Abstract
The effectiveness of autologous cell-based skeletal repair continues to be controversial in part because in vitro predictors of in vivo human bone formation by cultured human progenitor cells are not reliable. To assist in the development of in vivo assays of human osteoprogenitor potential, a fluorescence-based histology of nondecalcified mineralized tissue is presented that provides multiple criteria to distinguish human and host osteoblasts, osteocytes, and accumulated bone matrix in a mouse calvarial defect model. These include detection of an ubiquitously expressed red fluorescent protein reporter by the implanted human cells, antibodies specific to human bone sialoprotein and a human nuclear antigen, and expression of a bone/fibroblast restricted green fluorescent protein reporter in the host tissue. Using low passage bone marrow-derived stromal cells, robust human bone matrix formation was obtained. However, a striking feature is the lack of mouse bone marrow investment and osteoclasts within the human bone matrix. This deficiency may account for the accumulation of a disorganized human bone matrix that has not undergone extensive remodeling. These features, which would not be appreciated by traditional decalcified paraffin histology, indicate the human bone matrix is not undergoing active remodeling and thus the full differentiation potential of the implanted human cells within currently used mouse models is not being realized.
Keywords: bone cell transplantation, bone matrix, calvarial defect model, cryohistology, epifluorescence imaging, GFP, immunostaining, mineralized tissues, primary human MSCs, RFP, xenograft
Introduction
A reliable cell-based therapy for repair of the mineralized skeleton in humans is yet to be realized despite undocumented claims from commercial websites1–3 or less than critical review papers.4–7 Although radiographic or functional repair after an intervention can be demonstrated, it is not clear whether the mechanism was cell-dependent or cell non-autonomous. This distinction is crucial for the interpretation of a repair outcome to knowledgeably optimize the source and preparation of the transplanted cells. From a clinical perspective, the gold standard cell source for effective bone repair are autologous bone chips8 and an outcome that can be replicated in animal repair models.9–11 However, it is not firmly established that primary cells derived from a variety of autologous or heterologous human tissue sites contribute to new osteogenic cells within the repair site. The most current example of this problem is the use of adipose-derived stromal cells, which in primary culture can be induced to express some features of osteoblastic cells12 and appear to modify repair of a skeletal defect in animal models,13 but failed to be an effective approach in a clinical trial.14,15 Because there are no reliable cell surface markers or cell culture methods for predicting in vivo differentiation, investigators are forced to rely on transplantation models with the assumption that if osteogenic differentiation from either the host or donor component fails in the animal model, it will also fail in a human subject. Such a determination is reliant on a histological interpretation, which must be rigorous and independently verifiable by the unbiased observer.
This article presents three levels of a methodological approach to meet a standard of histological rigor and transparency. For rigor, the same histological section needs to be probed and imaged multiple times such that a number of histological criteria can be mapped to the same tissue section. For mineralized tissue, being able to retain the mineral signals within the tissue section as well as capture the major enzymatic signals that are unique to osteoblasts and osteoclasts is essential to the description of a repair field. Once the landscape is defined by these fundamental structures, the histological markers of levels of cellular differentiation and host or donor origin can be mapped to the repair process to provide the needed interpretation. Rigor also includes the use of high-resolution digital scanning of the entire repair field and adjacent host tissues so that a comprehensive interpretation of the transplantation outcome can be developed. Because each feature is recorded in a separate digital file, an image stack needs to be created and aligned to facilitate the mapping of various histological signals to the reference tissue. This process requires software with the ability to create image stacks and computers with sufficient memory and processing power to manage the gigabyte-sized files that are generated in this approach. For transparency, the entire image stack file that was used in the analysis needs to be archived in a manner that another investigator can examine the same repair process as the authors who provided the interpretation. The power of digital histology now makes it possible for independent assessment of a primary data set as is now required in microarray studies in which the primary data set is archived in a centralized source. By insisting on a histological standard for providing the information that was used in the interpretation of an experimental outcome, this approach will be helpful to the advocates of cell-based skeletal repair and the regulatory agencies that must adjudicate the effectiveness of a preclinical application.16
Materials and Methods
Overview of the Workflow
This histological approach to study skeletal tissues evolved from the initial observation that cryohistology of mineralized tissue provided a non-autofluorescent platform for detecting green fluorescent protein (GFP), fluorescent enzymatic substrates, and fluorescent-labeled antibodies. The quality of the section was obtained using tape transfer technology initially with the CryoJane system17 and subsequently with adhesive capture tape with minimal autofluorescence (cat. no C-FP092, Section Lab Co. Ltd., Hiroshima, Japan). This later proved effective in maintaining the mineral component of the section while adherence of the tape to the glass slide provided the opportunity for multiple rounds of staining and sectioning.18 Links to videos of the histological process and interpretation of the imaging files are provided on the website www.bonebase.org.
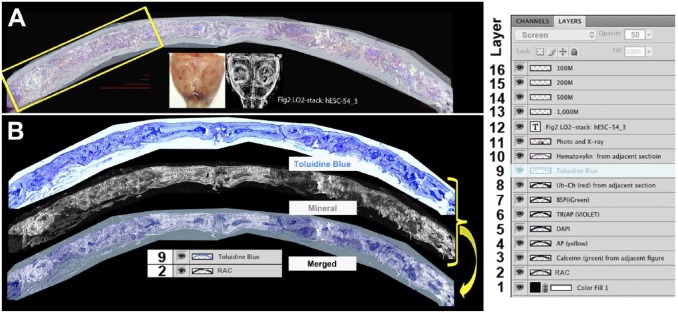
Once the section is cut, the capture tape is adhered to a glass slide with the tape side down. A sequence of steps is initiated to capture the fluorescent signals in a manner that is not compromised by the previous round of imaging. One adjacent section is captured just before and after the central section that can be used for additional staining protocols. Note that all the steps are performed under aqueous condition to prevent tissue shrinkage, which would undermine subsequent signal alignment. The first step in creation of an image stack as illustrated in Fig. 1A identifies the mineralized tissue using ring aperture contrast (RAC) and endogenous fluorescent signals that exist in the section before any manipulation. This includes the accumulated mineral (RAC, Fig. 1A, layer 2), fluorescent mineralization signals from administered fluorochromes (Fig. 1A, calcein green, layer 3), and any fluorescence-derived signals that were incorporated into the experimental protocol (Fig. 1A, ubiquitin cherry [Ub-Ch], layer 8). After scanning, step 2 is initiated with removal of the cover slip and equilibration with the acid environment needed for the tartrate resistant acid phosphatase (TRAP) reaction (Fig. 1A, TRAP, layer 6). This removes the mineral (RAC) and mineral labels with some diminution of the endogenous fluorescent signals. Step 3 begins by neutralizing the pH followed by fluorescent immunostaining using antibodies that do not require proteinase K pretreatment (Fig. 1A, bone sialoprotein [BSP] green, layer 7). It can also be performed on an adjacent section. Step 4 is the alkaline phosphatase (AP) reaction that uses a substrate that is both fluorescent and chromogenic (Fig. 1A, AP yellow, layer 4). It can be used simultaneously with nuclear stain (Fig. 1A, DAPI [4′,6-diamidino-2-phenylindole], layer 5) and staining for the Click-iT substrate (5-ethynyl-2′deoxyuridine) used for the EdU reaction (cell proliferation, not shown). Step 5 can be used for protease K pretreatment that is needed to expose chromatin-embedded antigens such as the human nuclear antigen or the Click-iT-based terminal transferase (TUNEL) technology that is the basis for detecting apoptotic cells. It is usually performed on one of the two adjacent sections. Step 6 is terminal aqueous chromogenic stain such as hematoxylin (Fig. 1A, layer 10), toluidine blue (TB; Fig. 1A, layer 9), or safranin-O/fast green (not shown), which obscures all the previous fluorescent signals. Because a single tissue section may not tolerate the complete sequence, the use of adjacent section preserves tissue integrity and still permits alignment of signals. For example, more than one chromogenic stain can be obtained by using the adjacent sections. Thus, in the image stack shown in Fig. 1A, steps 1, 2, 4, and 6 are performed on the same section and used to define the primary cellular landscape of the section while steps 3 and 5 can be performed on the adjacent section and mapped back to the central section.
Figure 1.
Creation and use of the image stack. (A) Root image stack: This file contains all the intensity-adjusted layers, x-rays, photographs, scale bars, and a file title stamp. To the right is a screen shot of the layers window that illustrates all the layers that the file contains. Layers 2 through 11 are superimposed. Layer 2 is the mineral layer and is adjusted to the normal image mode. Layers 3 to 7 are fluorescent signals that are adjusted to the screen mode at 100% opacity. Layers 8 and 9 are chromogenic images that are in the screen mode at 50% opacity. Layer 9 is highlighted and its properties (screen, 50% opacity) are shown in the first line of the layer file. This root file is maintained as a multilayered .pdf file and will provide the primary source for all subsequent figures. (B) Example of flat file used for data presentation. The toluidine blue image (9) and mineral layer (2) are placed in a new file, overlaid, aligned, and merged into a single (flat) .jpg file. The resolution (300 pixels/inch) and image size (~40″ × 19″) enables creation of enlarged subregions (e.g., yellow box in Fig. 1A) that can be presented in the context of the overall lower resolution flat image (Fig. 3, overview layer). Four scale bar lines ranging for 100 to 1000 µm are deposited within the stack for use in subsequent composite figure development. Abbreviations: RAC, ring aperture contrast; BSP, bone sialoprotein; TRAP, tartrate resistant acid phosphatase; DAPI, 4′,6-diamidino-2-phenylindole.
Critical to this protocol is the use of a computer controlled fluorescent microscope capable of repetitive sample scanning, auto focusing, and scan file labeling using predefined naming schema. Each scanning session will generate multiple gray scale images for each fluorochrome such that a complete analysis may have 10 to 15 files that need to be loaded into an image stack. Because the scanning microscope performs these steps in an automated fashion, the technician is free to perform other duties in the overall flow of the histological analysis. Once the image stack is assembled, the software will control the signal intensity for each file, which is then visually superimposed on the mineral (RAC) layer. It is from this basic image stack that the analysis is performed and presented as defined regions within the entire scanned field and compressed to a flat presentation file.
Preparation of Cells Before Transplantation
The cells were derived from primary explants of a total bone marrow aspirates using an approved human protocol (Institutional Review Board no. 06 577 2). Various concentration procedures and incubation conditions were evaluated and the results of those studies will be presented elsewhere. The following is a generic description of the method that can be applied to any source of freshly aspirated human bone marrow samples, which in this case employed a Magellan system (Isto Biologics, Hopkinton, MA) for enrichment of nucleated cells.19,20 Beginning with 30 to 35 ml of sample containing approximately 15 × 106 nucleated cells per milliliter, the first centrifugation is performed at 2800 rpm (~900 × g) and the majority of the pelleted red blood cells are discarded. Then the speed of centrifugation was increased to 3800 rpm (~1,500 × g), and the nucleated cells and platelets were collected in a volume of 5 ml containing ~6.0 × 106 cells per milliliter. From this sample, a 50 µl aliquot containing ~3.0 × 105 cells was seeded onto each of four 100 mm cell culture dishes containing α-minimum essential media (Life Technologies, Grand Island, NY), 10% v/v fetal bovine serum (Life Technologies), and 1% penicillin/streptomycin (cat. no 15140122, Life Technologies) and placed under humidified and low oxygen conditions (LO2: 5% O2, 5% CO2, Sanyo, O2/CO2 incubator) at 37C. The media was changed every 3 to 4 days to reveal small colonies of attached cells that gradually expand but are not allowed to become confluent. The attached cells were released after 5 min of incubation with 3 ml of Accutase (BD Biosceinces, cat. no 561527). Cells were resuspended in 7 ml of PBS containing 10% v/v FBS (Hyclone, GE Healthcare, Pittsburgh, PA) and pelleted by centrifugation at 280 × g for 5 min. The cells were resuspended in culture medium and seeded into the culture dish at a density of 2 × 105 cells/100 mm plate. After 3 to 5 days, the passage 1 cells reached 80% confluence at which time they were either harvested for implantation (passage 1, P1) or for subsequent passages at a 1:3 ratio and 3 to 5 days interval (P2–P6). Under these expansion conditions, the in vivo differentiation capacity (as described below) of these cultured cells, now identified as human bone marrow-derived stromal cells (hBMSC), will persist through P3 after which this property is gradually lost. In our hands, the cells cannot be frozen and recovered with any in vivo osteogenic potential. In practice, approximately 9 × 106 is the number of hBMSCs that can be produced at the P2 level from 1/100th of the original 35 ml bone marrow aspirate, which is sufficient to initiate eight mouse calvarial defect transplants.
Lentiviral Labeling of the hBMSCs
Delivery of a GFP reporter that is active in the majority of the lineages that arise from the culture is one technique that can be used to identify donor-derived cells after transplantation. In our hands, we have found that the ubiquitin C promoter is the most consistent and strongest driver of GFP within the cells of the skeletal lineages as previously described.21 The lentiviral vector that we have employed is called FURW and was derived from FUGW22 (Addgene plasmid 14883) by replacing GFP with the red fluorescent protein, cherry (RFPchry). A total of 10 µg DNA of transfection components, consisting of 4.5 µg of FURW plasmid, 3.5 µg of pMDLg/pRRE (RRE), 1 µg of pRSV-Rev (REV), and 1.5 µg of VSV(G), were diluted in 2.5 ml Opti-MEM (Life Technology) medium for each 100 mm dish. The transfection reagent was composed of 30 µl of Lipofectamine 2000 (Life Technology) and 2.5 ml Opti-MEM medium for each 100 mm dish. HEK293 cells were used as packaging cell. They were grown to 90% confluency, rinsed with PBS, and detached with 3 ml 0.05% trypsin/ethylenediaminetetraacetic acid (EDTA) (cat. no 25300062, Life Technology) for 5 min in a 37C incubator after which the trypsin was inactivated by addition of 7 ml of 10% v/v FBS/PBS buffer. Cells within the suspension was pelleted at 280 × g for 5 min and resuspended in 1 ml of Opti-MEM. The transfection reaction was initiated by addition of 2.5 ml of the transfection components and 2.5 ml of the transfection reagent to the 1.0 ml of the HEK293 cells. The cell suspension was placed in the cell incubator for 30 min followed by plating into one 100 mm dish previously coated for 1 hr with 3 ml of 0.1 µg/ml poly-D-lysine (Sigma) in PBS. The following day, the culture had attained full confluency and ~90% were RFPchry positive. The medium was changed into a viral production medium made from DMEM (Dulbecco’s Modified Eagle Medium) with 4.5 g/L glucose (Lonza) and 10% FBS (Hyclone, heat inactivated at 55C for 30 min). Viral medium was harvested 48 and 72 hr after transfection. Virus was concentrated 100 fold by centrifugation at 77,000 × g for 90 min and the viral pellet was gently resuspended in serum-free DMEM basal medium. Aliquots of the concentrated virus were kept in a −70C freezer. To titer the virus, hEK293 cells were plated into a 12 well plate with 5 × 104 cells/well 1 day before titering. Concentrated virus was serially diluted into 1 ml of the culture medium at concentration range of 102 to 109. There were 11 virus dilution points that were added into each culture well. The titer of the concentrated virus was evaluated by examining RFPchry positive foci, and the virus titer was determined by the dilution factor in the well that had only one positive foci. The titer for FURW packaged virus was usually around 107 to 109 IFU/ml.
The lentiviral transduction procedure uses the primary seeded bone marrow cells. When each colony contained about 100 cells (about 5 to 7 days), the cultured cells were exposed to the lentiviral-containing medium expressing RFPchry.21 For each 100 mm dish, the media was replaced with 5 ml of culture medium containing 50 µl of concentrated virus preparation and 6 µg/ml of polybrene (Sigma, cat. no 107689). After 4 hr of viral medium exposure, 5 ml of fresh culture medium was applied to the cells without removing viral medium. This protocol was repeated for two consecutive days (three applications) and resulted in transducing approximately 30% of the cells. In subsequent experiments, higher transduction efficiency was obtained by performing a single lentiviral exposure on suspended cells at the time of the primary culture passage. In this modified protocol, P-0 BMSCs that are detached and pelleted for primary passage are resuspended in the virus/polybrene-containing media as described above, incubated in the cell culture incubator for 30 min, and plated into one 100 mm dish for culture. We have obtained transduction efficiency up to 50% or 60% with this modified method.
Mouse Calvarial Defect Model
The surgical procedure was approved by the institutional animal use committee (ACC protocol 101111-0718). As described previously,23 a 3.5 mm diameter calvarial bone defect was created on one or both sides of the sagittal suture (SS) of the mice that were anesthetized with ketamine/xylazine. Hydroxyapatite (HA)/collagen matrix (HEALOS, DePuy Spine, Inc., Raynham, MA) scaffold disks (3.5 mm diameter × 0.5 mm thick) were cut and loaded with cell samples (1 × 106) and placed into the defect area. Postoperative analgesics were administered using isoflurane anesthesia to prevent dislodging of the implant secondary to animal resistance to the analgesic injection. In addition, male mice were separated for the first week to reduced implant shifting as a consequence of fighting. Implanted mice were maintained for 6 to 12 weeks. One day before sample harvesting, demeclocycline (30 µg/g) or calcein (10 µg/g) dissolved in 2% g/v NaHCO3 (pH = 7.4) was injected intraperitoneally (IP) to label newly deposited mineral.
During euthanasia, the intact calvaria was dissected free of the surrounding tissue and was fixed in 10% formalin (cat. no MFCD000003274, Sigma) at 4C for 2 to 3 days. Subsequently, digital photographic and X-ray (Faxitron LX 60, Lincolnshire, IL) images were taken followed by placement in cold 30% g/v sucrose/PBS for 12 to 24 hr in preparation for histological evaluation (see below). The enzymatic activity within the sample can be preserved for a few days at −20C or for an indefinite time period at −80C.
The surgical procedure was carried out in the immunodeficient NOD.Cg-Prkdcscid Il2rgtm1wj1/SzJ mice (NSG, The Jackson Laboratory, Bar Harbor, ME) that were purchased and maintained in our animal facility. This genetic background has proven to be tolerant to the long-term human cell engraftment that is necessary to interpret a bone transplantation experiment. To provide a visual histological marker that would identify mouse-derived bone and fibrous tissue within a surgical repair site, Col3.6GFPtpz transgenic mice in a CD1 background were crossed to NSG mice and then backcrossed with NSG for 10 generations. The resulting cross, called NSG/Col3.6GFPtpz, was tested to identify breeders that had regained their full parental immunosuppressed properties using antibodies suggested by The Jackson Laboratory: CD11b (macrophage/granulocyte marker) conjugated with Pacific Blue (eFluor 450, Bioscience, cat. no 48-0112), CD3e (T-cell receptor complex) conjugated with Phycoerythrin (PE-cy7, BD Pharmingen, cat. no 552774), B220 (a restricted isoform of mouse CD45 found on B cells) conjugated with Allophycocyanin (CD-45R(B220)APC eBioscience, cat. no 17-0452), and CD49b (an antigen expressed on majority of mouse killer cells and subset of T-cells) conjugated with R-Phycoerythrin (Clone Dx5, eBioscience, cat. no 12-5971). Blood samples (200 µl) were harvested by retro-orbital sinus bleeding, incubated with the antibody mixture for 45 min, and used for flow cytometry analysis on the BD LSR II (Flow Cytometry core, University of Connecticut Health, Farmington, CT). Mouse tail DNA was extracted for PCR to detect the Il2rgtm1wj1 mouse genotype using primers (oIMR5330: 5′ GTGGGTAGCCAGCTCTTCAG, oIMR5331: 5′ CCTGGAGCTGGACAACAAAT, and Oimr7415 5′ GCCAGAGGCCACTTGTGTAG) based on the sequences provided by The Jackson Laboratory. The annealing temperature was 67C and total cycle number was 35 cycles. Identification of mice carrying the Col3.6GFPtpz reporter was detected in neonatal mice using miner’s goggles and confirmed by fluorescence microscopy of a fresh tail snip.
The analysis of mice obtained after 10 generations of backcrossing used flow cytometry on peripheral blood to detect the markers of B (B220), T (CD3), natural killer (NK, DX5), and macrophage (CD11b) cells (Supplemental Fig. 1, Panel A). Normal numbers of these cell types were detected in the control blood (bottom panel), but negligible numbers of these cells were found in the NSG/Col3.6GFPtpz mice (top panel). Genomic DNA genotyping showed that both alleles of the IL2Rgamma gene were deleted (Supplemental Fig. 1, Panel B). The Col3.6GFPtpz reporter in skeletal cells in the NSG mouse line had a similar pattern and fluorescent intensity as the parental Col3.6GFPtpz line (data not shown). The NSG/Col3.6GFPtpz mouse line was established and used for further implantations.
Histology Sample Preparation and Staining Steps
The calvaria was trimmed to frontal strips that encompassed the implanted tissue and multiple strips from different calvaria (up to four) were embedded in a stacked orientation within a single block (Supplemental Fig. 2) using Neg-50 frozen section medium (Richard-Allan Scientific, Kalamazoo, MI). Cryosectioning was performed in a Leica cryotome, CM3050-S, using a non-autofluorescent adhesive film (Section Lab, Co., Ltd., Toyota-gun) to capture the section (5 µm). The tape was adhered, section side up, to a glass slide using a 0.2% g/v chitosan solution (Sigma, cat. no C3646) dissolved in 0.25% acetic acid followed by air drying overnight at 4C or ultraviolet (UV) adhesive glue (Norland Optical Adhesive, cat. no 6301, Cranbury, NJ). A set of three adjacent sections is collected at each level chosen for analysis and identified as the central and two adjacent sections. In the following protocols, a sample set consisting of four to eight slides each with three to four calvarial sections per tape and two tapes per slide (Supplemental Fig. 2) are processed and imaged together to ensure that the observed histological signals are produced under identical experimental conditions.
Step 1: Endogenous fluorescent signals. The slide is soaked in PBS for 10 min and a cover slip is mounted with 50% g/v glycerol in PBS before microscopy for detection of accumulated mineral, regions of active mineral deposition, and cellular GFP. In the subsequent staining steps of the same section, the cover slip was removed by brief soaking in PBS and then processed for additional stains.
Step 2: TRAP enzymatic activity. The cover slip was removed and placed in the TRAP reaction solution (112 mM sodium acetate, 76 mM sodium tartrate, 11 mM sodium nitrite, pH 4.1 to 4.3) for 10 min. The ELF 97 phosphatase substrate (1:20 to 1:60, Life Technologies, cat. no E-6588) was added to the section and exposed to UV blue light for 5 min (GE Lightening, Model 23301-A, black light tube, F15T8-BLB). The reaction was stopped by submerging the slides in three changes of PBS followed by mounting the cover slip in 50% glycerol in PBS. After the sections were imaged, the cover slip was removed as described above.
Step 3: AP. After scanning, the cover slip was removed and the section was incubated in the AP reaction buffer (100 mM Tris, pH 9.5, 50 mM MgCl2, 100 mM NaCl) for 10 min followed by substrate buffer containing 200 µg/ml Fast Red TR (Sigma, cat. no F8764-5G) and 100 µg/ml Naphthol AS-MX Phosphate (Sigma, cat. no N-4875) for 5 min. The slides were washed three times in PBS and mounted using 50% glycerol in PBS solution.
Step 4: Human BSP (hBSP) immunostaining: This step used an adjacent section that was decalcified using the TRAP buffer and neutralized. The slides are covered with 0.1% v/v Triton (Sigma, cat. no T8787) ×100 in PBS for 10 min followed by two PBS wash steps. Background suppression of the secondary antibody is initiated by addition of a 1:20 dilution of normal donkey serum (cat. no 017-000-121, Jackson ImmunoResearch Labs, West Grove, PA) for 1 hr at room temperature. After three washing steps in PBS, a 1:500 dilution of Fab Fragment Donkey anti-mouse IgG (cat. no 715-007-003, Jackson ImmunoResearch Labs) was incubated with the slides for 1 hr at room temperature. This step suppresses nonspecific staining of the mouse bone marrow compartment by the secondary antibody. After three PBS wash steps, the sections are covered with a 1:400 dilution of mouse anti-human BSP antibody (anti-hBSP, Millipore, cat. no MAB1061) made up in blocking solution and incubated overnight at 4C. The following day, the slides were rinsed with PBS three times and incubated with a 1:500 dilution of FITC conjugated donkey anti-mouse IgG (cat. no A31571, Invitrogen) at room temperature for I hr. It may be necessary to centrifuge the FITC antibody solution at ~10,000 × g for 15 min at 4C to remove fluorescent particulates. The slides were washed in PBS three times and mounted in 50% glycerol and 0.1% Hoechst 33342 (DAPI, Molecular Probes cat. no H-3570) in PBS.
Step 5: Human nuclear antigen (HuNucAg) antibody staining. This step is performed on an adjacent section that has been treated with the TRAP buffer step to remove mineral. The sensitivity of the HuNucAg staining step can be improved by partial proteolytic degradation of the chromatin proteins. From a 20 mg/ml stock solution of proteinase K (cat. no aB00925-00100, American Bio, Canton, MA), 1 µl was diluted to 1 ml in TE buffer. After the slide was washed in PBS for 5 min, the sections were covered with the diluted proteinase K and placed in a humidified incubator at 37C for 10 min, after which the slides were washed three times in PBS. Then the slides are covered with 0.1% Triton ×100 in PBS for 10 min followed by two PBS wash steps. Background suppression of the secondary antibody used the same combination of non-immune donkey serum as described for hBSP described above. Once the suppression steps are completed, HuNucAg antibody (cat. no MAB1281, Millipore) was diluted at 1:200 in blocking solution and placed on the slides for 1 hr at room temperature or overnight at 4C. The sample was washed in PBS three times. Donkey anti-mouse Cy3 (cat. no 715-166-150, Jackson ImmunoResearch Labs) was diluted at 1:500 in PBS and incubated with the slides for 1 hr at room temperature. The slides were washed three times in PBS and mounted in 0.1% DAPI in 50% glycerol for microscope scanning.
Step 6. Chromogenic staining. This step was performed on the central or adjacent section allowing for multiple chromogenic stains to be included in the histological analysis. For TB staining, sections were washed three times for 3 min in water, blot dried, stained with 0.025% g/v toluidine blue O (Sigma, cat. no T3260) for 5 min, rinsed in water three times for 1.5 min, placed in bluing solution (Shandon, cat. no 6769001) for 1 min, rinsed as before in water and mounted in 30% glycerol in water. For hematoxylin staining, sections were rinsed with distilled water, followed by Mayer’s modified hematoxylin solution (Polyscience, cat. no 3216) for 1 min. After a rinse in distilled water, the slide was placed in the bluing solution for 1 min and rinsed a second time. The sections were covered with a mounting solution of 50% glycerol in water and imaged as described above.
Fluorescent and Chromogenic Imaging and Image Processing
Imaging the sections was performed with an Axio Scan.Z1 microscope (Carl Zeiss, Thornwood, NY) and Colibri 2 LED light source. Accumulated mineral was assessed by RAC optics. DAPI was imaged using a near blue filter (Chroma 49000). Calcein, Col3.6GFPtpz, and FITC were detected with a yellow fluorescent protein filter (eYFP, Chroma cat. no 49003ET, EX: 500/20, EM: 535/30), while a custom RFPchry filter was used for detecting alizarin complexone staining, AP fast red staining, and Cy3 (EX: 577/20, bp595, EM: 640/40). The TRAP signal is segmented with a filter optimized for ELF97 and demeclocycline (Chroma Technology Custom HQ409sp, 425dcxr, HQ555/30). The fluorescent images were captured by an AxioCam MRm Rev.3 monochromatic camera while the chromogenic sections were recorded with a Hitachi HV-F202 brightfield camera. The scans were performed using the 10× objective and the resulting 100× image has a resolution of 0.44 µm/pixel.
Each fluorescent scanning step can produce three to five separate gray scale files that are assigned to a specific pseudocolored output and a single chromogenic file. The images are exported from the Zen 2.0 software as .jpg files and mounted into an image stack with Photoshop (v 11.0 or higher). Using the layer window, each layer will contain the label assigned by the nomenclature within the Zen program and will be in the normal layer mode. The signal strength of each layer is adjusted to fill 50% of the full digital spectrum as illustrated in Supplemental Fig. 3. With the exception of the lowest layer containing the accumulated mineral, each layer is adjusted to the screen mode to allow all the layers above the mineral level to be projected onto the base level after which the layers are manually aligned using the fluorescent microbeads and visual histological features. In addition, the chromogenic layer should be adjusted to about 50% setting using the opacity slider shown in Supplemental Fig. 4 to allow the deeper fluorescent signals to project into the chromogenic image.
Results
Image File Management
The root image stack consists of all the digital image files produced from the multiple rounds of staining and scanning that are assembled and aligned within the Photoshop environment in a vertical manner. In addition, it is useful to include the photographic and X-ray documentation of the dissected calvaria, scale bars, and the name of the computer file (Fig. 1A). It is regarded as the primary data source from which specific regions of interest can be isolated as a secondary stack that maintains all the layers of the parental stack. It is from the root and sub-stacks that flat files are created for publication or visual presentations (Supplemental Fig. 5). Examples of original high-resolution image stack file (~0.5 Gb) or sub-stack files can be downloaded from our laboratory image server (see http://ucsci.uchc.edu/cryomethod/) for analysis using Photoshop. The layer table in Fig. 1A illustrates how the stack is constructed and controlled using the layers feature of Photoshop. The stack consists of 16 layers of which layers 2 to 10 were obtained from the Axioscan microscope. Layers 11 to 16 add the calvarial photograph and X-ray, the bar graphs, and file name. Figure 1B illustrates how specific layers (9 and 2) are merged to produce a composite flat file that shows accumulated mineral over the decalcified TB layer. Note that in the layer table, the TB layer was set to the screen mode with an opacity setting of 50% to allow the mineral image to project into the TB image. Another example of merging TB with the fluorescent signals is provided in Supplemental Fig. 4. Once the alignment and image settings are optimized, the image stack is flattened and saved as a .jpg file, which prevents further alterations to the microscope image. The reader is encouraged to download each figure and increase it to its full image size to appreciate the cellular details described below.
Define the Cellular Landscape of the Mineralized Tissue Section
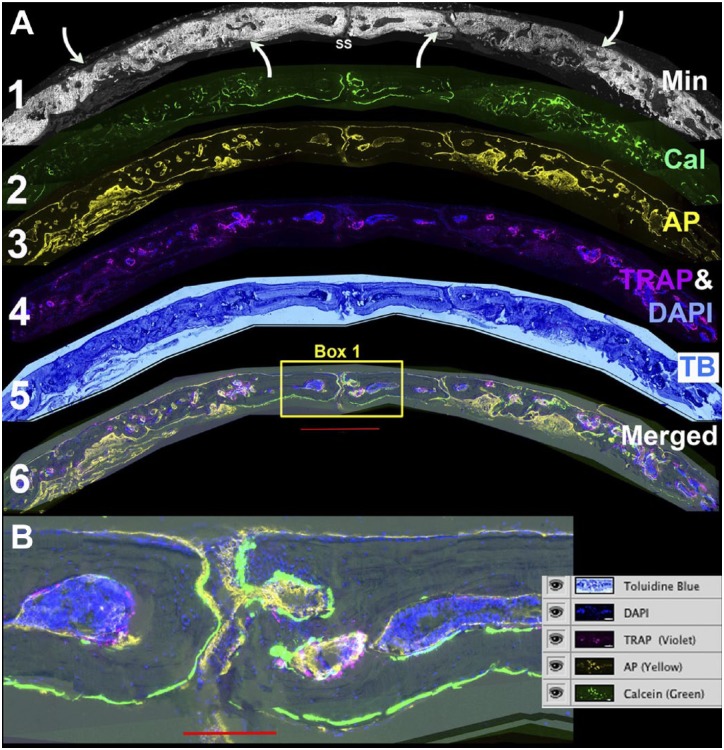
Figure 2 is derived from the root image (shown in Fig. 1) of a 12-week-old repair defect performed in a 13-week-old NSG male. The overall composition of the section is defined by merging the accumulated mineral and chromogenic layers as shown in Fig. 1B. However, the subtle difference of mineral accumulation is better appreciated in the mineral only layer shown in Fig. 2A-1. The accumulated mineral (gray color) in the bone surrounding the defect has a deeper intensity than the newly formed mineral and helps to distinguish the boundary between the host membranous bone and tissue that develops within the defect space (curved arrow) as well as the SS. The TB stain (Fig. 2A-5) provides the cellular context of the composite image, which is necessary to appreciate the marrow spaces and cell nuclei that are embedded into the mineralized tissues.
Figure 2.
Defining the osteogenic landscape of the repair field. (A) Individual layers of the stack are assembled to characterize the major features of the repair process. Layer 1: Accumulated mineral from which the overall orientation of the defect field is related to the sagittal suture (SS) and the boundaries of the repair are identified by the relative intensity of newly formed versus pre-existing mineralized bone (curved arrows). Layer 2: Calcein labeling of actively mineralizing bone surfaces. Both the accumulated mineral and mineral labeling signals are removed in the TRAP step. Layer 3: AP enzymatic activity. This step assesses the osteogenic activity of the repair process and is performed after the TRAP step. Layer 4. TRAP (violet) and DAPI (blue). The two are paired to emphasize the association of the endocortical bone surfaces with the bone marrow islands. The DAPI is also useful to identify cells with the bone matrix. Layer 5: TB. This stain provides a familiar chromogenic context for the fluorescent signals. Layer 6: Merging of all the cellular layers to show their relationship with active bone matrix forming surfaces. (B) Enlarged view of box 1 with a screen shot of the layer file. The green calcein label highlights the active periosteal and endosteal bone surfaces in the region of the SS. Most of the surfaces have an overlying AP (yellow signal). The bone marrow islands are intensely DAPI positive with TRAP-positive foci on the endocortical surface. The scale bar for Fig. 2A = 1000 µm and for Fig. 2B = 200 µm. Abbreviations: TRAP, tartrate resistant acid phosphatase; DAPI, 4′,6-diamidino-2-phenylindole; AP, alkaline phosphatase; TB, toluidine blue.
The next step selects image layers containing signals that represent regions of active bone formation as assessed by ongoing mineral deposition, osteoblast, and osteoclast activity. These features will be better appreciated by downloading and magnifying the image. The calcein signal (Fig. 2A-2, green), which labels recently deposited mineral on the bone surface, is best appreciated when the mineral channel is turned off. A strong sharp signal is seen on the outer surface of the host bone and the endocortical surface of the small bone marrow islands (see bone adjacent to the SS). The signal is weaker and more diffuse in the bone matrix within the defect area. Similarly, the AP signal (Fig. 2A-3, yellow), which arises from the surface of osteoblastic cells, is strongest above the same regions that have the strong calcein signal. The AP signal from cells in the repair area is weaker and less distinct but is still located on the bone surface. Cells from the osteoclast lineage identified by the TRAP stain (Fig. 2A-4, violet) are found primarily on the endocortical surface of the bone marrow, which is appreciated in fluorescence with the DAPI channel. The marrow spaces are located in the adjacent host bone and are not found in the repair region, although foci of TRAP activity are found in this region and will be discussed later. All of these features are merged (Fig. 2A-6) and an enlarged insert of host bone surrounding the SS (box 1) illustrates the relationship of the calcein, AP, and TRAP signals relative to the TB stained bone (Fig. 2B) as shown in the adjacent layer table.
Discriminating Host and Donor Formed Tissue Within the Repair Defect
Two different imaging sequences have been developed that are based on GFP-expressing cells (lentiviral-introduced ubiquitin-RFPchry that identifies donor cells and the Col3.6GFPtpz reporter carried by the NSG mice that identifies host cells) and human-specific antibody staining (hBSP and HuNucAg). Each choice has advantages and limitations.
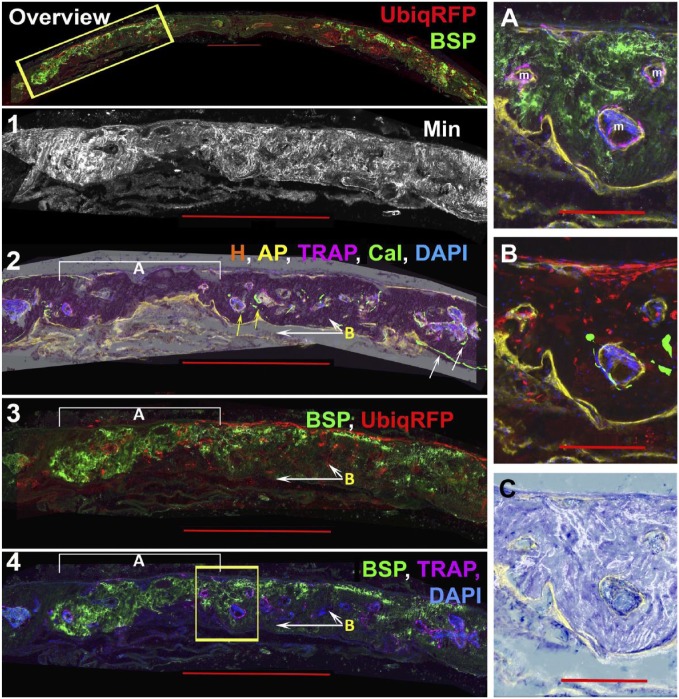
a. Identifying human tissue using ubiquitin-RFPchry and hBSP (Fig. 3): The overview panel shows the distribution of the ubiquitin-RFPchry (red) and hBSP (green) signals within the two repair fields. The hBSP antibody has proven to be exceptionally specific for staining the extracellular matrix formed by the human osteoblastic cells23 as demonstrated by the lack of signal in the host bone separating the two defects. Panel 1 (accumulated mineral) and Panel 2 (calcein, AP, TRAP, and DAPI mapped to the hematoxylin stain) are derived from the boxed area of the overview panel and demonstrate the osteogenic status of the repair field. Sharp calcein lines (green), overlying AP (yellow), and osteoclasts (violet) identify host bone on the bone surface and in marrow spaces (Panel 2, white arrows). In contrast, the region of bone underlying bracket A is mostly devoid of TRAP-lined marrow spaces or calcein-labeled bone surface (Panels 2 and 4). Panel 3 colocalizes the immunostains for hBSP and expression of the ubiquitin-RFPchry. The red signal is restricted to the cells within the defect space and can also be found in the adjacent marrow spaces. It is present on the external bone surface and extends into the bone matrix. The hBSP antibody is localized to the bone matrix and extends across the length of the repair defect. There is good alignment of the hBSP and ubiquitin-RFPchry fluorescent signals within the outer region of mineralized tissue of the repair field (bracket A). However the weaker hBSP and absence of RFP positive cells on the dural side of the tissue to the right side of bracket A indicates that an extension of host bone is projecting into the human repair tissue (arrows B). This interface of both cell sources (Panel 2, yellow arrows) complicates the interpretation of the repair process.
Figure 3.
Identification of human bone matrix by the colocalization of ubiq-RFPchry and anti-hBSP. The experiment used a 13-week-old NSG male mouse and the repair tissue was obtained 12 weeks after the surgical procedure. Panel 1 is an overall view of the ubiq-RFPchry (red) and anti-hBSP (green) signals from the same image stack shown in Figs. 1 and 2. These signals emanate primarily from the two repair defect regions. Panels 2 to 4 enlarge the region boxed in the overview panel and also the boxed regions shown in Figs. 1 and 2. Layers 1 and 2 describe the osteogenic landscape based on accumulated mineralized layer 1 and AP (yellow), TRAP (violet), calcein (green), and DAPI (blue) signals (layer 2). The fluorescent signals are layered over the hematoxylin (H) layer, which preserves regions of mineralized bone as a dense red stain. The white arrows point to the strong calcein labeling of the host periosteal and endocortical bone and the relatively rare calcein labeling within the repair region (yellow arrows). As described in layer 3, bracket A is a region that is exclusively human while the white arrows labeled B identify mouse bone that is extending forward from the dural side of host calvarial border. Layer 3 shows the merging of hBSP (green) and ubiq-RFPchry while layer 4 is TRAP (purple) and hBSP (green). Most of the region to the left of the boxed area which co-express hBSP and ubiq-RFPchry lacks TRAP activity, while regions to the right which are hBSP and ubiq-RFPchry negative show marrow islands with TRAP activity. The boxed region are enlarged in Panels (A–C) to better appreciate the interface between the human and mouse-derived bone. Panel A shows the hBSP and TRAP signals while Panel B is ubiq-RFPchry and calcein. This image has a minor stitching error, which misaligns the AP and calcein signals. The AP signal (yellow) is present in both panels. The two panels provide the impression of a gradual ingrowth of mouse cells extending from the dural side into the overlapping human bone. Marrow islands with osteoclasts develop at the interface but not in the interior of the human bone matrix. Panel C superimposes the mineral layer over toluidine blue staining to demonstrate the disordered pattern of mineral deposition as contrasted with the smooth pattern of host bone shown in Fig. 4, Panel C-3. The scale bar for the overview and Panels 1–4 = 1000 µm and for the subpanels A-C is 200 µm. Abbreviations: RFP, red fluorescent protein; hBSP, Human bone sialoprotein; TRAP, tartrate resistant acid phosphatase; DAPI, 4′,6-diamidino-2-phenylindole; AP, alkaline phosphatase.
The difficulty of interpreting a region containing a mixture of both human and mouse matrix is illustrated by the boxed area of Panel 4 and enlarged inserts (A–C). All three marrow spaces (m) within the boxed region contain endocortical TRAP stained cells (insert A), AP-positive osteoblasts, and some evidence of calcein deposition (insert B). However, the distribution of ubiquitin-RFPchry and hBSP around each marrow space is not uniform suggesting that the cellular mechanism for the formation of marrow space could be either host or donor cells in origin. Discerning this difference is important because of the recognition that competent osteoprogenitor cells have the ability to induce bone marrow formation, which in turn is important for bone remodeling.24 The disordered mineralization pattern of this region (see Fig. 3, insert C vs. Fig. 4 Panel C, insert 3) may reflect this lack of bone remodeling.
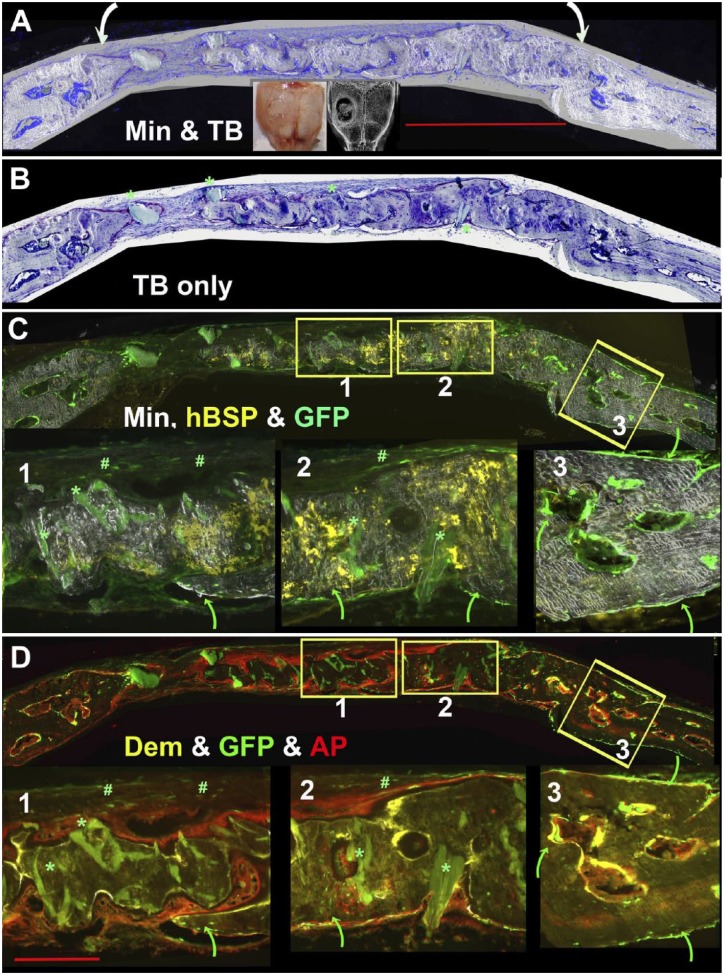
Figure 4.
Use of hBSP and host-derived Col3.6GFPtpz to distinguish human and mouse-derived bone matrix. The experiment used a 17-week-old NSG/Col3.6GFPtpz female mouse and the defect was harvested 8 weeks after surgery. (Panel A) Flattened view of the entire root file of a single hole calvarial defect in which the margins of the repair field are indicated by the curved arrows. (Panel B) Toluidine Blue layer. Some of the larger acellular and weak green hydroxyapatite (HA) deposits of the scaffold are indicated by the green asterisk. (C and D) Osteogenic landscape as defined by accumulated mineral, hBSP (yellow), and Col3.6GFPtpz (green) in Panel C and demeclocycline (DEM, yellow) and AP (red) staining in Panel D. Boxes 1, 2, and 3 are enlarged in this and subsequent panels to represent a region primarily composed of human bone (box 1), a region of interface of human and mouse bone (box 2) and adjacent host bone (box 3). Human bone in boxes 1 and 2 is identified by the hBSP signal, the broad layers of AP-positive cells on the bone surface, the unresorbed HA (green asterisk) and the disorganized mineral deposition. The mouse bone lacks the hBSP and HA staining while the forming mouse bone has a narrow layer of AP-positive osteoblasts and matrix shows a uniform pattern of mineral distribution. Strong expression of host-derived Col3.6GFPtpz cells lining the periosteal and endosteal bone are indicated by the curved green arrows in box 3. The reporter also is seen within the bone matrix external to the human bone (box 1) or intermingled with human bone (box 2). Weak expression of the Col3.6GFPtpz reporter is seen in the fibrous tissue overlying the external side of the repair (marked with a green pound sign). The scale bar for the overview panels is 1000 µm and for the subpanels is 200 µm.
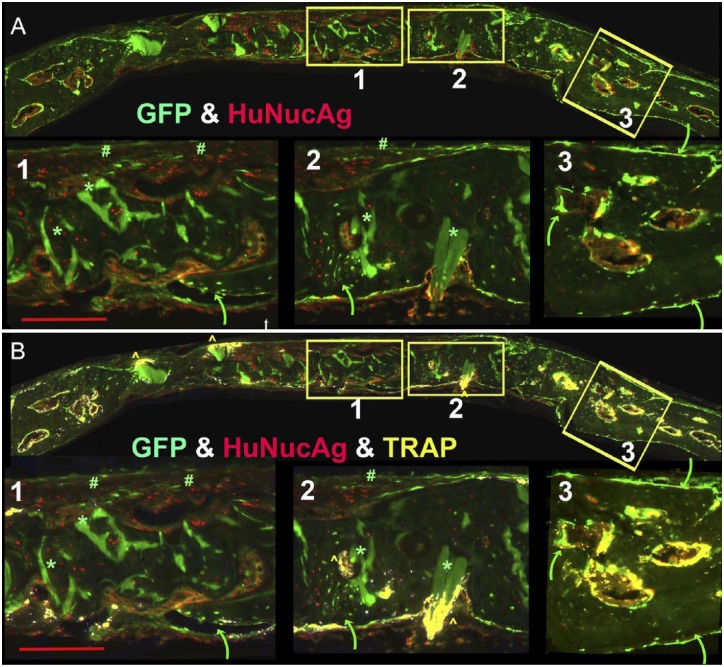
b. Discriminating human and mouse tissue using HuNucAg and host-expressed Col3.6GFPtpz.
Another test for human cell identification, which is useful when the ubiquitin-RFPchry vector is not available, is the antibody to HuNucAg. It is a more difficult antibody to use than hBSP because the epitope needs to be partially exposed using proteinase K and finding the right conditions for a particular section can be a problem. When the HuNucAg positive cells are mapped to the corresponding section showing the Col3.6GFPtpz reporter carried by the host tissues, the colocalization of the human and mouse-derived bone cells within bone matrix can be appreciated (see Fig. 5 Panel A).
Figure 5.
Use of HuNucAg (red) and host-derived Col3.6GFPtpz (green) to distinguish human and mouse-derived bone matrix. These images including the three boxed regions are derived from the same experiment described in Fig. 4A. Box 1 shows HuNucAg positive cells throughout the matrix and distinct from the Col3.6GFPtpz positive cell indicated by the curved green arrow. However, in box 2, the green arrow points to a region where the two populations of cells are intermingled with the bone matrix, although most of this hBSP positive matrix only shows HuNucAg positive cells. In box 3, all of the matrix is hBSP negative and Col3.6GFPtpz positive cells are distributed on the periosteal and endosteal surfaces. (B) The distribution of TRAP (yellow) is layered over the image shown in Panel A. Box 1 show a punctate distribution within the dural fibrous tissue in the region of unresorbed Hydroxyapatite (HA) and the small ingrowth of mouse bone (green arrow). Box 2 again show larger accumulation of TRAP-positive cells surrounding the HA deposit (yellow caret) and on the bone surface lined with Col3.6GFPtpz positive cells (green arrow). However, in both boxes, no TRAP signal is detected in regions where huNucAg positive cells predominate. In contrast, box 3 shows the expected strong TRAP positivity of cells lining the endocortical bone of the marrow islands. The scale bar for the overview panels is 1000 µm and for the subpanels is 200 µm. Abbreviations: hBSP, Human bone sialoprotein; GFP, green fluorescent protein; AP, alkaline phosphatase; TRAP, tartrate resistant acid phosphatase.
Figure 4 illustrates the use of these histological signals in an 8-week-old single hole repair defect performed in a 17-week-old female NSG-Col3.6GFPtpz mouse that did not completely fill the defect space. Panel A is from the root image and shows the overlay of the accumulated mineral (Min) and TB and the curved arrows define the limits of the repair field. Panel B (TB only) shows the cellular detail within the mineralized matrix and the lack of cells with deposits of unresorbed HA (green asterisk).
In Panel C, the overview image is separated into three enlarged regions that demonstrate different contribution from the host or donor cells: box 1, human distinct from mouse; box 2, human intermingled with mouse; box 3, mouse only. This panel overlays the hBSP (yellow), host Col3.6GFPtpz, and accumulated mineral signals to distinguish host and donor matrix. The curved green arrows point to the Col3.6GFPtpz positive cells while the green asterisk localize the deposits of unresorbed autofluorescent HA. The autofluorescent signals are distinguished from a specific fluorescent probe by positive fluorescence at every excitatory/emission setting, while the specific probe is active at one specific setting. Panel C, box 3 shows the host osteoblasts as a strong green signal on the periosteal and marrow endosteal surfaces, where it is usually associated with an underlying yellow demeclocycline (Dem) line (Panel D, box 3). Also note the ordered RAC pattern of accumulated mineral of host (box 3) versus the irregular pattern with the donor bone (boxes 1 and 2). Col3.6GFPtpz positive host cells are also present in Panel C boxes 1 and 2 but primarily on the dural side of the bone (green arrows). The loose connective tissue overlying the bone surface of boxes C1 and C2 reveal faint Col3.6GFP positive cells (green pound sign) characteristic of type I collagen-producing fibroblasts. However, within most of hBSP positive matrix (yellow) of boxes C1 and C2, no Col3.6GFPtpz cells are evident. The exception is an ingrowth of Co3.6GFPtpz positive mouse osteoblasts on the dural side (green arrows). Panel D contrasts the AP (red) signals and DEM (yellow) signals versus host Col3.6GFPtpz cells. The diffuse AP and DEM signal overlying the hBSP positive matrix (boxes D1 and D2) differs from the sharp signals lining the endosteal and periosteal surfaces (box D3).
Figure 5, Panel A contrasts the distribution of HuNucAg and Col3.6GFPtpz signals in the three zones. In box A1, HuNucAg positive cells are present throughout most of the matrix with the exception of the small tongue of mouse bone (green arrow) and the fibrous cellular layer (#) over the external surface of the bone. Box A2 also shows HuNucAg positive cells throughout most of the matrix with the exception of an area of mixing of Col3.6GFPtpz positive cells marked by the curved green arrow. In contrast, box A3 is mostly negative for HuNucAg in the bone matrix although positive cells are present in the marrow spaces. Panel B adds the TRAP stain to the signals in Panel A. The overview image of the entire calvaria shows strong TRAP activity in the adjacent host bone that outline the circular endocortical surfaces of the marrow spaces. With the exception of the TRAP surrounding deposits of the yet to be resorbed HA (*), there is minimal TRAP in the repair region. In box B1, the TRAP signal is external to regions of human bone, while in Panel B2 the TRAP is seen in an area composed of a mixture of HuNucAg and Col3.6GFPtpz positive cells (yellow caret). The overview panel shows an intense osteoclastic response to HA deposits that extend outward from the human bone matrix (yellow caret) but a lack of response to deposits that are internal to the human matrix (*). Panel B3 emphasizes the strong TRAP signal on the endocortical surface of the marrow islands within the host bone.
Interpretation of the Repair Process
Although the GFP and immunological data strongly indicates that the bone matrix that was formed in the repair space is of human origin, its character is different from the surrounding mouse bone. Specifically, it appears to lack formation of bone marrow islands and the accumulation of osteoclastic cells. The pattern of mineral deposition is sparse and disorganized as might be expected if there is a lack of osteoclastic remodeling. However, osteoclasts can invade the tissue at sites of unresorbed HA, which is a component of the HEALOS scaffold used in the implantation protocol. The small deposits of unresorbed HA are recognized in the histological sections as regions of intense autofluorescence that are detected in all filter settings, and the absence of cell nuclei within the deposit as detected in the chromogenic stains. Marrow spaces and osteoclasts can also be observed at the interface between the mouse and human bone but in most cases the endocortical bone will contain a few mouse osteoblasts as indicated by the GFP signal expressed by the NSG reporter mice (Fig. 5, box A2 and B2). The radiographic and histological interpretation of the repair strategy is that sufficient human bone was formed to fill the defects space, but because it did not get remodeled into a lamellar structure, it is unlikely to have sufficient mechanical properties to produce a functional skeletal repair.
Discussion
The cryohistological protocol presented here provides a number of operational and interpretive options that are not available for traditional paraffin or plastic embedded histological methods. The sample can be sectioned within a few days of harvesting, the mineralized sections can be imaged immediately after cutting and the mineral component can be removed from the section after a few minutes in the acidic conditions used for the TRAP stain. Most repair tissue sites can be cut with a disposable steel blade, but the method has been adapted to cutting mouse teeth using a titanium blade. Thus, the ability to begin the histological analysis can progress without the delays of tissue decalcification and dehydration that are required for other paraffin- and plastic-based methods.
Because the cryohistological sections do not have significant autofluorescent background, the interpretative power of fluorescence-based histology can be applied. Not only can these signals be designed to reflect a specific biological activity or marker of cell specificity, they can be easily mapped back to a familiar chromogenic image. The use of the two adjacent sections opens an opportunity of additional probes that provide regional signals that are informative to the central section. The formation of the image stack that is developed from whole-section imaging provides the opportunity for comprehensive analysis that can be electronically shared across all-interested observers. This capability is similar to the studies employing microarray technologies in which the raw microarray file and associated metadata are stored on central databank (see https://www.ncbi.nlm.nih.gov/geo/) available to any investigator who want to use their software package for data interpretation.
There are many options for probes that reflect the repair process and the source of the cells that mediate the repair. Our workflow begins by determining the primary tissue and cellular composition of the central section. The accumulated mineral is mapped to the chromogenic images and subsequently the location of the mineralizing surfaces, the surfaces that express a strong AP signal and the distribution of TRAP-positive cells both within the DAPI-intense marrow spaces and other sites are determined from the central section. Once this mineralized landscape is defined, the specific probes of cells or tissue matrix are added.
GFP-derived reporters provide the most unequivocal and distinct signal for cell of origin. Placing a reporter in a host mouse background such as NSG provides for consistency across many experimental models for discrimination of mouse-derived tissues and cells. Once produced and shared as a resource animal, greater consistency of experimental interpretation could be obtained across different laboratories. Introduction of a fluorescent reporter into primary human cells has to use a method that does not require cell selection because that process would require multiple cell passages and destroy the progenitor potential. Use of lentiviral system is probably the most suitable vector for introducing a stable reporter without compromising the progenitor properties of the target population. The ubiquitin promoter has proven to produce a strong fluorescent signal in many cell types including most mesenchymal-derived lineages.
In the absence of a source-restricted reporter, various species-specific antibodies have been used with various success. Because cryoembedded sections preserve tissue antigens better than other methods, the range of possibilities is wide. In addition, the acid-demineralization step may have a secondary antigen retrieval activity. In our hands, both human nuclear antigen and human mitochondrial antigens are useful but each has its advantages and difficulties. Human nuclear antigen produces a strong signal over the nucleus but performs better after the tissue is briefly exposed to proteinase K, while the signal from the human mitochondrial antigen is diffuse in the cytoplasm and is not detectable in osteocytes. Of the antibodies that detect extracellular matrix proteins, the human-specific BSP antibody has proven to be exceptionally reliable for detecting the antigen in the matrix produced by human osteoblasts and hypertrophic chondrocytes without a background activity in murine tissues. In situ hybridization for human-specific genomic sequences (Alu) or RNAs using RNAscope probes can be used on a Cryojane slide, which can tolerate the high incubation temperatures required for molecular hybridization.
Despite the formation of human bone tissue, the overall quality is significantly different from bone formed using mouse BMSCs. Instead of forming a cortical-like structure with ample internal bone marrow (Supplemental Fig. 6), the human cells produce a membranous-like bone tissue that is essentially devoid of bone marrow. Our findings with human cells are inconsistent with the report of Sacchetti et al.25 who emphasized the importance of marrow development at sites of human bone matrix formation. Other studies that used in vivo transplantation have made reference to variation in marrow or osteoclast enhancement of bone formation.26,27 This discrepancy may relate to differences between the BMSC culture protocol, the character of the matrix used to retain the transplanted BMSCs at the implant site, or differences in a model that uses calvaria, subcutaneous pouch, or the subrenal capsule. These difference in experimental outcome will have to be resolved with more informative histology.
Use of a host-specific GFP reporter is particularly helpful in demonstrating that where bone marrow is formed, cells with a Col3.6GFPtpz signal usually accompany it. Thus histology that does not distinguish between human and mouse-derived tissue may not reveal co-mingling of both tissue types. In addition, areas of mouse bone formation show a sharp mineralization line that is overlaid with a Col3.6GFPtpz reporter cell that has a strong AP signal, while human bone has a weak mineralization line and diffuse AP signal. In some regions, a wide osteoid seam separates the mineralization line from the AP signal (not shown). All of these features suggest that the bone matrix produced from the BMSC generated in our hands is not being adequately remodeled and invested with mouse-derived bone marrow cells. Specifically, the presence of osteoclasts remodeling the bone surfaces is noticeably absent within the human bone matrix except in areas adjacent to mouse bone or sites of un-reabsorbed HA incorporated into the Healos scaffold. In contrast, even membranous mouse bone formed from fetal calvarial cells shows intense osteoclast activity in the absence of bone marrow and complete resorption of the HA (Supplemental Fig. 7).
The lack of obvious remodeling and absence of osteoclasts within areas of human bone matrix accumulation suggests that human RANKL is relatively ineffective in interacting with mouse RANK receptor. Although in vitro interaction between human RANKL and mouse RANK is routinely used in cell culture, the concentration that is used far exceeds the biological levels. In vivo evidence for the ineffective mouse/human RANK/RANKL interaction was observed in mice engineered to replace mouse RANKL with human RANKL that was required for the development of an animal model responsive to anti-human RANKL therapy.28 These animals developed a high bone mass phenotype suggesting impaired mouse osteoclast activity. Thus, in a competitive environment, human RANKL expressed from the newly formed human osteoblasts will not be as an attractive target as osteoblasts expressing mouse RANKL. The importance of the RANKL/Rank interaction for bone remodeling is underscored by RANKL–/– mice that produce an immature bone matrix due to a lack of remodeling.29 It is also possible that signaling from other human cytokines secreted by osteoblasts such as M-CSF, G-CSF, IL-6, BMP, Jagged 1, CX3CL1, and CXCL12 that promote bone marrow formation/expansion may contribute to this histological feature. These problems may be overcome with the use of NSG mice that have been engineered to express the human cytokines that promote engraftment and the systemic distribution of human bone marrow cells including macrophages.30-33 To date, the peripheralization of osteoclasts has not been demonstrated, but it would be anticipated because of the close lineage relationship to macrophages and a recent report of human marrow investment of heterotopic bone formed by hBMSCs.34
The primary objective of this work is to develop histological criteria to identify a human osteogenic progenitor population that is capable of forming human bone tissue and heal a murine skeletal defect. Because existing cell surface markers or cell culture methods are not predictive of this property, the field needs an affordable in vivo model system that can distinguish between effective and ineffective progenitor cell source as a baseline for interpreting competing cell-based strategies for bone repair. The criteria of the in vivo model should imply that the human tissue is able to integrate into the host bone, undergo multiple rounds of osteoclastic-driven remodeling, and be capable of bearing a mechanical load. Although low passage human BMSCs produced in our hands do form human bone matrix, appear to integrate with mouse bone, and persist for an extended length of time, they do not remodel or produce the cortical bone and associated bone marrow characteristic of mouse BMSCs. This inability may be due to incompatibilities between human and mouse signaling similar to that encountered by human to mouse bone marrow transplantation. Thus, further modifications to either the donor or the host model system will have to be engineered before a murine platform is capable of reproducing the full developmental potential of human osteoprogenitor cells.
Supplemental Material
Supplemental material, DS_10.1369_0022155419836436 for Histological Criteria that Distinguish Human and Mouse Bone Formed Within a Mouse Skeletal Repair Defect by Xiaonan Xin, Xi Jiang, Liping Wang, Paiyz Mikael, Mary Beth McCarthy, Li Chen, Augustus D. Mazzocca, Syam Nukavarapu, Alexander C. Lichtler and David W. Rowe in Journal of Histochemistry & Cytochemistry
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: XX performed the surgical procedure and interpreted the histological outcome; PM processed the Lonza-derived bone marrow aspirate; MBM processed the University of Connecticut-derive bone marrow aspirate; LC and XJ developed and performed the histological processing, staining, and imaging; AM is the orthopedic surgeon who obtained the human bone marrow aspirate; SM provided the initial impetus for the project; AL developed the Ubiq-RFP lentiviral vector; and DR wrote the manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research reported in this publication was supported by the NIAMS/NIH under Award Number AR064381, and the DOD/USAMRMC under award number W81XWH-11-1-0262. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or DOD/USAMRMC.
ORCID iD: David W. Rowe  https://orcid.org/0000-0001-7852-7775
https://orcid.org/0000-0001-7852-7775
Contributor Information
Xiaonan Xin, Department of Reconstructive Sciences, University of Connecticut Health Center, Farmington, Connecticut.
Xi Jiang, Department of Reconstructive Sciences, University of Connecticut Health Center, Farmington, Connecticut.
Liping Wang, Department of Reconstructive Sciences, University of Connecticut Health Center, Farmington, Connecticut.
Paiyz Mikael, Department of Orthopedic Surgery, University of Connecticut Health Center, Farmington, Connecticut.
Mary Beth McCarthy, Department of Orthopedic Surgery, University of Connecticut Health Center, Farmington, Connecticut.
Li Chen, Department of Reconstructive Sciences, University of Connecticut Health Center, Farmington, Connecticut.
Augustus D. Mazzocca, Department of Orthopedic Surgery, University of Connecticut Health Center, Farmington, Connecticut
Syam Nukavarapu, Department of Orthopedic Surgery, University of Connecticut Health Center, Farmington, Connecticut; Department of Materials Science and Engineering, University of Connecticut, Storrs, Connecticut.
Alexander C. Lichtler, Department of Reconstructive Sciences, University of Connecticut Health Center, Farmington, Connecticut
David W. Rowe, Department of Reconstructive Sciences, University of Connecticut Health Center, Farmington, Connecticut.
Literature Cited
- 1. Bone Therapeutics: A Regenerative Therapy Company. Available from: www.bonetherapeutics.com
- 2. Novadip Biosciences. Available from: http://www.novadip.com
- 3. Bone Biologics Corp. Available from: http://bonebiologics.com
- 4. Derubeis AR, Cancedda R. Bone marrow stromal cells (BMSCs) in bone engineering: limitations and recent advances. Ann Biomed Eng. 2004;32(1):160–65. [DOI] [PubMed] [Google Scholar]
- 5. Panetta NJ, Gupta DM, Longaker MT. Bone regeneration and repair. Curr Stem Cell Res Ther. 2010;5(2):122–28. [DOI] [PubMed] [Google Scholar]
- 6. Gomez-Barrena E, Rosset P, Lozano D, Stanovici J, Ermthaller C, Gerbhard F. Bone fracture healing: cell therapy in delayed unions and nonunions. Bone. 2015;70:93–101. [DOI] [PubMed] [Google Scholar]
- 7. Grayson WL, Bunnell BA, Martin E, Frazier T, Hung BP, Gimble JM. Stromal cells and stem cells in clinical bone regeneration. Nat Rev Endocrinol. 2015;11(3):140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jakoi AM, Iorio JA, Cahill PJ. Autologous bone graft harvesting: a review of grafts and surgical techniques. Musculoskelet Surg. 2015;99(3):171–8. [DOI] [PubMed] [Google Scholar]
- 9. Toombs JP, Wallace LJ. Evaluation of autogeneic and allogeneic cortical chip grafting in a feline tibial nonunion model. Am J Vet Res. 1985;46(2):519–28. [PubMed] [Google Scholar]
- 10. Kroese-Deutman HC, Vehof JW, Spauwen PH, Stoelinga PJ, Jansen JA. Orthotopic bone formation in titanium fiber mesh loaded with platelet-rich plasma and placed in segmental defects. Int J Oral Maxillofac Surg. 2008;37(6):542–9. [DOI] [PubMed] [Google Scholar]
- 11. Liao JC, Chen WJ, Chen LH, Lai PL, Keorochana G. Low-intensity pulsed ultrasound enhances healing of laminectomy chip bone grafts on spinal fusion: a model of posterolateral intertransverse fusion in rabbits. J Trauma. 2011;70(4):863–9. [DOI] [PubMed] [Google Scholar]
- 12. Shahdadfar A, Fronsdal K, Haug T, Reinholt FP, Brinchmann JE. In vitro expansion of human mesenchymal stem cells: choice of serum is a determinant of cell proliferation, differentiation, gene expression, and transcriptome stability. Stem Cells. 2005;23(9):1357–66. [DOI] [PubMed] [Google Scholar]
- 13. Levi B, James AW, Nelson ER, Li S, Peng M, Commons GW, Lee M, Wu B, Longaker MT. Human adipose-derived stromal cells stimulate autogenous skeletal repair via paracrine Hedgehog signaling with calvarial osteoblasts. Stem Cells Dev. 2011;20(2):243–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ankrum JA, Ong JF, Karp JM. Mesenchymal stem cells: immune evasive, not immune privileged. Nat Biotechnol. 2014;32(3):252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thesleff T, Lehtimaki K, Niskakangas T, Huovinen S, Mannerstrom B, Miettinen S, Seppanen-Kaijansinkko R, Ohman J. Cranioplasty with adipose-derived stem cells, beta-tricalcium phosphate granules and supporting mesh: six-year clinical follow-up results. Stem Cells Transl Med. 2017;6(7):1576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Featherall J, Robey PG, Rowe DW. Continuing challenges in advancing preclinical science in skeletal cell-based therapies and tissue regeneration. J Bone Miner Res. 2018;33(10):1721–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang X, Kalajzic Z, Maye P, Braut A, Bellizzi J, Mina M, Rowe DW. Histological analysis of GFP expression in murine bone. J Histochem Cytochem. 2005;53(5):593–602. [DOI] [PubMed] [Google Scholar]
- 18. Dyment NA, Jiang X, Chen L, Hong SH, Adams DJ, Ackert-Bicknell C, Shin DG, Rowe DW. High-throughput, multi-image cryohistology of mineralized tissues. J Vis Exp. 2016;115:e54468. doi: 10.3791/54468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dohan Ehrenfest DM, Rasmusson L, Albrektsson T. Classification of platelet concentrates: from pure platelet-rich plasma (P-PRP) to leucocyte- and platelet-rich fibrin (L-PRF). Trends Biotechnol. 2009;27(3):158–67. [DOI] [PubMed] [Google Scholar]
- 20. DeLong JM, Russell RP, Mazzocca AD. Platelet-rich plasma: the PAW classification system. Arthroscopy. 2012;28(7):998–1009. [DOI] [PubMed] [Google Scholar]
- 21. Bakhtina A, Tohfafarosh M, Lichtler A, Arinzeh TL. Characterization and differentiation potential of rabbit mesenchymal stem cells for translational regenerative medicine. In Vitro Cell Dev Biol Anim. 2014;50(3):251–60. [DOI] [PubMed] [Google Scholar]
- 22. Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–72. [DOI] [PubMed] [Google Scholar]
- 23. Xin X, Jiang X, Wang L, Stover ML, Zhan S, Huang J, Goldberg AJ, Liu Y, Kuhn L, Reichenberger EJ, Rowe DW, Lichtler AC. A site-specific integrated Col2.3GFP reporter identifies osteoblasts within mineralized tissue formed in vivo by human embryonic stem cells. Stem Cells Transl Med. 2014;3(10):1125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riminucci M, Remoli C, Robey PG, Bianco P. Stem cells and bone diseases: new tools, new perspective. Bone. 2015;70:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, Bianco P. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–36. [DOI] [PubMed] [Google Scholar]
- 26. Jeon OH, Panicker LM, Lu Q, Chae JJ, Feldman RA, Elisseeff JH. Human iPSC-derived osteoblasts and osteoclasts together promote bone regeneration in 3D biomaterials. Sci Rep. 2016;6:26761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan CKF, Gulati GS, Sinha R, Tompkins JV, Lopez M, Carter AC, Ransom RC, Reinisch A, Wearda T, Murphy M, Brewer RE, Koepke LS, Marecic O, Manjunath A, Seo EY, Leavitt T, Lu WJ, Nguyen A, Conley SD, Salhotra A, Ambrosi TH, Borrelli MR, Siebel T, Chan K, Schallmoser K, Seita J, Sahoo D, Goodnough H, Bishop J, Gardner M, Majeti R, Wan DC, Goodman S, Weissman IL, Chang HY, Longaker MT. Identification of the human skeletal stem cell. Cell. 2018;175(1):43–56.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kostenuik PJ, Nguyen HQ, McCabe J, Warmington KS, Kurahara C, Sun N, Chen C, Li L, Cattley RC, Van G, Scully S, Elliott R, Grisanti M, Morony S, Tan HL, Asuncion F, Li X, Ominsky MS, Stolina M, Dwyer D, Dougall WC, Hawkins N, Boyle WJ, Simonet WS, Sullivan JK. Denosumab, a fully human monoclonal antibody to RANKL, inhibits bone resorption and increases BMD in knock-in mice that express chimeric (murine/human) RANKL. J Bone Miner Res. 2009;24(2):182–95. [DOI] [PubMed] [Google Scholar]
- 29. Schena F, Menale C, Caci E, Diomede L, Palagano E, Recordati C, Sandri M, Tampieri A, Bortolomai I, Capo V, Pastorino C, Bertoni A, Gattorno M, Martini A, Villa A, Traggiai E, Sobacchi C. Murine RANKL-/- mesenchymal stromal cells display an osteogenic differentiation defect improved by a RANKL-expressing lentiviral vector. Stem Cells. 2017;35(5):1365–77. [DOI] [PubMed] [Google Scholar]
- 30. Miller PH, Cheung AM, Beer PA, Knapp DJ, Dhillon K, Rabu G, Rostamirad S, Humphries RK, Eaves CJ. Enhanced normal short-term human myelopoiesis in mice engineered to express human-specific myeloid growth factors. Blood. 2013;121(5):e1–4. [DOI] [PubMed] [Google Scholar]
- 31. Rathinam C, Poueymirou WT, Rojas J, Murphy AJ, Valenzuela DM, Yancopoulos GD, Rongvaux A, Eynon EE, Manz MG, Flavell RA. Efficient differentiation and function of human macrophages in humanized CSF-1 mice. Blood. 2011;118(11):3119–28. [DOI] [PubMed] [Google Scholar]
- 32. Tanaka S, Saito Y, Kunisawa J, Kurashima Y, Wake T, Suzuki N, Shultz LD, Kiyono H, Ishikawa F. Development of mature and functional human myeloid subsets in hematopoietic stem cell-engrafted NOD/SCID/IL2rgammaKO mice. J Immunol. 2012;188(12):6145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M, Mulloy JC. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24(10):1785–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reinisch A, Thomas D, Corces MR, Zhang X, Gratzinger D, Hong WJ, Schallmoser K, Strunk D, Majeti R. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med. 2016;22(7):812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1369_0022155419836436 for Histological Criteria that Distinguish Human and Mouse Bone Formed Within a Mouse Skeletal Repair Defect by Xiaonan Xin, Xi Jiang, Liping Wang, Paiyz Mikael, Mary Beth McCarthy, Li Chen, Augustus D. Mazzocca, Syam Nukavarapu, Alexander C. Lichtler and David W. Rowe in Journal of Histochemistry & Cytochemistry