Abstract
Background
Clubroot, caused by Plasmodiophora brassicae Woronin, is a very important disease of Brassica species. Management of clubroot relies heavily on genetic resistance. In a cross of Brassica nigra lines PI 219576 (highly resistant, R) × CR2748 (highly susceptible, S) to clubroot, all F1 plants were resistant to clubroot. There was a 1:1 ratio of R:S in the BC1 and 3R:1S in the F2, which indicated that a single dominant gene controlled clubroot resistance in PI 219576. This gene was designated Rcr6. Mapping of Rcr6 was performed using genome sequencing information from A-genome of B. rapa and B-genome of B. nigra though bulked segregant RNA sequencing (BSR-Seq) and further mapping with Kompetitive Allele Specific PCR (KASP) analysis.
Results
Reads of R and S bulks from BSR-Seq were initially aligned onto B. rapa (A-genome; B. nigra has the B-genome) where Rcr6 was associated with chromosome A08. KASP analysis showed that Rcr6 was flanked by SNP markers homologous to the region of 14.8–15.4 Mb of chromosome A08. There were 190 genes annotated in this region, with five genes (Bra010552, Bra010588, Bra010589, Bra010590 and Bra010663) identified as encoding the toll-interleukin-1 receptor / nucleotide-binding site / leucine-rich-repeat (TIR-NBS-LRR; TNL) class of proteins. The reads from BSR-Seq were then aligned into a draft B-genome of B. nigra, where Rcr6 was mapped on chromosome B3. KASP analysis indicated that Rcr6 was located on chromosome B3 in a 0.5 Mb region from 6.1–6.6 Mb. Only one TNL gene homologous to the B. rapa gene Bra010663 was identified in the target region. This gene is a likely candidate for Rcr6. Subsequent analysis of the Rcr6 equivalent region based on a published B. nigra genome was performed. This gene is located into chromosome B7 of the published B-genome, homologous to BniB015819.
Conclusion
Rcr6 was the first gene identified and mapped in the B-genome of Brassica species. It resides in a genomic region homologous to chromosome A08 of A-genome. Based on this finding, it could possibly integrate into A08 of B. napus using marker assisted selection with SNP markers tightly linked to Rcr6 developed in this study.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1844-5) contains supplementary material, which is available to authorized users.
Keywords: Brassica nigra, Brassica rapa, Clubroot, Plasmodiophora brassicae, Bulked segregant RNA-sequencing
Background
Brassica species are important oilseed and vegetable crops worldwide. The genomic relationships among these important crop species (diploid and amphidiploid) have been summarized in the ‘Triangle of U’ [1]. The diploid species are B. rapa (L.) (AA; n = 10), B. nigra (L.) (genome BB; n = 8), and B. oleracea (L.) (CC; n = 9) and the amphidiploid species are B. juncea (L.) (AABB; n = 18), B. napus (L.) (AACC; n = 19) and B. carinata (L.) (BBCC; n = 17). The amphidiploid species were derived from interspecific hybridization between the corresponding pairs of the diploid species.
Species containing the B-genome are a useful source of genes when breeding for biotic and abiotic stress tolerance, disease resistance and oil quality [2, 3]. In Canada, transferring disease resistance from B. nigra to canola (B. napus) has potential to expand the genetic base in canola germplasm [4].
Clubroot, caused by Plasmodiophora brassicae Woronin, attacks a wide range of Brassica species. In western Canada, it was discovered in canola fields for the first time in 2003, but has spread rapidly [5] and currently poses a serious threat to canola production in Canada. Genetic resistance is the most widely used method for clubroot management. However, resistant sources for canola are limited, and no resistance is available in mustard species (B. juncea and B. carinata). Interestingly, lines effectively resistant to a broad range of pathotypes of P. brassicae have been identified in ancestral diploid species [6, 7], which could greatly broaden the genetic base of clubroot resistance (CR) in B. napus, B. juncea and B. carinata.
Several CR genes have been identified and mapped in B. rapa, B. oleracea and B. napus. For example, more than 10 loci on five chromosomes have been identified in the A-genome of B. rapa; Crr1, Crr2, Crr4, and CRc mapped to chromosomes A08, A01, A06, and A02, respectively [8, 9], and CRa, CRb, CRk, CRbkato, Rcr1 and Rcr2 were mapped onto linkage group A03 [8–19]. CRa, CRbkato and Crr1, which have been cloned from B. rapa [20–22], encode toll-interleukin-1 receptor / nucleotide binding site / leucine-rich repeat (TIR-NBS-LRR or TNL) proteins. In recent studies, six QTLs residing in five CR QTL regions of chromosomes A01, A03, and A08 were identified in B. rapa [23] and three strong QTLs (Rcr4, Rcr8 and Rcr9) were identified in chromosomes A03, A02 and A08, respectively [24].
In the C-genome of B. oleracea, one major resistance gene, Rcr7 originating from B. rapa [25], and over 20 QTLs [26, 27] have been identified. So far, at least 20 QTLs involving CR have been identified in the A and C-genomes of B. napus. A major gene, Pb-Bn1, which confers resistance to two P. brassicae collections, has been mapped to chromosome A03, and a minor QTL for each collection was mapped to linkage groups C2 and C9 [28]. Also, 19 QTLs involving race-specific resistance were mapped on eight chromosomes: A02, A03, A08, A09, C3, C5, C6 and C9 [29]. From these 19 QTLs, four were closely linked on A03 and three were linked on A08. However, mapping and identification of CR genes has not been carried out in Brassica species containing the B-genome.
Next-generation sequencing (NGS) has been used to develop the genome sequences of B. rapa [30], B. oleracea [31], B. nigra [32], B. napus [33] and B. juncea [32]. The genomes of all Brassica species underwent a lineage-specific whole-genome triplication, followed by diploidization that involved substantial genome reshuffling and gene losses [32]. Another application of NGS is gene mapping through mapping by sequencing, which has been used to map important genes in the genomes of organisms such as Arabidopsis thaliana [34–36] and pok choy [19]. In addition, the CR gene Rcr7 was successfully mapped in cabbage through analysis of percentage of polymorphic variants (PPV) based on bulked segregant RNA sequencing (BSR-Seq) [25].
A previous study identified B. nigra accession PI 219576 as highly resistant to clubroot [7]. The objectives of the current study were to: i) identify the resistance gene in PI 219576; ii) map the resistance gene using genome sequencing information from the A-genome of B. rapa and B-genome of B. nigra; iii) develop SNP markers tightly linked to the resistance gene; and iv) identify the most probable candidates for Rcr6.
Results
Inheritance of CR in line PI 219576
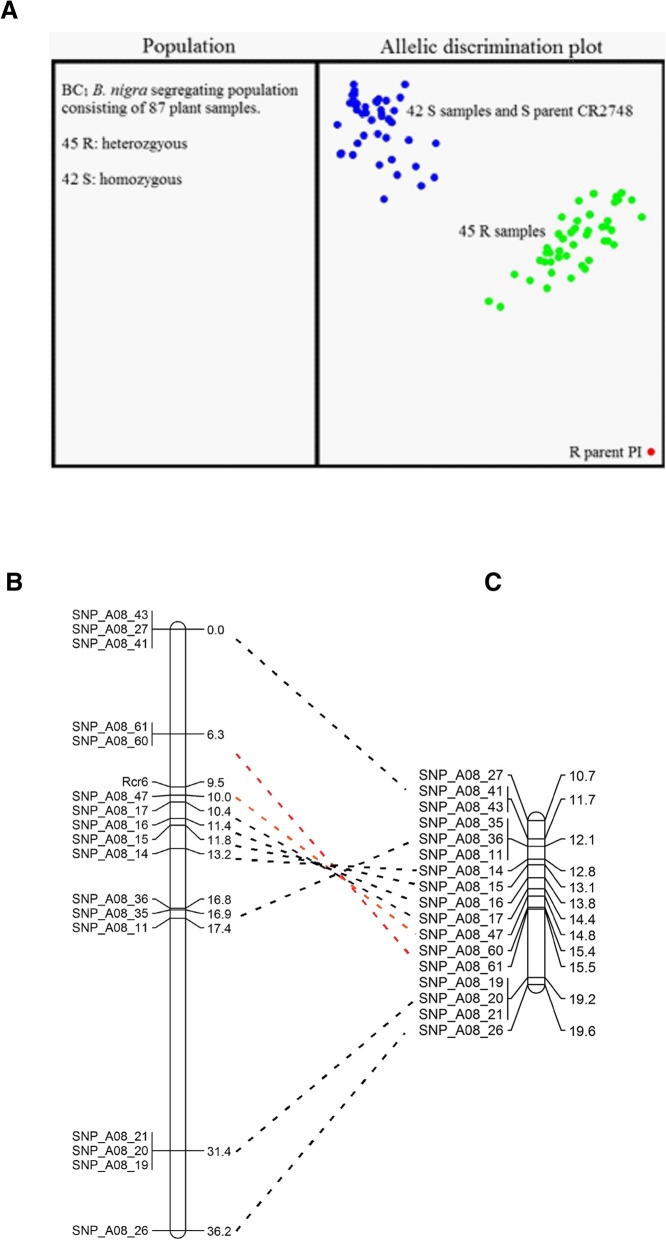
The parental line PI 219576 was highly resistant to pathotype 3 and inoculation produced no clubroot symptoms. Line CR2748 was highly susceptible with large galls (Fig. 1a). The F1 generation derived from the cross of CR2748 with PI 219576 was highly resistance to pathotype 3 (Fig. 1b), indicating that PI 219576 was likely a homozygous resistant line and the resistance was dominant. The F2 plants from the cross showed a 3:1 segregation ratio for R and S (Fig. 1b, Table 1). Evaluation of BC1 plants from PI 219576 showed a 1:1 segregation ratio (Fig. 1b, Table 1), indicating that CR in PI 219576 is controlled by a single dominant gene designated as Rcr6 (Resistance to club root 6).
Fig. 1.
Evaluation of clubroot disease reaction to inoculation with pathotype 3 under controlled conditions: (a). phenotypes of resistant line PI 2195760 and susceptible (CR2748) line; (b). distribution of phenotypes in Brassica nigra parental lines (PI 219576 and CR2748) and the F1, F2 and BC1 populations
Table 1.
Genetic analysis of resistance to clubroot in the segregating populations derived from CR2748 (Susceptible, S) x PI 219576 (Resistant, R) with pathotype 3 of Plasmodiophora brassicae
| Phenotype | ||||||
|---|---|---|---|---|---|---|
| Population | Total | R | S | Expected ratio (R:S) | χ2 | P-value |
| BC1 | 234 | 123 | 111 | 1:1 | 0.62 | 0.43 |
| F2 | 57 | 42 | 15 | 3:1 | 0.05 | 0.82 |
Assembly of RNA-seq short reads into the reference genomes of B. rapa
The pooled sample assembly (PSA) of the three R bulks and three S bulks was carried out. A total of 32.0 million (M) sequences, 2062.2 Mb in length, with 8-fold coverage of the reference A-genome were assembled into B. rapa chromosomes from the pool of three R bulks, and 39.5 M sequences, 2523.4 Mb in length, with 10-fold coverage were assembled from the pool of three S bulks (Table 2). The sequence counts assembled into the genome for each chromosome were significantly correlated to chromosome length for the R bulks (r = 0.90, P = 0.003) and S bulks (r = 0.89, P = 0.0005).
Table 2.
Short reads from the resistant (R) and susceptible (S) bulks were assembled into chromosomes of the Brassica rapa and B. nigra reference genomes
| Chromosome number | Chromosome size (bases × 106) | Number of sequences (× 106) | Accumulated length of sequences (bases × 106) | ||
|---|---|---|---|---|---|
| R | S | R | S | ||
| B. rapa | |||||
| A01 | 26.8 | 3.0 | 3.7 | 191.4 | 233.8 |
| A02 | 27.0 | 2.8 | 3.4 | 176.4 | 215.3 |
| A03 | 31.8 | 4.4 | 5.4 | 284.8 | 342.2 |
| A04 | 19.3 | 2.6 | 3.3 | 170.4 | 210.7 |
| A05 | 25.3 | 3.0 | 3.6 | 192.5 | 230.7 |
| A06 | 25.2 | 3.4 | 4.4 | 219.2 | 285.0 |
| A07 | 25.9 | 3.3 | 4.0 | 211.6 | 255.5 |
| A08 | 20.8 | 3.0 | 3.6 | 192.9 | 234.6 |
| A09 | 38.9 | 4.5 | 5.6 | 292.2 | 356.2 |
| A10 | 16.4 | 2.0 | 2.5 | 130.8 | 159.4 |
| Total | 257.4 | 32.0 | 39.5 | 2062.2 | 2523.4 |
| B. nigra | |||||
| B1 | 42.3 | 5.0 | 5.7 | 345.6 | 395.1 |
| B2 | 52.7 | 5.3 | 6.2 | 362.6 | 426.7 |
| B3 | 46.9 | 5.6 | 6.4 | 385.9 | 449.1 |
| B4 | 43.4 | 4.8 | 5.6 | 334.6 | 393.3 |
| B5 | 51.8 | 5.0 | 6.0 | 347.5 | 415.9 |
| B6 | 36.7 | 5.9 | 6.9 | 407.7 | 481.9 |
| B7 | 41.7 | 4.1 | 4.8 | 281.6 | 331.0 |
| B8 | 54.2 | 5.4 | 6.3 | 375.8 | 439.9 |
| Total | 369.7 | 41.1 | 47.9 | 2841.3 | 3332.9 |
Mapping of Rcr6 with PPV against the A-genome
In total, 120.2 K polymorphic (poly) variants (SNPs and InDels) were identified when aligned with the A-genome. Variants in the B. rapa genome generally consisted of 85–88% monomorphic (mono) variants and 12–15% poly variants. The only exception was chromosome A08, which carried more ploy variants (31%) and fewer mono variants (69%) than the other chromosomes (Fig. 2a). This difference relative to A08 indicated that Rcr6 may reside in a genomic region in the B-genome of B. nigra homologous to A-genome chromosome A08. The PPV on chromosome A08 was further analyzed. The highest PPV was located in the physical range from approximately 10–20 Mb (Fig. 2b), indicating that Rcr6 is located in a genomic region homologous to 10–20 Mb of chromosome A08.
Fig. 2.
Analysis of BSR-Seq to map Rcr6 based on the reference genome of B. rapa: (a). the percentage (%) of monomorphic and polymorphic variants on each chromosome; and (b). % polymorphic variants on chromosome A08
Defining Rcr6 location relative to the A-genome
The 221 plants in the BC1 population were assessed using KASP analysis of 17 poly SNP sites against chromosome A08; these sites spanned a distance of 10.7 Mb to 19.8 Mb (Fig. 3a). A linkage map of 36.2 cM was constructed (Fig. 3b), which confirmed that Rcr6 lay in a region homologous to 10–20 Mb of chromosome A08. Homology between chromosome A08 of B. rapa in the region and the corresponding region in B. nigra genome was confirmed through genetic mapping. However, the DNA fragment from the markers SNP_A08_61 to SNP_A08_11 in the Rcr6 region (Fig. 2c) was reversed comparing with the B. rapa chromosome (Fig. 3c). Rcr6 was flanked by SNP_A08_60 & 61 and SNP_A08_47, in an interval of 3.7 cM. The flanked segment was homologous to the region between 15.4 Mb and 14.8 Mb of chromosome A08 (Fig. 3c), spanning about 0.6 Mb region. There were 190 genes annotated in this region of reference genome v1.5, with five genes (Bra010552, Bra010588, Bra010589, Bra010590 and Bra010663) identified as encoding TNL class of proteins (Additional file 4: Table S1).
Fig. 3.
Genetic mapping of Rcr6: (a). genotyping of SNP markers using KASP. The R parent (homozygous resistant) segregated to the lower right quadrant, the S parent (homozygous recessive) and S individuals (homozygous recessive) from BC1 population formed a cluster in the upper left quadrant, and the R individuals (heterozygous resistant) from the BC1 population were positioned between the S cluster and R parent; (b). genetic map of the region in which the Rcr6 gene is located (genetic distance on right); and (c). physical location of the Rcr6 region (in bases, on right), with SNP markers connected with a broken line between the maps
Identification of the Rcr6 region in the draft B. nigra genome
The short reads from the BSR-Seq project were assembled into Dr. Parkin’s draft B. nigra genome sequence after mapping of Rcr6 with the B. rapa reference genome was completed. A total of 41.1 M sequences, 2841.3 Mb in length, with 8-fold coverage of the reference B-genome were assembled into B. nigra chromosomes from the pool of three R bulks; and 47.9 M sequences, 3332.9 Mb in length, with 9-fold coverage were assembled from the pool of three S bulks (Table 2). More sequences were aligned into chromosomes B3 and B6; fewer sequences were aligned into chromosomes B4 and B7 than in the shortest chromosome B6 (Table 2). However, the sequence counts assembled into the genome for each chromosome were not correlated to chromosome length for the R bulks (r = 0.07, P = 0.87) and S bulks (r = 0.11, P = 0.79).
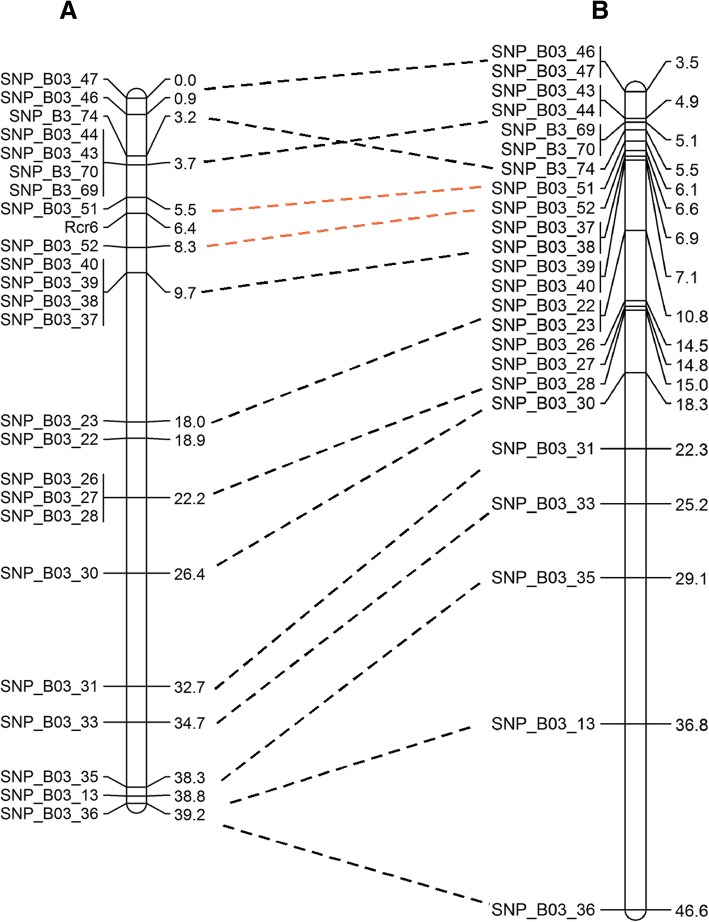
The variants identified across the B. nigra usually consisted of 63–67% mono variants and 33–37% poly variants, except for chromosome B3, which carried fewer mono variants (51%) and more poly variants (49%) than the other chromosomes (Additional file 1: Figure S1A). This indicated that Rcr6 was located in B. nigra chromosome B3. Also, there was a peak in PPV in the 36–37 Mb region of B3 (Additional file 1: Figure S1B), but there were very few poly variants in the region (Additional file 1: Figure S1C). A SNP marker, SNP_B03_13, located in 36.8 Mb of chromosome B3 was genotyped using KASP. However, it was not closely associated with Rcr6 (Fig. 4a), indicating that Rcr6 was not in the region.
Fig. 4.
Mapping of Rcr6 into chromosome B3 using the draft genome of B. nigra from Canada: (a). genetic map of the region in which the Rcr6 gene was located (genetic distance on right); and (b). physical location of the Rcr6 region in bases (on right) and genetic location, with each SNP marker connected between the two maps with a broken line
The BC1 population consisting of 221 plants was analyzed with 23 SNP markers on chromosome B3 and a linkage map covering 39.2 cM was constructed (Fig. 4a). Rcr6 was flanked by SNP_B03_51 and SNP_B03_52, in an interval of 2.8 cM. The flanked segment is homologous to the region of 6.1–6.6 Mb on chromosome B3 (Fig. 4b), spanning about 0.5 Mb. A joint map was also constructed with SNP markers developed based on both chromosomes A08 and B3 (Additional file 2: Figure S2).
There were 109 genes predicted in the Rcr6 target region of 6.1–6.6 Mb on chromosome B3. Analysis with Blast2Go indicated that only one gene (FGENESH_B3_72) encoded the TNL class of proteins (Additional file 4: Table S2). This B. nigra TNL gene is homologous to the B. rapa gene Bra010663, which is located from 15,082,118 to 15,087,789 bases of chromosome A08, based on a blast search at http://brassicadb.org/brad against the B. rapa reference genome.
Determining the location of Rcr6 in the published B. nigra genome
The short reads from the BSR-Seq project were also assembled into the published B. nigra reference genome v1.1 at http://brassicadb.org/brad. Analysis of PPV was also performed following the identification of genome wide DNA variants. The variants identified across the B. nigra genome usually comprised of 61–63% mono variants and 37–39% poly variants. Unexpectedly, chromosome B7 [32] carried fewer mono variants (49%) and more poly variants (51%) than the other chromosomes (Additional file 3: Figure S3A). This indicated that Rcr6 was located in the B. nigra chromosome B7 (designated as B7_China in the current study). This result also indicated that chromosome B3 sequenced by the research group in Canada (designated as B3_Canada) is equivalent to B7_China. Pairwise chromosome alignments of B3_Canada with the reference genome of B. nigra confirmed that B3_Canada was the same chromosome as B7_China. However, the orientation is opposite (Additional file 3: Figure S3B).
Blast search in the B. nigra reference genome v1.1 was performed using the DNA fragment in the Rcr6 target region defined by SNP_B03_51 and SNP_B03_52 at http://brassicadb.org/brad. This fragment hit B7_China with the highest score of 2.427e+ 04. The TNL gene of B3_Canada in the Rcr6 interval homologous to the B. rapa TNL gene Bra010663 hit the B. nigra TNL gene BniB015819 in B7_China with a score of 729.0. This gene is located into 36,680,688 to 36,684,758 bases of B7_China.
Discussion
Brassica species with the B-genome contain a large reservoir of genes conferring resistance to important diseases of canola such as clubroot and blackleg. Resistance to clubroot in B. nigra could be transferred into B-genome species such as B. juncea and B. carinata by interspecific hybridization or re-synthesis of these species. Due to differentiation among the Brassica genomes caused by genome rearrangements and gene deletions, the level of homology between B and A/C chromosomes is low [37], which makes introgression of genes from the B-genome into the A or C-genomes of canola (B. napus) difficult through conventional breeding. To facilitate the transfer of resistance available in the B-genome into canola, it is necessary to accurately map genes in B. nigra, so that molecular markers tightly linked to these resistance genes can be identified for use in marker-assisted selection (MAS). Alternatively, the genes could be isolated from B. nigra so that the genes could be delivered into canola through gene transformation.
When the project to map Rcr6 was initiated, no reference genome for B. nigra was available. Therefore, Rcr6 was initially mapped against a reference A-genome of B. rapa [30]. The genetic location of Rcr6 was further defined using SNP markers identified though BSR-Seq based on the B. rapa reference genome. This demonstrated that mapping by sequencing was possible in a species using the genome sequence from a close relative if its own genome sequence was not available. In the current study, Rcr6 was mapped into chromosome A08 using the reference genome from B. rapa.
A draft genome of B. nigra sequenced in Canada subsequently became available. Rcr6 was then mapped into B. nigra chromosome B3 (designated here as B3_Canada), which indicated that the target region of Rcr6 in B3_Canada of B. nigra was homologous to the corresponding region in chromosome A08 of B. rapa. Rcr6 was also mapped using a published B. nigra sequence from http://brassicadb.org/brad in China [32] into chromosome B7_China. The Canadian group used a nomenclature published by Lagercrantz and Lydiate [38], while the Chinese group accepted a proposal for nomenclature described by Panjabi et al. [39]. Therefore, it is not surprising that Rcr6 was mapped into different chromosomes using these two separately derived sources of the B. nigra genome.
In a previous study, RNA-Seq and block homoeology analysis had identified synteny between the A and B-genomes in B. juncea [40]. Based on the current study, we believe that Rcr6 could possibly integrate into A08 of B. napus in inter-specific crosses between B. napus and B. nigra, but this speculation has not yet been confirmed. Despite the challenges involved in transferring agronomically important traits from the B-genome species into B. napus, a blackleg resistance gene from the B-genome of B. juncea, Rlm6, has been successfully introgressed into B. napus [2].
Substantial effort has been made to identify genes or QTLs for CR in Brassica species containing A- or C-genome. In contrast, Rcr6 is the first CR gene to be finely mapped in a B-genome species. It was located on chromosome B3_Canada, homologous to 14.8–15.4 Mb of Mb of chromosome A08. Five genes (Bra010552, Bra010588, Bra010589, Bra010590 and Bra010663) were identified as encoding the TNL-class of proteins in this interval. The CR gene Crr1 has previously been mapped into chromosome A08 [8] and cloned [20]. It is highly homologous to gene Bra020861, which located in the 10.8 Mb region of chromosome A08. In addition, a strong QTL, Rcr9, was also mapped into chromosome A08 [24]. The nearest SNP to Rcr9 was A08_10272562. The current study indicated that Rcr6 was not located on the corresponding genomic region of Rcr9 or Crr1 on chromosome A08. In the current study, the only TNL gene identified in the Rcr6 interval is FGENESH_B3_72 (Additional file 4: Table S2), which is highly homologous to the B. rapa TNL gene Bra010663 and the B. nigra TNL gene BniB015819. The availability of closely linked SNP markers will facilitate molecular cloning of Rcr6 from the donor line. Although the cloned CR genes encode the TNL-class of proteins [20–22], it is possible that clubroot resistance is encoded by other classes of disease resistance genes. However, further clarification of the relationship between classes of disease resistance proteins and Rcr6 was beyond the scope of the present study.
Breeding for CR can be severely constrained when the pathotype of interest does not occur near the site of established breeding institutions [19], so the use of molecular markers in marker-assisted selection can be extremely important. More than 10 robust SNP markers developed in this study were shown to be tightly linked to Rcr6 using KASP. These markers could facilitate introgression of Rcr6 into canola.
Until recently, only five pathotypes of P. brassicae (2, 3, 5, 6 and 8, based on Williams’ differentials) had been identified in Canada and pathotype 3 was the most prevalent pathotype on canola. Line PI 219576 of B. nigra, which was the resistant parent in the cross examined in the current study, was highly resistant to all of these pathotypes [7]. Recently, 12 new pathotypes of P. brassicae virulent on canola were identified using a new Canadian Clubroot Differential set [41]. PI 219576 also showed resistance to all of these new pathotypes (data not shown). Rcr6 from PI 219576 was identified through testing for disease reaction to pathotype 3 only, so studies to confirm that Rcr6 confers resistance to other pathotypes are needed.
RNA-Seq produces a large amount of short DNA sequence reads from random places in the transcriptome. The RNA-Seq data generated from the R and S bulks in the BC1 population was aligned into the A-genome of B. rapa and B-genome of B. nigra. This study characterized genome-wide variants in the B. nigra population that carried Rcr6 and demonstrated that the sequence counts assembled into the A-genome of B. rapa for each chromosome were correlated to chromosome length from the R and S bulks, which was consistent with a previous study [19]. However, there was no correlation between assembled sequence lengths and the B-genome of B. nigra, which was not anticipated and requires additional study.
A previous study identified a high proportion of PPV on chromosome A03 of B. rapa adjacent to the CR gene Rcr1 [19] and Rcr7 was mapped into chromosome C7 of B. oleracea through identification of PPV [25]. In the current study, Rcr6 was mapped into 10–20 Mb of A08 of B. rapa and B. nigra B3_Canada. However, the peak region based on PPV on B3 was not closely associated with Rcr6. One reason for this could be the low number of variants identified in the region due to a relatively low depth of RNA-Seq reads (Additional file 1: Figure S1). Greater depth of sequencing for genetic mapping via identification of PPV is recommended for future research.
Conclusion
We aimed to identify novel CR genes from B. nigra that can be used in canola and mustard crops in the Canadian prairies. Rcr6 was the first gene for resistance to clubroot identified and mapped in the B-genome of Brassica species. The sequencing information from B. rapa and B. nigra was used for BRS-Seq to map the gene. It resides in the genomic region of B. nigra homologous to chromosome A08 of B. rapa. Based on this funding, we believe that it could possibly integrate into A08 of canola crop B. napus. SNP markers tightly linked to the gene were developed, facilitating canola and mustard breeders for use of MAS in introgression of the CR gene into their cultivars. This gene was mapped into a small interval with one TNL gene (BniB015819) identified. This gene can be the candidate of Rcr6 for gene cloning.
Methods
Plant materials
Nutrien Ag Solutions (201–407 Downey Rd., Saskatoon, SK, Canada) provided seed of the clubroot-resistant line PI 219576. The susceptible line CR2748 was provided by IPK (Leibniz Institute of Plant Genetics and Crop Plant Research, OT Gatersleben, Corrensstrasse 3, D-06466 Seeland, Germany). The resistant line (male) was crossed with the susceptible line (female) to produce the F1. Self-pollination of F1 plants produced the F2. A resistant F1 plant (male) was backcrossed with the susceptible line (female) to produce a BC1 population.
Assessing clubroot resistance
Seed was planted in Sunshine #3 soil-less planting mix (SunGro Horticulture, Vancouver, BC) in tall, narrow plastic pots (5-cm diameter, 20-cm height, Steuwe & Sons, Corvalis, OR). The soil mix was treated with 1% (w/v) 16–8-12 (N-P-K) control-released fertilizer. Plants were maintained in a greenhouse (~ 22°/18 °C, day/night) with a 14-h photoperiod (230 μmol/m2/s at the canopy level). After inoculation with P. brassicae, plants were transferred to a growth room at 23°/20 °C and 14-h photoperiod (512 μmol/m2/s).
Pathotype 3 of P. brassicae (Williams’ differential system), which was originated from a canola field infected with clubroot in Alberta Canada, was used for inoculation throughout the study. The inoculum was prepared as a spore suspension with the concentration adjusted to 1 × 107 resting spores mL− 1. For inoculation, 5 mL of spore suspension was applied adjacent to the seed, resulting in about 1 × 106 spores g− 1 growth medium. Inoculated seedlings in the growth room were maintained at a high soil moisture level for 2 weeks by retaining an excessive amount of water in the tray under the pots.
At 5 weeks post-inoculation, clubroot severity was assessed using a 0–3 scale where 0 = no clubbing; 1 = a few small clubs; 2 = moderate clubs; and 3 = severe clubs [42]; a rating of 0 was considered resistant (R) and 1–3 were susceptible (S). The F2 populations were analyzed to confirm the expected genetic analysis ratio of 3R: 1S and the BC1 populations (expected 1R: 1S ratio) were used for genetic mapping. The goodness of fit for the segregation was analyzed using a Chi-square (X2) Test [43].
DNA extraction
DNA was extracted from young leaves using the CTAB method [44] with the following modifications: freeze-dried leaf samples were incubated with extraction buffer (2% CTAB; pH 8.0) at 65 °C, followed by extraction in chloroform-isoamylalcohol (24:1, v/v) and alcohol precipitation. RNA was eliminated by adding 1/10 volume of 10 mg/mL RNase A. The DNA concentration was estimated using a NanoDrop ND-2000c spectrophotometer (Thermo Scientific, Wilmington, DE).
RNA-Seq
The BC1 population was used for RNA-Seq analysis. At 15 days post-inoculation, leaf tissues from 30 R plants were combined to form an R bulk, and tissues from 30 S plants were combined to form an S bulk. Together, the two bulks comprised one biological replicate. There were three replicates, with a total of 90 R and 90 S plants assessed. Total RNA from each bulk was extracted using the RNeasy Plant Mini Kit (Qiagen; Toronto, ON) with on-column deoxyribonuclease (DNAse) digestion using a Qiagen RNase-Free DNase Set, following the manufacturer’s instruction. RNA concentration and purity were checked using a NanoDrop ND-2000c spectrophotometer. The Experion RNA StdSens analysis kit (Bio-Rad Laboratories, Inc., Montreal, QC) was used for RNA quality analysis, with the Experion automated electrophoresis system. RNA quality was analyzed to ensure that the RNA integrity number (RIN) was > 8 for each sample.
cDNA libraries were prepared following the TruSeq RNA Sample Preparation v2 Guide (Illumina; San Diego, CA). The NanoDrop ND-2000c spectrophotometer was used to check cDNA concentrations and purity. Quality control and qPCR analysis were carried out to validate the cDNA libraries. The Experion DNA 1 K Analysis Kit (Bio-Rad Laboratories, Inc.) was used to confirm the size and purity of the cDNA libraries, based on a band at approximately 260 bps. The KAPA Library Quantification Kit v4.11 was used to perform qPCR of cDNA libraries.
RNA-Seq was carried out on samples from each inoculated R and S bulk using the Illumina MiSeq platform at the University of Saskatchewan (Saskatoon, SK, Canada).
Sequence alignment, SNP discovery and mapping of the causal gene
No DNA sequence of the B-genome of B. nigra was available when mapping of the CR gene in B. nigra resistant line PI 219576 was initiated. Therefore, high-quality reads from BSR-Seq were aligned to the A-genome of B. rapa ssp. pekinesis (Chinese cabbage) cv. Chiifu genome v1.5 (http://brassicadb.org/brad) following PSA method described by Yu et al. [19] using SeqMan NGen 12 software (DNASTAR, Madison, WI). The reference A-genome consists of 10 chromosomes, with total combined length of about 257 million bases (Mb). Subsequently, a draft B-genome of B. nigra (developed by Dr. I. Parkin) became available for this study. The total length of the eight chromosomes in the draft genome was about 370 Mb. The short reads from the BSR-Seq project were aligned to the chromosomes of the draft genome. SNP discovery for marker development was carried out using SeqMan Pro 12 software (DNASTAR). The SNP discovery parameter was set at default, with Q call ≥15 and depth ≥ 5. Mapping of the causal gene was performed using PPV [25].
Gene prediction and annotation
Gene prediction from the target region of the draft B-genome was performed at http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind using “Dicot, Arabidopsis (generic)” as the organism-specific finding parameter. Gene annotation for DNA fragments in the target region was analyzed with Blast2GO [45].
SNP genotyping and linkage analysis
Selected SNPs identified through BSR-Seq were confirmed using the Kompetitive Allele Specific PCR (KASP) method (http://www.lgcgroup.com/), following the manufacturer’s instruction. PCR reactions were performed using a StepOne Plus Real Time PCR System (Applied Biosystem, Mississauga, ON, Canada). Genetic linkage maps were constructed by JoinMap 4.1 software, with distance measured in centimorgans (cM).
Locating the causal gene in a published B. nigra genome
A B. nigra reference genome (v1.1) from China [32] became available at http://brassicadb.org/brad in 2017. Since this version had already been published and so was publically available, this was the version that was selected for the final version of alignment and analysis. The short reads from the BRS-Seq project were aligned to B. nigra v1.1 using SeqMan NGen 12 to identify the corresponding chromosome related to the causal gene. Additional confirmation was provided from pairwise chromosome alignments using two methods; i) the Blastn command line tool and the top hits with ≥98% to identity the query sequence in the reference genome, and ii) use LASTZ version 1.04.00 [46] to carry out pairwise chromosome alignments. Results were visualized using the Artemis comparison tool ACT [47]. The target region for the causal gene was searched at http://brassicadb.org/brad using the Blast tool.
Additional files
Figure S1. Mapping Rcr6 based on BSR-Seg using a draft genome of B. nigra from Canada: A. The percentage of monomorphic and polymorphic variants on each chromosome; B. distribution of the percentage of polymorphic variants on chromosome B3; and C. distribution of polymorphic variants on chromosome B3. (JPG 434 kb)
Figure S2. The genetic map of Rcr6 with SNP markers identified through BSR-Seq from both chromosomes A08 and B3. (JPG 493 kb)
Figure S3. Mapping of Rcr6 into chromosome B7 using the published B-genome of B. nigra from China: A. the percentage (%) of monomorphic and polymorphic variants on each chromosome; and B. comparison of B3_Canada and B7_China. The dot plot was created using R (https://cran.r-project.org/). (JPG 1126 kb)
Table S1. Blast2GO result in the Rcr6 target region of chromsome A08. Table S2. Blast2GO result in the Rcr6 target region of chromsome B3. (XLSX 59 kb)
Acknowledgements
We thank L. McGregor and M. Kehler for technical assistance, Dr. Qilin Chen for KASP analysis, C. Franke for providing the B. nigra resistant line and Dr. M. Hecker for providing the MiSeq platform for RNA-Seq.
Abbreviations
- BSR-Seq
Bulked segregant RNA sequencing
- CR
Clubroot resistance
- MAS
Marker-assisted selection
- PPV
Percentage of polymorphic variants
- PSA
Pooled sample assembly
- R
Resistant
- Rcr6
Resistance to club root 6
- S
Susceptible
- SNP
Single-nucleotide polymorphism
- TNL
TIR-NBS-LRR, toll-interleukin-1 receptor / nucleotide-binding site / leucine-rich-repeat
Authors’ contributions
FY and YW conceived of and designed the study; AC conducted the experiments; AC, ML, FY and HH analyzed data; IAPP, BDG and GP provided important information and materials. AC and FY drafted the manuscript. All authors reviewed the manuscript and approved the final draft.
Funding
This work was funded by a competitive grant from Agriculture and Agri-Food Canada. The funding body played no role in the design of the study and collection, analysis, and.interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Adrian Chang, Email: adrian.chang.ca@gmail.com.
Mebarek Lamara, Email: m.lamara2@gmail.com.
Yangdou Wei, Email: yangdou.wei@usask.ca.
Hao Hu, Email: hao.hu@canada.ca.
Isobel A. P. Parkin, Email: isobel.parkin@canada.ca
Bruce D. Gossen, Email: bruce.gossen@canada.ca
Gary Peng, Email: gary.peng@canada.ca.
Fengqun Yu, Email: Fengqun.yu@canada.ca.
References
- 1.Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot. 1935;7(7):389–452. [Google Scholar]
- 2.Chevre A, Eber F, Barret P, Dupuy P, Brace J. Identification of the different Brassica nigra chromosomes from both sets of B. oleracea-B. nigra and B. napus-B. nigra addition lines with a special emphasis on chromosome transmission and self-incompatibility. Theor Appl Genet. 1997;94(5):603–611. doi: 10.1007/s001220050457. [DOI] [Google Scholar]
- 3.Struss D, Bellin U, Röbbelen G. Development of B-genome chromosome addition lines of B. napus using different interspecific Brassica hybrids. Plant Breed. 1991;106(3):209–214. doi: 10.1111/j.1439-0523.1991.tb00503.x. [DOI] [Google Scholar]
- 4.Gerdemann-Knörck M., Nielen S., Tzscheetzsch C., Iglisch J., Schieder O. Developments in Plant Breeding. Dordrecht: Springer Netherlands; 1995. Transfer of disease resistance within the genus Brassica through asymmetric somatic hybridization; pp. 247–253. [Google Scholar]
- 5.Gossen BD, Deora A, Peng G, Hwang S-F, McDonald MR. Effect of environmental parameters on clubroot development and the risk of pathogen spread. Can J Plant Pathol. 2014;36(sup1):37–48. doi: 10.1080/07060661.2013.859635. [DOI] [Google Scholar]
- 6.Jakir Hasan M, Strelkov SE, Howard RJ, Rahman H. Screening of Brassica germplasm for resistance to Plasmodiophora brassicae pathotypes prevalent in Canada for broadening diversity in clubroot resistance. Can J Plant Sci. 2012;92(3):501–515. doi: 10.4141/cjps2010-006. [DOI] [Google Scholar]
- 7.Peng G, Falk KC, Gugel RK, Franke C, Yu F, James B, Strelkov SE, Hwang S-F, McGregor L. Sources of resistance to Plasmodiophora brassicae (clubroot) pathotypes virulent on canola. Can J Plant Pathol. 2014;36(1):89–99. doi: 10.1080/07060661.2013.863805. [DOI] [Google Scholar]
- 8.Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 2006;173(1):309–319. doi: 10.1534/genetics.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakamoto K, Saito A, Hayashida N, Taguchi G, Matsumoto E. Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis) Theor Appl Genet. 2008;117(5):759–767. doi: 10.1007/s00122-008-0817-0. [DOI] [PubMed] [Google Scholar]
- 10.Chu M, Song T, Falk KC, Zhang X, Liu X, Chang A, Lahlali R, McGregor L, Gossen BD, Yu F. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics. 2014;15(1):1166. doi: 10.1186/1471-2164-15-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirai M, Harada T, Kubo N, Tsukada M, Suwabe K, Matsumoto S. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor Appl Genet. 2004;108(4):639–643. doi: 10.1007/s00122-003-1475-x. [DOI] [PubMed] [Google Scholar]
- 12.Huang Z, Peng G, Liu X, Deora A, Falk KC, Gossen BD, McDonald MR, Yu F. Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front Plant Sci. 2017;8:1448. doi: 10.3389/fpls.2017.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato T, Hatakeyama K, Fukino N, Matsumoto S. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed Sci. 2013;63(1):116–124. doi: 10.1270/jsbbs.63.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuginuki Y, Ajisaka H, Yui M, Yoshikawa H, Hida K-i, Hirai M. RAPD markers linked to a clubroot-resistance locus in Brassica rapa L. Euphytica. 1997;98(3):149–154. doi: 10.1023/A:1003147815692. [DOI] [Google Scholar]
- 15.Matsumoto E, Yasui C, Ohi M, Tsukada M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis) Euphytica. 1998;104(2):79. doi: 10.1023/A:1018370418201. [DOI] [Google Scholar]
- 16.Piao Z, Deng Y, Choi S, Park Y, Lim Y. SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis) Theor Appl Genet. 2004;108(8):1458–1465. doi: 10.1007/s00122-003-1577-5. [DOI] [PubMed] [Google Scholar]
- 17.Saito M, Kubo N, Matsumoto S, Suwabe K, Tsukada M, Hirai M. Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor Appl Genet. 2006;114(1):81. doi: 10.1007/s00122-006-0412-1. [DOI] [PubMed] [Google Scholar]
- 18.Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, Fukuoka H, Matsumoto S, Hirai M. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor Appl Genet. 2003;107(6):997–1002. doi: 10.1007/s00122-003-1309-x. [DOI] [PubMed] [Google Scholar]
- 19.Yu F, Zhang X, Huang Z, Chu M, Song T, Falk KC, Deora A, Chen Q, Zhang Y, McGregor L. Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PLoS One. 2016;11(4):e0153218. doi: 10.1371/journal.pone.0153218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatakeyama K, Suwabe K, Tomita RN, Kato T, Nunome T, Fukuoka H, Matsumoto S. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One. 2013;8(1):e54745. doi: 10.1371/journal.pone.0054745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatakeyama K, Niwa T, Kato T, Ohara T, Kakizaki T, Matsumoto S. The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol Gen Genomics. 2017;292(2):397–405. doi: 10.1007/s00438-016-1281-1. [DOI] [PubMed] [Google Scholar]
- 22.Ueno H, Matsumoto E, Aruga D, Kitagawa S, Matsumura H, Hayashida N. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol Biol. 2012;80(6):621–629. doi: 10.1007/s11103-012-9971-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Jing J, Zhan Z, Zhang T, Zhang C, Piao Z. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS One. 2013;8(12):e85307. doi: 10.1371/journal.pone.0085307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu F, Zhang X, Peng G, Falk KC, Strelkov SE, Gossen BD. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci Rep. 2017;7(1):4516. doi: 10.1038/s41598-017-04903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dakouri A, Zhang X, Peng G, Falk KC, Gossen BD, Strelkov SE, Yu F. Analysis of genome-wide variants through bulked segregant RNA sequencing reveals a major gene for resistance to Plasmodiophora brassicae in Brassica oleracea. Sci Rep. 2018;8(1):17657. doi: 10.1038/s41598-018-36187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocherieux J, Glory P, Giboulot A, Boury S, Barbeyron G, Thomas G, Manzanares-Dauleux M. Isolate-specific and broad-spectrum QTLs are involved in the control of clubroot in Brassica oleracea. Theor Appl Genet. 2004;108(8):1555–1563. doi: 10.1007/s00122-003-1580-x. [DOI] [PubMed] [Google Scholar]
- 27.Nagaoka T, Doullah M, Matsumoto S, Kawasaki S, Ishikawa T, Hori H, Okazaki K. Identification of QTLs that control clubroot resistance in Brassica oleracea and comparative analysis of clubroot resistance genes between B. rapa and B. oleracea. Theor Appl Genet. 2010;120(7):1335–1346. doi: 10.1007/s00122-010-1259-z. [DOI] [PubMed] [Google Scholar]
- 28.Manzanares-Dauleux M, Delourme R, Baron F, Thomas G. Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theor Appl Genet. 2000;101(5–6):885–891. doi: 10.1007/s001220051557. [DOI] [Google Scholar]
- 29.Werner S, Diederichsen E, Frauen M, Schondelmaier J, Jung C. Genetic mapping of clubroot resistance genes in oilseed rape. Theor Appl Genet. 2008;116(3):363. doi: 10.1007/s00122-007-0674-2. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 31.Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, Zhao M, Ma J, Yu J, Huang S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Liu G, Zhao N, Chen S, Liu D, Ma W, Hu Z, Zhang M. Comparative mitochondrial genome analysis reveals the evolutionary rearrangement mechanism in Brassica. Plant Biol. 2016;18(3):527–536. doi: 10.1111/plb.12414. [DOI] [PubMed] [Google Scholar]
- 33.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 34.Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW. Next-generation mapping of Arabidopsis genes. Plant J. 2011;67(4):715–725. doi: 10.1111/j.1365-313X.2011.04619.x. [DOI] [PubMed] [Google Scholar]
- 35.Mokry M, Nijman IJ, van Dijken A, Benjamins R, Heidstra R, Scheres B, Cuppen E. Identification of factors required for meristem function in Arabidopsis using a novel next generation sequencing fast forward genetics approach. BMC Genomics. 2011;12(1):256. doi: 10.1186/1471-2164-12-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen J-E, Weigel D, Andersen SU. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6(8):550. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 37.Attia T, Busso C, Röbbelen G. Digenomic triploids for an assessment of chromosome relationships in the cultivated diploid Brassica species. Genome. 1987;29(2):326–330. doi: 10.1139/g87-053. [DOI] [Google Scholar]
- 38.Lagercrantz U, Lydiate DJ. Comparative genome mapping in Brassica. Genetics. 1996;144(4):1903–1910. doi: 10.1093/genetics/144.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V, Pradhan AK, Pental D. Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the a, B and C Brassica genomes. BMC Genomics. 2008;9(1):113. doi: 10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paritosh K, Gupta V, Yadava SK, Singh P, Pradhan AK, Pental D. RNA-seq based SNPs for mapping in Brassica juncea (AABB): synteny analysis between the two constituent genomes a (from B. rapa) and B (from B. nigra) shows highly divergent gene block arrangement and unique block fragmentation patterns. BMC Genomics. 2014;15(1):396. doi: 10.1186/1471-2164-15-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strelkov SE, Hwang S-F, Manolii VP, Cao T, Fredua-Agyeman R, Harding MW, Peng G, Gossen BD, Mcdonald MR, Feindel D. Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can J Plant Pathol. 2018;40(2):284–298. doi: 10.1080/07060661.2018.1459851. [DOI] [Google Scholar]
- 42.Strelkov S, Tewari J, Smith-Degenhardt E. Characterization of Plasmodiophora brassicae populations from Alberta, Canada. Can J Plant Pathol. 2006;28(3):467–474. doi: 10.1080/07060660609507321. [DOI] [Google Scholar]
- 43.Sokal RR, Rohlf FJ. The principles and practice of statistics in biological research. San Francisco: WH freeman and company; 1969. [Google Scholar]
- 44.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 46.Harris RS. Improved pairwise Alignmnet of genomic DNA. 2007. [Google Scholar]
- 47.Carver Tim, Berriman Matthew, Tivey Adrian, Patel Chinmay, Böhme Ulrike, Barrell Barclay G., Parkhill Julian, Rajandream Marie-Adèle. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24(23):2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mapping Rcr6 based on BSR-Seg using a draft genome of B. nigra from Canada: A. The percentage of monomorphic and polymorphic variants on each chromosome; B. distribution of the percentage of polymorphic variants on chromosome B3; and C. distribution of polymorphic variants on chromosome B3. (JPG 434 kb)
Figure S2. The genetic map of Rcr6 with SNP markers identified through BSR-Seq from both chromosomes A08 and B3. (JPG 493 kb)
Figure S3. Mapping of Rcr6 into chromosome B7 using the published B-genome of B. nigra from China: A. the percentage (%) of monomorphic and polymorphic variants on each chromosome; and B. comparison of B3_Canada and B7_China. The dot plot was created using R (https://cran.r-project.org/). (JPG 1126 kb)
Table S1. Blast2GO result in the Rcr6 target region of chromsome A08. Table S2. Blast2GO result in the Rcr6 target region of chromsome B3. (XLSX 59 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.