Abstract
Objectives
The purpose of this study was to explore the association of miRNA-146 and miRNA-499 polymorphisms with inflammatory arthritis.
Methods
A systematic search of studies on the association of miRNA-146 and miRNA-499 polymorphisms with inflammatory arthritis susceptibility was conducted in PubMed, Web of science, Elsevier ScienceDirect, and Cochrane Library. Eventually, 18 published studies were included. The strength of association between miRNA-146/499 polymorphisms and inflammatory arthritis susceptibility was assessed by odds ratios (ORs) with its 95% confidence intervals (CIs).
Results
A total of 18 case-control studies, consisting of 3385 inflammatory arthritis patients and 4584 controls, were included in the meta-analysis. This meta-analysis showed significant association between miRNA-499 rs3746444 polymorphism and inflammatory arthritis susceptibility in overall population (C vs T, OR: 1.422, 95% CI= 1.159-1.745, P=0.001). Similar results were found in subgroup analysis by region. But we did not find association between miRNA-146 rs2910164 polymorphism and inflammatory arthritis susceptibility in overall population (C vs T, OR: 1.061, 95% CI= 0.933-1.207, P=0.365).
Conclusions
The present study indicates that miRNA-499 rs3746444 polymorphism is associated with inflammatory arthritis susceptibility. However, there is lack of association between miRNA-146 rs2910164 polymorphism and inflammatory arthritis susceptibility. But, we also find miRNA-146 rs2910164 and miRNA-499 rs3746444 polymorphism are associated with inflammatory arthritis in Middle East. Therefore, more large-scale studies are warranted to replicate our findings.
1. Introduction
Inflammatory arthritis is a group of complex diseases, including rheumatoid arthritis (RA), psoriatic arthritis (PsA), juvenile idiopathic arthritis (JIA), etc. And it is characterized by inflammatory cell infiltration into the joints [1, 2]. There is no doubt that environmental factors and genetic factors play an important role in the development of inflammatory arthritis.
MicroRNAs (miRNAs) are small noncoding RNAs, which are about 22 nucleotides long. They play an important role in regulating transcription and translation [3–6]. Some studies indicated that they also participate in cell proliferation, differentiation, and apoptosis [7, 8]. And, miRNAs can cause the degradation or translation repression of their target miRNA by binding to the 3′-untranslated region of specific mRNAs (messenger RNAs) [9–12]. Recently, some studies reported that miRNA may associate with inflammatory arthritis, such as miRNA-146 rs2910164 and miRNA-499 rs3746444. Therefore, we chose miRNA-146 rs2910164 and miRNA-499 rs3746444 to explore the association between its polymorphisms and inflammatory arthritis.
MiRNA-146 is located on human chromosome 5q34 and related to immune regulation, inflammatory signaling pathway, because it can regulate the expression of IL-1 receptor-associated kinase (IRAK1) and IRAK2 and targets TNF receptor-associated factor 6 (TRAF6) [13]. And, miRNA-499 is related to the expression of Interleukin-17 receptor B (IL-17RB), IL-2RB, IL-6, B and T lymphocyte attenuator (BTLA), and peptidyl arginine deiminase 4 (PADI4) [14]. Both miRNA-146 and miRNA-499 are thought to be involved in autoimmune and inflammatory diseases [15].
Considering the results of miRNA-146 and miRNA-499 polymorphisms with inflammatory arthritis are inconsistent [16–20], this discrepancy might be caused by some studies that have small sample size, low statistical power, ethnicity differences, and publication bias. It is necessary to conduct meta-analysis to explore this association.
2. Methods
2.1. Search Strategy
A systematic search of studies on the association of miRNA-146 and miRNA-499 polymorphisms with inflammatory arthritis susceptibility was conducted in PubMed, Web of science, Elsevier ScienceDirect, and Cochrane Library. Keywords for the search were as follows: (“microRNA” or “miRNA” or “microRNAs”) and (“Inflammatory arthritis”) and (“polymorphism” or “variant” or “genotype” or “SNP” or “mutations”). All relevant studies were retrieved. We screened those studies by reading its titles, abstracts, and contents carefully.
2.2. Eligibility Criteria
The inclusion criteria were (i) studies involving the association between miRNA gene polymorphisms and inflammatory arthritis; (ii) case-control study; (iii) studies based on human; (iv) studies providing the detailed relevant genotype data of both case group and control group. Studies were excluded if (i) the study was a review, editorial, abstract, case report, or unpublished article; (ii) studies were nonhuman studies or animal experiments or cell experiments; (iii) studies had no controls or no detailed relevant genotype data.
2.3. Data Extraction
The data of the eligible studies were extracted by one investigator (Mr.Wang). The following information were collected: first author's name; year of publication; type of disease; country; region; genotyping methods; number of cases and controls in each study; characteristics of controls; category of polymorphisms and other additional information.
2.4. Quality Assessment
The included studies in this meta-analysis were evaluated by another inspector (Pan). The quality assessment was based on the modified Newcastle-Ottawa Quality Assessment Scale (NOS). The scale consists of eight multiple-choice questions that involve subjects' selection, comparability in cases and controls, and assessment of exposure. High-quality response earns a point, totaling up to nine points (the comparability question earns up to two points). The higher score indicates better quality.
2.5. Statistical Analysis
All statistical analyses were performed by Stata 12.0 (StataCorp, College Station, TX, USA) software. OR and 95% CI were used to evaluate the strength of association between miRNA-146/499 polymorphisms with inflammatory arthritis susceptibility under different genetic models. The effect of heterogeneity was evaluated using I2 statistics. Heterogeneity was recognized as statistically significant when I2 > 50%. According to the value of I2, we chose fixed-effects or random-effects model. All subgroups were analyzed. Sensitivity analysis was used to evaluate the influence of individual study on the overall OR. Publication bias was assessed by Funnel's plot, Begg's test and Egger's test. An asymmetric plot and the P value of Egger's test or Begg's test less than 0.05 were considered as significant publication bias.
3. Results
3.1. Literature Search
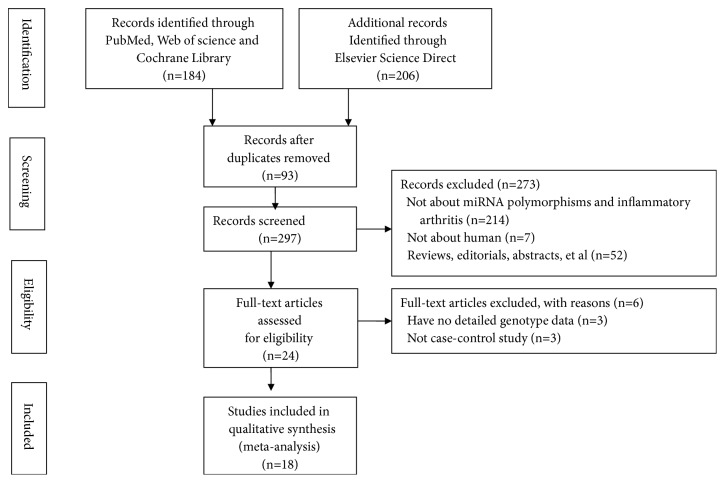
A total of 390 studies were retrieved from PubMed, Web of science, Elsevier Science Direct and Cochrane Library. Finally, 18 eligible studies were included in this meta-analysis. A flowchart of the included and excluded studies was shown in Figure 1.
Figure 1.
A flowchart of the included and excluded studies.
3.2. Characteristics of the Included Studies
Table 1 showed the main features of those included studies. Those studies were all English articles, which have been published from 2010 to 2018. All studies included in this meta-analysis were case-control studies. Eighteen studies involving 3385 inflammatory arthritis patients and 4584 controls were included in this meta-analysis.
Table 1.
The main features of included studies.
| Author | Year | Disease | Country | Region | Genotyping methods | Number of Cases | Number of controls | Characteristics of controls | Polymorphisms | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| Chatzikyriakidou A | 2010 | RA | Greece | Europe | PCR–RFLP | 136 | 147 | Healthy | rs2910164(miRNA-146) | 6 |
|
| ||||||||||
| Yang XK | 2016 | RA | China | Asia | TaqMan | 386 | 576 | Healthy | rs3746444(miRNA-499) | 7 |
|
| ||||||||||
| Singh S | 2014 | JIA-ERA | India | Asia | PCR–RFLP | 150 | 216 | Healthy | rs2910164(miRNA-146) | 7 |
|
| ||||||||||
| El-Shal AS | 2013 | RA | Egypt | Middle East | PCR–RFLP | 217 | 245 | Healthy | rs2910164(miRNA-146), rs3746444(miRNA-499) |
6 |
|
| ||||||||||
| Shaker OG | 2018 | RA | Egypt | Middle East | TaqMan | 104 | 112 | Healthy | rs2910164(miRNA-146), rs3746444(miRNA-499) |
7 |
|
| ||||||||||
| Hashemi M | 2012 | RA | Iran | Middle East | T-ARMS-PCR | 104 | 110 | Healthy | rs2910164(miRNA-146), rs3746444(miRNA-499) |
6 |
|
| ||||||||||
| Yang B | 2010 | RA | China | Asia | PCR–RFLP | 208 | 240 | Healthy | rs2910164(miRNA-146), rs3746444(miRNA-499) |
7 |
|
| ||||||||||
| Aleman-Avila I | 2017 | RA | Mexico | North America | TaqMan | 412 | 486 | Healthy | rs2910164(miRNA-146), rs3746444(miRNA-499), rs11614913(miRNA-196) |
7 |
|
| ||||||||||
| Zhou X | 2015 | RA | China | Asia | NA | 598 | 821 | NA | rs2910164(miRNA-146) | 5 |
|
| ||||||||||
| Hassine HB | 2017 | RA | Tunisia | Middle East | PCR–RFLP | 165 | 150 | Healthy | rs2910164(miRNA-146) | 6 |
|
| ||||||||||
| Maharaj AB(1) | 2018 | PsA | South Africa | Africa | PCR–RFLP | 84 | 62 | Healthy | rs2910164(miRNA-146) | 6 |
|
| ||||||||||
| Maharaj AB(2) | 2018 | PsA | South Africa | Africa | PCR–RFLP | 32 | 38 | Healthy | rs2910164(miRNA-146) | 6 |
|
| ||||||||||
| Jimenez-Morales S | 2012 | JRA | Mexico | North America | TaqMan | 210 | 531 | Healthy | rs2910164(miRNA-146) | 7 |
|
| ||||||||||
| Ciccacci C | 2016 | RA | Italy | Europe | TaqMan | 192 | 298 | Healthy | rs2910164(miRNA-146) | 7 |
|
| ||||||||||
| Ayeldeen G | 2018 | RA | Egypt | Middle East | real-time PCR | 52 | 56 | Healthy | rs2910164(miRNA-146), rs3746444(miRNA-499) |
6 |
|
| ||||||||||
| Toraih EA | 2016 | RA | Egypt | Middle East | TaqMan | 95 | 200 | Healthy | rs3746444(miRNA-499), rs11614913(miRNA-196) |
6 |
|
| ||||||||||
| Fattah SA | 2018 | RA | Egypt | Middle East | PCR–RFLP | 100 | 100 | Healthy | rs3746444(miRNA-499) | 6 |
|
| ||||||||||
| Bogunia-Kubik K | 2016 | RA | Poland | Europe | PCR–RFLP | 111 | 130 | Healthy | rs2910164(miRNA-146) | 6 |
|
| ||||||||||
| Chatzikyriakidou A | 2010 | PsA | Greece | Europe | PCR–RFLP | 29 | 66 | NA | rs2910164(miRNA-146) | 6 |
HWE: Hardy–Weinberg equilibrium; NOS: Newcastle-Ottawa Quality Assessment Scale.
3.3. Meta-Analysis of Association between miRNA-146 rs2910164 Polymorphism and Inflammatory Arthritis
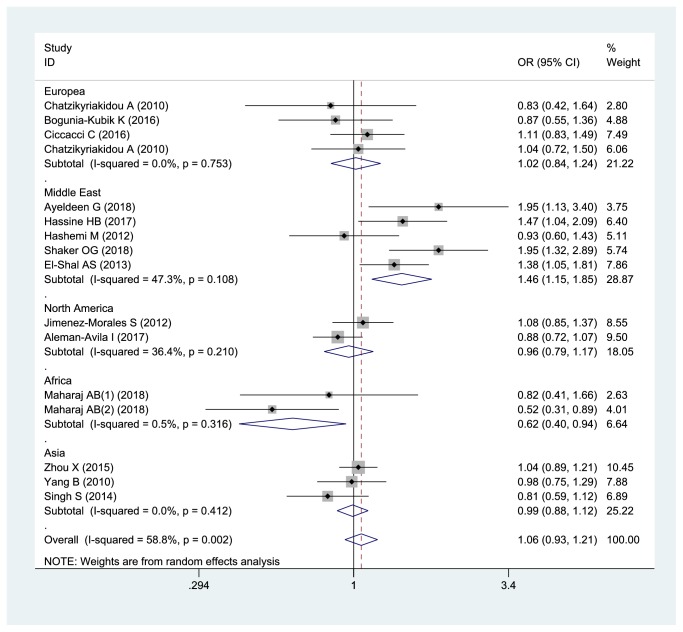
Meta-analysis indicated that there is lack of association between miRNA-146 rs2910164 polymorphism and inflammatory arthritis susceptibility in the overall population (G vs C, OR: 1.061, 95% CI= 0.933-1.207, P=0.365; GC+CC vs GG, OR: 0.919, 95% CI=0.762-1.110, P=0.381; CC vs GG+GC, OR: 0.935, 95% CI=0.821-1.065, P=0.310). After stratifying by region, it only showed the association between miRNA-146 rs2910164 polymorphism and inflammatory arthritis susceptibility in Middle East (Figure 2). This may be due to the differences in geographic region, ethnicity, and sample size.
Figure 2.
The forest plot about association between miRNA-146 rs2910164 and inflammatory arthritis under allelic model (G vs C).
3.4. Meta-Analysis of Association between miRNA-499 rs3746444 Polymorphism and Inflammatory Arthritis
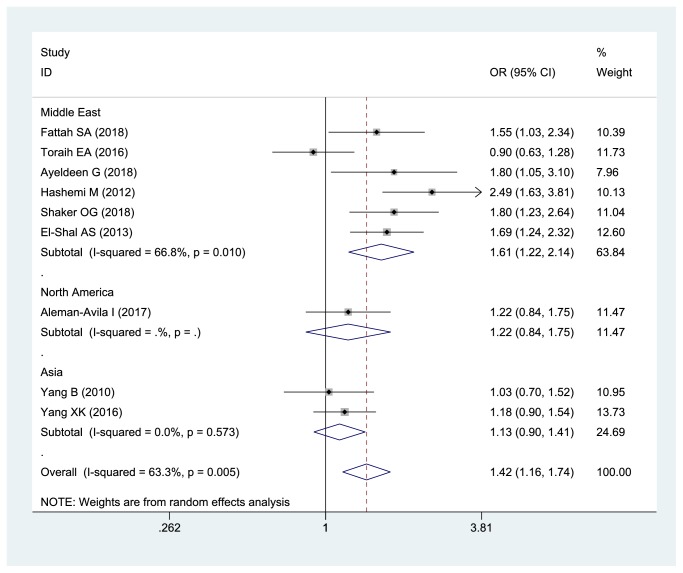
Meta-analysis indicated significant association between miRNA-499 rs3746444 polymorphism and inflammatory arthritis susceptibility in the overall population (C vs T, OR: 1.422, 95% CI= 1.159-1.745, P=0.001; TC +CC vs TT, OR: 1.409, 95% CI=1.072-1.852, P=0.014; CC vs TT+TC, OR: 1.872, 95% CI=1.420-2.467, P<0.001). After stratifying by region, the association between miRNA-499 rs3746444 polymorphism and inflammatory arthritis susceptibility was still extant in Middle East (Figure 3). But, we did not find this association in Asia and North America.
Figure 3.
The forest plot about association between miRNA-499 rs3746444 and inflammatory arthritis under allelic model (C vs T).
3.5. Heterogeneity and Publication Bias
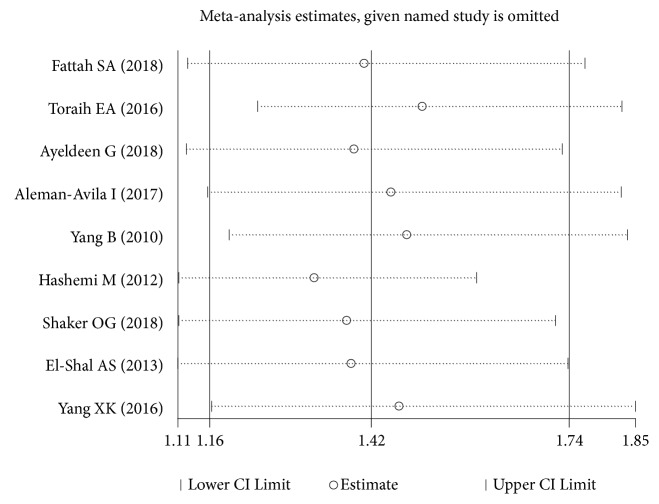
The results of heterogeneity test were shown in Table 2. And the results of Begg's test and Egger's test were also shown in Table 2. As for some studies that had heterogeneity (I2 > 50%), the random-effects models were performed; others were analyzed by fixed-effects model. As for miRNA-499 rs3746444 polymorphism, the plot of sensitivity analysis was shown in Figure 4. And there was no significant evidence of publication bias found in the meta-analysis.
Table 2.
The results of meta-analysis, heterogeneity test, and publication bias.
| polymorphism | Number of studies | Comparable genotype | No. of Cases/Controls | Test of association | Test of heterogeneity | Test of publication bias | Model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Begg's test | Egger's test | ||||||||||||
| OR (95%CI) | P | I 2 | P H | Z | P | t | P | ||||||
| rs2910164 (miRNA-146) |
|||||||||||||
| Overall | 16 | G vs C | 2802/3708 | 1.061(0.933-1.207) | 0.365 | 58.80% | 0.002 | 0.14 | 0.893 | 0.60 | 0.559 | R | |
| Region | |||||||||||||
| Europe | 4 | G vs C | 468/641 | 1.017(0..836-1.236) | 0.868 | 0.00% | 0.753 | 1.02 | 0.308 | -2.93 | 0.100 | F | |
| Asia | 3 | G vs C | 956/1277 | 0.991(0.877-1.120) | 0.890 | 0.00% | 0.412 | 1.04 | 0.296 | -1.89 | 0.309 | F | |
| Middle East | 5 | G vs C | 642/673 | 1.449(1.229-1.709) | 0.001 | 47.30% | 0.108 | 0.24 | 0.806 | 0.53 | 0.630 | F | |
| Africa | 2 | G vs C | 116/100 | 0.615(0.405-0.935) | 0.023 | 0.50% | 0.316 | 0.00 | 1.000 | NA | NA | F | |
| North America | 2 | G vs C | 620/1017 | 0.955(0.820-1.111) | 0.551 | 36.40% | 0.210 | 0.00 | 1.000 | NA | NA | F | |
| HWE | |||||||||||||
| Y | 13 | G vs C | 2524/3359 | 1.053(0.916-1.211) | 0.469 | 61.90% | 0.002 | 0.18 | 0.855 | 0.99 | 0.344 | R | |
| Overall | 16 | GC+CC vs GG |
2802/3708 | 0.919(0.762-1.110) | 0.381 | 57.20% | 0.002 | -0.05 | 1.000 | 0.05 | 0.963 | R | |
| Region | |||||||||||||
| Europe | 4 | GC+CC vs GG |
468/641 | 1.000(0.784-1.276) | 0.998 | 0.00% | 0.606 | 1.70 | 0.089 | 13.66 | 0.005 | F | |
| Asia | 3 | GC+CC vs GG |
956/1277 | 0.980(0.795-1.209) | 0.851 | 0.00% | 0.869 | 0.00 | 1.000 | 0.16 | 0.896 | F | |
| Middle East | 5 | GC+CC vs GG | 642/673 | 0.530(0.320-0.877) | 0.014 | 64.30% | 0.024 | 0.73 | 0.462 | -0.99 | 0.394 | R | |
| Africa | 2 | GC+CC vs GG | 116/100 | 1.915(1.107-3.314) | 0.020 | 49.40% | 0.160 | 0.00 | 1.000 | NA | NA | F | |
| North America | 2 | GC+CC vs GG | 620/1017 | 1.009(0.735-1.385) | 0.956 | 57.20% | 0.127 | 0.00 | 1.000 | NA | NA | R | |
| HWE | |||||||||||||
| Y | 13 | GC+CC vs GG | 2524/3359 | 0.942(0.780-1.137) | 0.532 | 55.80% | 0.007 | 0.06 | 0.951 | 0.02 | 0.982 | R | |
| Overall | 16 | CC vs GG+GC | 2802/3708 | 0.935(0.821-1.065) | 0.310 | 34.60% | 0.086 | 1.22 | 0.224 | 0.98 | 0.343 | F | |
| Region | |||||||||||||
| Europe | 4 | CC vs GG+GC | 468/641 | 0.914(0.581-1.439) | 0.699 | 0.00% | 0.839 | -0.34 | 1.000 | 0.23 | 0.837 | F | |
| Asia | 3 | CC vs GG+GC | 956/1277 | 1.149(0.803-1.645) | 0.447 | 59.10% | 0.087 | 1.04 | 0.296 | 2.69 | 0.227 | R | |
| Middle East | 5 | CC vs GG+GC | 642/673 | 0.631(0.483-0.823) | 0.001 | 0.00% | 0.431 | 0.24 | 0.806 | -0.60 | 0.590 | F | |
| Africa | 2 | CC vs GG+GC | 116/100 | 2.211(0.674-7.256) | 0.191 | 0.00% | 0.581 | 0.00 | 1.000 | NA | NA | F | |
| North America | 2 | CC vs GG+GC | 620/1017 | 1.151(0.837-1.582) | 0.387 | 0.00% | 0.739 | 0.00 | 1.000 | NA | NA | F | |
| HWE | |||||||||||||
| Y | 13 | CC vs GG+GC | 2524/3359 | 0.959(0.834-1.103) | 0.556 | 40.20% | 0.066 | 0.55 | 0.583 | 0.37 | 0.722 | F | |
| rs3746444 (miRNA-499) |
|||||||||||||
| Overall | 9 | C vs T | 1678/2125 | 1.422(1.159-1.745) | 0.001 | 63.30% | 0.005 | 1.15 | 0.251 | 1.30 | 0.234 | R | |
| Region | |||||||||||||
| Asia | 2 | C vs T | 594/816 | 1.128(0.904-1.406) | 0.286 | 0.00% | 0.573 | 0.00 | 1.000 | NA | NA | F | |
| Middle East | 6 | C vs T | 672/823 | 1.614(1.219-2.137) | 0.001 | 66.80% | 0.010 | 0.75 | 0.452 | 0.77 | 0.485 | R | |
| North America | 1 | C vs T | 412/486 | 1.215(0.845-1.748) | 0.294 | NA | NA | NA | NA | NA | NA | F | |
| HWE | |||||||||||||
| Y | 7 | C vs T | 1479/1815 | 1.390(1.214-1.591) | 0.001 | 31.00% | 0.191 | 0.30 | 0.764 | 0.99 | 0.368 | F | |
| Overall | 9 | TC+CC vs TT | 1678/2125 | 1.409(1.072-1.852) | 0.014 | 67.50% | 0.002 | 0.10 | 0.917 | 1.03 | 0.335 | R | |
| Region | |||||||||||||
| Asia | 2 | TC+CC vs TT | 594/816 | 1.138(0.891-1.454) | 0.301 | 0.00% | 0.374 | 0.00 | 1.000 | NA | NA | F | |
| Middle East | 6 | TC+CC vs TT | 672/823 | 1.634(1.062-2.513) | 0.026 | 73.40% | 0.002 | 0.75 | 0.452 | 0.10 | 0.924 | R | |
| North America | 1 | TC+CC vs TT | 412/486 | 1.188(0.809-1.742) | 0.379 | NA | NA | NA | NA | NA | NA | F | |
| HWE | |||||||||||||
| Y | 7 | TC+CC vs TT | 1479/1815 | 1.399(1.190-1.645) | 0.001 | 38.20% | 0.138 | 0.00 | 1.000 | 1.11 | 0.319 | F | |
| Overall | 9 | CC vs TT+TC | 1678/2125 | 1.872(1.420-2.467) | 0.001 | 0.00% | 0.588 | 0.10 | 0.917 | 0.42 | 0.686 | F | |
| Region | |||||||||||||
| Asia | 2 | CC vs TT+TC | 594/816 | 1.224(0.551-2.717) | 0.619 | 0.00% | 0.495 | 0.00 | 1.000 | NA | NA | F | |
| Middle East | 6 | CC vs TT+TC | 672/823 | 1.960(1.455-2.641) | 0.001 | 0.00% | 0.462 | 0.38 | 0.707 | 0.66 | 0.545 | F | |
| North America | 1 | CC vs TT+TC | 412/486 | 3.557(0.369-34.331) | 0.273 | NA | NA | NA | NA | NA | NA | F | |
| HWE | |||||||||||||
| Y | 7 | CC vs TT+TC | 1479/1815 | 1.910(1.334-2.736) | 0.001 | 0.00% | 0.696 | 0.00 | 1.000 | -0.21 | 0.840 | F | |
R: random-effects model; F: fixed-effects model; Y: yes (studies that suitable for HWE); PH: the value of P in the heterogeneity test.
Figure 4.
The plot of sensitivity analysis about association between miRNA-499 rs3746444 and inflammatory arthritis.
4. Discussion
Both environmental factors and genetic factors are related to the development of inflammatory arthritis. As for miRNAs, it is reported that they are involved in cancers and autoimmune diseases [15, 21]. But some recent studies about miRNA-146/499 polymorphisms and inflammatory arthritis are inconsistent; it needs us to explore the real association between miRNA-146/499 polymorphisms and inflammatory arthritis by conducting this meta-analysis.
Present meta-analysis included 18 relevant studies, 9 only about miRNA-146 rs2910164, 3 only about miRNA-499 rs3746444, and 6 about both miRNA-146 rs2910164 and miRNA-499 rs3746444. There were 5 studies not met Hardy–Weinberg equilibrium; we did not exclude those 5 studies. We conducted subgroup analysis according to HWE and found that the results are similar (Table 2).
Of course, there were some limitations in this meta-analysis. First, we searched literature based on English; it may cause language bias. Second, the number of included studies was not sufficient, which could not conduct comprehensive analysis. Third, our data of meta-analysis were from retrospective research, which may be related to the methodological deficiencies. Finally, our meta-analysis did not take the interactions between environmental factors and genetic factors into account. Therefore, the results should be interpreted with caution.
In conclusion, the present study indicates that miRNA-499 rs3746444 polymorphism is associated with inflammatory arthritis susceptibility. However, there is lack of association between miRNA-146 rs2910164 polymorphism and inflammatory arthritis susceptibility. But, we find miRNA-146 rs2910164 and miRNA-499 rs3746444 polymorphism are associated with inflammatory arthritis in Middle East. Therefore, it is necessary to conduct more large-sample and more high-quality studies to further validate our findings.
Acknowledgments
This study was supported by grants from the Anhui Medical University Doctoral Research Foundation (XJ201414), Anhui Province Postdoctoral Foundation (2017B237), and the National Natural Science Foundation of China (11801010; 11526034).
Conflicts of Interest
All members declare they have no conflicts of interest.
References
- 1.Miyabe Y., Miyabe C., Luster A. D. LTB4 and BLT1 in inflammatory arthritis. Seminars in Immunology. 2017;33:52–57. doi: 10.1016/j.smim.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Song G., Bae S., Seo Y., et al. The association between susceptibility to inflammatory arthritis and miR-146a, miR-499 and IRAK1 polymorphisms a meta-analysis. Zeitschrift für Rheumatologie. 2015;74(7):637–645. doi: 10.1007/s00393-014-1493-x. [DOI] [PubMed] [Google Scholar]
- 3.Chatzikyriakidou A., Voulgari P. V., Georgiou I., Drosos A. A. The role of microRNA-146a (miR-146a) and its target IL-1R-associated kinase (IRAK1) in psoriatic arthritis susceptibility. Scandinavian Journal of Immunology. 2010;71(5):382–385. doi: 10.1111/j.1365-3083.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 4.Chatzikyriakidou A., Voulgari P. V., Georgiou I., Drosos A. A. A polymorphism in the 3'-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine. 2010;77(5):411–413. doi: 10.1016/j.jbspin.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Yang X.-K., Li P., Zhang C., et al. Association between IRAK1 rs3027898 and miRNA-499 rs3746444 polymorphisms and rheumatoid arthritis: a case control study and meta-analysis. Zeitschrift für Rheumatologie. 2017;76(7):622–629. doi: 10.1007/s00393-016-0169-0. [DOI] [PubMed] [Google Scholar]
- 6.Bogunia-Kubik K., Wysoczańska B., Piątek D., Iwaszko M., Ciechomska M., Świerkot J. Significance of polymorphism and expression of miR-146a and NFkB1 genetic variants in patients with rheumatoid arthritis. Archivum Immunologiae et Therapia Experimentalis. 2016;64(suppl. 1):131–136. doi: 10.1007/s00005-016-0443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaker O. G., El Boghdady N. A., El Sayed A. E.-D. Association of MiRNA-146a, MiRNA-499, IRAK1 and PADI4 polymorphisms with rheumatoid arthritis in Egyptian population. Cellular Physiology and Biochemistry. 2018;46(6):2239–2249. doi: 10.1159/000489592. [DOI] [PubMed] [Google Scholar]
- 8.Yang B., Chen J., Li Y., et al. Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Human Immunology. 2012;73(1):101–106. doi: 10.1016/j.humimm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Singh S., Rai G., Aggarwal A. Association of microRNA-146a and its target gene IRAK1 polymorphism with enthesitis related arthritis category of juvenile idiopathic arthritis. Rheumatology International. 2014;34(10):1395–1400. doi: 10.1007/s00296-014-3001-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X., Zhu J., Zhang H., Zhou G., Huang Y., Liu R. Is the microRNA-146a (rs2910164) polymorphism associated with rheumatoid arthritis? Association of microRNA-146a (rs2910164) polymorphism and rheumatoid arthritis could depend on gender. Joint Bone Spine. 2015;82(3):166–171. doi: 10.1016/j.jbspin.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Hassine H. B., Boumiza A., Sghiri R., et al. Micro RNA-146a but not IRAK1 is associated with rheumatoid arthritis in the tunisian population. Genetic Testing and Molecular Biomarkers. 2017;21(2):92–96. doi: 10.1089/gtmb.2016.0270. [DOI] [PubMed] [Google Scholar]
- 12.Fattah S. A., Ghattas M. H., Saleh S. M., Abo-Elmatty D. M. Pre-micro RNA-499 gene polymorphism rs3746444 T/C is associated with susceptibility to rheumatoid arthritis in Egyptian population. Indian Journal of Clinical Biochemistry. 2018;33(1):96–101. doi: 10.1007/s12291-017-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maharaj A. B., Naidoo P., Ghazi T., et al. MiR-146a G/C rs2910164 variation in South African Indian and Caucasian patients with psoriatic arthritis. BMC Medical Genetics. 2018;19(1):p. 48. doi: 10.1186/s12881-018-0565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayeldeen G., Nassar Y., Ahmed H., Shaker O., Gheita T. Possible use of miRNAs-146a and -499 expression and their polymorphisms as diagnostic markers for rheumatoid arthritis. Molecular and Cellular Biochemistry. 2018;449(1-2):145–156. doi: 10.1007/s11010-018-3351-7. [DOI] [PubMed] [Google Scholar]
- 15.Alemán-Ávila I., Jiménez-Morales M., Beltrán-Ramírez O., et al. Functional polymorphisms in pre-miR146a and pre-miR499 are associated with systemic lupus erythematosus but not with rheumatoid arthritis or Graves’ disease in Mexican patients. Oncotarget. 2017;8(54):91876–91886. doi: 10.18632/oncotarget.19621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Shal A. S., Aly N. M., Galil S. M. A., Moustafa M. A., Kandel W. A. Association of microRNAs genes polymorphisms with rheumatoid arthritis in Egyptian female patients. Joint Bone Spine. 2013;80(6):626–631. doi: 10.1016/j.jbspin.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Hashemi M., Eskandari-Nasab E., Zakeri Z., et al. Association of pre-miRNA-146a rs2910164 and pre miRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Molecular Medicine Reports. 2013;7(1):287–291. doi: 10.3892/mmr.2012.1176. [DOI] [PubMed] [Google Scholar]
- 18.Yang B., Zhang J. L., Shi Y. Y., et al. Association study of single nucleotide polymorphisms in premiRNA and rheumatoid arthritis in a Han Chinese population. Molecular Biology Reports. 2011;38(8):4913–4919. doi: 10.1007/s11033-010-0633-x. [DOI] [PubMed] [Google Scholar]
- 19.Ciccacci C., Conigliaro P., Perricone C., et al. Polymorphisms in STAT-4, IL-10, PSORS1C1, PTPN2 and MIR146A genes are associated differently with prognostic factors in Italian patients affected by rheumatoid arthritis. Clinical & Experimental Immunology. 2016;186(2):157–163. doi: 10.1111/cei.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiménez-Morales S., Gamboa-Becerra R., Baca V., et al. MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens. 2012;80(4):317–321. doi: 10.1111/j.1399-0039.2012.01929.x. [DOI] [PubMed] [Google Scholar]
- 21.Toraih E. A., Ismail N. M., Toraih A. A., Hussein M. H., Fawzy M. S. Precursor miR-499a variant but not miR-196a2 is associated with rheumatoid arthritis susceptibility in an Egyptian population. Molecular Diagnosis & Therapy. 2016;20(3):279–295. doi: 10.1007/s40291-016-0194-3. [DOI] [PubMed] [Google Scholar]