Abstract
Secretoglobins (SCGBs) are cytokine-like small molecular weight secreted proteins with largely unknown biological functions. Three SCGB proteins, SCGB1A1, SCGB3A1, and SCGB3A2, are predominantly expressed in lung airways. To gain insight into the possible functional relationships among the SCGBs, their protein and mRNA expression patterns were examined in lungs during gestation and in adult mice, using Scgb3a1-null and Scgb3a2-null mice as negative controls, by immunohistochemistry and by qRT-PCR analysis, respectively. The three SCGBs exhibited unique spatiotemporal expression patterns during embryogenesis. The lack of Scgb3a1 or Scgb3a2 did not affect expression of the other Scgb genes as determined by mRNA measurements. Moreover, the lack of Scgb3a1 or Scgb3a2 did not affect development of the pulmonary neuroepithelial bodies during embryogenesis, while the lack of Scgb3a2 may have resulted in slightly fewer ciliated cells than in the wild-type. These results suggest that SCGB1A1, SCGB3A1, and SCGB3A2 each may possess its own unique biological function.
Keywords: CC10, CCSP, HIN-1, immunomodulatory, lung, secretoglobin, UGRP1, UGRP2, NEB
Introduction
The secretoglobin (Scgb) gene superfamily consists of cytokine-like secreted proteins of small molecular weight (~10 kDa).1–3 Most SCGB proteins are found in secretions such as those of lung, lacrimal gland, salivary gland, prostate gland, and uterus.1 They share a common four helical bundle subunit structure and exist as dimers.2,4 SCGBs are thought to be involved in immunomodulatory activities, however, their biological functions and their modes of action are not understood.
Three SCGB proteins, SCGB1A1, SCGB3A1, and SCGB3A2, are expressed in lung airway epithelial cells.3,5,6 SCGB1A1, also called uteroglobin, CC10 (Club cell 10-kDa protein), and CCSP (Club cell secretory protein), is the most widely studied member of the SCGB proteins, and it is thought to be involved in modulation of inflammation, tissue repair, and tumorigenesis.4,7–10 SCGB3A2, also called UGRP1,6 appears to share activities similar to those of SCGB1A1. It was demonstrated to exhibit antiinflammatory,11–15 growth factor,16,17 and antifibrotic activities.17–20 SCGB3A2 also serves as a marker for lung adenocarcinomas.21,22 The antifibrotic activity involves the increased phosphorylation of STAT1 and increased levels of inhibitory SMAD7, which suppresses the TGFβ signaling.18 SCGB3A1, also called UGRP2 or high in normal–1 (HIN-1), was described as a tumor suppressor in various human tumors including breast, prostate, lung, and pancreatic carcinomas.23,24 However, the biological functions of SCGB3A1 remain largely unknown.
This study was initiated to examine the expression patterns of SCGB1A1, SCGB3A1, and SCGB3A2 in mouse airways during development and in the adult to determine their cell-specific and spatial relationships to gain insights into their biological functions.
Materials and Methods
Animals
Scgb3a1(–/–) mice (unpublished results, manuscript in preparation), Scgb3a2(–/–) mice,14 and their respective wild-type littermates were used in this study. Scgb3a1(–/–) mice are healthy and fertile, and do not show any lung or other abnormal phenotypes. In some experiments, mice on the C57BL/6N background were used (the number of backcross is provided in each figure legend). Embryos were collected from wild-type C57BL/6N mice at embryonic day (E) 14.5, 15.5, 16.5, 17.5, and 18.5. Noon on the day in which the vaginal plug was observed was considered as E0.5. For qRT-PCR analysis, each lung was considered an n=1 for individual mouse and embryos older than E16.5, while for embryos of E12.5 and 14.5, two lungs were combined, which was considered as an n=1. All animal studies were carried out after approval by the National Cancer Institute Animal Care and Use Committee.
Histological Analysis
Resected lungs and embryos were fixed in 4% buffered paraformaldehyde, pH 7.2 to 7.4, overnight at 4C, followed by dehydration in a series of 30–50–70% ethanol (1–2 hr each). Tissues in 70% ethanol were sent to American HistoLabs (Gaithersburg, MD) for paraffin embedding, sectioning, and mounting.
Immunohistochemistry
Dewaxed and rehydrated sections were immunostained by three methods: (1) double immunofluorescence using secondary antibody coupled Alexa Fluor dyes (Molecular Probes, Eugene, OR) 488 (green) and 594 (red), applied together (dilution 1:250) and recorded as digital photos, (2) avidin-biotin-complex (ABC kit, Vector Laboratories, Burlingame, CA) conjugated with peroxidase and using diaminobenzidine (DAB) as chromogen, counterstained with Light Green (Sigma-Aldrich, St. Louis, MO), or (3) DAB immunostaining (as above) followed by costaining with ABC kit conjugated with alkaline phosphatase and using Vector Red (Vector Laboratories) as a chromogen, counterstained with hematoxylin. Antigen retrieval was accomplished either by 10 min microwaving in 0.05 M citrate buffer, pH 6, or by 50 min steaming in Tris-EDTA buffer, pH 9. In all procedures, primary antibody application was overnight at 4C, followed by secondary antibody (45 min at room temperature) the next day. Primary antibodies used were specific for SCGB3A1 (dilution 1:100, AF2954, R&D Systems, Minneapolis, MN), SCGB3A2 (dilution 1:100, as described in Niimi et al.6), SCGB1A1 (dilution 1:1000, obtained from Francesco DeMayo, NIEHS), and β-tubulin (dilution 1:100, obtained from Charles Bevins, Cleveland Clinic). Sections were evaluated visually by brightfield or epi-fluorescence microscopy, and digital images were captured using MetaMorph software (Universal Imaging, Bedford Hills, NY) or Keyence microscope BZ-X800. Airways with columnar and cuboidal epithelial cells were defined, respectively, as large and small airways; typically, large airways were 1.5 to two times larger in lumen diameter than small airways.
Pulmonary neuroepithelial bodies (NEBs) were detected by DAB immunostaining (as above) for a NEB-specific marker, CGRP (calcitonin gene-related peptide) (primary antibody dilution 1:3000, C-8198, Sigma-Aldrich). Digital images were captured of all detectable NEB foci in lung sections from eight mice in each of the four experimental groups (Scgb3a1(–/–), Scgb3a2(–/–), and their respective littermate wild-type mice), a total of 32 mice; average NEB areas were calculated by MetaMorph software after manually circling every NEB’s digital image. Foci/airway ratios were determined after visually counting all NEB foci and all airways (both large and small) in these same 32 mouse lung specimens. For β-tubulin analysis, 10 randomly selected visual fields of large airways (×400 magnification) from three each of Scgb3a1(–/–) and Scgb3a2(–/–) mice, and two respective littermate wild-type mice were used. Percentages of ciliated cells were obtained by manually counting the β-tubulin positive cells and negative club cells, followed by the β-tubulin positive cell numbers being divided by the combined numbers of β-tubulin positive and negative cells, and the results were multiplied by 100. Each field was counted twice. Note that these two types of cells (ciliated cells and nonciliated club cells) constitute almost the entire population of airway epithelial cells in mice under normal condition.25,26 The statistical differences were compared within a group by unpaired t-test for NEB data and by one-way ANOVA for β-tubulin percentage.
Quantitative RT-PCR
Total RNA was extracted from whole left lung lobes of adult knockout or wild-type mice by TRIzol (Life Technologies, Carlsbad, CA) and reverse transcribed into cDNA by using SuperScript III reverse transcriptase (Life Technologies) according to the manufacturer’s protocol. Analysis of mRNA levels was performed on a 7900 Fast Real-Time PCR System (Life technologies) with SYBR Green-based real-time PCR. The primer sequences used for real-time PCR are as follows: Scgb1a1 (Forward) 5′-AGAGACTGGTGGATACCCTC-3′, (Reverse) 5′-TCTTGCTTACACAGAGGACTTG-3′; Scgb3a1 (Forward) 5′-ATGTCCCCACAATCAGCAAG-3′, (Reverse) 5′-CTCTGCAGCTGGAGCAAGG-3′; Scgb3a2 (Forward) 5′-AAGCTGGTATCTATCTTTCTGC-3′, (Reverse) 5′-ACAGGGAGACGGTTGATGAG-3′.
Results
Coexpression of SCGB1A1, SCGB3A1, and SCGB3A2 in Nonciliated Airway Club Cells
Specificity of the anti-SCGB3A1 and anti-SCGB3A2 antibodies used in this study was examined using immunohistochemistry. They were highly specific to SCGB3A1 and SCGB3A2, respectively, as demonstrated by using Scgb3a2(–/–) and Scgb3a1(–/–) mouse lungs as negative controls (Fig. 1A and B). SCGB1A1 was known to be specifically expressed in club cells, the principal secretory cell type in the airways,27,28 to which the antibody was specifically reacted29 (Fig. 2A–C). In these cells, SCGB3A1 is coexpressed with SCGB3A2 and SCGB1A1 (Fig. 1C and D).
Figure 1.
Antibody specificity and expression of SCGB1A1, SCGB3A1, and SCGB3A2 in airway epithelial cells. Immunofluorescence analysis was carried out using Scgb3a1(–/–) (A) and Scgb3a2(–/–) (B) mouse lungs with SCGB3A1 (green) and SCGB3A2 (red) antibodies, and using wild-type littermate mouse lungs with SCGB3A1 (green) and SCGB3A2 (red) antibodies (C), and SCGB3A1 (green) and SCGB1A1 (red) antibodies together (D). Co-immunohistochemistry for β-tubulin (brown, representative indicated by black arrow) and SCGB3A2 (burgundy red, representative indicated by red arrow) (E), and β-tubulin (brown, representative indicated by black arrow) and SCGB3A1 (burgundy red, representative indicated by red arrow) (F) using wild-type mice lungs. (E, F) Counterstained with hematoxylin. Airways shown are large airways. Scale bars: 25 µm for A to D, and E and F. Abbreviations: Alv, alveolar area; Lu, lung airway lumen; WT, wild-type.
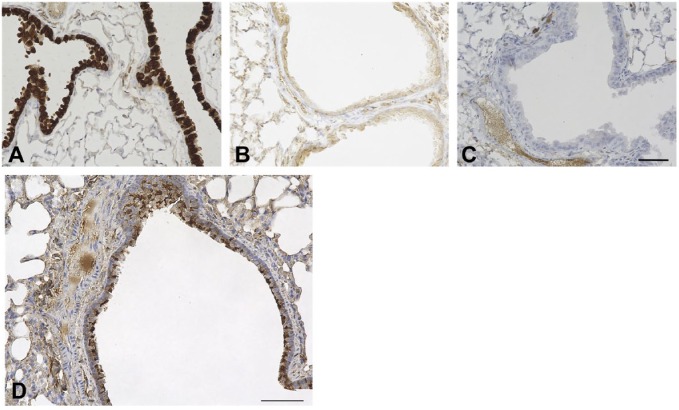
Figure 2.
Anti-mouse SCGB1A1 antibody specificity and airway β-tubulin expression. Immunohistochemistry was carried out using lungs of wild-type (A) and Scgb1a1(–/–) mice (kindly provided by Dr. Anil Mukherjee, NICHD) (B), and wild-type without primary antibody (C). Airways shown are large airways. Counterstained with hematoxylin. Scale bar: 50 µm for all panels. (D) Abundance of ciliated cells in mouse lung airway using C57BL/6N mouse. The airway shown is a large airway. Counterstained with hematoxylin. Scale bar: 50 µm.
Immunohistochemistry for ciliated cells was also carried out using β-tubulin as a marker (Fig. 2D). Club cells and ciliated cells are the two abundantly found epithelial cells (~50% each depending on the airway location) while other cell types such as goblet cells and basal cells are few that line the conducting airways of the respiratory tract in rodents under normal condition.25,26 Our results on the expression of SCGB proteins and β-tubulin are in agreement with these earlier reported results. Double immunostaining confirmed that cells expressing SCGB3A1 or SCGB3A2 are not ciliated cells (Fig. 1E and F).
Differential Expression of SCGB1A1, SCGB3A1, and SCGB3A2 During Lung Development
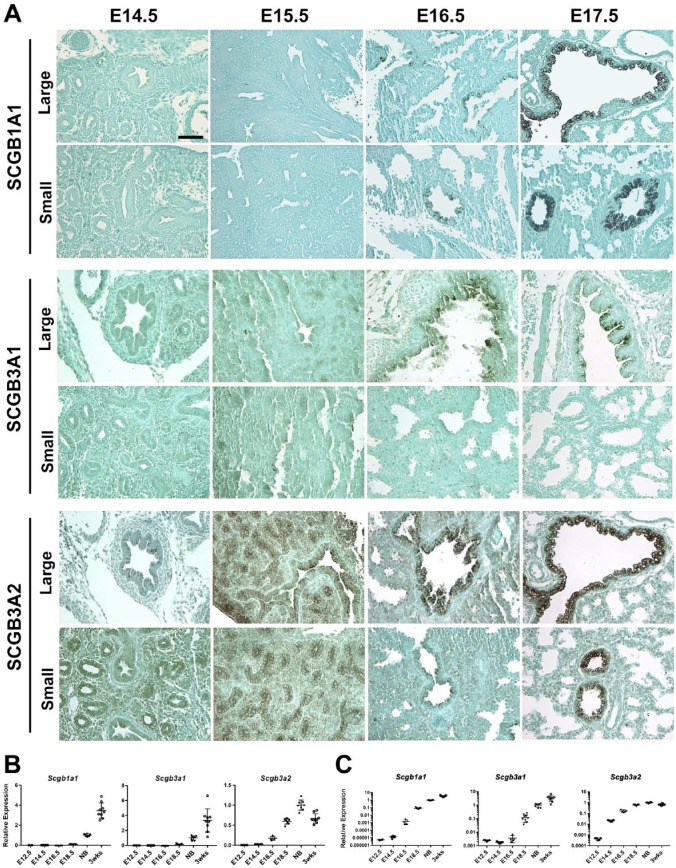
To determine the timing and sites of expression of these three SCGB proteins during lung development, mouse embryonic lungs of E14.5, 15.5, 16.5, and 17.5 were subjected to immunohistochemistry for SCGB1A1, SCGB3A1, and SCGB3A2 (Fig. 3A and Table 1). Epithelial cells started expressing SCGB1A1 at E16.5 in both large (bronchus) and small (bronchiole) airways, which became intense at E17.5. SCGB3A1 expression was observed only in large airways at E16.5 and thereafter, but not in small airways. In contrast, SCGB3A2 expression was already found at E14.5, albeit weak in both the large and small airways, which gradually became intense as gestation proceeded. The increase in expression of these three proteins during the gestational period was in good agreement with the results obtained by mRNA analysis (Fig. 3B and C and Table 2). The expression of Scgb1a1 and Scgb3a1 mRNAs continued to increase up to 3 weeks after birth, while that of Scgb3a2 peaked around birth. The latter is in good agreement with previous results.30 These results clearly demonstrate that the spatiotemporal expressions of the SCGB proteins differ during lung development, suggesting their possible unique as well as cooperative functions in this process.
Figure 3.
Spatiotemporal expression of SCGB1A1, SCGB3A1, and SCGB3A2 during mouse lung gestation. (A) Immunohistochemistry for the expression of the three SCGBs in large and small airways at various mouse embryonic stages. Counterstained with light green. Scale bar: 50 µm for all panels. (B, C) qRT-PCR analysis for the expression of Scgb1a1, Scgb3a1, and Scgb3a2 mRNAs at various embryonic days through 3 weeks old. Relative expression plotted by logarithmic scale is shown in C. C57BL/6N mice were used for these analyses. A dot indicates an individual lung (N>5) for embryos/mice older than E16.5, and two combined lungs as one lung for embryos of E12.5 and 14.5. qRT-PCR was carried out in triplicate per sample. The average ± SE is shown. Abbreviations: SCGBs, Secretoglobins; NB, newborn.
Table 1.
Spatiotemporal Expression of Three SCGB Proteins During Embryogenesis.
| Embryo (Days) | |||||
|---|---|---|---|---|---|
| 14.5 | 15.5 | 16.5 | 17.5 | ||
| SCGB1A1 | L. Airway | − | − | + | ++ |
| S. Airway | − | − | + | ++ | |
| SCGB3A1 | L. Airway | − | − | + | + |
| S. Airway | − | − | − | − | |
| SCGB3A2 | L. Airway | + | + | + | ++ |
| S. Airway | +/– | + | + | ++ | |
Abbreviations: SCGB, secretoglobin; L. Airway, large airway; S. Airway, small airway; –, no expression found; +, modest expression found; ++, strong expression found.
Table 2.
qRT-PCR Ct Values for Scgb1a1, Scgb3a1, and Scgb3a2.
| Scgb1a1 | Scgb3a1 | Scgb3a2 | |
|---|---|---|---|
| E12.5 | 34 | 31 | 30 |
| E14.5 | 32 | 31 | 22 |
| E16.5 | 25 | 29 | 19 |
| E18.5 | 19 | 25 | 17 |
| Newborn | 16 | 21 | 16 |
| 3 weeks | 15 | 20 | 17 |
Expression of Scgb1a1, Scgb3a1, and Scgb3a2 in Adult Mouse Airways
The effect of the lack of Scgb3a1 or Scgb3a2 on the expression of Scgb1a1, Scgb3a1, and/or Scgb3a2 was examined in Scgb3a1(–/–) and Scgb3a2(–/–) mouse lungs (Fig. 4). Except in the corresponding knockout mice, the expressions of Scgb1a1, Scgb3a1, and Scgb3a2 mRNAs were not affected by the lack of Scgb3a1 or Scgb3a2 and remained at similar levels, suggesting that these three genes do not compensate for the loss of expression of the other genes, at least at mRNA levels.
Figure 4.

Effect of the lack of Scgb3a1 or Scgb3a2 on the expression of Scgb1a1, Scgb3a1, or Scgb3a2. The mRNA levels were measured by qRT-PCR using a mouse adult whole left lung lobe. The average ± SE is shown. N=3–4, each in triplicate. Statistical analysis was performed using one-way ANOVA. WT: wild-type (C57BL/6N), S1KO: Scgb3a1(–/–) mice, S2KO: Scgb3a2(–/–) mice. For this analysis, the knockout mice used were 10 times backcrossed to C57BL/6N.
Impact of SCGB3A1 or SCGB3A2 Expression on the Differentiation of Neuroendocrine and Ciliated Cells
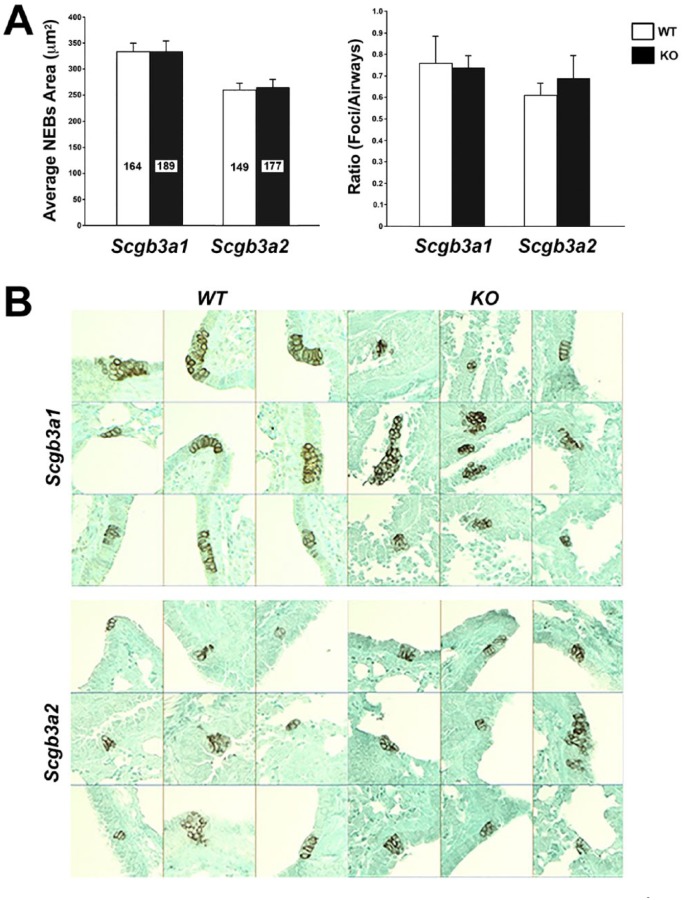
Previously, SCGB1A1 deficiency was shown to attenuate pulmonary neuroendocrine differentiation in mice lacking Scgb1a1.31 Furthermore, it was reported that Scgb3a2 expression is enriched in clusters in the pulmonary NEB microenvironment, juxtaposed to Ascl1-expressing cells during lung development.32 Ascl1(–/–) mice have no detectable neuroendocrine cells.33 Thus, to understand the possible roles for SCGB3A1 and SCGB3A2 in NEB differentiation, CGRP immunostaining that specifically detects NEBs was carried out to determine the size of NEBs and the number of NEB foci per airway as indicators for neuroendocrine differentiation using Scgb3a1(–/–) and Sgb3a2(–/–) mice31 (Fig. 5A and B). No differences were found in the differentiation of NEBs between Scgb3a1(–/–) and Sgb3a2(–/–) mice as compared with their corresponding wild-type littermate mice.
Figure 5.
Analysis of NEB development in Scgb3a1(–/–) and Scgb3a2(–/–) mouse lungs. (A) Average NEB areas (µm2) and foci/airway ratio are shown using Scgb3a1(–/–) mice in mixed background and Scgb3a2(–/–) mice that were three times backcrossed to C57BL/6N. Values for Scgb3a1(–/–) (black column) and corresponding wild-type littermates (white column) are indicated by Scgb3a1. Similarly, those of Scgb3a2(–/–) (black column) and corresponding wild-type littermates (white column) are indicated by Scgb3a2. For each genotype, eight mice were used, and the total number of NEBs analyzed is shown inside each column. The average ± SEM (standard error of the mean) is shown. No significant difference was observed between WT and KO littermates of Scgb3a1 or Scgb3a2 genotype as determined by t-test within each group. The differences seen between Scgb3a1(–/–) corresponding WT littermates and those of Scgb3a2(–/–) are detailed in the Discussion. (B) Representative immunohistochemistry for CGRP (calcitonin gene-related peptide, a neuroendocrine cell marker) in Scgb3a1(–/–) (KO), Scgb3a2(–/–) (KO), and corresponding WT littermates mouse lungs. The CGRP-positivity was used to define NEB foci, which were used to calculate the average NEB areas and the foci/airways ratios shown in A.29 NEB locations shown include both large and small airways. Counterstained with light green. Abbreviation: NEB, neuroepithelial body; WT, wild-type; KO, knockout.
Furthermore, the distribution of ciliated cells in the airway was compared between Scgb3a1(–/–), Scgb3a2(–/–), and wild-type mice using immunostaining for β-tubulin, a marker for ciliated cells (Fig. 6A and B). There was a slight but statistically significant reduction in ciliated cells in the large airways of Sgb3a2(–/–) mice as compared with their wild-type littermate mice. A tendency toward reduced ciliated cells was also found in Scgb3a1(–/–) mice as compared with wild-type mice, but with no significant difference between Scgb3a1(–/–) and Scgb3a2(–/–) mice.
Figure 6.
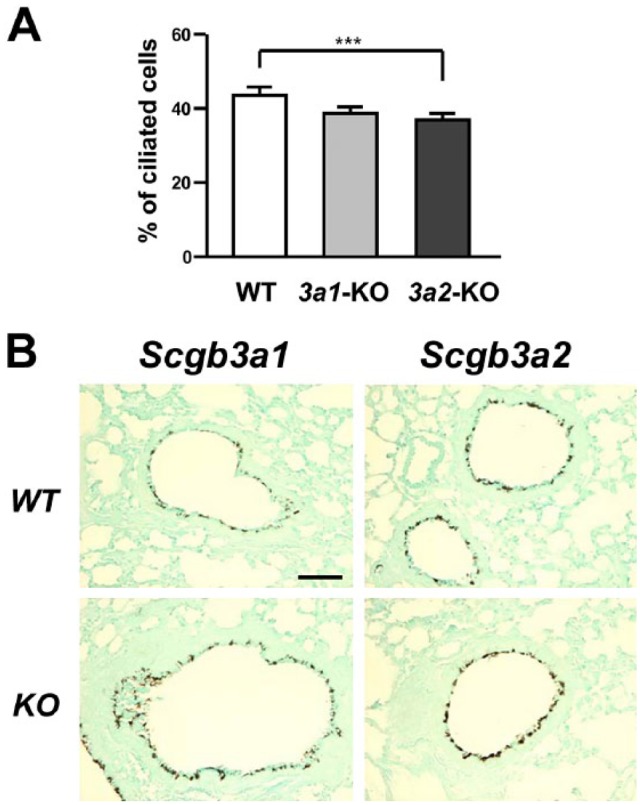
Analysis of ciliary development in Scgb3a1(–/–) and Scgb3a2(–/–) mouse lungs. (A) Percentage of ciliated cells in large airways of Scgb3a1(–/–) (3a1-KO), Scgb3a2(–/–) (3a2-KO), and their WT littermates mice. Mice used for this analysis were those 10 times backcrossed to C57BL/6N mice. Statistical difference between WT and 3a2-KO was obtained using one-way ANOVA. (B) Representative β-tubulin immunostaining using lungs of Scgb3a1(–/–) (KO), Scgb3a2(–/–) (KO), and their respective WT littermates. Airways shown include both large and small airways. Counterstained with light green. Scale bar: 50 µm for all panels. Abbreviation: WT, wild-type; KO, knockout.
Discussion
In this study, spatial and/or temporal differences were demonstrated in the expression of three SCGB proteins in airway epithelial club cells, namely, SCGB1A1, SCGB3A1, and SCGB3A2, during embryonic development and in adult mice. Previously, a similar comparison of the expression pattern for these SCGB proteins was carried out mainly at the mRNA level.3 The current results confirmed the previous observations at protein levels. Among the three, SCGB3A2 is the earliest protein expressed in both large and small airways of embryos at E14.5, followed by expression of SCGB1A1 in large and small airways starting at E16.5. In contrast, SCGB3A1 is expressed only in large airways after E16.5. Expressions of all three Scgb mRNAs markedly increase toward the end of gestation. The earlier onset of expression of SCGB3A2 suggests that this protein may play a role in lung development. Indeed, our previous work demonstrated that SCGB3A2 is a growth factor promoting lung branching morphogenesis16 and that SCGB3A2 together with Notch signaling plays a role in the determination of secretory cell fate in the NEB microenvironment.32 However, SCGB1A1 was used as a marker for airway epithelial Club cells during lung development and regeneration.34,35 The differential use of SCGB1A1 and SCGB3A2 as lung epithelial markers may provide a tool to more precisely determine the events and their roles occurring between E14.5 and 16.5 during embryonic lung development, due to the delayed expression of SCGB1A1 as opposed to SCGB3A2.
Both SCGB1A1 and SCGB3A2 exhibit similar antiinflammatory, growth stimulatory, and antifibrotic activities,4,7–20 while the function of SCGB3A1 in the lung remains unknown. It is not clear whether the similar activities of SCGB1A1 and SCGB3A2 mean that at least these two SCGB proteins share redundant functions to sustain homeostasis of lung function. In this regard, the present results using Scgb3a1(–/–) and Scgb3a2(–/–) mouse lungs showed no compensatory increase in expression among the three SCGBs, at least at mRNA level, suggesting that each SCGB protein may possess a distinct function. However, in the Scgb1a1(–/–) mouse lung, Scgb3a2 (reported as a previously nonannotated expressed sequence tag, EST W82219) was identified as a highly upregulated gene based on microarray data.36 It is interesting to note that Scgb1a1 deficiency results in Scgb3a2 upregulation, but there is no Scgb1a1 upregulation in Scgb3a1 or Scgb3a2 deficiency. This apparent discrepancy might suggest the presence of different mechanism(s) to regulate expression of mRNAs between Scgb1a1 and Scgb3a2 and/or Scgb3a1, which could be related to their distinct functions. However, it is possible that the three genes share redundant/overlapping functions without apparent compensation at the mRNA expression level. Further studies are required to address these questions.
In the current study, neither SCGB3A1 nor SCGB3A2 was demonstrated to play a role in pulmonary NEB differentiation, which is opposite to what was reported for SCGB1A1 using Scgb1a1(–/–) mice; lack of SCGB1A1 attenuated pulmonary neuroendocrine differentiation.31 Yet, the expression pattern of Scgb3a2, enriched in clusters in the NEB microenvironment, is reminiscent of that of SCGB1A1.32,37 These results may again suggest that SCGB1A1 has a different function from SCGB3A2 and/or SCGB3A1. When a percentage of ciliated cells in the large airways was calculated, Scgb3a2(–/–) and Scgb3a1(–/–) mice had, respectively, slightly reduced and a tendency of reduced numbers of ciliated cells in their lungs as compared with wild-type mice. It is possible that at least SCGB3A2 may affect the development of ciliated cells, although at small degree. In fact, ciliated cells are known to be produced from club cells.38,39 Understanding the exact roles for these three SCGB proteins in lung development and function requires further studies.
Finally, it is important to note that the values for two factors used to describe NEB development, namely, average NEB areas and foci/airway ratios, differed in the corresponding wild-type littermate control lungs between Scgb3a1(–/–) and Scgb3a2(–/–) mice. The Scgb3a1(–/–) mice used in this analysis were on a mixed background of C57BL/6N and 129Sv, whereas Scgb3a2(–/–) mice were at least three times backcrossed with C57BL/6N mice, by which the mice were >85% congenic to C57BL/6N.40 Care must be taken to consider the background of mice when interpreting the results.
In conclusion, SCGB1A1, SCGB3A1, and SCGB3A2 are expressed in their unique spatiotemporal patterns in mouse airways during development and in adult mice, suggesting a possible unique biological function for each protein.
Acknowledgments
We thank Florent Suau and Yan Li for their help in collection of mouse embryos used in the immunohistochemical studies, and Francesco DeMayo (National Institute of Environmental Health Sciences [NIEHS]) and Charles Bevins (Cleveland Clinic) for providing antibodies.
Footnotes
Competing Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: The authors prepared this work within the scope of their employment with National Cancer Institute/National Institutes of Health.
Author Contributions: All authors have contributed to this article as follows: XN, RIL, and SK conceived the project and designed the experiments; XN performed immunohistochemical, and pulmonary neuroepithelial body and β-tubulin analyses and interpreted the results; TK and SY performed qRT-PCR analyses; XN and SK wrote the manuscript; XN, TK, SY, and SK edited the manuscript; SK provided overall supervision; and all authors have read and approved the manuscript as submitted.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by an Intramural Research Program of the National Cancer Institute, Center for Cancer Research (ZIA BC010449 for S.K.).
ORCID iD: Shioko Kimura  https://orcid.org/0000-0001-9627-6818
https://orcid.org/0000-0001-9627-6818
Contributor Information
Xu Naizhen, Cell and Cancer Biology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland; Laboratory of Pathology, National Cancer Institute, Bethesda, Maryland.
Taketomo Kido, Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland; Laboratory of Stem Cell Therapy, Institute for Quantitative Biosciences, The University of Tokyo, Tokyo, Japan.
Shigetoshi Yokoyama, Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland.
R. Ilona Linnoila, Cell and Cancer Biology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland.
Shioko Kimura, Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland.
Literature Cited
- 1. Jackson BC, Thompson DC, Wright MW, McAndrews M, Bernard A, Nebert DW, Vasiliou V. Update of the human secretoglobin (SCGB) gene superfamily and an example of “evolutionary bloom” of androgen-binding protein genes within the mouse Scgb gene superfamily. Hum Genomics. 2011;5(6):691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klug J, Beier HM, Bernard A, Chilton BS, Fleming TP, Lehrer RI, Miele L, Pattabiraman N, Singh G. Uteroglobin/Clara cell 10-kDa family of proteins: nomenclature committee report. Ann N Y Acad Sci. 2000;923:348–54. [DOI] [PubMed] [Google Scholar]
- 3. Reynolds SD, Reynolds PR, Pryhuber GS, Finder JD, Stripp BR. Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am J Respir Crit Care Med. 2002;166(11):1498–509. [DOI] [PubMed] [Google Scholar]
- 4. Mukherjee AB, Zhang Z, Chilton BS. Uteroglobin: a steroid-inducible immunomodulatory protein that founded the secretoglobin superfamily. Endocr Rev. 2007;28(7):707–25. [DOI] [PubMed] [Google Scholar]
- 5. Niimi T, Copeland NG, Gilbert DJ, Jenkins NA, Srisodsai A, Zimonjic DB, Keck-Waggoner CL, Popescu NC, Kimura S. Cloning, expression, and chromosomal localization of the mouse gene (Scgb3a1, alias Ugrp2) that encodes a member of the novel uteroglobin-related protein gene family. Cytogenet Genome Res. 2002;97(1–2):120–7. [DOI] [PubMed] [Google Scholar]
- 6. Niimi T, Keck-Waggoner CL, Popescu NC, Zhou Y, Levitt RC, Kimura S. UGRP1, a uteroglobin/Clara cell secretory protein-related protein, is a novel lung-enriched downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor. Mol Endocrinol. 2001;15(11):2021–36. [DOI] [PubMed] [Google Scholar]
- 7. Saha A, Lee YC, Zhang Z, Chandra G, Su SB, Mukherjee AB. Lack of an endogenous anti-inflammatory protein in mice enhances colonization of B16F10 melanoma cells in the lungs. J Biol Chem. 2010;285(14):10822–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang Y, Zhang Z, Mukherjee AB, Linnoila RI. Increased susceptibility of mice lacking Clara cell 10-kDa protein to lung tumorigenesis by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a potent carcinogen in cigarette smoke. J Biol Chem. 2004;279(28):29336–40. [DOI] [PubMed] [Google Scholar]
- 9. Lee YC, Zhang Z, Mukherjee AB. Mice lacking uteroglobin are highly susceptible to developing pulmonary fibrosis. FEBS Lett. 2006;580(18):4515–20. [DOI] [PubMed] [Google Scholar]
- 10. Harrod KS, Mounday AD, Stripp BR, Whitsett JA. Clara cell secretory protein decreases lung inflammation after acute virus infection. Am J Physiol. 1998;275(5, Pt 1):L924–30. [DOI] [PubMed] [Google Scholar]
- 11. Chiba Y, Kurotani R, Kusakabe T, Miura T, Link BW, Misawa M, Kimura S. Uteroglobin-related protein 1 expression suppresses allergic airway inflammation in mice. Am J Respir Crit Care Med. 2006;173(9):958–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiba Y, Kusakabe T, Kimura S. Decreased expression of uteroglobin-related protein 1 in inflamed mouse airways is mediated by IL-9. Am J Physiol Lung Cell Mol Physiol. 2004;287(6):L1193–8. [DOI] [PubMed] [Google Scholar]
- 13. Chiba Y, Srisodsai A, Supavilai P, Kimura S. Interleukin-5 reduces the expression of uteroglobin-related protein (UGRP) 1 gene in allergic airway inflammation. Immunol Lett. 2005;97(1):123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kido T, Yoneda M, Cai Y, Matsubara T, Ward JM, Kimura S. Secretoglobin superfamily protein SCGB3A2 deficiency potentiates ovalbumin-induced allergic pulmonary inflammation. Mediators Inflamm. 2014;2014:216465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoneda M, Xu L, Kajiyama H, Kawabe S, Paiz J, Ward JM, Kimura S. Secretoglobin superfamily protein SCGB3A2 alleviates house dust mite-induced allergic airway inflammation in mice. Int Arch Allergy Immunol. 2016;171(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kurotani R, Tomita T, Yang Q, Carlson BA, Chen C, Kimura S. Role of secretoglobin 3A2 in lung development. Am J Respir Crit Care Med. 2008;178(4):389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cai Y, Winn ME, Zehmer JK, Gillette WK, Lubkowski JT, Pilon AL, Kimura S. Preclinical evaluation of human secretoglobin 3A2 in mouse models of lung development and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2014;306(1):L10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurotani R, Okumura S, Matsubara T, Yokoyama U, Buckley JR, Tomita T, Kezuka K, Nagano T, Esposito D, Taylor TE, Gillette WK, Ishikawa Y, Abe H, Ward JM, Kimura S. Secretoglobin 3A2 suppresses bleomycin-induced pulmonary fibrosis by transforming growth factor beta signaling down-regulation. J Biol Chem. 2011;286(22):19682–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cai Y, Kimura S. Secretoglobin 3A2 exhibits anti-fibrotic activity in bleomycin-induced pulmonary fibrosis model mice. PLoS One. 2015;10(11):e0142497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cai Y, Yoneda M, Tomita T, Kurotani R, Okamoto M, Kido T, Abe H, Mitzner W, Guha A, Kimura S. Transgenically-expressed secretoglobin 3A2 accelerates resolution of bleomycin-induced pulmonary fibrosis in mice. BMC Pulm Med. 2015;15:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurotani R, Kumaki N, Naizhen X, Ward JM, Linnoila RI, Kimura S. Secretoglobin 3A2/uteroglobin-related protein 1 is a novel marker for pulmonary carcinoma in mice and humans. Lung Cancer. 2011;71(1):42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tachihara-Yoshikawa M, Ishida T, Watanabe K, Sugawara A, Kanazawa K, Kanno R, Suzuki T, Niimi T, Kimura S, Munakata M. Expression of secretoglobin3A2 (SCGB3A2) in primary pulmonary carcinomas. Fukushima J Med Sci. 2008;54(2):61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krop IE, Sgroi D, Porter DA, Lunetta KL, LeVangie R, Seth P, Kaelin CM, Rhei E, Bosenberg M, Schnitt S, Marks JR, Pagon Z, Belina D, Razumovic J, Polyak K. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc Natl Acad Sci U S A. 2001;98(17):9796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krop I, Player A, Tablante A, Taylor-Parker M, Lahti-Domenici J, Fukuoka J, Batra SK, Papadopoulos N, Richards WG, Sugarbaker DJ, Wright RL, Shim J, Stamey TA, Sellers WR, Loda M, Meyerson M, Hruban R, Jen J, Polyak K. Frequent HIN-1 promoter methylation and lack of expression in multiple human tumor types. Mol Cancer Res. 2004;2(9):489–94. [PubMed] [Google Scholar]
- 25. Harkema JR, Nikula KJ, Haschek WM. Respiratory system. Haschek and Rousseaux’s handbook of toxicologic pathology. 3rd ed. San Diego: Elsevier; 2013. p. 1935–2003. [Google Scholar]
- 26. Plopper CG, Hyde DM. Epithelial cells of the bronchiole. In: Parent RA, editor. Comparative biology of the normal lung. 2nd ed. San Diego: Academic Press; 2015. p. 83–92. [Google Scholar]
- 27. Hicks SM, Vassallo JD, Dieter MZ, Lewis CL, Whiteley LO, Fix AS, Lehman-McKeeman LD. Immunohistochemical analysis of Clara cell secretory protein expression in a transgenic model of mouse lung carcinogenesis. Toxicology. 2003;187(2–3):217–28. [DOI] [PubMed] [Google Scholar]
- 28. Ryerse JS, Hoffmann JW, Mahmoud S, Nagel BA, deMello DE. Immunolocalization of CC10 in Clara cells in mouse and human lung. Histochem Cell Biol. 2001;115(4):325–32. [DOI] [PubMed] [Google Scholar]
- 29. Naizhen X, Linnoila RI, Kimura S. Co-expression of achaete-scute homologue-1 and calcitonin gene-related peptide during NNK-induced pulmonary neuroendocrine hyperplasia and carcinogenesis in hamsters. J Cancer. 2016;7(14):2124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomita T, Kido T, Kurotani R, Iemura S, Sterneck E, Natsume T, Vinson C, Kimura S. CAATT/enhancer-binding proteins alpha and delta interact with NKX2-1 to synergistically activate mouse secretoglobin 3A2 gene expression. J Biol Chem. 2008;283(37):25617–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Castro CM, Yang Y, Zhang Z, Linnoila RI. Attenuation of pulmonary neuroendocrine differentiation in mice lacking Clara cell secretory protein. Lab Invest. 2000;80(10):1533–40. [DOI] [PubMed] [Google Scholar]
- 32. Guha A, Vasconcelos M, Cai Y, Yoneda M, Hinds A, Qian J, Li G, Dickel L, Johnson JE, Kimura S, Guo J, McMahon J, McMahon AP, Cardoso WV. Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways. Proc Natl Acad Sci U S A. 2012;109(31):12592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Borges M, Linnoila RI, van de Velde HJ, Chen H, Nelkin BD, Mabry M, Baylin SB, Ball DW. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature. 1997;386(6627):852–5. [DOI] [PubMed] [Google Scholar]
- 34. Herriges M, Morrisey EE. Lung development: orchestrating the generation and regeneration of a complex organ. Development. 2014;141(3):502–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng D, Soh BS, Yin L, Hu G, Chen Q, Choi H, Han J, Chow VT, Chen J. Differentiation of club cells to alveolar epithelial cells in vitro. Sci Rep. 2017;7:41661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Watson TM, Reynolds SD, Mango GW, Boe IM, Lund J, Stripp BR. Altered lung gene expression in CCSP-null mice suggests immunoregulatory roles for Clara cells. Am J Physiol Lung Cell Mol Physiol. 2001;281(6):L1523–30. [DOI] [PubMed] [Google Scholar]
- 37. Khoor A, Gray ME, Singh G, Stahlman MT. Ontogeny of Clara cell-specific protein and its mRNA: their association with neuroepithelial bodies in human fetal lung and in bronchopulmonary dysplasia. J Histochem Cytochem. 1996;44(12):1429–38. [DOI] [PubMed] [Google Scholar]
- 38. Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106(31):12771–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watson JK, Rulands S, Wilkinson AC, Wuidart A, Ousset M, Van Keymeulen A, Gottgens B, Blanpain C, Simons BD, Rawlins EL. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. 2015;12(1):90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cowin RM, Bui N, Graham D, Green JR, Grueninger S, Yuva-Paylor LA, Syed AU, Weiss A, Paylor R. Onset and progression of behavioral and molecular phenotypes in a novel congenic R6/2 line exhibiting intergenerational CAG repeat stability. PLoS One. 2011;6(12):e28409. [DOI] [PMC free article] [PubMed] [Google Scholar]