Abstract
Hepatitis B virus (HBV) has a 3.2 kb circular DNA genome. It employs four promoters in conjunction with a single polyadenylation signal to generate 3.5, 2.4, 2.1 and 0.7 kb co-terminal RNAs. The 3.5 kb RNA is subdivided into the precore RNA for e-antigen expression and pregenomic RNA for genome replication. When introduced to a genotype A clone, several core promoter mutations markedly enhanced HBV genome replication, but suppressed e-antigen expression through up-regulation of pregenomic RNA at the expense of precore RNA. In this study, we found such mutations also diminished envelope proteins and hepatitis B surface antigen, products of the 2.1 and 2.4 kb subgenomic RNAs. Indeed, Northern blot analysis revealed overall increase in 3.5 kb RNA, but reduction in all subgenomic RNAs. To validate transcriptional interference, we subcloned 1.1×, 0.7× and 0.6× HBV genome, respectively, to a vector with or without a cytomegalovirus (CMV) promoter at the 5′ end, so as to produce the pregenomic RNA, 2.4 kb RNA, and 2.1 kb RNA in large excess or not at all. Parallel transfection of the three pairs of constructs into a human hepatoma cell line confirmed the ability of pregenomic RNA to suppress all subgenomic transcripts and established the ability of the 2.4 and 2.1 kb RNAs to suppress the 0.7 kb RNA. Consistent with our findings, pregenomic RNA of the related duck HBV has been reported to interfere with transcription of the subgenomic RNAs. Transcriptional interference might explain why HBV produces so little 0.7 kb RNA and HBx protein despite a strong X promoter.
Keywords: core promoter, genetic variants, hepatitis B virus, hepatitis B e antigen, hepatitis B surface antigen, transcriptional interference
Introduction
Chronic infection with hepatitis B virus (HBV) is a leading cause of liver cirrhosis and hepatocellular carcinoma (HCC) (Trépo et al., 2014). HBV is an enveloped virus with a small DNA genome of 3.2 kb (Fig. 1a) (Seeger & Mason, 2015). The two structural genes specify core and envelope proteins, which are required for the assembly of capsids and formation of infectious virions, respectively. The P gene encodes the P protein, which during genome replication serves as both the primer and enzyme of reverse transcription, and also as RNase and DNA-dependant DNA polymerase. HBx, the X gene product, is a weak transcriptional transactivator implicated in hepatocarcinogenesis (Slagle & Bouchard, 2016). Alternative translation initiation from in-frame AUG codons in the envelope gene generates three co-terminal envelope proteins: large (L), middle (M) and small (S), with the M protein having an extra preS2 domain than the S protein and the L protein having an extra preS1 domain than the M protein. Majority of the S and M proteins are released as noninfectious subviral particles greatly exceeding virions. They are detected by ELISA as hepatitis B surface antigen (HBsAg). Another serological marker is hepatitis B e-antigen (HBeAg), a processed form of the precore/core protein. Therefore, alternative translation initiation from the two structural genes generates five of the seven viral proteins.
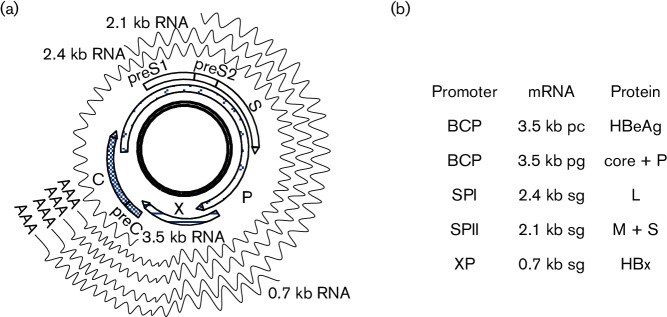
Fig. 1.
HBV genome, genes, transcripts and proteins. (a) The cccDNA template, four ORFs, and 3.5, 2.4, 2.1 and 0.7 kb HBV RNAs. These RNAs are co-terminal at the 3′ end, with the shorter ones completely overlapped by the longer ones. (b) Correspondence between the four size forms of HBV RNAs and seven viral proteins. The longer and shorter versions of the 3.5 kb RNA are called precore RNA (pcRNA) and pregenomic RNA (pgRNA), respectively.
Expression of the seven proteins is made possible by transcription of four size forms of viral RNAs from the cccDNA template in the nucleus (Nassal, 2015) (Fig. 1a). The four unidirectional promoters (core, SPI, SPII and X) scattered on the cccDNA drive transcriptional initiation at different sites, while a single polyadenylation signal at the 5′ end of the core gene ensures transcriptional termination at the same position. Consequently, the 3.5, 2.4, 2.1 and 0.7 kb HBV RNAs are unidirectional, overlapping and co-terminal at the 3′ end (Fig. 1a). The longer version of the 3.5 kb RNA, or precore RNA (pcRNA), is responsible for HBeAg expression, while the pregenomic RNA (pgRNA) drives core and P protein expression (Fig. 1b). The pgRNA is also the HBV RNA to be packaged inside core particles for converting into dsDNA, and hence the only RNA required for genome replication. L and M/S proteins are translation products of the 2.4 and 2.1 kb subgenomic mRNAs, respectively, while the 0.7 kb subgenomic RNA directs HBx expression. The overlapping nature of the HBV mRNAs raises the possibility of transcriptional interference by promoter occlusion (Shearwin et al., 2005).
Both HBeAg and HBsAg could suppress the host immune surveillance mechanisms to promote persistent infection. With the break of immune tolerance at the late stage of chronic infection, mutations that reduce or abolish HBeAg and HBsAg expression are gradually selected (Kramvis, 2014; Tong et al., 2013). The most common mutations to reduce HBeAg expression are A1762T and G1764A in the basal core promoter (BCP), which together with the upstream regulatory region constitutes the core promoter responsible for transcription of the 3.5 kb RNAs (Kramvis & Kew, 1999; Okamoto et al., 1994). We previously introduced the T1753C, A1762T, G1764A and C1766T BCP mutations in different combinations into a genotype A clone (Parekh et al., 2003; Tsai et al., 2009). The 1762/1764/1766 and 1753/1762/1764/1766 mutations were much more effective at suppressing HBeAg expression than the double mutation alone. They also markedly enhanced genome replication (by >10-fold). Primer extension assay revealed marked reduction in pcRNA but increase in pgRNA, thus explaining differential regulation of genome replication versus HBeAg expression (Tsai et al., 2009). In this study, we discovered their suppression of envelope protein expression at the transcriptional level. Overproduction of the 3.5, 2.4 or 2.1 kb RNA through the CMV promoter could repress transcription of the shorter co-terminal HBV RNA(s), most likely through promoter occlusion.
Results
Combined BCP mutations reduced S protein and HBsAg even in the absence of genome replication
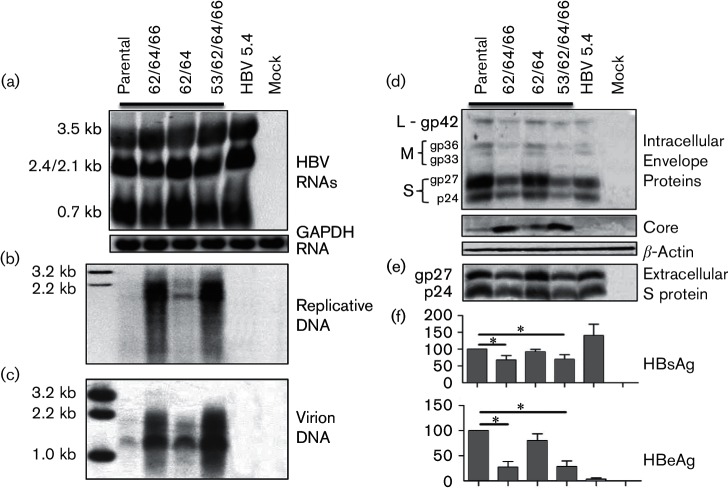
As we previously reported (Parekh et al., 2003), the 1762/1764/1766 and 1753/1762/1764/1766 BCP mutations introduced into clone 2A of genotype A markedly enhanced DNA replication and core protein expression (Fig. 2b, d), but significantly reduced HBeAg production (Fig. 2f). A muchuch milder effect was produced by the 1762/1764 double mutation. Unexpectedly, the triple and quadruple BCP mutations, but not the double mutation, also reduced S protein and HBsAg levels (Fig. 2d–f). One possible explanation for the reduced intracellular pool of S protein was its depletion through virion release, which was increased due to enhanced genome replication (Fig. 2c). We therefore introduced a C2044A nonsense mutation into the core gene to prevent genome replication and consequently virion secretion (Fig. 3b, c). However, the triple BCP mutant was still associated with less S protein and HBsAg, than clone 2A (Fig. 3d–f).
Fig. 2.
Impact of different combinations of BCP mutations on HBV biological properties. The T1753C (simplified as 53), A1762T (62), G1764A (64) and C1766T (66) mutations were introduced to clone 2A of genotype A in various combinations, and tandem EcoRI dimers were transiently transfected to Huh7 cells. HBV 5.4 was defective in genome replication and HBeAg expression. (a) Northern blot analysis of HBV RNAs using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. Southern blot analysis of (b) intracellular replicative DNA and (c) extracellular virion DNA. (d) Western blot analysis of intracellular envelope and core proteins using β-actin as a loading control. (e) Western blot analysis of secreted S protein following polyethylene glycol (PEG) precipitation. (f) HBsAg and HBeAg values averaged from seven transfection experiments, with those from clone 2A set at 100 %. *P<0.05.
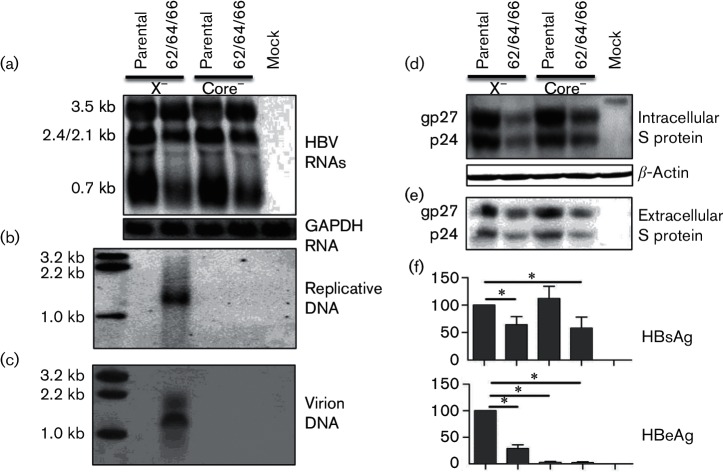
Fig. 3.
Biological impact of the 1762/1764/1766 BCP mutations in the absence of HBx or core protein expression. The X-minus or core-minus mutant of clone 2A and its 1762/1764/1766 (62/64/66) mutant in the form of 1.3-mer constructs were transiently transfected to Huh7 cells. (a) Intracellular HBV RNAs using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. (b) Intracellular replicative HBV DNA. (c) Extracellular virion DNA. (d) Intracellular S protein with β-actin serving as a loading control. (e) Extracellular S protein following polyethylene glycol (PEG) precipitation. (f) Extracellular HBsAg and HBeAg averaged from six transfection experiments. Values of clone 2A were set at 100 %. *P<0.05.
Mutated BCP rather than HBx diminished subgenomic RNAs
Northern blot analysis revealed that both 1762/1764/1766 and 1753/1762/1764/1766 mutations increased 3.5 kb RNA, which was accompanied by reduced levels of 2.4/2.1 kb RNAs (these two RNA species could not be resolved into separate bands) and 0.7 kb RNA (Fig. 2a). The transcriptional difference between clone 2A and the triple BCP mutant was reproducible by their core-minus mutants (Fig. 3a). Therefore, reduced S protein and HBsAg stemmed from a reduction in transcript level. BCP overlaps with the X gene, and the A1762T/G1764A/C1766T mutations introduce K130M and V131I substitutions in HBx protein, a transcriptional transactivator. Possible involvement of mutant HBx protein in altering the HBV transcriptional profile was investigated by introducing a C1684G nonsense mutation into the X gene upstream of the BCP mutations. However, the mutation altered neither the transcriptional difference between clone 2A and its 1762/1764/1766 mutant (Fig. 3a), nor the difference in S protein expression/HBsAg secretion (Fig. 3d–f).
Overproduction of pgRNA could down-regulate 2.4/2.1 kb and 0.7 kb RNAs
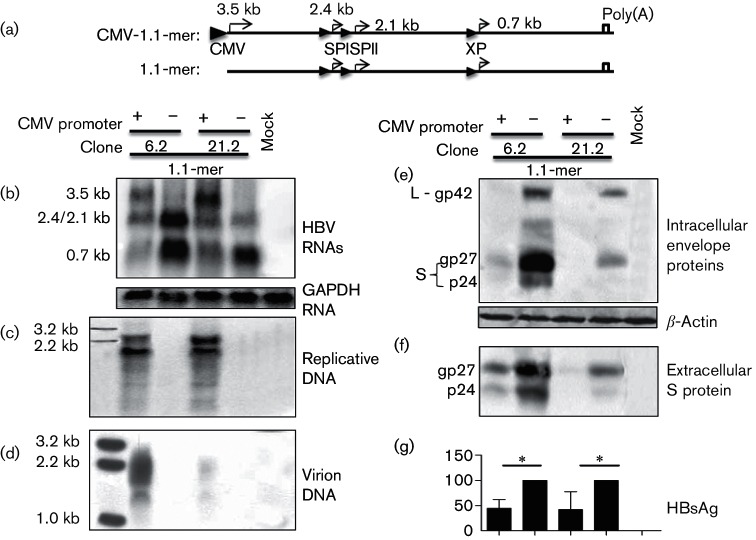
The BCP mutations that diminished both 2.4/2.1 kb and 0.7 kb RNAs strongly suggested transcriptional interference. To unequivocally establish the ability of the 3.5 kb pgRNA to repress transcription of the subgenomic RNAs, its cDNA equivalent (1.1 copies of the HBV genome) was inserted into the pcDNA3.1zeo(−) vector. This will ensure CMV promoter-driven overproduction of pgRNA, while maintaining transcription of subgenomic RNAs under endogenous HBV promoters (Fig. 4a). Deleting the CMV promoter will generate a 1.1-mer construct, which is unable to transcribe the pgRNA, and its comparison with the CMV-1.1-mer construct should provide the best chance to reveal transcriptional interference, if it does exist. To reach reliable conclusions, we employed a genotype A clone (6.2) and a genotype D clone (21.2) for parallel analysis. Genotype D isolates are characterized by reduced levels of HBsAg and especially intracellular S protein than genotype A isolates (Zhang et al., unpublished). As expected, only the CMV-1.1-mer constructs supported transcription of the 3.5 kb RNA (Fig. 4b), leading to HBV genome replication and virion secretion (Fig. 4c, d). Lack of a CMV promoter not only prevented transcription of the 3.5 kb RNA but also increased 2.4/2.1 kb and especially 0.7 kb RNA, which was more striking for the genotype A clone (Fig. 4b). Western blot analysis and ELISA confirmed increased levels of envelope proteins and HBsAg for the 1.1-mer construct than CMV-1.1-mer construct, whether for genotype A or D (Fig. 4e–g). Besides the pgRNA, we found the 3.5 kb pcRNA driven by the CMV promoter could also down-regulate the subgenomic RNAs (data not shown).
Fig. 4.
Overproduction of the 3.5 kb pgRNA diminished transcription of subgenomic RNAs. (a) 1.1 copies of clone 6.2 or 21.2 were inserted to pcDNA3.1zeo(−) vector, with or without the CMV promoter removed to overproduce or not to produce the 3.5 kb pg RNA. These 1.1-mer HBV DNA constructs were transiently transfected to Huh7 cells. (b) Intracellular HBV RNAs using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. (c) Intracellular replicative HBV DNA. (d) Extracellular virion DNA. (e) Intracellular envelope proteins with β-actin serving as a loading control. (f) Extracellular S protein following polyethylene glycol (PEG) precipitation. (g) Extracellular HBsAg averaged from 10 transfection experiments. For both clones 6.2 and 21.2, value of the construct without the CMV promoter was set at 100 %. *P<0.05.
The 2.4 kb RNA has the potential to suppress the 0.7 kb RNA
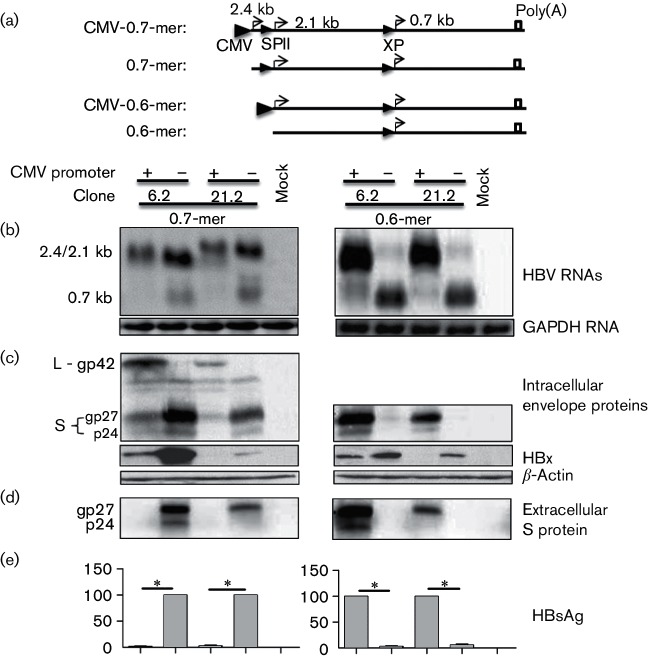
Having established transcriptional interference by the 3.5 kb pgRNA, we next employed the same approach to examine the impact of overproducing the 2.4 kb subgenomic RNA. Its cDNA equivalent (0.7× genome length or 0.7-mer) with or without CMV promoter was inserted to a modified pUC18 vector (Fig. 5a). Whether preventing transcription of the 2.4 kb RNA increased the 2.1 kb RNA could not be established by Northern blot analysis (Fig. 5b, left panel), but Western blot analysis revealed increased S protein expression by the 0.7-mer construct than CMV-0.7-mer construct, for both genotypes (Fig. 5c, d, left panels). Complete lack of HBsAg secretion by the CMV-0.7-mer construct (Fig. 5e, left panel) was most likely attributed to dramatically increased L/S protein ratio, because the L protein is known to suppress S protein secretion in a dose-dependant manner (Garcia et al., 2009; Ou & Rutter, 1987; Persing et al., 1986; Standring et al., 1986). The 0.7 kb RNA and HBx protein were clearly up-regulated in the absence of transcription of the 2.1 kb RNA, although the antibody appeared less efficient at recognizing HBx of genotype D (Fig. 5b, c, left panels). Taken together, up-regulation of the 2.4 kb RNA reduced 0.7 kb RNA and possibly also the 2.1 kb RNA.
Fig. 5.
Overproduction of either the 2.4/2.1 kb RNA diminished 0.7 kb RNA and HBx protein. (a) A 0.7-mer (2.4 kb) or 0.6-mer (2.1 kb) HBV sequence of clone 6.2 or 21.2, with or without a CMV promoter at the 5′ end, was inserted to a modified pUC18 vector. This will ensure overproduction or no production of the 2.4 kb RNA (0.7-mer construct), or overproduction or no production of the 2.1 kb RNA (0.6-mer construct). The 0.7-mer and 0.6-mer constructs were separately transfected to Huh7 cells. (b) Northern blot of HBV RNAs using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. Note that for the 0.7-mer constructs, the 2.4 and 2.1 kb RNAs cannot be resolved into separate bands. (c) Western blot analysis of intracellular envelope and HBx proteins, using β-actin as a loading control. (d) Western blot analysis of extracellular S protein following polyethylene glycol (PEG) precipitation. (e) Secreted HBsAg averaged from seven transfection experiments. Both, for the 0.7-mer and 0.6-mer constructs, the values for 6.2 and 21.2 constructs without a CMV promoter were set at 100 %. *P<0.05.
Overproduction of the 2.1 kb RNA reduced HBx protein at the transcriptional level
Finally, we examined transcriptional impact of the 2.1 kb RNA. A 0.6-mer DNA sequence for the 2.1 kb RNA with or without an upstream CMV promoter was inserted to the modified pUC18 vector (Fig. 5a). Overproduction of the 2.1 kb RNA was accompanied by a near loss of the 0.7 kb RNA for both genotypes (Fig. 5b, right panel). Western blot analysis confirmed that only the CMV-0.6-mer construct could express S protein and secrete HBsAg, but the 0.6-mer construct expressed much higher level of HBx protein than CMV-0.6-mer construct (Fig. 5c–e, right panels). Therefore, similar to the 3.5 and 2.4 kb RNAs, the 2.1 kb RNA also has the potential to suppress its downstream transcription unit.
Discussion
BCP mutations alter both the ratio and total amounts of pcRNA and pgRNA to augment core protein expression, DNA replication while suppressing HBeAg expression (Baumert et al., 1996; Buckwold et al., 1996; Jammeh et al., 2008; Moriyama et al., 1996; Parekh et al., 2003; Tsai et al., 2009). In this study, we found those with the strongest effect (TA1762T/G1764A/C1766T and T1753C/A1762T/G1764A/C1766T) also reduced intracellular and extracellular levels of envelope proteins, whether according to Western blot or ELISA (in the form of HBsAg). The impact on viral envelope proteins did not require DNA replication and was independent of concomitant mutations (K130M/V131I) in the HBx protein. Northern blot analysis revealed that the triple and quadruple mutations, but not the double mutation, increased 3.5 kb RNA while diminishing both 2.4/2.1 kb and 0.7 kb RNAs (Fig. 2a). These findings suggested that increased transcription of the 3.5 kb RNAs could interfere with transcription of the shorter co-terminal HBV RNAs, possibly by promoter occlusion (Boussadia et al., 1997; Greger et al., 1998; Proudfoot, 1986; Shearwin et al., 2005). This conclusion was reinforced by a more drastic approach, in which the pgRNA was either produced at very high level through the CMV promoter or not produced at all (Fig. 4). Compared with the CMV-1.1-mer construct of clone 6.2, the corresponding 1.1-mer construct without any promoter at its 5′ end showed marked increase in both 2.4/2.1 kb RNAs and 0.7 kb RNA (Fig. 4b). It will be very interesting to introduce a polyadenylation signal upstream of the SPI, SPII or X promoter to prematurely terminate pgRNA transcription. If pgRNA diminishes the subgenomic RNAs by transcriptional interference, then premature termination of transcription will relieve interference on a downstream transcription unit.
While much more study is needed to clarify the mechanism whereby the longer HBV transcripts diminish levels of the shorter co-terminal transcripts, a similar observation has been made with duck hepatitis B virus (DHBV). HBV, DHBV, woodchuck hepatitis virus and ground squirrel hepatitis are founding members of Hepadnaviridae (hepatotropic DNA viruses). Studies from Summers laboratory found that in either a 1.5-mer DHBV DNA construct or circularized DHBV genome, preventing pgRNA transcription by deleting its promoter increased levels of the two mRNA species for envelope proteins (Beckel-Mitchener & Summers, 1997; Huang & Summers, 1994). Deleting a so-called ‘PET’ (positive effector of transcription) sequence located at the 5′ end of the pgRNA could also prevent transcription of the pgRNA, and likewise increase the subgenomic RNAs. Therefore, ability of the pgRNA to suppress transcription of the mRNAs for envelope proteins appears to be a shared feature between distant members of hepadnaviruses.
Employing the 0.7-mer and 0.6-mer HBV sequences with or without 5′ fusion with the CMV promoter, we further demonstrated ability of both the 2.4 and 2.1 kb RNAs to suppress transcription of the 0.7 kb RNA (Fig. 5b–f). Western blot analysis confirmed concomitant reduction in the HBx protein (Fig. 5c–g). Due to the close spacing between the 2.4 and 2.1 kb RNAs, we could not establish by Northern blot transcription interference of the 2.4 kb RNA against the 2.1 kb RNA. Nevertheless, higher S protein level by the 0.7-mer construct than CMV-0.7-mer construct was consistent with such transcriptional suppression. It is worth mentioning that the total amount of the 2.4/2.1 kb HBV RNAs was higher with the 0.7-mer construct without a CMV construct, yet such a construct produced higher level of the 0.7 kb RNA than the CMV-0.7-mer construct (Fig. 5b). This observation suggests that the 2.4 kb RNA driven by the CMV promoter is a more potent suppressor of the transcription of the 0.7 kb RNA than is the 2.1 kb RNA driven by the endogenous SPII promoter. Certainly, during the natural course of HBV infection, the 2.4 kb RNA is produced at much a lower level than the 2.1 kb RNA to exert strong transcriptional interference. Comparison of the HBV RNA patterns between CMV-1.1-mer construct of clone 6.2 and corresponding 1.1-mer construct suggests that pgRNA driven by the CMV promoter is a more potent inhibitor of the 0.7 kb RNA than the 2.4/2.1 kb RNAs driven by SPI and SPII promoters (Fig. 4b, left). Whether the use of a CMV promoter enhances transcriptional interference is unknown, but ability of the 3.5, 2.4 and 2.1 kb RNAs to suppress transcription of the 0.7 kb RNA may partly explain why the shortest HBV RNA is transcribed at low level despite a strong X promoter (Antonucci & Rutter, 1989). Nevertheless, it should be pointed out that during the natural course of HBV infection, the cccDNA is the template for HBV RNA transcription. On such a transcriptional template, upstream or downstream is relative, and the 0.7 kb RNA also has the potential to diminish transcription of the 3.5 kb RNAs. Therefore, further studies are needed to validate the extent of transcriptional interference in the context of a circular HBV genome without any foreign sequence.
Transcription of HBV RNAs is further augmented by two enhancer elements, EnI and EnII. As an enhancer works in an orientation- and position-independent manner, mutations affecting EnI or EnII are expected to simultaneously affect the transcription of multiple HBV RNAs. On the other hand, mutations within one particular HBV promoter are not predicted to affect transcription of other HBV RNAs. Demonstration of transcriptional interference among HBV RNAs challenges such a prediction. In this regard, HCC development has been linked to HBx protein (Diamantis et al., 1992; Hsia et al., 1996; Paterlini et al., 1995), BCP mutations (Baptista et al., 1999; Kao et al., 2003; Kuang et al., 2004) and preS deletions (Chen et al., 2008; Fang et al., 2008; Huang et al., 2010; Wang et al., 2006). The amino acid substitutions in the HBx protein as a consequence of BCP mutations have been found to alter HBx function from growth inhibitory to growth promoting (Huang et al., 2011, 2013). However, according to the current work, BCP mutations will reduce HBx protein expression at the transcript level. As some of the preS deletions remove the SPII promoter, it will be interesting to examine whether such deletions could augment expression of the mutant HBx protein at the transcriptional level. This would cause synergistic effects between BCP mutations and preS deletions.
Methods
EcoRI dimers and 1.3-mer constructs of genotype A.
EcoRI dimers of HBV clones 2A, 5.4 and 6.2 of genotype A have been described (Parekh et al., 2003), with clone 5.4 being defective in HBV DNA replication and HBeAg expression due to a single nucleotide deletion in the core gene. Different combinations of BCP mutations were introduced to clone 2A to generate mu1 (A1762T/G1764A), mu2a (A1762T/G1764A/C1766T) and Ex 2 (T1753C/A1762T/G1764A/C1766T) (Parekh et al., 2003; Tsai et al., 2009). A 1.3-mer construct of clone 2A and its 1762/1764/1766 mutant was generated by inserting nucleotide sequence 980–3221/1–1948 to the SacI and SalI sites of pBluescript SK(−) vector in two cloning steps [Table S1 (available in the online Supplementary Material) for sequences of the two pairs of primers]. Introduction of the C1684G and C2044A nonsense mutations (Table S1) to such 1.3-mer constructs prevented translation of full-length HBx and core protein, respectively.
Genotype A and D constructs with or without CMV promoter-driven transcription of the 3.5, 2.4 and 2.1 kb RNAs.
The 1.1×, 0.7× and 0.6× genome length of clone 6.2 and clone 21.2 (genotype D, unpublished) were amplified by PCR (Table S1 for primer sequences) and inserted to a vector downstream of the CMV promoter to drive efficient transcription of the 3.5, 2.4 and 2.1 kb HBV RNAs, respectively. In addition, the same constructs without the 5′ CMV promoter were generated for comparison. The 1.1-mer HBV genome (nt 1818–3221/1–1960 for clone 6.2 and nt 1818–3182/1–1960 for clone 21.2) was inserted to pcDNA3.1zeo(−) vector. Since an MluI site was present on the vector upstream of the CMV promoter, and another MluI site was introduced to the 5′ end of the HBV sequence (Table S1), the CMV promoter was removed by MluI digestion followed by self-ligation of the large DNA fragment. The 0.7-mer (nt 2807–3221/1–1960 or 2807–3182/1–1960) and 0.6-mer (nt 4–1960) genomes with or without upstream CMV promoter were constructed in pcDNA3.1zeo(−) vector in a similar manner, and the CMV promoter–HBV sequence or HBV sequence was subsequently transferred to a modified pUC18 vector. To modify pUC18, two copies of the polyadenylation signal of bovine growth hormone were inserted between the AatI and PciI sites, with several unique restriction sites introduced between the two copies. Next, the HBV sequence was inserted between two copies using such unique sites.
Transient transfection and protein analysis.
The human hepatoma cell line Huh7 was cultured in Dulbecco’s Modified Eagle’s Medium (GIBCO) supplemented with 10 % FBS (Sigma). Transient transfection was performed on cells seeded in six-well plates using Mirus reagent (Tsai et al., 2009). Cells were lysed 3 days later in 80 µl of lysis buffer (10 mM HEPES, pH 7.5; 100 mM NaCl; 1 mM EDTA and 1 % NP40), and one-fourth was subject to Western blot analysis by incubating at 4 °C overnight with a 1 : 4000 dilution of rabbit polyclonal anti-HBs (Novus). After further incubation with HRP-conjugated goat anti-rabbit antibody at 1 : 10 000 dilution, signals were revealed by enhanced chemiluminescence (PerkinElmer) and visualized by chemiluminescent imaging system (Tanon). The antibodies were removed by the stripping buffer (CWBIO), and the blot was incubated sequentially with mouse anti-actin antibody (1 : 3000 dilution; Proteintech) and HRP-conjugated goat anti-mouse antibody (1 : 10 000 dilution). HBx was detected by a mouse mAb (16F9) at 1 : 2000 dilution and HRP-conjugated goat anti-mouse antibody. A polyclonal rabbit anti-core antibody (Dako) at 1 : 3000 dilution was used for the detection of core protein.
HBsAg and HBeAg secreted to culture supernatant were measured by ELISA kits (KHB) with proper dilution to prevent signal saturation. To detect secreted HBsAg by Western blot, 650 µl of culture supernatant was mixed with 250 µl of 36 % polyethylene glycol 8000 (dissolved in PBS). After rotating at 4 °C overnight, the samples were centrifuged at 18 620 g for 1 h and the pellets were resuspended in 20 µl of lysis buffer.
Northern blot analysis.
Huh7 cells were lysed by TRI reagent (Sigma) at day 2 post-transfection, and RNA was extracted according to the manufacturer′s protocol. Total RNA (10 µg) in 1× RNA loading buffer was denatured at 65 °C for 10 min and separated in a 1.5 % agarose gel with morpholinepropanesulfonic acid and formaldehyde. After transfer to the positively charged nylon membrane (Roche), the blot was hybridized using a DIG-Northern Starter kit (Roche) according to the manufacturer’s instructions. To generate an HBV probe, a 0.7 kb HBV DNA fragment covering positions 1266–1950 was cloned to the KpnI–XhoI sites of pcDNA3 vector (Table S1). A DIG-labelled, negative-stranded HBV RNA probe was generated by in vitro transcription of the plasmid linearized at the KpnI site, using SP6 RNA polymerase. For the loading control, a 970 bp RNA probe of glyceraldehyde-3-phosphate dehydrogenase was generated in a similar manner (Table S1 for primer sequences).
Southern blot analysis of HBV genome replication and virion secretion.
Huh7 cells seeded in six-well plates were harvested at day 5 post-transfection. Core particles were precipitated from half of the cell lysate, followed by nuclease treatment, proteinase K digestion and DNA extraction as detailed elsewhere (Guarnieri et al., 2006). DNA was separated in 1.2 % agarose gel, transferred to the nylon membrane, followed by hybridization with 32P-labelled full-length HBV DNA probe (Parekh et al., 2003). For unbiased detection of both genotypes A and D, probes of genotype A and D were mixed at 1 : 1 ration. The blots were washed at 65 °C in 2× SSC/0.1 % SDS solution.
Protein G beads were conjugated with a mixture of rabbit anti-S (Novus) and custom-made rabbit anti-preS1 antibodies against peptide MGTNLSVPNPLGFFPDHQLDPAFGANSNNPDWDFN (GenScript), at a ratio of 10 µl bed volume of beads with 3 µl of preS1 antibody and 1 µl of HBs antibody. The preconjugated beads (10 µl bed volume) were incubated with 1.5 ml of culture supernatant of transfected Huh7 cells to immunoprecipitate virus particles. The immunoprecipitate was treated sequentially with nucleases and proteinase K, followed by DNA extraction for Southern blot analysis.
Statistical analysis.
Data were expressed as mean±sd. Statistical analysis was performed by IBM SPSS Statistics (version 23). One-way ANOVA was chosen for comparing the means, and Duncan’s multiple range test was used to analyse the difference among the multiple groups. P<0.05 were considered as statistically significant.
Supplementary Data
Acknowledgement
This work was supported by National Institutes of Health grant numbers AI103648, AI107618, AI113394 and AI116639, and also by a grant from National Science Foundation of China (81371822).
Footnotes
One supplementary table is available with the online Supplementary Material.
References
- Antonucci T. K., Rutter W. J.(1989). Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J Virol 63579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista M., Kramvis A., Kew M. C.(1999). High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology 29946–953. 10.1002/hep.510290336 [DOI] [PubMed] [Google Scholar]
- Baumert T. F., Rogers S. A., Hasegawa K., Liang T. J.(1996). Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J Clin Invest 982268–2276. 10.1172/JCI119037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckel-Mitchener A., Summers J.(1997). A novel transcriptional element in circular DNA monomers of the duck hepatitis B virus. J Virol 717917–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussadia O., Amiot F., Cases S., Triqueneaux G., Jacquemin-Sablon H., Dautry F.(1997). Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett 42020–24. [DOI] [PubMed] [Google Scholar]
- Buckwold V. E., Xu Z., Chen M., Yen T. S., Ou J. H.(1996). Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol 705845–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Changchien C. S., Lee C. M., Hung C. H., Hu T. H., Wang J. H., Wang J. C., Lu S. N.(2008). Combined mutations in pre-S/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: a case-control study. J Infect Dis 1981634–1642. 10.1086/592990 [DOI] [PubMed] [Google Scholar]
- Diamantis I. D., McGandy C. E., Chen T. J., Liaw Y. F., Gudat F., Bianchi L.(1992). Hepatitis B X-gene expression in hepatocellular carcinoma. J Hepatol 15400–403. 10.1016/0168-8278(92)90077-3 [DOI] [PubMed] [Google Scholar]
- Fang Z. L., Sabin C. A., Dong B. Q., Wei S. C., Chen Q. Y., Fang K. X., Yang J. Y., Huang J., Wang X. Y., Harrison T. J.(2008). Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: a matched nested case-control study. J Gen Virol 892882–2890. 10.1099/vir.0.2008/002824-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia T., Li J., Sureau C., Ito K., Qin Y., Wands J., Tong S.(2009). Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J Virol 8311152–11165. 10.1128/JVI.00905-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger I. H., Demarchi F., Giacca M., Proudfoot N. J.(1998). Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res 261294–1301. 10.1093/nar/26.5.1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri M., Kim K. H., Bang G., Li J., Zhou Y., Tang X., Wands J., Tong S.(2006). Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J Virol 80587–595. 10.1128/JVI.80.2.587-595.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia C. C., Yuwen H., Tabor E.(1996). Hot-spot mutations in hepatitis B virus X gene in hepatocellular carcinoma. Lancet 348625–626. 10.1016/S0140-6736(05)64851-9 [DOI] [PubMed] [Google Scholar]
- Huang H. P., Hsu H. Y., Chen C. L., Ni Y. H., Wang H. Y., Tsuei D. J., Chiang C. L., Tsai Y. C., Chen H. L., Chang M. H.(2010). Pre-S2 deletions of hepatitis B virus and hepatocellular carcinoma in children. Pediatr Res 6790–94. 10.1203/PDR.0b013e3181c1b0b7 [DOI] [PubMed] [Google Scholar]
- Huang M., Summers J.(1994). pet, a small sequence distal to the pregenome cap site, is required for expression of the duck hepatitis B virus pregenome. J Virol 681564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Tong S., Tai A. W., Hussain M., Lok A. S.(2011). Hepatitis B virus core promoter mutations contribute to hepatocarcinogenesis by deregulating SKP2 and its target, p21. Gastroenterology 1411412–1421. 10.1053/j.gastro.2011.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Tai A. W., Tong S., Lok A. S.(2013). HBV core promoter mutations promote cellular proliferation through E2F1-mediated upregulation of S-phase kinase-associated protein 2 transcription. J Hepatol 581068–1073. 10.1016/j.jhep.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammeh S., Tavner F., Watson R., Thomas H. C., Karayiannis P.(2008). Effect of basal core promoter and pre-core mutations on hepatitis B virus replication. J Gen Virol 89901–909. 10.1099/vir.0.83468-0 [DOI] [PubMed] [Google Scholar]
- Kao J. H., Chen P. J., Lai M. Y., Chen D. S.(2003). Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 124327–334. 10.1053/gast.2003.50053 [DOI] [PubMed] [Google Scholar]
- Kramvis A., Kew M. C.(1999). The core promoter of hepatitis B virus. J Viral Hepat 6415–427. 10.1046/j.1365-2893.1999.00189.x [DOI] [PubMed] [Google Scholar]
- Kramvis A.(2014). Genotypes and genetic variability of hepatitis B virus. Intervirology 57141–150. 10.1159/000360947 [DOI] [PubMed] [Google Scholar]
- Kuang S. Y., Jackson P. E., Wang J. B., Lu P. X., Muñoz A., Qian G. S., Kensler T. W., Groopman J. D.(2004). Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci U S A 1013575–3580. 10.1073/pnas.0308232100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K., Okamoto H., Tsuda F., Mayumi M.(1996). Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology 226269–280. 10.1006/viro.1996.0655 [DOI] [PubMed] [Google Scholar]
- Nassal M.(2015). HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 641972–1984. 10.1136/gutjnl-2015-309809 [DOI] [PubMed] [Google Scholar]
- Okamoto H., Tsuda F., Akahane Y., Sugai Y., Yoshiba M., Moriyama K., Tanaka T., Miyakawa Y., Mayumi M.(1994). Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol 688102–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J. H., Rutter W. J.(1987). Regulation of secretion of the hepatitis B virus major surface antigen by the preS-1 protein. J Virol 61782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh S., Zoulim F., Ahn S. H., Tsai A., Li J., Kawai S., Khan N., Trépo C., Wands J., Tong S.(2003). Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol 776601–6612. 10.1128/JVI.77.12.6601-6612.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini P., Poussin K., Kew M., Franco D., Brechot C.(1995). Selective accumulation of the X transcript of hepatitis B virus in patients negative for hepatitis B surface antigen with hepatocellular carcinoma. Hepatology 21313–321. [PubMed] [Google Scholar]
- Persing D. H., Varmus H. E., Ganem D.(1986). Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science 2341388–1391. 10.1126/science.3787251 [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J.(1986). Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature 322562–565. 10.1038/322562a0 [DOI] [PubMed] [Google Scholar]
- Seeger C., Mason W. S.(2015). Molecular biology of hepatitis B virus infection. Virology 479-480672–686. 10.1016/j.virol.2015.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin K. E., Callen B. P., Egan J. B.(2005). Transcriptional interference – a crash course. Trends Genet 21339–345. 10.1016/j.tig.2005.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagle B. L., Bouchard M. J.(2016). Hepatitis B virus X and regulation of viral gene expression. Cold Spring Harb Perspect Med 6a021402. 10.1101/cshperspect.a021402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring D. N., Ou J. H., Rutter W. J.(1986). Assembly of viral particles in Xenopus oocytes: pre-surface-antigens regulate secretion of the hepatitis B viral surface envelope particle. Proc Natl Acad Sci U S A 839338–9342. 10.1073/pnas.83.24.9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Li J., Wands J. R., Wen Y. M.(2013). Hepatitis B virus genetic variants: biological properties and clinical implications. Emerg Microbes Infect 2,e10. 10.1038/emi.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trépo C., Chan H. L., Lok A.(2014). Hepatitis B virus infection. Lancet 3842053–2063. 10.1016/S0140-6736(14)60220-8 [DOI] [PubMed] [Google Scholar]
- Tsai A., Kawai S., Kwei K., Gewaily D., Hutter A., Tong D. R., Li J., Wands J. R., Tong S.(2009). Chimeric constructs between two hepatitis B virus genomes confirm transcriptional impact of core promoter mutations and reveal multiple effects of core gene mutations. Virology 387364–372. 10.1016/j.virol.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. C., Huang W., Lai M. D., Su I. J.(2006). Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci 97683–688. 10.1111/j.1349-7006.2006.00235.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.