Abstract
Purpose:
Glioma is identified as a broad category of brain and spinal cord tumors. MiR-32 is important in regulating the genesis of different cancers; however, the underlying mechanisms of miR-32 in glioma still largely unknown. This study aimed to elucidate pathobiological functions of miR-32 in glioma and verify its effect on the regulation of enhancer of zeste homolog 2.
Methods:
The expression of miR-32 and enhancer of zeste homolog 2 was detected by quantitative real-time polymerase chain reaction and Western blot in glioma tissues and cells. Cell Counting Kit-8 (CCK-8) assay was used to examine the effects of miR-32 on human glioma cells proliferation. Transwell assay was used to examine cell metastasis, respectively. Two bioinformatics analysis software and luciferase reporter assay were chosen to confirm targeting association between miR-32 and enhancer of zeste homolog 2.
Results:
MiR-32 was downregulated in glioma tissues and cells. Furthermore, enhancer of zeste homolog 2 expression was upregulated and negatively correlated with miR-32 in clinical tissues. Ectopic expression of miR-32 inhibited glioma cell proliferation, migration, and invasion. Enhancer of zeste homolog 2 was identified as direct target gene of miR-32 in glioma. Overexpression of enhancer of zeste homolog 2 ablated the inhibitory effects of miR-32.
Conclusion:
In summary, our finding suggests that miR-32 acts an important role in inhibiting glioma cell proliferation and metastasis and suppresses the expression of ABCC4 by directly targeting its 3′-untranslated region. The miR-32/enhancer of zeste homolog 2 axis may provide new insights to the treatment for glioma.
Keywords: miR-32, EZH2, glioma, proliferation, migration, invasion
Instruction
Glioma is the most untreatable and aggressive type of primary neoplasm of the central nervous system in adults, with significant mortality, increasing relapse, and poor survival rate.1 The gliomas are classified into grades I to IV by the World Health Organization (WHO) classification according to the degree of malignancy.2 Glioblastoma multiforme (WHO grade IV) has been the most aggressive primary brain tumor,3 and about 10 000 cases of glioma are diagnosed every year.4 Although the availability of multiple strategies such as maximal surgical excision, radiation therapy, or chemotherapy for the glioma therapy in recent years,5 the strong metastasis ability of malignant gliomas cells impeded the surgical resection and radiotherapy effect of glioma treatment.6 The molecular mechanisms of glioma propagation or metastasis require further studies, it is imperative to clarify the precise mechanism and find out novel therapeutic treatments for glioma patients.
MicroRNAs (miRNAs), a kind of noncoding RNA, were 20 to 24 nucleotide in length in eukaryotic organisms.7 MicroRNAs negatively modulate gene expression transcriptionally or post-transcriptionally by binding to their 3′-untranslated regions (3′-UTRs), thereby inducing their degradation and/or translational repression.8,9 Functional studies have suggested the involvement of miRNAs in various physiological and pathological processes, such as the cell cycle, cell division, cellular development, apoptosis, metastasis, and angiogenesis.10–12 MiR-32 is a miRNA which located on chromosome band Xq26.2 works differently in tumors. For example, its expression was reduced in gastric cancer13 and osteosarcoma,14 and upregulated in colorectal cancer15 and prostate cancer.16 A variety of miRNAs including miR-32, miR-29b, and miR-222 have been found being downregulated in glioma patients.17 Therefore, we want to investigate the mechanism of miR-32 in glioma patients from the clinical perspective.
Enhancer of zeste homolog 2 (EZH2) is the core structural composition of polycomb group gene family, which is involved in cell cycle regulation and carcinogenesis through methylating H3K27.18 Enhancer of zeste homolog 2, as a tumor-promoting gene, might play important role in tumorigenesis on account of its alterations of a great range of tumors such as colorectal cancer,19 clear cell renal cell carcinoma,20 non-small cell lung cancer,21 and breast cancer.22 Intriguingly, miR-32 participates in the regulating of EZH2 expression in human oral squamous cell carcinoma,23 but the potential effect of miR-32 on EZH2 remains unconfirmed in glioma.
We examined miR-32 was frequently downregulated in glioma tissues and cells. MiR-32 abnormal expression could inhibit glioma cell proliferation and metastasis. Enhancer of zeste homolog 2 is a novel target gene of miR-32. Additionally, miR 32 protein level was higher in glioma tissues, and EZH2 was negatively associated with miR-32 in glioma. The newly identified miR-32/EZH2 axis partially represents a novel potential therapeutic target for glioma treatment.
Materials and Methods
Tissues Samples
From Cangzhou Central Hospital (Cangzhou, Hebei Province, China), a total of 70 cases of tissue specimens including human glioma tissues and paired normal adjacent tissues were obtained from spinal glioma who underwent surgery between March 2016 and September 2017. Liquid nitrogen was used to flash-frozen all the tissue specimens, and then the specimens stored at −80°C. None of spinal glioma patients had received chemotherapy or radiotherapy before surgery. The use of human specimens was approved by the Ethics Committee of Cangzhou Central Hospital (Approval No. 2016-14). All patients provided written informed consent prior to enrollment in the study. The patients’ information is summarized in Table 1.
Table 1.
The MiR-32 Expression and Clinicopathological Characteristics in Patients With Glioblastoma.
| Clinicopathological Features | Cases (n = 70) | MiR-32 Expression | P Valuea | |
|---|---|---|---|---|
| High (%) | Low (%) | |||
| Gender | ||||
| Male | 30 | 10 (33.3) | 20 (66.7) | .719 |
| Female | 40 | 15 (37.5) | 25 (62.5) | |
| Age (years) | ||||
| ≤41 | 34 | 15 (44.1) | 19 (55.9) | .657 |
| >41 | 36 | 14 (38.9) | 22 (61.1) | |
| KPS score | ||||
| <80 | 34 | 22 (66.7) | 12 (33.3) | .009b |
| >80 | 36 | 12 (35.7) | 24 (64.3) | |
| WHO grade | ||||
| I + II | 30 | 18 (60.0) | 12 (40.0) | .022a |
| III + IV | 40 | 13 (32.5) | 27 (67.5) | |
Abbreviations: KPS score, Karnofsky Performance Score; WHO, World Health Organization.
aχ2 test.
b P < 0.01.
Cell Lines
The 4 cell lines (U87, U251, A172, and U118) and primary normal human astrocytes (NHA) were obtained from ATCC (Manassas, Virginia). All cells were cultured in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, California), 2 mM L-glutamine (Invitrogen), 100 U/mL penicillin (Invitrogen), and 0.1 mg/mL streptomycin (Invitrogen) at 37°C and 5% CO2.
Tumor cells were plated onto a 6-well plate at a density of 3 × 105 cells/well for about 24 hours prior to transfection. MiR-32 mimics/inhibitor and miRNA negative control (NC) were transfected into cells using Lipofectamine 2000 Reagent (Invitrogen). Small interfering RNA targeting EZH2 and small interfering NC were synthesized by RiboBio (Guangzhou, China). Then, cells were used for cell proliferation and metastasis after transfection. Enhancer of zeste homolog 2 overexpressed plasmid (pCDNA3.1-EZH2) and blank plasmid were obtained from Genepharma (Shanghai, China).
Quantitative Real-Time PCR
Total RNA of glioma cells and tissues were extracted using Trizol (Invitrogen). quantitative real-time polymerase chain reaction (qRT-PCR) was conducted with SYBR Premix Ex Taq (TaKaRa, China). U6 and glyceraldehyde phosphate dehydrogenase (GAPDH) acted as the endogenous control for the expressions of miR-32 and EZH2. The expression was tested using the 2− ΔΔCt method. The conditions for PCR were as follows: 95°C for 5 minutes, 40 cycles of denaturation at 95°C (15 seconds), 50°C (30 seconds), and 72°C (30 seconds). Primers were as follows: miR-32 forward, 5′-GCGGCGTATTGCACATTACT-3′, reverse, 5′-TCGTATCCAGTGCAGGGTC-3′; U6 forward, 5′-CTC GCTTCGGCAGCACA-3′, reverse, 5′-AACGCTTCACGAATTTGCGT-3′; EZH2 forward, 5′-TTCATGCAACACCCAACACT-3′, reverse, 5′-GAGAGCAGCAGCAAACTCCT-3′; GAPDH forward, 5′-CTGGGCTACACTGAGCACC-3′, reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′.
Western Blot
Cultured cells were collected and lysed with (Radio Immunoprecipitation Assay [RIPA]) lysis Buffer (Beyotime, China). The measurement of total protein concentration used BCA Assay Kit (Beyotime, China). Protein (20 μg) was resolved on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and then transferred to polyvinylidene fluoride membrane (Millipore, Darmstadt, Germany). The membranes were blocked by 5% skimmed milk in tris buffered saline containing 0.1% Tween-20 (TBST) and incubated with specific primary antibodies rabbit polyclonal anti-EZH2 antibody (ab186006, 1:1000; Abcam, Cambridge, United Kingdom) or rabbit polyclonal anti-GAPDH (ab9485, 1:1000; Abcam) at 4°C overnight. Subsequently, the membranes were incubated in the goat polyclonal antirabbit immunoglobulin G H&L secondary antibody (ab150077, 1:2000; Abcam) at room temperature for 1 hour. GAPDH was used as internal control. Image J software (National Institutes of Health, Bethesda, Mary Land) was used to quantify the protein bands.
CCK-8 Assay
Cell Counting Kit-8 (CCK-8) assay (Dojindo, Japan) was chosen to detect cell proliferative capacity. Briefly, cells (1 × 105) transfected with miR-32 mimics/inhibitor were reseeded into 96-well plates. Subsequently, the cells were incubated for 0, 24, 48, 72, and 96 hours at 37°C with 5% CO2. CCK-8 reagent (10 μL) was added into each plate for 2 hours incubation at 37°C. The supernatant was removed and dissolved the formazan crystals by dimethyl sulfoxide (DMSO) (150 μL/well). The absorbance at 450 nm was detected using microplate (Bio-Rad, San Jose, California).
Transwell Assay
The ability of glioma cell metastasis was measured by transwell assay with or without Matrigel (Clontech, CA). For the migration assay, 1 × 105 glioma cells were seeded into the top of 24-well transwell chambers (BD Biosciences). The lower chamber was added with DMEM medium with 20% FBS. Following 2-day incubation, the nonmigrated cells were carefully removed using cotton swabs. Then, migrated cells were fixed with methanol, stained with 0.05% crystal violet at room temperature for 15 minutes, and washed with phosphate-buffered saline and dried in air. Finally, migrated cells were photographed using inverted microscope (Olympus Corp, Tokyo, Japan). Invasion assay was conducted in the similar manner except that transwell chambers were precoated with Matrigel.
Luciferase Assay
Potential targets of miR-32 were predicted by choosing the bioinformatics analysis software miRanda (www.microrna.org/microrna/home.do) and TargetScan (www.targetscan.org). The wild type (WT) and mutant type (Mut) 3′-UTR of EZH2 were cloned into pGL3-REPORT luciferase vector (Ambion; Thermo fisher Scientific, Inc., California). For the luciferase assay, the cells were seeded into a 24-well plate at a density of 1.5 × 105 cells/well and cultured overnight at 37°C prior to cotransfected with a reporter plasmid, along with miR-32 mimics and WT or Mut 3′-UTR of EZH2 using Lipofectamine 2000 transfection reagent at 37°C. Following 48 hours of incubation, the cells were harvested, and then detected by using Dual-Luciferase Reporter Assay System (Promega).
Statistical Analysis
All the results were present as the mean (standard deviation, SD). Each assay was repeated 3 times independently. P < .05 was considered to be significantly different. Data were analyzed with 2-tailed Student t test or 1-way analysis of variance using SPSS (version 19.0; IBM Corp, Armonk, New York). Student-Newman-Keuls method was used to compare differences between 2 groups in multiple comparison analyses following 1-way analysis of variance.
Results
MiR-32 Is DownRegulated and EZH2 Expression Is Upregulated in Glioma
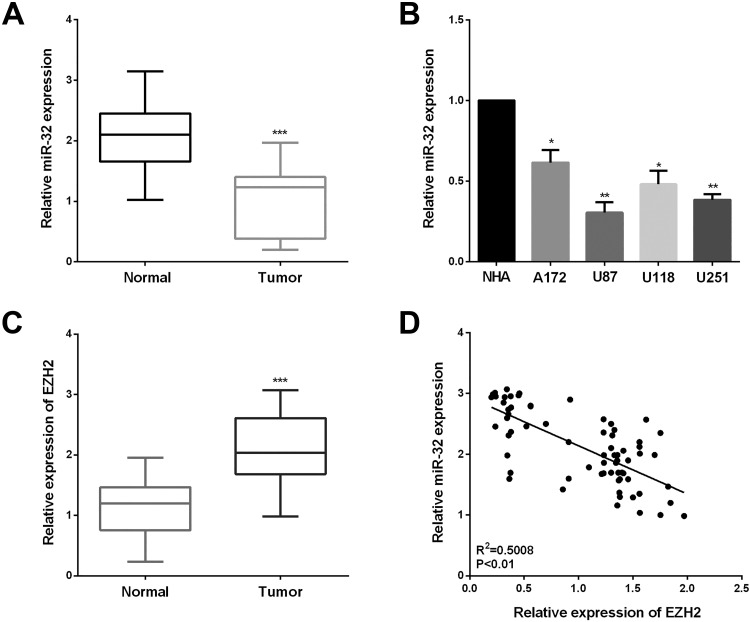
The miR-32 expression was detected in 70 cases of tissue specimens including human glioma tissues and paired tissues by qRT-PCR. In Figure 1A, the miR-32 expression was decreased in glioma tissues than that in paired tissues. Analogously, lower miR-32 expression was observed in 4 glioma cell lines compared with NHA (Figure 1B). These results presented that miR-32 was downregulated in both the glioma tissues and cell lines. Table 1 presents the results of the correlation analysis between clinicopathological characteristics and the relative miR-32 expression in glioma patients. There were significant differences in Karnofsky Performance Score and WHO grade between the high miR-32 expression group and the low group (P = .009; P = .022, respectively). However, we did not find any significant differences in the age and gender group of miR-32 expression (Table 1).
Figure 1.
MiR-32 was downregulated and inversely connected with enhancer of zeste homolog 2 (EZH2). A, miR-32 expression in 70 pairs of samples was detected by quantitative real-time polymerase chain reaction (qRT-PCR). B, MiR-32 levels in 4 glioma cell lines (U87, U251, A172, and U118) compared with normal human astrocytes (NHA). C, Relative mRNA expression of EZH2 in glioma tissues. D, Spearman correlation analysis of miR-32 and EZH2 expression in glioma tissues. ***P < .001, **P < .01, and *P < .05.
Furthermore, the EZH2 expression in the glioma tissues was also determined. Enhancer of zeste homolog 2 mRNA was increased in the glioma tissues compared with the paired tissues (Figure 1C). Then, miR-32 expression was inversely correlated with the EZH2 level in these clinical specimens (Figure 1D). There was an inverse correlation between miR-32 and EZH2 mRNA expression by Spearman correlation analysis in glioma tissues (r = −0.7077; P < .01). The above results indicated that the reduced expression of miR-32 and increased EZH2 level may play an essential part in the progression of glioma.
MiR-32 Inhibits Cell Proliferation, Migration, and Invasion of Glioma
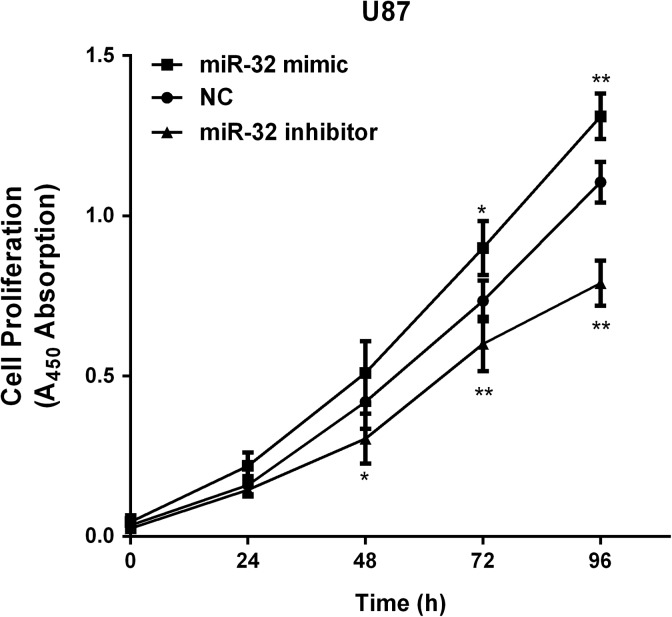
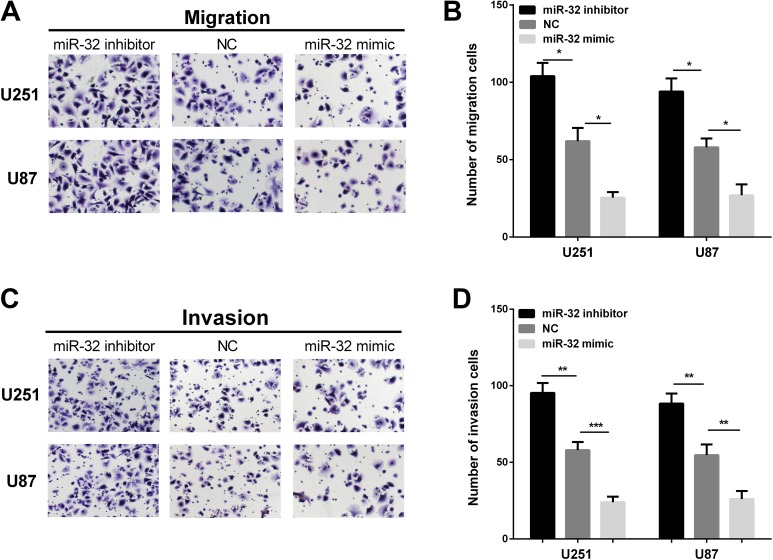
We established stable cells to examine the functions of miR-32 in glioma in both U251 and U87 cells, miR-32 mimic/inhibitor was transiently transfected into both cell lines. The miR-32 expression was higher after transfecting with miR-32 mimic, while the miR-32 expression was reduced when transfected with miR-32 inhibitor (Figure 2A and B). Firstly, we used CCK-8 assay to measure glioma cells proliferation. MiR-32 overexpression inhibited glioma cell proliferation, and miR-32 inhibitor could promote proliferative abilities (Figure 2C and D). Transwell assays were performed to assess whether miR-32 influenced invasive and migration abilities of glioma cells. High expression of miR-32 significantly inhibited cell migration in 2 glioma cell lines, whereas inhibiting miR-32 expression promoted cell migration (Figure 3A and B). Overexpression of miR-32 inhibited cell invasion, and conversely, transfection with miR-32 inhibitor promoted cell invasion ability (Figure 3C and D). Overall, these results revealed that miR-32 may serve as tumor suppressor in glioma progression.
Figure 2.
The overexpression of miR-32 induced growth inhibition in glioma. A and B, MiR-32 was reexpressed in U251 and U87 cell lines, and miR-211 levels were determined by quantitative real-time polymerase chain reaction (qRT-PCR) after transfected miR-32 mimic/inhibitor and NC. C and D, CCK-8 assay showed that miR-32 suppressed viability of U251 and U87 cells lines after transfection of miR-32 mimic. ***P < .001, **P < .01, *P < .05.
Figure 3.
MiR-32 inhibits glioma cell migration and invasion. A and B, Ectopic expression of miR-32 significantly suppressed U251 and U87 cell migration, whereas inhibiting miR-32 expression promoted U251 and U87 cell migration. C and D, The overexpression of miR-32 dramatically inhibited U251 and U87 cell invasion, whereas inhibition of miR-32 expression significantly promoted cell invasion. ***P < .001, **P < .01, and *P < .05.
MiR-32 Directly Targets EZH2
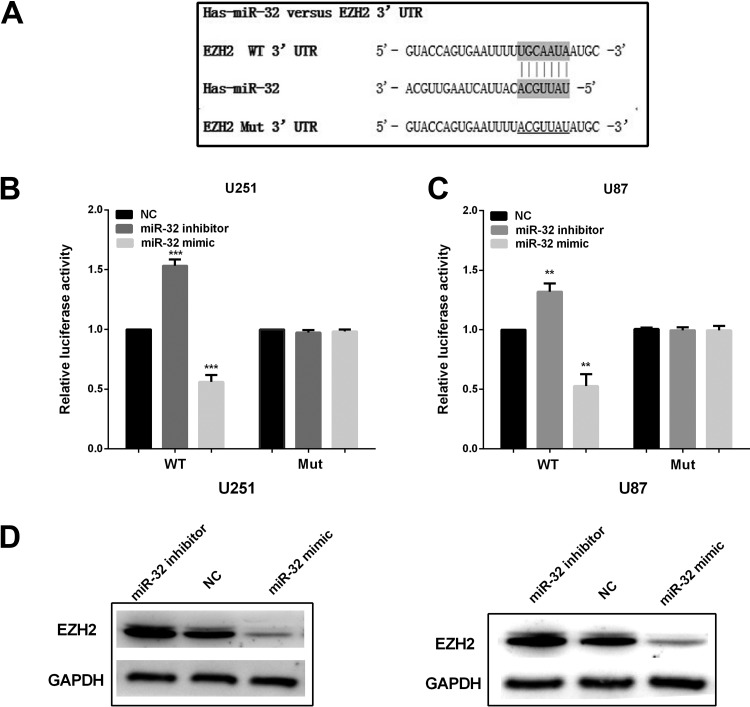
To further explore the detailed mechanism of miR-32-regulated glioma progression, prediction program of TargetScan Human software was chosen to analyze the miR-32 putative targets. The results showed that EZH2, a key transcription factor involved in development of a diversity of tumors, was a potential target of miR-32. Figure 4A showed putative binding site of miR-32 in the 3′-UTR region of EZH2 mRNA. To further confirm whether EZH2 is the target of miR-32, the dual luciferase reporter assay was conducted with U251 and U87 cells. Results showed that the high expression of miR-32 inhibited the luciferase activity in cells transfected with 3′-UTR-WT luciferase plasmid, and low expression of miR-32 enhanced the luciferase activity in U251 and U87 cells; however, mutation of this putative binding site abrogated the inhibitory effect by miR-32 (Figure 4B and C). The EZH2 expression was decreased after overexpression of miR-32 in U251 and U87 cells (Figure 4D). Moreover, knockdown of miR-32 had the opposite influence on EZH2 expression to that of miR-32 overexpression. These upon results indicate that EZH2 is a direct target gene of miR-32.
Figure 4.
MiR-32 suppressed enhancer of zeste homolog 2 (EZH2) expression by targeting its 3′-untranslated region (UTR). A, The binding sites of miR-32 on EZH2 3′-UTR. B and C, Luciferase reporter assay with the pGL3-EZH2-3′-UTR-WT or pGL3-EZH2-3′-UTR-Mut were cotransfected with miR-32 mimics/inhibitor/NC. D, EZH2 expression in cells transfected with miR-32 mimic/inhibitor/NC. ***P < .001 and **P < .01.
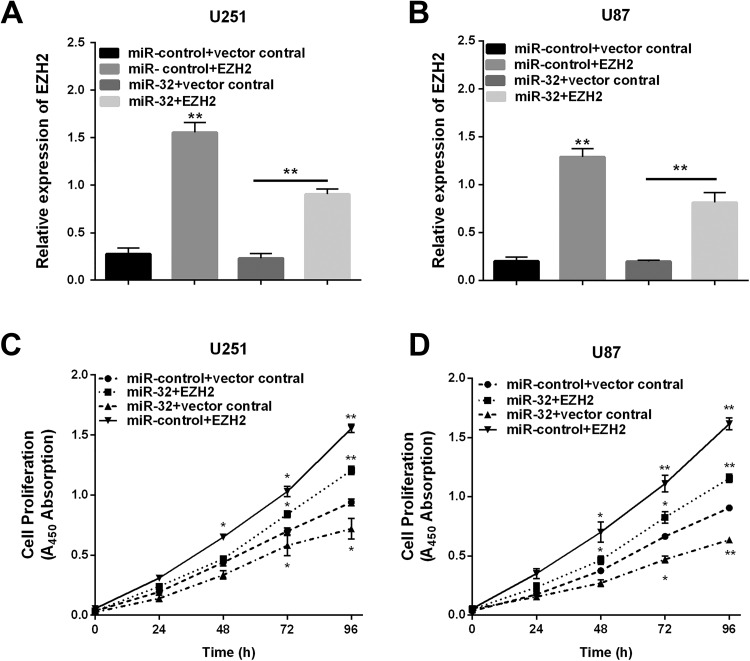
Overexpression of EZH2 Ablated the Inhibitory Effects of miR-32
Given evidence has indicated EZH2 was the direct target of miR-32 in the U251 and U87 cells, EZH2 might take part in miR-32-mediated inhibition of glioma cells proliferation. To detect whether overexpression of EZH2 would simulate miR-32-mediated effects, overexpression of EZH2 was transfected into U251 and U87 cells. The qRT-PCR suggested the EZH2 mRNA level was reduced by miR-32 mimic, and the levels were restored after con-transfected EZH2 and miR-32 mimic (Figure 5A and B), and then we calculated glioma cell proliferation. Subsequently, the effect of EZH2 on relative cell viability regulated by miR-32 was investigated by CCK-8 assay. The results showed that the overexpression of EZH2 increased viability of glioma cells, and EZH2 may reverse the miR-32-inhibiting effect on glioma cell proliferation (Figure 5C and D). These results demonstrated that miR-32 functioned as a tumor suppressor through regulating EZH2.
Figure 5.
Upregulation of enhancer of zeste homolog 2 (EZH2) reverses the inhibitory effects of miR-32. A and B, EZH2 was abnormally expressed in U251 and U87 and EZH2 levels were determined by quantitative real-time polymerase chain reaction (qRT-PCR). C and D, CCK-8 assay was conducted in 2 cells lines. **P < .01.
Discussion
Dysregulation of miRNAs might lead to uncontrolled and progressive tumorigenesis and has been thoroughly reported in almost all kinds of human malignancies, such as glioma.24–26 Mounting evidence has demonstrated miR-32 could act as an inhibitor or oncogene in malignant biological behaviors such as cell growth, invasion, and migration in different cancers.13–15 But the molecular mechanism in which miR-32 regulates the progression and development of glioma has not been totally investigated.
More and more data indicated that miR-32 has different expression in different tumors. For example, the miR-32 expression was significantly decreased in lung cancer,27 on the contrary, was raised in renal and prostate cancer tissue.28,29 Moreover, miR-32 has divergent effects on different cancers, such as miR-32 could inhibit gastric cancer and osteosarcoma cell proliferation, migration, and invasion13,14 and promote progression of CRC cells.15 We firstly demonstrated the synthesized analysis of miR-32 effects on glioma in this report. This research demonstrated that the miR-32 expression was decreased in glioma tissues and cells. We hypothesized that miR-32 may play as tumor suppressor of gliomas based on these results. In order to prove this hypothesis, we examine the miR-32 functions in the progression of glioma. We found that miR-32 inhibited proliferation and metastasis of glioma. MicroRNAs has been demonstrated to be involved in modulating the tumor physiological process or pathogenesis by negatively regulating target genes, and miRNA could target to about 1/3 of human genes.30 We predicted that EZH2 acts as one target of miR-32 with microRNA Targetscan software.
MiRs exert their functions through inhibiting their target genes’ expression31; therefore, it is important to elucidate their target genes. As a tumor-promoting gene, EZH2 has a significant influence in cellular proliferation, apoptosis, migration, and invasion of tumor.32,33 The levels of EZH2 in glioma,34 oral squamous cell carcinoma,35 and prostate cancer36 tissues are higher than their normal adjacent tisssues, and overexpression of EZH2 was confirmed as specific predictor of poor prognostic role for patients. Chang and Hung reported that EZH2 plays important role in tumor progression.37 Furthermore, Lu et al EZH2 could regulate tumor angiogenesis by epigenetic activation of specific pathways.38 Further analysis corroborated EZH2 expression was raised in OSCC and could affect OSCC carcinogenesis.35,39 Previous study showed that miR-32 proliferation, migration, and invasion and promoted cell apoptosis of human oral squamous cell carcinoma cells via targeting EZH2,23 which was largely consistent with our findings of the miR-32 effect. The results presented that EZH2 was a direct target of miR-32 in glioma by luciferase reporter assay. MiR-32 expression inversely correlated with EZH2 in glioma tissues, and the luciferase reporter and Western blot also showed that overexpression of miR-32 inhibited 3′-UTR luciferase report activity, and this influence was abolished by mutation of the miR-32 seed binding site. Given evidence has indicated EZH2 was the direct target of miR-32 in the U251 and U87 cells, EZH2 might take part in miR-32-mediated inhibition of glioma cell proliferation. The results showed that the overexpression of EZH2 increased viability of glioma cells, and EZH2 may reverse the miR-32-inhibiting effect on glioma cell proliferation. These results suggested that EZH2 is a direct gene of miR-32, miR-32 functioned as a tumor suppressor via regulating EZH2. The above results suggested that miR-32 may function as tumor inhibitor by repressing EZH2 expression in glioma progression.
In summary, our findings identified the specific molecular mechanisms of miR-32 in human glioma proliferation, and verified EZH2, a multifaceted oncogene, as the downstream target gene of miR-32, whose expression was suppressed by miR-32. MiR-32 is able to inhibit EZH2 expression by directly binding to its 3′-UTR. Thus, miR-32 may be considered as a tumor suppressor and may develop to be a novel strategy against gliomas.
Abbreviations
- DMEM
Dulbecco Modified Eagle Medium
- EZH2
Enhancer of zeste homolog 2
- FBS
fetal bovine serum
- miRNA
microRNAs
- NC
negative control
- NHA
normal human astrocytes
- qRT-PCR
quantitative real-time polymerase chain reaction
- WHO
World Health Organization
- WT
wild type
- 3′-UTR
3′-untranslated region
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by Key Research and Development Plan of Shandong Province (2016GSF201063).
ORCID iD: Wei Zheng, PhD  https://orcid.org/0000-0003-0747-7146
https://orcid.org/0000-0003-0747-7146
References
- 1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2. Komori T, Sasaki H, Yoshida K. Revised WHO classification of tumours of the central nervous system: summary of the revision and perspective [in Japanese]. No Shinkei Geka. 2016;44(8):625–635. doi:10.11477/mf.1436203347. [DOI] [PubMed] [Google Scholar]
- 3. Easaw JC, Mason WP, Perry J, et al. Canadian recommendations for the treatment of recurrent or progressive glioblastoma multiforme. Curr Oncol. 2011;18(3):e126–e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9(7):717–726. doi:10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40(1):1–14. doi:10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 6. Wang X, Jia Y, Wang P, Liu Q, Zheng H. Current status and future perspectives of sonodynamic therapy in glioma treatment. Ultrason Sonochem. 2017;37:592–599. doi:10.1016/j.ultsonch.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 7. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi:10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 9. Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92. [PMC free article] [PubMed] [Google Scholar]
- 10. Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25(46):6176–6187. doi:10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 11. Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15(5):563–568. doi:10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 12. Magee P, Shi L, Garofalo M. Role of microRNAs in chemoresistance. Ann Transl Med. 2015;3(21):332 doi:10.3978/j.issn.2305-5839.2015.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Kuai X, Song M, et al. microRNA-32 inhibits the proliferation and invasion of the SGC-7901 gastric cancer cell line in vitro. Oncol Lett. 2014;7(1):270–274. doi:10.3892/ol.2013.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu JQ, Zhang WB, Wan R, Yang YQ. MicroRNA-32 inhibits osteosarcoma cell proliferation and invasion by targeting Sox9. Tumour Biol. 2014;35(10):9847–9853. doi:10.1007/s13277-014-2229-x. [DOI] [PubMed] [Google Scholar]
- 15. Wu W, Yang J, Feng X, et al. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013;12:30 doi:10.1186/1476-4598-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jalava SE, Urbanucci A, Latonen L, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31(41):4460–4471. doi:10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 17. Munthe S, Halle B, Boldt HB, et al. Shift of microRNA profile upon glioma cell migration using patient-derived spheroids and serum-free conditions. J Neurooncol. 2017;132(1):45–54. doi:10.1007/s11060-016-2356-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chinaranagari S, Sharma P, Chaudhary J. EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4 in prostate cancer. Oncotarget. 2014;5(16):7172–7182. doi:10.18632/oncotarget.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song-Bing H, Hao Z, Jian Z, et al. Inhibition of EZH2 expression is associated with the proliferation, apoptosis and migration of SW620 colorectal cancer cells in vitro. Exp Biol Med (Maywood). 2015;240(4):546–555. doi:10.1177/1535370215573463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu ZQ, Zhang L, Gao BS, et al. EZH2 promotes tumor progression by increasing VEGF expression in clear cell renal cell carcinoma. Clin Transl Oncol. 2015;17(1):41–49. doi:10.1007/s12094-014-1195-5. [DOI] [PubMed] [Google Scholar]
- 21. Geng J, Li X, Zhou Z, Wu CL, Dai M, Bai X. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015;359(2):275–287. doi:10.1016/j.canlet.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 22. Wang X, Hu B, Shen H, et al. Clinical and prognostic relevance of EZH2 in breast cancer: a meta-analysis. Biomed Pharmacother. 2015;75:218–225. doi:10.1016/j.biopha.2015.07.038. [DOI] [PubMed] [Google Scholar]
- 23. Zhang D, Ni Z, Xu X, Xiao J. MiR-32 functions as a tumor suppressor and directly targets EZH2 in human oral squamous cell carcinoma. Med Sci Monit. 2014;20:2527–2535. doi:10.12659/MSM.892636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pandima Devi K, Rajavel T, Daglia M, Nabavi SF, Bishayee A, Nabavi SM. Targeting miRNAs by polyphenols: novel therapeutic strategy for cancer. Semin Cancer Biol. 2017;46:146–157. doi:10.1016/j.semcancer.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 25. Muluhngwi P, Klinge CM. Identification of miRNAs as biomarkers for acquired endocrine resistance in breast cancer. Mol Cell Endocrinol. 2017;456:76–86. doi:10.1016/j.mce.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 26. Wang ZY, Xiong J, Zhang SS, Wang JJ, Gong ZJ, Dai MH. Up-regulation of microRNA-183 promotes cell proliferation and invasion in glioma by directly targeting NEFL. Cell Mol Neurobiol. 2016;36(8):1303–1310. doi:10.1007/s10571-016-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dacic S, Kelly L, Shuai Y. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod Pathol. 2010;23(12):1577–1582. doi:10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- 28. Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, Teh BT. MicroRNA profiling of human kidney cancer subtypes. Int J Oncol. 2009;35(1):109–114. [DOI] [PubMed] [Google Scholar]
- 29. Bandres E, Cubedo E, Agirre X, et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29 doi:10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31. Liu GF, Tang D, Li P, et al. S-1-based combination therapy vs S-1 monotherapy in advanced gastric cancer: a meta-analysis. World J Gastroenterol. 2014;20(1):310–318. doi:10.3748/wjg.v20.i1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou X, Ren Y, Kong L, et al. Targeting EZH2 regulates tumor growth and apoptosis through modulating mitochondria dependent cell-death pathway in HNSCC. Oncotarget. 2015;6(32):33720–33732. doi:10.18632/oncotarget.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zingg D, Debbache J, Schaefer SM, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051 doi:10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 34. Zhang J, Chen L, Han L, et al. EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in glioblastoma. Cancer Lett. 2015;356(2 Pt B):929–936. doi:10.1016/j.canlet.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 35. Kidani K, Osaki M, Tamura T, et al. High expression of EZH2 is associated with tumor proliferation and prognosis in human oral squamous cell carcinomas. Oral Oncol. 2009;45(1):39–46. doi:10.1016/j.oraloncology.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 36. Melling N, Thomsen E, Tsourlakis MC, et al. Overexpression of enhancer of zeste homolog 2 (EZH2) characterizes an aggressive subset of prostate cancers and predicts patient prognosis independently from pre- and postoperatively assessed clinicopathological parameters. Carcinogenesis. 2015;36(11):1333–1340. doi:10.1093/carcin/bgv137. [DOI] [PubMed] [Google Scholar]
- 37. Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106(2):243–247. doi:10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu C, Han HD, Mangala LS, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18(2):185–197. doi:10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao L, Yu Y, Wu J, et al. Role of EZH2 in oral squamous cell carcinoma carcinogenesis. Gene. 2014;537(2):197–202. doi:10.1016/j.gene.2014.01.006. [DOI] [PubMed] [Google Scholar]