sets Used in the Sequential Binary Classification Modeling.
| transport inhibition data | total | inhibitor | noninhibitor |
|---|---|---|---|
| all inhibitors + general noninhibitors (training set) | 324 | 262 | 62 |
| all inhibitors + general noninhibitors (test set) | 139 | 113 | 26 |
| OATP1B1 training set | 937 | 232 | 705 |
| OATP1B1 test set | 403 | 100 | 303 |
| OATP1B3 training set | 875 | 139 | 736 |
| OATP1B3 test set | 375 | 59 | 316 |
| OATP2B1 training set | 161 | 43 | 118 |
| OATP1B1 test set | 69 | 19 | 50 |
Descriptor Calculation and Feature Selection
Twenty-six two-dimensional descriptors representing interpretable physicochemical properties were calculated using the “RDKit Descriptor Calculation node” in KNIME. The most relevant descriptors for the respective data set were selected using the “CfsSubsetEval” algorithm implemented in Weka27 with the “BestFirst” search method. Weka is an open-source tool comprising different machine learning algorithms. The exact list of descriptors is given in Tables S13–S16.
Machine Learning Models
Weka27 nodes implemented in KNIME28 were used to train binary classification models for inhibitors of OATP1B1, OATP1B3, and OATP2B1. “Random tree”29,30 (with default parameters) was used as the base classifier. In order to overcome the problem of data imbalance, two different meta-classifiers were used: a cost-sensitive classifier31 and stratified bagging.32,33 In case of the cost-sensitive classifier, the misclassification was applied in accordance with the imbalance ratio. For stratified bagging, the number of bags was adjusted to 64, as a previous study33,34 suggested that generation of 64 models provides satisfactory results without exponentially increasing the computational cost.
Evaluation Method
All of the models were validated by 10-fold cross-validation and by their performances on the external test sets. In both validation schemes, the confusion matrix, sensitivity, specificity, balanced accuracy, and Matthews correlation coefficient (MCC) are reported as measures of the predictive power of the models.
Analyzing Important Molecular Features for OATP Inhibition
The features appearing as most relevant for hepatic OATP inhibition (as selected by the feature selection methodology) were further analyzed by plotting the distribution of their values for inhibitors versus noninhibitors for the three hepatic OATPs and the level 1 (general inhibitors) data set. These analyses as well as the calculations of the statistical significance of the pairwise comparisons of the distributions using the Wilcoxon test were done in R version 1.0.143. The R Project is a software for statistical analysis and data visualization and is freely available at https://www.r-project.org/.
Results and Discussion
Semiautomatic Integration of Pharmacological Data from Different Sources
Compound bioactivity data on human OATP1B1, OATP1B3, and OATP2B1 were collected, mapped, and integrated from five different data sources openly available in the public domain: ChEMBL,21 Metrabase,24 DrugBank,23 the UCSF–FDA TransPortal database,22 and IUPHAR/Guide to Pharmacology.25 The motivation for curating data sets from such a large number of different sources was the wish to enhance the particular data sets not only in terms of their unique enumerated compounds but also in terms of chemical space. Since the different data sources focus on different aspects of bioactivity data (e.g., ChEMBL contains literature data from primarily SAR series, Metrabase has a focus on transporter substrates, and DrugBank contains a collection of marketed or withdrawn drugs), it can be expected that a greater variety in some molecular properties of pharmaceutical interest (e.g., lipophilicity, molecular weight, topological polar surface area, and the number of rotatable bonds) would be introduced by integrating these various sources. As shown in Figure S2, all four features are significantly different in the other databases (DrugBank, Metrabase, IUPHAR, TransPortal) compared with ChEMBL (the Wilcoxon test revealed p < 0.05 in all pairwise comparisons; data not shown), which illustrates the different constitution of the five considered data sources.
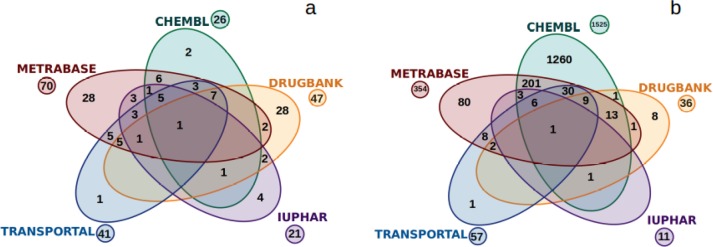
A major goal in this study was the generation of the most comprehensive data sets for hepatic OATPs available from the open domain. These data sets should reflect both the state of the art of available inhibitor and substrate compound spaces, and there was a particular attempt to separate the two sets. This objective was achieved by classifying compounds according to different types of activity end points (Km and EC50 for substrates; IC50, Ki, and percentage inhibition for inhibitors) and activity annotations (substrate, nonsubstrate, inhibitor, or noninhibitor). Interestingly, in terms of the increase in the size of the data sets achieved by integrating data from different sources, the situation looks strikingly different for inhibitor data sets versus substrate data sets for hepatic OATPs (see Figure 1).
Figure 1.
Venn diagrams showing the contributions from the different data sources (in terms of the numbers of unique compounds extracted and curated from them) to the final data sets for (a) (non)substrates and (b) (non)inhibitors.
Whereas ChEMBL accounts for the largest collection of compounds contributing to the inhibitor data set (1525 unique compounds; 94% of all unique inhibitors/noninhibitors), for substrates, Metrabase (70 unique compounds; 69% of all unique substrates/nonsubstrates) and DrugBank (47 unique compounds; 46%) were identified as the most useful resources. Interestingly, just 25% (26 unique compounds) of all substrates/nonsubstrates could be retrieved from ChEMBL, which indeed justifies the integration of data from various sources, especially when it comes to investigations on transporter substrates.
Metrabase24 was originally created to serve as a large open source for transporter ligand data with a special focus on substrates. In total, 631 substrates, 183 nonsubstrates, 1256 inhibitors, and 370 noninhibitors of hepatic OATPs are currently reported in Metrabase. Nevertheless, only a minority of the data entries in Metrabase also report distinct bioactivity values; instead, mostly the data are presented with activity annotations only (e.g., substrate, nonsubstrate, inhibitor, or noninhibitor). However, it is unclear how the data curators decided upon the particular annotations in certain cases. To give an example, primovist was defined as an OATP1B3 substrate, having Km = 4.1 mM.35 On the other hand, clarithromycin was classified as an OATP1B3 nonsubstrate on the basis of its reported Km value of 1 μM.36 In order to further assess the confidence of Metrabase entries, activity annotations from Metrabase were compared with annotations that were assigned to bioactivity measurements from CHEMBL (for the chosen cutoff for classifying actives/inactives, see below). Strikingly, we found conflicting annotations for up to 74% of the compounds retrieved from Metrabase (see Table S9). Thus, only Metrabase entries including numerical bioactivity values were included in our final data sets. Consequently, only 60 substrates/nonsubstrates (7% of the available substrates in Metrabase) and 350 inhibitors/noninhibitors (22% of the available inhibitors in Metrabase) from Metrabase are part of our final data sets for hepatic OATPs.
DrugBank is a comprehensive repository comprising detailed descriptions of small-molecule drugs and their associated targets. Drug activity linked to a respective target is expressed in the form of activity annotations (e.g., substrate, inhibitor, unknown, stimulator, activator, or reducer). Interestingly, DrugBank provided quite a balanced number of both (non)substrates (47 unique compounds) and (non)inhibitors (36 unique compounds) for our final data sets. A similar number of total compounds was included from the UCSF–FDA TransPortal database, but with a predominance of (non)inhibitors (57 unique compounds) over (non)substrates (27 unique compounds). Providing data about FDA-approved drugs linked to pharmaceutically relevant targets, UCSF–FDA TransPortal comprises numerical bioactivity measurements (e.g., Km, IC50, Ki) for hepatic OATPs. The source with the lowest number of compounds for hepatic OATPs [21 unique (non)substrates, 11 unique (non)inhibitors] turned out to be IUPHAR, which provides both real activity measurements and/or annotations for all licensed drugs and other ligands of biologically relevant targets, including transporters. It mainly provided additional information about the hepatic OATP natural substrates. Finally, three novel OATP2B1 inhibitors/noninhibitors recently reported by Giacomini and co-workers18 and 10 novel OATP1B1 and OATP1B3 inhibitors/noninhibitors reported by the group of Ecker17 (just one compound, sirolimus, has been annotated to be a OATP1B1 inhibitor in DrugBank before) were also manually added to the data sets.
In addition to enrichment in terms of chemical space and data set size, we sought to increase the confidence in the final data annotations (as actives or inactives) by collecting multiple independently measured bioactivities or activity annotations for compound/target pairs. Box plots showing the distributions of the number of bioactivities/annotations per single compound and transporter are shown in Figure S3.
For the sake of establishing quantitative SAR (QSAR) models, it is not advisible to mix data from different bioactivity end points or different assay setups.3738 When it comes to binary classification (e.g., into actives and inactives), however, the final label (e.g., inhibitor or noninhibitor) should be independent of the specific experimental protocol.39 Combining data from different activity end points can thus provide a more accurate perception of the OATP pharmacological profiles since measurement errors will be detected and sorted out to a higher extent.
Data Curation
Once the small-molecule bioactivity data had been successfully fetched, the compound data had to be mapped across the various sources in order to identify all assays/bioactivity measurements for a particular compound against one particular target but also across the three different transporters. Hereby, the availability of encoded chemical structures (in the form of InChIKeys, InChIs, or SMILES) was a great advantage. However, this information is not implicitly included in all of the databases used herein (e.g., the UCSF–FDA TransPortal provides only generic names for the compounds). In such cases, the Chemical Identifier Resolver (CIR) web service provided by NIH (available at https://cactus.nci.nih.gov/chemical/structure) can be used in order to assign chemical structural information (SMILES, InChI, InChIKey, etc.) to a compound’s generic name.40 Since for our data sets this procedure failed for 132 compounds, we generated in house a fit-for-purpose “name-to-structure” conversion workflow that retrieves standard InChIKeys from the PubChem database. The majority of these compounds could be mapped by this procedure (68%); however, for 41 compounds the mapping failed because of the wide range of compound expressions and associated synonyms. InChIKeys were manually added in these cases.
All of the precurated entries were subjected to Atkinson’s standardization procedure. To account for consistency during mapping of data from different sources, unified standard InChIKeys were calculated from standardized compounds.
The selected cutoff for separating actives from inactives at 10 μM appears as a good choice upon inspection of the distribution of the median bioactivities for each target since we can observe a certain plateau when looking at the density plots (see Figure S4).
Setting the cutoff for percentage inhibition values resulted in a more complicated procedure. As can be seen from Table 2, percentage inhibition values account for approximately 77% of entries from the overall inhibitor data set. Interestingly, the interpretation of percentage inhibition values is highly inconsistent in different data sets originating from different articles. In the case of CHEMBL entries, three out of 11 integrated data sets reported percentage inhibition values in the form of the inhibitory effect, i.e., the higher the value, the stronger the inhibitor. However, the remaining eight data sets present inhibition as a percentage of control (also expressed as “residual activity”), i.e., the lower the value, the stronger the inhibitor. Interpretation of CHEMBL data gets even more complicated, as some of the data (e.g., the data set reported by Nozawa et al.41) were converted to the opposite form of percentage inhibition values prior to being uploaded to CHEMBL. Since a strict removal of entries with percentage inhibition values would have resulted in a tremendous reduction in the compound numbers of the inhibitor data sets, we manually curated these data sets and transformed the data into a uniform representation of the activity end point “percentage inhibition”. For the ∼150 data sets with percentage inhibition data provided by Metrabase, this curation exercise was alleviated by the availability of activity comments [“Uptake/Inhibition (% of control)” or “Inhibition”]. Cutoffs for separating inhibitors and noninhibitors were set individually on the basis of recommendations given in the primary literature (Tables S1 and S2). The assignment of activity labels was done prior to the creation of a pharmacological overlap matrix. Consequently, compounds with conflicting activity measurements (i.e., equivalent frequencies of the active and inactive binary labels) could be sorted out during this important step of mapping standard InChIKeys in order to represent the whole data set together with their activity labels toward the three transporters. Activity labels for more than 65% of the compounds of the final data set were assessed on basis of more than a single bioactivity measurement. To give an example, we retrieved 59 independent data points (measured bioactivities and/or pure annotations) for cyclosporine from all of the integrated databases, including 19 values from CHEMBL (14 Ki/IC50 and five percentage inhibition values), 26 values from Metrabase (22 Ki/IC50 and four percentage inhibition values), 12 Ki/IC50 values from the UCSF–FDA TransPortal, and two IC50 values from IUPHAR.
Table 2. Numbers of Unique Compounds for Different Activity End Points.
| Ki | IC50 | Km | EC50 | % inhibition | manual annotation | |
|---|---|---|---|---|---|---|
| (non)substrates | – | – | 74 | 4 | – | 63 |
| (non)inhibitors | 170 | 236 | – | – | 1526 | 45 |
For the subsequent analyses on chemical fragments and features, two different data sets were generated. The “sparse hepatic OATP data set” comprises the whole data matrix (including missing annotations for one or two of the transporters) and is made up of 102 unique substrates/nonsubstrates and 1630 unique inhibitors/noninhibitors (see Table 3 for the respective data subset compositions). The “dense hepatic OATP data set”, however, comprises only 13 substrates and 163 inhibitors whose bioactivities have been measured against all three hepatic OATPs [see Table S10 for (non)substrates and Table S11 for (non)inhibitors]. Data from the latter data set provide information about general (i.e., completely overlapping), partially overlapping, and selective substrates/inhibitors. Both data sets are useful sources for studying features that are potentially important for hepatic OATP ligand activity or selectivity.
Table 3. Constitution of the “Sparse Hepatic OATP Data Set”: Numbers of Compounds Per Annotation and Transporter Are Shown (Compounds Might Appear Annotated to More than One Target).
| activity | OATP1B1 | OATP1B3 | OATP2B1 |
|---|---|---|---|
| substrates | 53 | 45 | 26 |
| nonsubstrates | 19 | 16 | 6 |
| inhibitors | 332 | 198 | 62 |
| noninhibitors | 1008 | 1052 | 168 |
Scaffold Clustering and Analysis
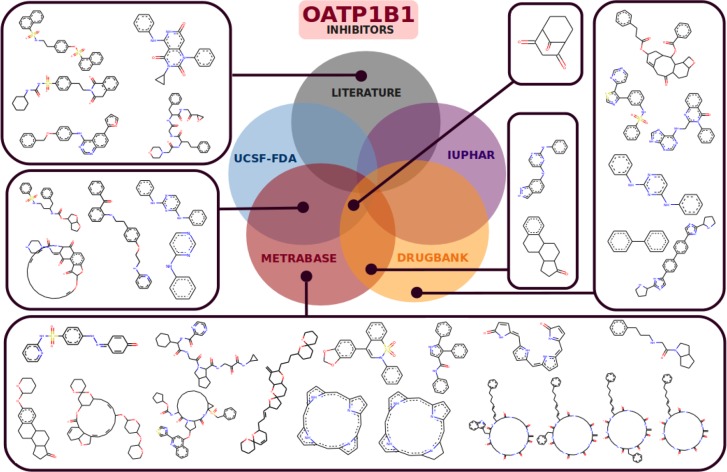
First, the analysis on structural determinants for ligand interaction and selectivity among hepatic OATPs was conducted at the scaffold level. As demonstrated previously by looking at the distributions of certain chemical features in the different data sources (Figure S2), adding data sources led to an increase in chemical space. In terms of new scaffolds, the addition of data from the UCSF–FDA TransPortal database, DrugBank, Metrabase, IUPHAR, and the literature to the data from ChEMBL also led to a gain in terms of new chemical scaffolds (as demonstrated for OATP1B1 inhibitors in Figure 2). Visualizations of new chemical scaffolds for OATP1B3 and OATP2B1 inhibitors are included in Figures S5 and S6, respectively.
Figure 2.
Murcko Scaffolds for OATP1B1 inhibitors retrieved from databases other than CHEMBL.
In order to analyze the frequencies of scaffolds across the different transporters, compounds were grouped by their Murcko scaffolds26 for each transporter. We have to point out that although these analyses were carried out for inhibitors and substrates separately, the majority of the results discussed here were derived from inhibitor data because of data sparseness for substrates in that domain.
The large number of different scaffolds (reflected by the scaffold-to-compound ratio; Table 4)42 strongly indicates that OATP ligands are structurally highly diverse compounds. However, a few scaffolds (23 for inhibitors) were significantly enriched in actives versus inactives (Fisher’s exact test, p < 0.05; see Figure 3).
Table 4. Numbers of Unique Scaffolds in Substrate and Inhibitor Data Sets and (in Parentheses) Their Scaffold-to-Compound Ratios.
| OATP1B1 | OATP1B3 | OATP2B1 | |
|---|---|---|---|
| substrates | 43 (0.86) | 39 (0.86) | 23 (0.88) |
| inhibitors | 250 (0.75) | 155 (0.78) | 54 (0.87) |
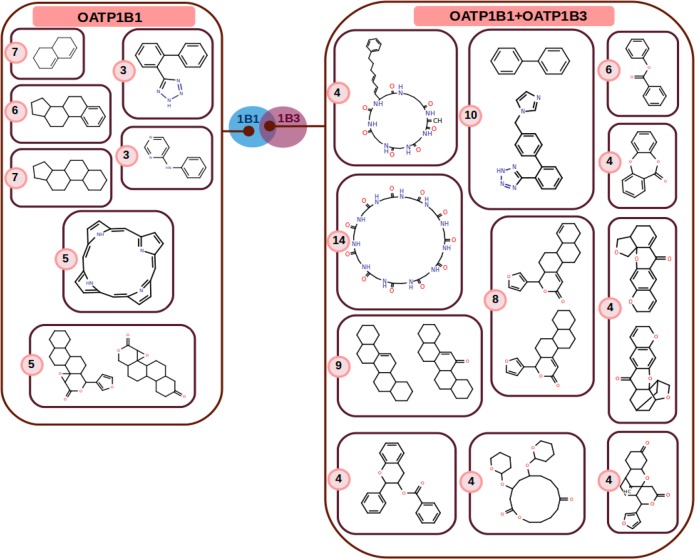
Figure 3.
Enriched scaffolds (p < 0.05) for hepatic OATP inhibitors grouped by their pharmacological profiles with respect to hepatic OATPs. Numbers in pink circles are the numbers of associated compounds for the respective scaffold clusters.
One limitation of the scaffold algorithm of Bemis and Murcko26 is the fact that adding (an) additional ring(s) leads to a new Murcko scaffold. Therefore, for detecting congeneric SAR series of compounds sharing a common scaffold within a data set, the grouping by scaffolds should be combined with additional substructure searches.43 In our case, this strategy has proven useful, e.g., in order to find additional structural analogues of pravastatin-like compounds in the inhibitor data set. In the first instance, only three compounds sharing a hexahydronaphthalene scaffold were detected in the 1B1 inhibitor data set, with pravastatin being a selective inhibitor for OATP1B1 (lovastatin acid and tenivastatin are OATP1B1 inhibitors but have unknown activity toward the other two transporters). By the subsequent substructure search, we could retrieve seven additional compounds with a hexahydronaphthalene substructure but with some variation in their activity profiles (see Table S12). While six compounds show activity against OATP1B1, some do possess additional activity against one of the other two transporters. A closer look at their structures revealed that potentially the addition of more rings, leading to three- or four-ring systems, is responsible for the shift in activity, turning them into unselective hepatic OATP inhibitors (also see the discussion on steroidal scaffolds below).
After enrichment of the scaffold series with additional compounds (by substructure searches), their pharmacological profiles were inspected in order to identify scaffolds with a pronounced activity for only one OATP, for two OATPs (dual inhibitors), or for all three OATPs (pan inhibitors). Furthermore, hierarchical scaffold clustering was applied in order to group structurally similar scaffolds with the same selectivity profile. Within the inhibitor data set, this procedure led to seven enriched scaffold clusters for OATP1B1 (eight scaffolds) and 11 enriched scaffold clusters for both OATP1B1 and OATP1B3 (15 scaffolds) (see Figure 3). Of course, this analysis is influenced by data availability/sparseness and by no means reflects a complete picture of the pharmacological profiles (which especially accounts for the less investigated target OATP2B1).
In order to be able to sort out scaffolds where a real selectivity claim can be made (compared with just enriched scaffolds without a complete pharmacological profile for hepatic OATPs) we applied the scaffold frequency analysis to the dense data set as well. This analysis delivered two scaffolds with indications for OATP1B1 selectivity (pravastatin-like scaffold, estrone-like) and one scaffold with an indication for OATP1B subfamily selectivity (cyclosporin-like scaffold) (Figure S7). In these cases, the available full pharmacological profiles indicate inactivity toward the other targets.
We were also interested in whether some of the trends in enriched scaffolds would remain if the analysis were repeated with full dose–response curve data only. As can be seen from Figure S8, upon exclusion of percentage inhibition data points, most of the enriched scaffolds persisted (20 scaffolds out of 23).
Enriched Scaffolds for OATP1B1 Inhibitors
As shown in Figure 3, frequently occurring scaffolds among the OATP1B1 inhibitors (eight scaffolds) can be grouped into seven different clusters with the available data. Some of the most populated clusters are those comprising steroid derivatives (estrone derivatives and cholate derivatives), with 13 associated compounds in total (six and seven compounds, respectively). The scaffold made up of pravastatin-like compounds, as already discussed above, is also among the most frequent ones for OATP1B1. The seven member compounds have been detected as either OATP1B1-selective inhibitors (pravastatin, simvastatin, and mevinolin) or as OATP1B1 inhibitors (e.g., cyproterone and lovastatin acid; no measurements against OATP1B3 and OATP2B1) in our data sets. Another cluster is derived from porphyrin (five associated compounds). This scaffold has been suggested for the design of new tool compounds for therapeutic applications, mainly because of its photodynamic effects against ovarian cancer. Current findings show that porphyrin and its derivatives exert inhibitory activity against OATP1B1.44 There is also evidence from activity measurements for OATP1B3, suggesting that protoporphyrin acts as a noninhibitor against OATP1B3.15 However, measurements for all porphyrin-associated compounds are needed to confirm the selectivity of this scaffold toward OATP1B1. The remaining three scaffold clusters represent gedunin- and khivorin-associated scaffolds (five associated compounds), N-phenylpyrimidin-4-amine (three associated compounds), and the valsartan-like scaffold (three compounds).
Enriched Scaffolds for Dual OATP1B1/OATP1B3 Inhibitors
In contrast to OATP1B1, inhibitors for OATP1B3 and OATP2B1 do not constitute enriched scaffolds that are specific for these transporters, since the number of respective enumerated compounds does not exceed two in these cases (data not shown).
Interestingly, the group of compounds and scaffolds with the highest occupied clusters belong to the class of compounds showing a pronounced activity against both OATP1B1 and OATP1B3 (dual inhibitors) (15 scaffolds and 11 scaffold clusters; depicted in Figure 3). This can be rationalized by the high sequence similarity between these two targets (∼80%). The largest scaffold cluster with this activity annotation (14 compounds) is derived from cyclosporine and other associated macrocyclic compounds. There are two more clusters possessing macrocyclic scaffolds (four associated compounds each). Macrocyclic compounds in many cases show peptidomimetic properties and will be interesting candidates for future structure-based in silico studies, since it is likely that they accommodate different binding pockets than the smaller molecules.
Enriched Scaffolds for Pan Inhibitors of Hepatic OATPs
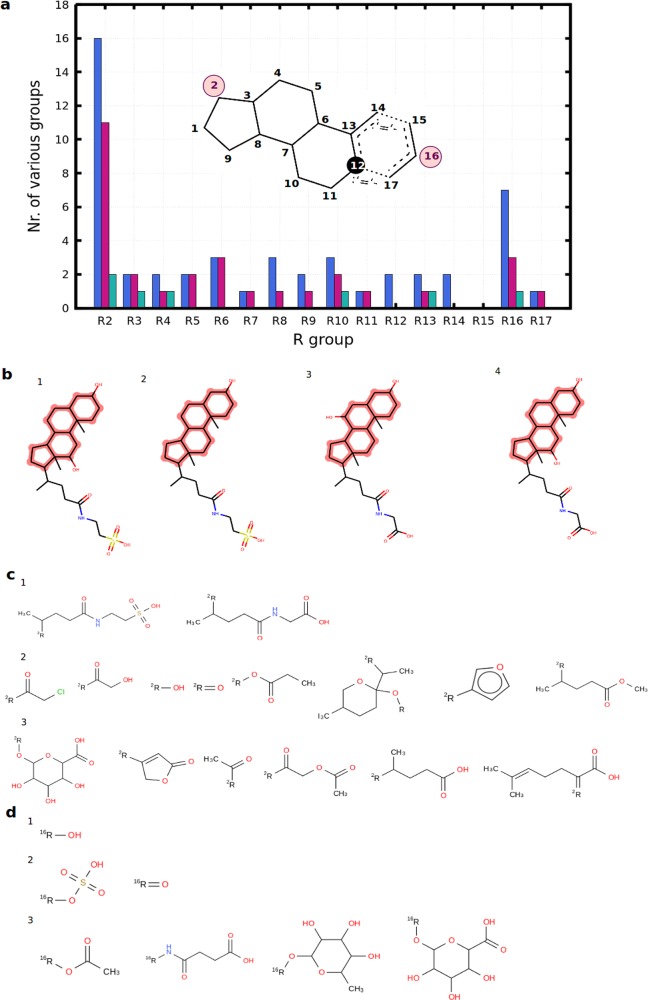
As a result of the scaffold frequency analysis undertaken for hepatic OATP inhibitors, no enriched scaffolds for pan inhibitors were detected as significantly enriched at p < 0.05. However, when the analysis was repeated at a bit weaker significance level (p < 0.1), we found the cholate-like steroidal scaffold to be enriched for all three hepatic OATPs (13 compounds in the sparse data set, four compounds in the dense data set; Figure S9). This is not surprising since the steroidal scaffold also occurs in natural substrates (e.g., cholate and taurocholate) and was already found to be enriched in the OATP1B1 inhibitor set. We applied an R-group decomposition procedure and analyzed the frequency of various R groups at certain positions in a targetwise manner. Positions 2 and 16 show the largest variety in terms of the numbers of functional groups. For substitutions at position 2, hydrophilic flexible side chains (e.g., N-sulfethylpropionamide-4-yl) occur in ligands for all three hepatic OATPs, while, e.g., dihydrofuran or tetrahydropyran groups were detected only among OATP1B1 inhibitors at position 2 (Figure 4). At position 16, substitutions in general appear to be of hydrophilic nature, with tetrahydropyran rings with hydroxyl groups attached to the ring occurring only among OATP1B1 ligands (Figure 4). Looking at compounds with a proven pan-inhibitory effect for hepatic OATPs (four compounds from the dense data set; Figure 4b), we can see that the trends that we found among the sparse data set are verified for pan-inhibitory activity. In order to be able to make real selectivity claims here, more data with measurements on all three transporters will need to be investigated in the future.
Figure 4.
R-group decomposition of steroidal inhibitors. (a) Stacked bar plot showing the distribution of the number of various functional groups at certain R-group positions (blue bar plots, OATP1B1 inhibitors; purple bar plots, OATP1B3 inhibitors; green bar plots, OATP2B1 inhibitors). The maximum common substructure of all of the steroidal inhibitors is shown to highlight the R-group positions. (b) Steroidal ligands with proven pan-inhibitory effect: (1) taurodeoxycholic acid; (2) lithocholyltaurine; (3) glycoursodeoxycholic acid; (4) glycodeoxycholic acid. (c) Functional groups identified at position 2 for (1) pan inhibitors, (2) dual OATP1B inhibitors, and (3) OATP1B1 inhibitors. (d) Functional groups identified at position 16 for (1) pan inhibitors, (2) dual OATP1B inhibitors, and (3) OATP1B1 inhibitors.
OATP Substrates
An analogous analysis of scaffold frequency was also performed for OATP substrates. Because of the considerably lower number of known substrates for hepatic OATPs compared with inhibitors (see Table 3), this analysis could not retrieve any statistically significantly enriched scaffolds. It will be interesting to repeat this analysis when more data become available for hepatic OATP substrates.
In terms of side chains of steroid-associated substrates, we observed consistent trends, as positions 2 and 16 also show the largest variety of different side chains (data not shown).
Important Molecular Features for Inhibitory Activity
After the investigation of molecular determinants for ligand profiles at the scaffold level, it appeared interesting to look at a more abstract representation of structural features: molecular features/descriptors. Such representations might capture commonalities among ligand sets of different hepatic OATPs that would not at first sight appear obvious at the level of scaffolds. The implemented strategy for retrieving important molecular features for the different data sets included the generation of binary classification models for hepatic OATP inhibitors. In more detail, we followed a sequential binary classification approach in which the first level comprised a machine learning model for general noninhibitors (compounds with annotations as “noninhibitors” for all three transporters) versus all inhibitors (OATP1B1 or/and OATP1B3 or/and OATP2B1 inhibitors). At the second level, three models for OATP inhibition (separately for OATP1B1, OATP1B3, and OATP2B1) were generated. It has to be pointed out that the major aim of this modeling approach was the extraction of relevant molecular descriptors and their careful analysis with respect to the transporters and already existing knowledge in that domain. The use of these models for screening purposes and the subsequent identification of novel compounds/scaffolds (potentially active on hepatic OATPs) is not the focus of this investigation but will be conducted in follow-up studies.
A similar approach was used by Karlgren et al.19 in order to describe hepatic OATP inhibitors in terms of chemical features. One of the motivations to repeat this analysis was our curiosity to check whether our models built on basis of the chemically enhanced data sets would still prioritize the same chemical features or if we could retrieve other or additional features that likely better describe the data added since then.
We performed attribute selection (“CfsSubsetEval”45) as implemented in the “BestFirst” search method in Weka27 before model building. For each inhibitor data set, significant molecular features that would aid in distinguishing between inhibitors and noninhibitors could thus be retrieved. On basis of these “relevant” features, classification models were built assuring that highly correlated features were eliminated in order to get rid of redundant information. To account for difficulties due to imbalanced data sets (imbalance ratios between 1:2.5 and 1:4.5 for the different models), which usually affect model accuracies, two different meta-classifiers were used on top of “random tree” as the base classifier: a cost-sensitive classifier31 and stratified bagging.32,33 In a recent study by Jain et al.,34 these two meta-classifiers were found to be the best-performing ones when dealing with imbalanced data sets. Assessing the performances of the final models, stratified bagging outperformed the cost-sensitive classifier. The balanced accuracies of the final models were in the range of 0.73 to 0.79, and the MCC values were between 0.43 and 0.5 (Table 5; model accuracies of all models built are given in Tables S13–S16).
Table 5. Results on Level 1 (All Inhibitors + General Noninhibitors) and Level 2 (OATP1B1, OATP1B3, and OATP2B1 Inhibition Models) in Stratified Bagging for All Calculated Statistical Metrics: Sensitivity, Specificity, Balanced Accuracy, and MCC (The Performance Is Given for Both 10-Fold Cross-Validation and on the External Test Set).
| validation | sensitivity | specificity | balanced accuracy | MCC | |
|---|---|---|---|---|---|
| level 1 | training set | 0.760 | 0.790 | 0.775 | 0.455 |
| level 1 | test set | 0.796 | 0.769 | 0.783 | 0.477 |
| level 2—OATP1B1 | training set | 0.703 | 0.799 | 0.751 | 0.462 |
| level 2—OATP1B1 | test set | 0.730 | 0.809 | 0.769 | 0.497 |
| level 2—OATP1B3 | training set | 0.748 | 0.834 | 0.791 | 0.486 |
| level 2—OATP1B3 | test set | 0.746 | 0.829 | 0.787 | 0.476 |
| level 2—OATP2B1 | training set | 0.698 | 0.771 | 0.734 | 0.434 |
| level 2—OATP2B1 | test set | 0.632 | 0.840 | 0.736 | 0.464 |
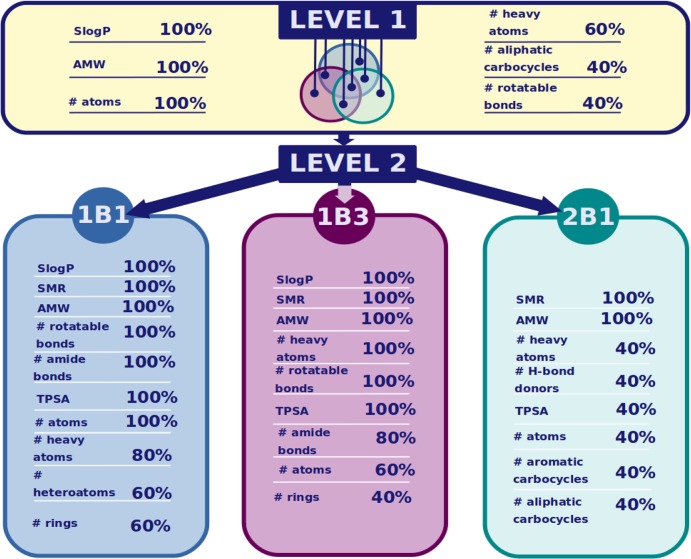
Figure 5 shows the list of important features for each level and category of our sequential modeling approach. Since some of the descriptors were correlated, the final models were constructed with only a selection of those features (available in Tables S13–S16). Upon inspection of the relevant features given in Figure 5 and comparison of them across level 1 and to the models from level 2, it becomes clear that the general inhibitor model (level 1) broadly reflects the important features from the three individual models at level 2. This is not unexpected but shows that our methodology can capture differences and commonalities in the data sets.
Figure 5.
List of relevant features extracted from four different binary classification models with percentage of descriptor importance: level 1 model (any inhibitor vs general noninhibitors); level 2 models (separate models for OATP1B1 inhibition, OATP1B3 inhibition, and OATP2B1 inhibition).
For all four models, average molecular weight (AMW) (100% descriptor importance), the number of atoms (100–40%), and the number of heavy atoms (100–40%) are among the most important features for separating hepatic OATP inhibitors from noninhibitors (Figure 5). Since these three features are highly correlated, for building the final models only AMW was considered.
Lipophilicity (SlogP) was found to be an important descriptor (100% descriptor importance) for all of the models except the OATP2B1 model (Figure 5). It was therefore not taken into account for building the OATP2B1 model. For topological polar surface area (TPSA), we observe that it plays a role for the individual models but not for the general level 1 model. In addition, it seems to be less important in the case of OATP2B1 (40% descriptor importance; Figure 5). Thus, TPSA was not considered for building the final level 1 and OATP2B1 models.
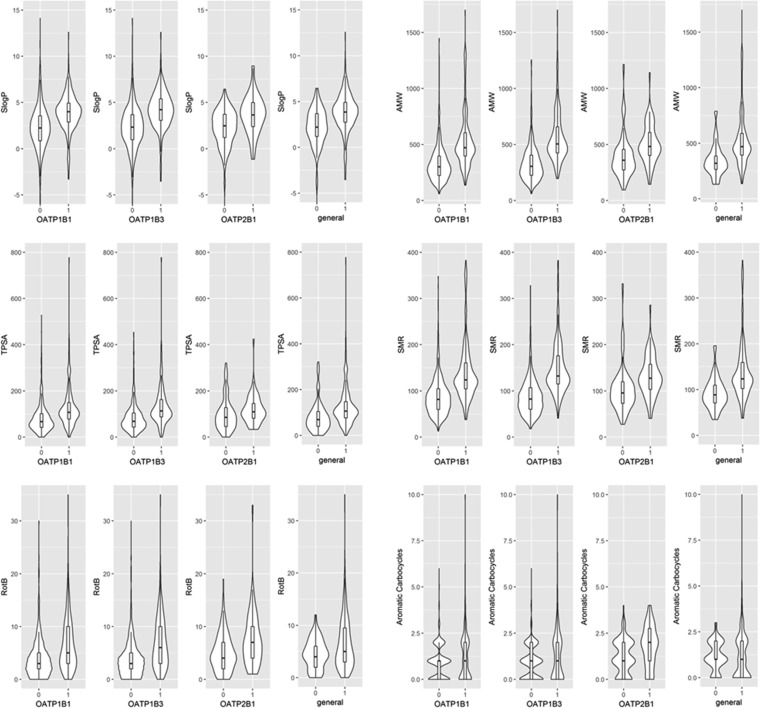
Upon examination of the distribution of those features within the individual data sets (Figure 6 and Table S17) it becomes obvious that in general hepatic OATP inhibitors do possess a higher lipophilicity, molecular weight, and polarity than noninhibitors. These findings are in accordance with the findings of Karlgren et al.,19 but in addition, we were able to prioritize a few other important features, one of which is the molecular refractivity or polarizability (SMR), which reflects the charge distribution on a molecules’ surface. Since in the case of OATPs an inwardly directed pH gradient likely drives the transport,46 a generally higher polarizability in the case of inhibitors versus noninhibitors together with a higher polarity seems very plausible (Figure 6). Interestingly, SMR appears with 100% descriptor importance for all of the individual level 2 models but does not contribute to the general level 1 model.
Figure 6.
Violin and box plots showing the distributions of different molecular discriptors, namely, lipophilicity (SlogP), average molecular weight (AMW), topological polar surface area (TPSA), molecular refractivity (SMR), the number of rotatable bonds (RotB), and the number of aromatic carbocycles (Aromatic Carbocycles), for inhibitors vs noninhibitors within four different data sets. Labeling on abscissae: 0, inactives; 1, actives.
Other important parameters that were not discussed before by Karlgren et al.19 include the influence of flexibility (expressed by the number of rotatable bonds) and counts of different ring systems (especially aromatic rings). The number of rotatable bonds has previously been described as a discriminating factor for OATP1B1 inhibitors versus noninhibitors by van de Steeg et al.16 Our analysis suggests an important role of this feature for all hepatic OATP inhibitors (Figure 6 and Table S17). The number of rings was previously described as a discriminative molecular property by van de Steeg et al.16 for OATP1B1 inhibitors. De Bruyn et al.15 correlated a number of rings < 4 with OATP1B inactivity, which could be confirmed by our analysis and was also observed here for OATP2B1 (see Table S17). We found the number of rings to be discriminative for OATP1B1 and OATP1B3 inhibitors versus the respective noninhibitors (60–40% descriptor importance). However, for OATP2B1 inhibitors, more specific descriptors—namely, the numbers of aliphatic and aromatic carbocycles—were among the list of selected features. Since aromaticity can be linked to molecular complexity or 3D-ness, we were interested in how the feature “number of aromatic carbocycles” was distributed among the four inhibitor data sets. From Figure 6 and Table S17 it becomes obvious that only for OATP2B1 inhibitor data there is a significant difference in the distribution of this feature for inhibitors versus noninhibitors (for OATP1B1/OATP1B3, p > 0.05 in the Wilcoxon test; for OATP2B1, p = 0.0004).
Although the feature “FractionCSP3” (Fsp3), i.e., the fraction of sp3-hybridized carbons, was not among the prioritized ones for any model, one would expect to observe a similar trend in the distribution of this feature across the different transporters. Indeed, it was observed that for all of the data sets except the OATP2B1 data set, the inhibitors show a significantly higher Fsp3 than the respective noninhibitors. For OATP2B1, it can be observed that inhibitors on average possess lower Fsp3 values than inhibitors from the OATP1 subfamiles, which correlates with higher aromaticity and therefore higher planarity (Figure S10). Here again, a lack of data might be the reason for a tendency of planar molecules to inhibit OATP2B1. As is also visible from Figure 3, inhibitors of the OATP1B family do include large, flexible ring systems (e.g., cyclosporine, antamanide, microcystin, caspofungin), which were mostly not tested against OATP2B1.
Finally, the number of amide bonds was highlighted in cases of OATP1B inhibition models but not for the OATP2B1 and the general inhibition model. This can again be explained by the availability of large ring systems containing up to 11 amide bonds (e.g., cyclosporin) in the OATP1B data sets preferentially.
Summary, Conclusions, and Outlook
The main aim of this study was to investigate potential structural determinants responsible for ligand activity or selectivity among hepatic OATPs on the basis of data available from the open domain. In this first study, we focused merely on ligand information as a rich source of chemical structures and bioactivities (pharmacological data).
Emphasis was put on data integration and data curation during the course of this study, as well as on semiautomatic processing of the data. All of the workflows have been made openly available to the scientific community so that they can be reused for other case studies. In addition, since hepatic OATPs are transporters of emerging interest for the research field of hepatotoxicity47 and also in relation to cancer48 and drug resistance,49,50 the current knowledge in this domain is expected to constantly increase in the near future. Therefore, our data integration, curation, and substructure analysis workflows will especially prove useful when a substantial amount of new data become available since in that case the whole analyses can be repeated and refined efficiently and swiftly.
As a side effect of this study, we collected six high-quality curated data sets, for substrates and inhibitors of OATP1B1, OATP1B3, and OATP2B1. Although data sparseness does not always allow delivery of a full ligand profile for all three hepatic OATPs, this analysis exemplifies that nonetheless commonalities and differences among related transporters can be determined by using the methods of data mining, cheminformatics, and ligand-based modeling.
These data sets as well as the information gained on enriched scaffolds and ligand properties of individual and general hepatic OATP inhibitors will serve as a basis for future investigations on ligand interactions and selectivity of hepatic OATPs. Especially the scaffold analyses delivered interesting scaffold series that will be exploited further in terms of their selectivity profiles with the help of structure-based in silico studies exploring individual ligand–protein binding events at the molecular level.
Acknowledgments
We are grateful for technical support by Jana Gurinova when fetching the data from different sources. Gratitude is further expressed to Jennifer Hemmerich for providing us with the Atkinson standardization protocol as a KNIME node. This work received funding from the Austrian Science Fund (FWF) (Grant P 29712).
Glossary
Abbreviations
- OATP
organic anion transporting polypeptide
- SLC
solute carrier
- KNIME
Konstanz Information Miner
- WEKA
Waikato Environment for Knowledge Analysis
- MCC
Matthews correlation coefficient
- MW
molecular weight
- SMR
molecular refractivity
- AMW
average molecular weight
- TPSA
topological polar surface area
- RotB
number of rotatable bonds
- Fsp3
fraction of sp3-hybridized carbons
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jcim.8b00466.
Data sets in (a) CHEMBL and (b) Metrabase annotated with bioactivity end point “inhibition”; lists of removed substrates and inhibitors with conflicting annotations; percentages of conflicting compound activities based on comparison of the data from CHEMBL and Metrabase; dense data sets for hepatic OATP substrates and inhibitors; 10 detected compounds with the hexahydronaphthalene-associated scaffold with pharmacological profiles included; results from level 1 models (all inhibitors + general noninhibitors) for all calculated statistical metrics; results from OATP1B1, OATP1B3, and OATP2B1 inhibition models (level 2) for all calculated statistical metrics; summary statistics for molecular descriptors calculated for inhibitors of OATP1B1, OATP1B3, and OATP2B1; schematic workflow for integrative data mining and curation; box-and-whisker plots showing the distribution of molecular properties for compounds measured against human OATP1B1, OATP1B3, and OATP2B1 originating from five different data sources (ChEMBL, Metrabase, DrugBank, IUPHAR, TransPortal); box plot with number of bioactivities/annotations per unique compound; histograms showing the distributions of median bioactivities for OATP1B1, OATP1B3, and OATP2B1; Murcko scaffolds for OATP1B3 and OATP2B1 inhibitors retrieved from databases other than CHEMBL; enriched scaffolds (p < 0.05) for hepatic OATP inhibitors considering the dense data set (with complete pharmacological profile); enriched scaffolds (p < 0.05) for hepatic OATP inhibitors, excluding percentage inhibition data; enriched scaffolds (p < 0.1) for hepatic OATP inhibitors; violin plots showing the distribution feature “FractionCSP3” (Fsp3) for inhibitors versus noninhibitors within four different data sets (PDF)
Supplementary data files with sparse substrate/nonsubstrate and inhibitor/noninhibitor data sets in CSV format (ZIP)
The authors declare no competing financial interest.
Supplementary Material
References
- Lin L.; Yee S. W.; Kim R. B.; Giacomini K. M. SLC Transporters as Therapeutic Targets: Emerging Opportunities. Nat. Rev. Drug Discovery 2015, 14 (8), 543–560. 10.1038/nrd4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullak-ublick G. A.; Stieger B.; Meier P. J. Enterohepatic Bile Salt Transporters in Normal Physiology and Liver Disease. Gastroenterology 2004, 126 (1), 322–342. 10.1053/j.gastro.2003.06.005. [DOI] [PubMed] [Google Scholar]
- Keppler D. The Roles of MRP2, MRP3, OATP1B1, and OATP1B3 in Conjugated Hyperbilirubinemia. Drug Metab. Dispos. 2014, 42 (4), 561–565. 10.1124/dmd.113.055772. [DOI] [PubMed] [Google Scholar]
- Seithel A.; Eberl S.; Singer K.; Auge D.; Heinkele G.; Wolf N. B.; Dörje F.; Fromm M. F.; König J. The Influence of Macrolide Antibiotics on the Uptake of Organic Anions and Drugs Mediated by OATP1B1 and OATP1B3. Drug Metab. Dispos. 2007, 35 (5), 779–786. 10.1124/dmd.106.014407. [DOI] [PubMed] [Google Scholar]
- Kunze A.; Huwyler J.; Camenisch G.; Gutmann H. Interaction of the Antiviral Drug Telaprevir with Renal and Hepatic Drug Transporters. Biochem. Pharmacol. 2012, 84 (8), 1096–1102. 10.1016/j.bcp.2012.07.032. [DOI] [PubMed] [Google Scholar]
- Obaidat A.; Roth M.; Hagenbuch B. The Expression and Function of Organic Anion Transporting Polypeptides in Normal Tissues and in Cancer. Annu. Rev. Pharmacol. Toxicol. 2012, 52 (1), 135–151. 10.1146/annurev-pharmtox-010510-100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu P.; Lee W.; Xu X.; Leake B. F.; Yamazaki M.; Stone J. A.; Lin J. H.; Pearson P. G.; Kim R. B. Hepatic Uptake of the Novel Antifungal Agent Caspofungin. Drug Metab. Dispos. 2005, 33 (5), 676–682. 10.1124/dmd.104.003244. [DOI] [PubMed] [Google Scholar]
- Kim R. B. 3-Hydroxy-3-methylglutaryl–Coenzyme A Reductase Inhibitors (Statins) and Genetic Variability (Single Nucleotide Polymorphisms) in a Hepatic Drug Uptake Transporter: What’s It All About?. Clin. Pharmacol. Ther. 2004, 75 (5), 381–385. 10.1016/j.clpt.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Cvetkovic M.; Leake B.; Fromm M. F.; Wilkinson G. R.; Kim R. B. OATP and P-Glycoprotein Transporters Mediate the Cellular Uptake and Excretion of Fexofenadine. Drug Metab. Dispos. 1999, 27 (8), 866–871. [PubMed] [Google Scholar]
- Bachmakov I.; Glaeser H.; Fromm M. F.; König J. Interaction of Oral Antidiabetic Drugs With Hepatic Uptake Transporters: Focus on Organic Anion Transporting Polypeptides and Organic Cation Transporter 1. Diabetes 2008, 57 (6), 1463–1469. 10.2337/db07-1515. [DOI] [PubMed] [Google Scholar]
- Mikkaichi T.; Suzuki T.; Tanemoto M.; Ito S.; Abe T. The Organic Anion Transporter (OATP) Family. Drug Metab. Pharmacokinet. 2004, 19 (3), 171–179. 10.2133/dmpk.19.171. [DOI] [PubMed] [Google Scholar]
- Kindla J.; Müller F.; Mieth M.; Fromm M. F.; König J. Influence of Non-Steroidal Anti-Inflammatory Drugs on Organic Anion Transporting Polypeptide (OATP) 1B1-and OATP1B3-Mediated Drug Transport. Drug Metab. Dispos. 2011, 39 (6), 1047–1053. 10.1124/dmd.110.037622. [DOI] [PubMed] [Google Scholar]
- Shitara Y.; Maeda K.; Ikejiri K.; Yoshida K.; Horie T.; Sugiyama Y. Clinical Significance of Organic Anion Transporting Polypeptides (OATPs) in Drug Disposition: Their Roles in Hepatic Clearance and Intestinal Absorption. Biopharm. Drug Dispos. 2013, 34 (1), 45–78. 10.1002/bdd.1823. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B.; Stieger B. The SLCO (Former SLC21) Superfamily of Transporters. Mol. Aspects Med. 2013, 34 (2–3), 396–412. 10.1016/j.mam.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyn T.; van Westen G. J. P.; IJzerman A. P.; Stieger B.; de Witte P.; Augustijns P. F.; Annaert P. P. Structure-Based Identification of OATP1B1/3 Inhibitors. Mol. Pharmacol. 2013, 83 (6), 1257–1267. 10.1124/mol.112.084152. [DOI] [PubMed] [Google Scholar]
- van de Steeg E.; Venhorst J.; Jansen H. T.; Nooijen I. H. G.; DeGroot J.; Wortelboer H. M.; Vlaming M. L. H. Generation of Bayesian Prediction Models for OATP-Mediated Drug–Drug Interactions Based on Inhibition Screen of OATP1B1, OATP1B1*15 and OATP1B3. Eur. J. Pharm. Sci. 2015, 70, 29–36. 10.1016/j.ejps.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Kotsampasakou E.; Brenner S.; Jäger W.; Ecker G. F. Identification of Novel Inhibitors of Organic Anion Transporting Polypeptides 1B1 and 1B3 (OATP1B1 and OATP1B3) Using a Consensus Vote of Six Classification Models. Mol. Pharmaceutics 2015, 12 (12), 4395–4404. 10.1021/acs.molpharmaceut.5b00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuri N.; Zur A. A.; Wittwer M. B.; Lin L.; Yee S. W.; Sali A.; Giacomini K. M. Computational Discovery and Experimental Validation of Inhibitors of the Human Intestinal Transporter OATP2B1. J. Chem. Inf. Model. 2017, 57 (6), 1402–1413. 10.1021/acs.jcim.6b00720. [DOI] [PubMed] [Google Scholar]
- Karlgren M.; Vildhede A.; Norinder U.; Wisniewski J. R.; Kimoto E.; Lai Y.; Haglund U.; Artursson P. Classification of Inhibitors of Hepatic Organic Anion Transporting Polypeptides (OATPs): Influence of Protein Expression on Drug–Drug Interactions. J. Med. Chem. 2012, 55 (10), 4740–4763. 10.1021/jm300212s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold M. R.; Cebron N.; Dill F.; Gabriel T. R.; Kötter T.; Meinl T.; Ohl P.; Thiel K.; Wiswedel B. KNIME—the Konstanz Information Miner: Version 2.0 and Beyond. SIGKDD Explor. Newsl. 2009, 11 (1), 26–31. 10.1145/1656274.1656280. [DOI] [Google Scholar]
- Bento A. P.; Gaulton A.; Hersey A.; Bellis L. J.; Chambers J.; Davies M.; Krüger F. A.; Light Y.; Mak L.; McGlinchey S.; Nowotka M.; Papadatos G.; Santos R.; Overington J. P. The ChEMBL Bioactivity Database: An Update. Nucleic Acids Res. 2014, 42 (D1), D1083–D1090. 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey K. M.; Wen C. C.; Johns S. J.; Zhang L.; Huang S.-M.; Giacomini K. M. The UCSF–FDA TransPortal: A Public Drug Transporter Database. Clin. Pharmacol. Ther. 2012, 92 (5), 545–546. 10.1038/clpt.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; Maciejewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le D.; Pon A.; Knox C.; Wilson M. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46 (D1), D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak L.; Marcus D.; Howlett A.; Yarova G.; Duchateau G.; Klaffke W.; Bender A.; Glen R. C. Metrabase: A Cheminformatics and Bioinformatics Database for Small Molecule Transporter Data Analysis and (Q)SAR Modeling. J. Cheminf. 2015, 7, 31. 10.1186/s13321-015-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson A. J.; Sharman J. L.; Benson H. E.; Faccenda E.; Alexander S. P. H.; Buneman O. P.; Davenport A. P.; McGrath J. C.; Peters J. A.; Southan C.; Spedding M.; Yu W.; Harmar A. J. The IUPHAR/BPS Guide to PHARMACOLOGY: An Expert-Driven Knowledgebase of Drug Targets and Their Ligands. Nucleic Acids Res. 2014, 42 (D1), D1098–D1106. 10.1093/nar/gkt1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis G. W.; Murcko M. A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39 (15), 2887–2893. 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- Frank E.; Hall M.; Trigg L.; Holmes G.; Witten I. H. Data Mining in Bioinformatics Using Weka. Bioinformatics 2004, 20 (15), 2479–2481. 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- Beisken S.; Meinl T.; Wiswedel B.; de Figueiredo L. F.; Berthold M.; Steinbeck C. KNIME-CDK: Workflow-Driven Cheminformatics. BMC Bioinf. 2013, 14, 257. 10.1186/1471-2105-14-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColm G. L. An Introduction to Random Trees. Res. Lang. Comput. 2003, 1 (3–4), 203–227. 10.1023/A:1024691507511. [DOI] [Google Scholar]
- Le Gall J.-F. Random Trees and Applications. Probab. Surveys 2005, 2, 245–311. 10.1214/154957805100000140. [DOI] [Google Scholar]
- López V.; Fernández A.; Moreno-Torres J. G.; Herrera F. Analysis of Preprocessing vs. Cost-Sensitive Learning for Imbalanced Classification. Open Problems on Intrinsic Data Characteristics. Expert Syst. Appl. 2012, 39 (7), 6585–6608. 10.1016/j.eswa.2011.12.043. [DOI] [Google Scholar]
- He H.; Garcia E. A. Learning from Imbalanced Data. IEEE Trans. Knowledge Data Eng. 2009, 21 (9), 1263–1284. 10.1109/TKDE.2008.239. [DOI] [Google Scholar]
- Tetko I. V.; Novotarskyi S.; Sushko I.; Ivanov V.; Petrenko A. E.; Dieden R.; Lebon F.; Mathieu B. Development of Dimethyl Sulfoxide Solubility Models Using 163 000 Molecules: Using a Domain Applicability Metric to Select More Reliable Predictions. J. Chem. Inf. Model. 2013, 53 (8), 1990–2000. 10.1021/ci400213d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S.; Kotsampasakou E.; Ecker G. F. Comparing the Performance of Meta-Classifiers—a Case Study on Selected Imbalanced Data Sets Relevant for Prediction of Liver Toxicity. J. Comput.-Aided Mol. Des. 2018, 32 (5), 583–590. 10.1007/s10822-018-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt M.; Keiser M.; Oswald S.; Kühn J.; Jia J.; Grube M.; Kroemer H. K.; Siegmund W.; Weitschies W. Hepatic Uptake of the Magnetic Resonance Imaging Contrast Agent Gd-EOB-DTPA: Role of Human Organic Anion Transporters. Drug Metab. Dispos. 2010, 38 (7), 1024–1028. 10.1124/dmd.110.032862. [DOI] [PubMed] [Google Scholar]
- Peters J.; Eggers K.; Oswald S.; Block W.; Lütjohann D.; Lämmer M.; Venner M.; Siegmund W. Clarithromycin Is Absorbed by an Intestinal Uptake Mechanism That Is Sensitive to Major Inhibition by Rifampicin: Results of a Short-Term Drug Interaction Study in Foals. Drug Metab. Dispos. 2012, 40 (3), 522–528. 10.1124/dmd.111.042267. [DOI] [PubMed] [Google Scholar]
- Kramer C.; Kalliokoski T.; Gedeck P.; Vulpetti A. The Experimental Uncertainty of Heterogeneous Public Ki Data. J. Med. Chem. 2012, 55 (11), 5165–5173. 10.1021/jm300131x. [DOI] [PubMed] [Google Scholar]
- Kalliokoski T.; Kramer C.; Vulpetti A.; Gedeck P. Comparability of Mixed IC50 Data—A Statistical Analysis. PLoS One 2013, 8 (4), e61007. 10.1371/journal.pone.0061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanari F.; Ecker G. F. BCRP Inhibition: From Data Collection to Ligand-Based Modeling. Mol. Inf. 2014, 33 (5), 322–331. 10.1002/minf.201400012. [DOI] [PubMed] [Google Scholar]
- Lowe D. M.; Corbett P. T.; Murray-Rust P.; Glen R. C. Chemical Name to Structure: OPSIN, an Open Source Solution. J. Chem. Inf. Model. 2011, 51 (3), 739–753. 10.1021/ci100384d. [DOI] [PubMed] [Google Scholar]
- Nozawa T.; Tamai I.; Sai Y.; Nezu J.-I.; Tsuji A. Contribution of Organic Anion Transporting Polypeptide OATP-C to Hepatic Elimination of the Opioid Pentapeptide Analogue [d-Ala2,d-Leu5]-Enkephalin. J. Pharm. Pharmacol. 2003, 55 (7), 1013–1020. 10.1211/0022357021440. [DOI] [PubMed] [Google Scholar]
- Jasial S.; Hu Y.; Bajorath J. Assessing the Growth of Bioactive Compounds and Scaffolds over Time: Implications for Lead Discovery and Scaffold Hopping. J. Chem. Inf. Model. 2016, 56 (2), 300–307. 10.1021/acs.jcim.5b00713. [DOI] [PubMed] [Google Scholar]
- Zdrazil B.; Hellsberg E.; Viereck M.; Ecker G. F. From Linked Open Data to Molecular Interaction: Studying Selectivity Trends for Ligands of the Human Serotonin and Dopamine Transporter. MedChemComm 2016, 7 (9), 1819–1831. 10.1039/C6MD00207B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Guo Z.; Wang Y.; Chen X.; Liu J.; Zhong D. Potential Role of Organic Anion Transporting Polypeptide 1B1 (OATP1B1) in the Selective Hepatic Uptake of Hematoporphyrin Monomethyl Ether Isomers. Acta Pharmacol. Sin. 2015, 36 (2), 268–280. 10.1038/aps.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. A.Correlation-Based Feature Selection for Machine Learning. Ph.D. Thesis, University of Waikato, Hamilton, New Zealand, 1999. [Google Scholar]
- Leuthold S.; Hagenbuch B.; Mohebbi N.; Wagner C. A.; Meier P. J.; Stieger B. Mechanisms of PH-Gradient Driven Transport Mediated by Organic Anion Polypeptide Transporters. Am. J. Physiol.: Cell Physiol. 2009, 296 (3), C570–C582. 10.1152/ajpcell.00436.2008. [DOI] [PubMed] [Google Scholar]
- Kotsampasakou E.; Escher S. E.; Ecker G. F. Linking Organic Anion Transporting Polypeptide 1B1 and 1B3 (OATP1B1 and OATP1B3) Interaction Profiles to Hepatotoxicity - The Hyperbilirubinemia Use Case. Eur. J. Pharm. Sci. 2017, 100, 9–16. 10.1016/j.ejps.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Thakkar N.; Lockhart A. C.; Lee W. Role of Organic Anion-Transporting Polypeptides (OATPs) in Cancer Therapy. AAPS J. 2015, 17 (3), 535–545. 10.1208/s12248-015-9740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster C. S.; Sprowl J. A.; Walker A. L.; Hu S.; Gibson A. A.; Sparreboom A. Modulation of OATP1B-Type Transporter Function Alters Cellular Uptake and Disposition of Platinum Chemotherapeutics. Mol. Cancer Ther. 2013, 12 (8), 1537–1544. 10.1158/1535-7163.MCT-12-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S.; Riha J.; Giessrigl B.; Thalhammer T.; Grusch M.; Krupitza G.; Stieger B.; Jäger W. The Effect of Organic Anion-Transporting Polypeptides 1B1, 1B3 and 2B1 on the Antitumor Activity of Flavopiridol in Breast Cancer Cells. Int. J. Oncol. 2015, 46 (1), 324–332. 10.3892/ijo.2014.2731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.