Abstract
Crossover recombination is essential for generating genetic diversity and promoting accurate chromosome segregation during meiosis. The process of crossover recombination is tightly regulated and is initiated by the formation of programmed meiotic DNA double-strand breaks (DSBs). The number of DSBs is around ten-fold higher than the number of crossovers in most species, since only a limited number of DSBs is repaired as crossovers during meiosis. Moreover, crossovers are not randomly distributed. Most crossovers are located on chromosomal arm regions and both centromeres and telomeres are usually devoid of crossovers. Either loss or mislocalization of crossovers frequently results in chromosome nondisjunction and subsequent aneuploidy, leading to infertility, miscarriages and birth defects such as Down syndrome. Here, we will review aspects of crossover regulation observed in most species and then focus on crossover regulation in the nematode Caenorhabditis elegans in which both the frequency and distribution of crossovers are tightly controlled. In this system, only a single crossover is formed, usually at an off-centered position, between each pair of homologous chromosomes. We have identified C. elegans mutants with deregulated crossover distribution and we are analyzing crossover control by using an inducible single DSB system with which a single crossover can be produced at specific genomic positions. These combined studies are revealing novel insights into how crossover position is linked to accurate chromosome segregation.
INTRODUCTION
Meiosis is a specialized cell division process that generates haploid gametes from diploid parental germ cells. This reduction in the number of chromosomes is achieved by following a single round of DNA replication with two consecutive cell divisions (meiosis I and II). Homologous chromosomes are separated at meiosis I and sister chromatids are separated at meiosis II. There are unique chromosomal events that need to take place during prophase to ensure that homologs segregate properly at meiosis I (Fig. 1). Homologous chromosomes need to find each other and pair, these pairing interactions need to be stabilized via the formation of a scaffold known as the synaptonemal complex, which assembles at the interface between paired homologs, and interhomolog recombination needs to take place in order to produce crossovers. Crossover formation is one of the sources of genetic diversity in the population. Moreover, crossovers result in physical attachments (chiasmata) between homologs, which underpinned by cohesion, confer the tension required to properly align the attached homologs (bivalents) at the metaphase plate and then orient them toward opposite poles of the meiosis I spindle. Errors in crossover formation result in chromosome nondisjunction leading to aneuploidy, which causes infertility, miscarriages, birth defects and cancers.
Figure 1. Meiosis and crossover formation.
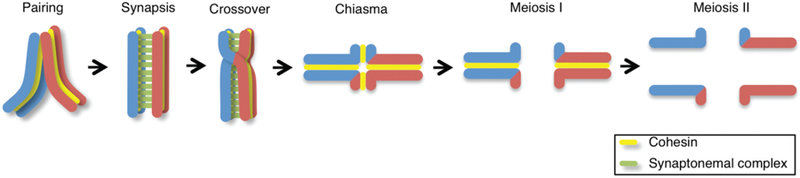
Chromosome dynamics during meiosis. After premeiotic DNA replication, homologous chromosomes find each other (pairing) during the leptotene-zygotene stages. The synaptonemal complex assembles, aligning and holding homologs together throughout their full lengths (synapsis) at the pachytene stage. Repair of DNA double-strand breaks (DSBs) via crossover formation involves the reciprocal exchange of genetic information between homologs. A chiasma is the cytologically visible manifestation of an earlier crossover event underpinned by flanking sister chromatid cohesion and is observed as a cruciform configuration during the diplotene to diakinesis transition. Homologous chromosomes are segregated at the metaphase I to anaphase I transition and sister chromatids are separated at the metaphase II to anaphase II transition. Paternal chromatids are blue and maternal chromatids are red. Sister chromatid cohesion is depicted in yellow and the synaptonemal complex is depicted in green.
Given the impact of crossover formation on human health and reproductive biology it is therefore not surprising that crossovers are tightly regulated. For example, crossover formation is not frequently observed near centromere and telomeres, suggesting they may be repressed in these regions. Crossovers at centromere regions lead to aneuploidy in female meiosis and crossovers at telomeres increase azoospermia (Ottolini et al. 2015; Ren et al. 2016). However, direct testing of how a crossover positioned near centromeres or telomeres might lead to increased errors in chromosome segregation has been challenging in metazoans.
Caenorhabditis elegans is an ideal model organism to study crossover control because crossover formation is tightly regulated in comparison to other known model organisms. A single off-centered crossover is formed on each of the six pairs of homologous chromosomes in C. elegans compared to the one to four crossovers per pair of homologs observed in other species (Barnes et al. 1995; Martinez-Perez and Colaiacovo 2009; Rockman and Kruglyak 2009). Surprisingly, a single DNA double-strand break (DSB) is sufficient to make a crossover in C. elegans (Rosu et al.2011). This property, coupled with the use of a system in which a single DSB can be induced at defined genomic positions, allows us to analyze how crossover position affects meiotic chromosome segregation in C. elegans. Here we review what is known for crossover control from studies in different organisms, our novel findings regarding regulation of crossover position using the single inducible DSB system in C. elegans and the future directions of research using this system aimed at understanding the origin of aneuploidies.
MOLECULAR STEPS IN CROSSOVER FORMATION
Crossover formation starts with the formation of DSBs by a topoisomerase-like protein present from yeast to humans known as Spo11 (Fig. 2; (Keeney et al. 1997)). DSBs then undergo 5’ end resection to produce 3’ overhangs through the activity of the Mre11/Rad50/Xrs2 exonuclease complex. Rad51 associates with the 3’ single-stranded DNA overhangs producing a DNA-protein filament that then engages in a search for homologous DNA sequences. The 3’end invades the homologous template (single strand invasion resulting in D-loop formation), followed by DNA synthesis. At this point, repair can proceed through different pathways resulting in either the displacement and annealing of the newly synthesized strand to its complementary strand (synthesis-dependent strand annealing; SDSA; Fig. 2A) or in second end capture to produce a double Holliday junction (dHJ) intermediate (Fig. 2B). The SDSA pathway results only in non-crossover products. Meanwhile, the asymmetric resolution of dHJs by structure-specific endonucleases (Slx1-Slx4, Mus81-Mms4 and Yen1) produces crossover products (Fig. 2 C), whereas their symmetric resolution results in non-crossovers (Fig. 2D). It has been proposed that designated and non-designated DSBs are converted to dHJs and resolved by different structure-specific endonucleases in yeast to assure and limit the number of crossovers (Zakharyevich et al. 2012). dHJs can also be processed through dissolution mediated by the Sgs1-Top3-Rmi1 complex during which the two Holliday junctions branch migrate toward one another until they form a hemicatenated intermediate that can be decatenated by topoisomerase III, resulting in non-crossovers (Fig. 2E). Finally, all DSBs undergoing intersister repair result in non-crossovers. This detailed blueprint of the molecular requirements for crossover formation allows for assessment of how these may differ during DSB repair depending on the location of the DSB in a metazoan.
Figure 2. Model of homologous recombination.
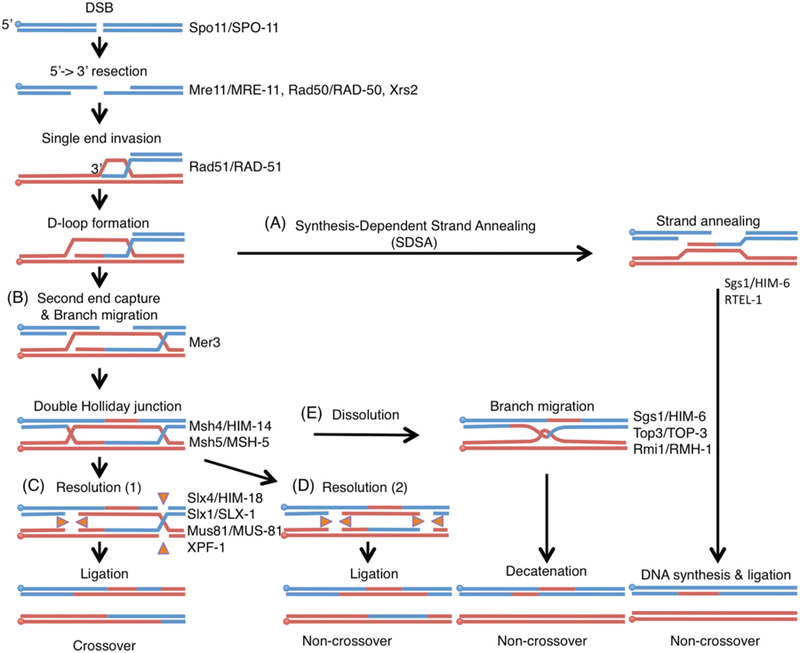
DNA double strand breaks are generated by the topoisomerase-like protein Spo11. The MRN/X complex (Mre11-Rad50-Nbs1/Xrs2) resects the 5’ends to expose 3’overhangs. Single end invasion (SEI) is mediated by Rad51. Homologous recombination can then proceed through the following pathways: (A) synthesis-dependent strand annealing resulting in non-crossover products or (B) double Holliday junction (dHJ) formation by Mer3 and Msh4-Msh5 resulting in crossover (CO) formation. The DNA helicase ReqQ homologs Sgs1 and RTEL-1 disrupt D-loops to anneal both ends of the DSB. Once double Holliday junctions are formed, they are resolved by the structure-specific endonucleases SLX-1-SLX-4/HIM-18, MUS-81-EME1 and XPF-ERCC1. (C) Asymmetric resolution of the dHJ produces crossovers and (D) symmetric resolution results in non-crossovers. (E) dHJs can also be processed by the dissolution pathway through the BTR complex (BLM-TOP3-RMI1/2) to make non-crossover products. Paternal DNAs are blue and maternal DNAs are red. Circles indicate 5’side of DNA. Orange triangles indicate the direction of catalytic activities of Holliday junction resolvases. Key proteins acting at each step are indicated on the right and both yeast and worm names are indicated.
CLASS I and CLASS II CROSSOVERS
Two different classes of crossovers have been identified, namely class I and class II. In yeast, around 90 crossovers are observed in 16 bivalents. 70% (range from 60–90%) of crossovers are class I crossovers, which are dependent on the meiosis-specific ZMM proteins (Zip1, Zip2, Zip3, Msh4, Msh5, Mer3, Spo16, and Spo22/Zip4). Zip1 is a structural component of the synaptonemal complex that holds pairs of homologous chromosomes together. Zip2 is a XPF-like Helix-hairpin-helix containing protein (Chua and Roeder 1998; Macaisne et al. 2008) and Zip3 is a SUMO E3 ligase, whereas Msh4 and Msh5 are homologs of the E. coli mismatch repair protein MutS implicated in stabilizing dHJ intermediates (Snowden et al. 2004). The number of class I crossovers (~60) matches the number of Zip2, Zip3, and Msh4-Msh5 foci observed in yeast pachytene nuclei. Mer3 is a DNA helicase required for Holliday junction branch migration (Nakagawa and Ogawa 1999; Mazina et al. 2004). Finally, Spo16 and Spo22/Zip4 are involved in synaptonemal complex assembly, and they are unique because, in contrast to the other ZMM proteins, they are not essential for crossover interference (Shinohara et al. 2008) (see the section of crossover interference). The remaining ~30% of crossovers fall into class II. Class II crossovers depend on double Holliday junction resolution executed by the structure-specific endonucleases Mus81-Mms4, Slx1-Slx4 and Yen1 in yeast (Zakharyevich et al. 2012) (Fig. 2C,D).
MULTIPLE LAYERS OF CROSSOVER REGULATION
Crossover assurance/Obligate crossover
Because crossover formation is essential for proper chromosome segregation at meiosis I (Fig. 1), at least one crossover (obligate crossover) has to be formed between each pair of homologous chromosomes. This phenomenon is called “crossover assurance” (Fig. 3A). A notable exception to this regulation is observed in Drosophila where both male meiosis and female chromosome 4 are devoid of crossover formation (Cooper 1949; Hartmann and Sekelsky 2017) and chromosome segregation occurs randomly at meiosis II. In contrast, recombinant chromatids are preferentially segregated to oocytes and not into the second polar body in human female meiosis (Ottolini et al. 2015). Robust crossover assurance is also observed in C. elegans, where only one DSB per homologous chromosome pair is sufficient to make a crossover (Rosu et al. 2011). Moreover, chromosomes that fail to undergo crossover formation, as a result of either impaired homologous pairing or due to lack of synapsis in the case of the extra chromosome present in trisomies, are preferentially segregated into the polar bodies during both anaphase I and anaphase II of C. elegans female meiosis (Cortes et al. 2015; Muscat et al. 2015; Vargas et al. 2017). Therefore, crossover formation is not only important for accurate homolog separation at meiosis I but also acts as a driving force during sister chromatid separation at meiosis II.
Figure 3. Different types of crossover control.
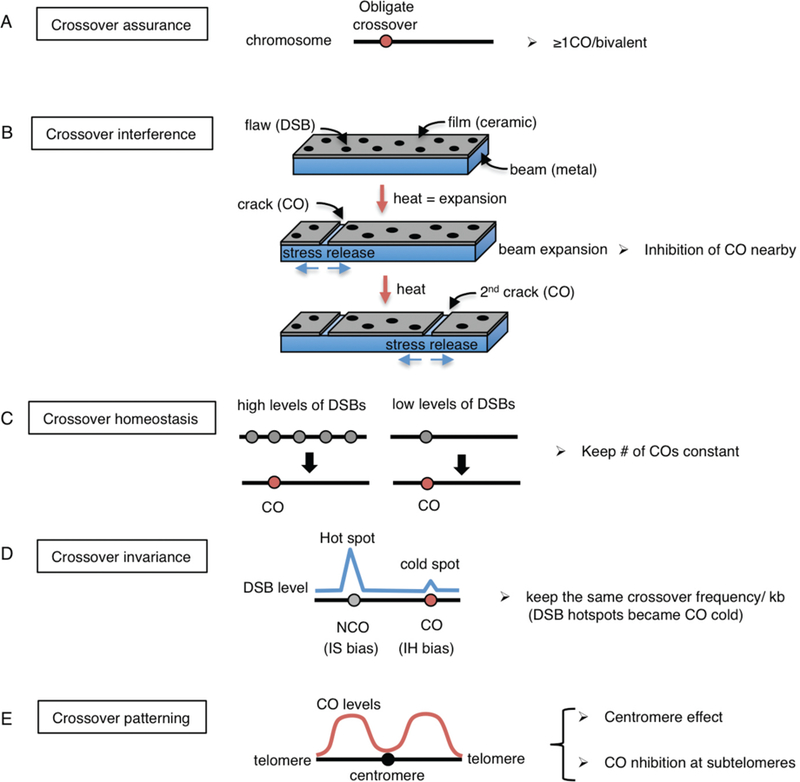
Five known forms of crossover control are depicted. (A) Crossover assurance. At least one crossover per homologous chromosome pair is essential for chiasma formation and proper chromosome segregation at meiosis I. (B) Crossover interference. The beam-film model (modified from Kleckner et al. 2004) is represented. Chromosome axes and chromatin loops are likened as metallic beams and ceramic films which are tightly bonded to the beam, respectively. Heating the beam results in a flaw (DSB) being converted into a crack (CO formation), and the release of stress then propagates in both directions. Continued heating generates a 2nd crack away from the 1st crack resembling interference. (C) Crossover homeostasis. Either high or low levels of DSBs per homologous chromosome pair result in the same number of crossovers. Gray circles are DSBs and red circles are crossovers. (D) Crossover invariance. DSBs at hot spots tend to undergo intersister bias, resulting in a non-crossover outcome, while DSBs at cold spots undergo interhomolog bias leading to crossover products in S. pombe. (E) Crossover distribution/centromere effect. Crossovers near centromeres and telomeres are suppressed. Crossovers are also suppressed at the center regions in the holocentric organism C. elegans.
Crossover interference
Crossover interference is a phenomenon in which a crossover at one location reduces the probability of a second crossover nearby such that when there are two or more crossovers along a bivalent these crossovers are separated away from each other (Fig. 3B). This phenomenon was first described more than 100 yeas ago (Sturtevant 1915) and is only observed for class I crossover events. A beam-film model has been proposed for crossover interference which simulates establishment and propagation of a mechanical stress along the chromosome axis depicted by an elastic beam plate (metal) covered with a thin brittle film (ceramic) with COs being seen as cracks that release the stress locally and thus abrogate COs nearby (Kleckner et al. 2004). Topoisomerase II and the meiosis-specific chromosome axis protein Red1 are suggested to be involved in crossover interference through ubiquitination by the Histone deacetylase Sir2 and the SUMO-targeted ubiquitin ligases Slx5-Slx8 (Zhang et al. 2014). Synaptonemal complex-dependent crossover interference is observed in C. elegans (Libuda et al. 2013) and in at least one yeast strain (Sym and Roeder 1994; Chen et al. 2008). The biological function of crossover interference is largely unknown but it may confer a selective advantage due to co-segregation of functionally related linked genes (Wang et al. 2015; Sun et al. 2017).
Crossover homeostasis
In general, DSBs, which are introduced at the leptotene/zygotene stage of prophase of meiosis I, occur at levels that are 10-fold higher than the number of crossovers detected. The number of crossovers is maintained constant at the expense of non-crossover events even when DSBs levels are reduced. This phenomenon is termed “crossover homeostasis” and has been observed in yeast (Martini et al. 2006), mice (Cole et al. 2012), worms (Yokoo et al. 2012) and plants (Varas et al. 2015)(Fig. 3C).
Crossover invariance
The choice of repair template is very important to make an interhomolog crossover. In addition, levels of DSBs vary largely across the genome as evidenced by the presence of both hot spots and cold regions of DSBs along chromosomes. In S. pombe, which lacks crossover interference, a nearly constant level of crossing over is maintained per unit physical distance across the genome by control of partner choice for DSB repair, a phenomenon referred to as “crossover invariance” (Hyppa and Smith 2010; Fowler et al. 2014) (Fig. 3D). At a DSB hotspot, intersister repair is predominant, whereas at a DSB cold region, interhomolog repair is more prevalent. This phenomenon may serve as an alternative mechanism of crossover homeostasis in other organisms to maintain crossover levels constant.
Crossover patterning/Centromere effect
Crossover formation is inhibited near centromeres and telomeres in many species including humans (Fig. 3E) (Ottolini et al. 2015; Ren et al. 2016). Crossovers at centromeres disrupt cohesion in the pericentric region and affect kinetochore orientation. Interestingly, even gene conversion near centromeres is associated with 60% of segregation errors at meiosis I in S. cerevisiae due primarily to premature sister chromatid separation (Sears et al. 1995). The “centromere effect” was first described in Drosophila as an inhibition of crossovers at centromeres and pericentromeric euchromatic regions (Beadle 1932). A combination of the centromere effect and crossover suppression by the Blm helicase results in the absence of crossovers on chromosome 4 in Drosophila (Hartmann and Sekelsky 2017; Hatkevich and Sekelsky 2017). In plants, cytosines at centromeric regions are highly methylated, thereby suppressing expression of repetitive sequences including transposons. Pollen-typing revealed that crossover frequency is increased in centromeric regions in the DNA methyltransferase met1 mutants, suggesting that DNA methylation at centromeres is important to suppress crossovers at those regions (Yelina et al. 2012). Yeast Slx4, a regulatory subunit of the structure-specific endonuclease Slx1, is also required for crossover suppression near centromeres in an Slx1-independent manner (Higashide and Shinohara 2016). A similar suppression mechanism was observed in C. elegans, where normally crossovers are located on the arms but not at the center of the chromosomes (Fig. 4). However, the crossover suppression observed at the center of the chromosomes in this worm is SLX-1-dependent and SLX4/HIM-18-independent (Saito et al. 2009; Saito et al. 2012; Saito et al. 2013). Taken together, these various layers of crossover control, underscore the importance of crossover formation during meiosis and the need for tightly regulating this process throughout species.
Figure 4. Tight regulation of crossover formation in C. elegans.
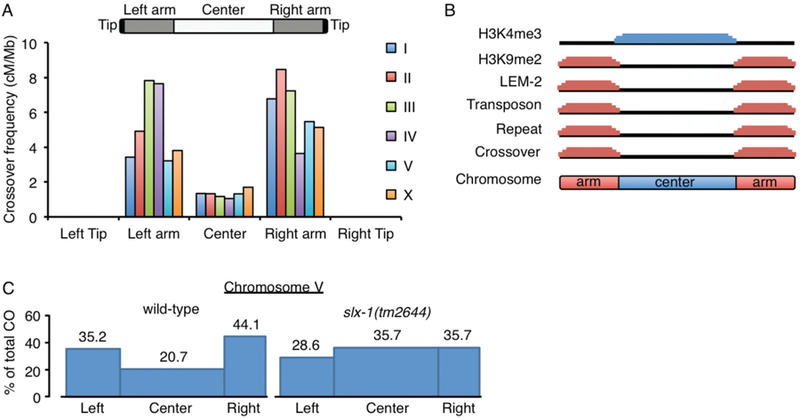
(A) Crossovers are enriched at arm regions but suppressed at center regions in both autosomes and the X chromosome in C. elegans. No crossovers are observed at subtelomeric regions (average <614kb from telomeres). Data was adapted from (Rockman and Kruglyak 2009). (B) Unique features divide chromosome domains in C. elegans. Although up to ~10 DSBs are distributed in a non-biased manner along chromosomes (Saito et al. 2012), crossovers occur at the arm regions where the heterochromatin marker histone H3K9me2, the nuclear membrane protein (LEM-2) binding sequences, transposons, and repeat sequences, are enriched. Crossover formation is suppressed at the center region where the euchromatic marker histone H3K4me3 is enriched. (C) Crossover suppression at the center region of autosomes is lost in slx-1(tm2644) null mutants. Blue boxes indicate crossover frequencies (Saito et al. 2012). While the overall crossover frequency is not altered, crossover distribution is altered by increasing at the center region and decreasing at the arms in slx-1 mutants compared to wild type.
CROSSOVER PATTERNING IN C. ELEGANS
Although studies of crossover control started a century ago, the molecular mechanisms underlying these processes are still largely unknown, in part because the various layers of crossover control are not independent phenomena. The regulation of obligatory crossover, interference and homeostasis are thought to be a cooperative process (Wang et al. 2015). To understand these complex phenomena, it is important to use a simple model.
In C. elegans, crossovers tend to occur in an off-centered position such that crossover frequencies are 1.3 cM/Mb at the central region of the chromosome, which encompasses 49% of the total length of the autosomes and 36% of the X chromosomes, and 4.7 cM/Mb and 6.1 cM/Mb on the left and right arms, respectively (Fig. 4A) (Rockman and Kruglyak 2009). There are some clear differences between the central and arm regions of the chromosomes in this organism. For example, essential genes and the euchromatin marker histone H3K4me3 are enriched at the central region. In contrast, the arm regions are gene poor, they are enriched for the heterochromatin marker H3K9me2/3, they are transposon- and repeat-rich and interact with the nuclear membrane via the LEM (LAP2, emerin, MAN1) domain-containing protein LEM-2 (Fig 4B) (Consortium 1998; Gerstein et al. 2010; Ikegami et al. 2010; Liu et al. 2011; Ho et al. 2014). Different mutants have been identified that shift crossover distribution to the center region of the chromosomes in C. elegans (Zetka and Rose 1995; Wagner et al. 2010; Meneely et al. 2012; Saito et al. 2012; Chung et al. 2015; Hong et al. 2016; Jagut et al. 2016). However, the molecular mechanism underlying the regulation of crossover position is largely unknown. We found that in mutants for SLX-1, a structure-specific endonuclease that cleaves 5’-flaps, replication forks and Holliday junctions, crossovers shifted to the center regions, while overall crossover frequency was preserved (Fig. 4C) (Saito et al. 2012; Saito et al. 2013). This was not due to alterations in DSB distribution since similar frequencies of markers for DSB repair sites were observed cytologically along the arms and at the center of the chromosomes in slx-1 mutants compared to wild type (Saito et al. 2012). This observation led us to hypothesize that there are mechanisms either inhibiting crossover formation or promoting non-crossover formation after DSB induction at the center region of the chromosomes, such as SDSA, double Holliday junction dissolution, “same sense resolution” of double Holliday junctions or intersister repair. Based on its Holliday junction resolvase activity, one possibility is that SLX-1 produces noncrossovers at the center region by same sense resolution of double Holliday junctions. An alternative, albeit non exclusive, possibility is that SLX-1 may act as an epigenetic reader given that it has a PHD/RING-like zinc finger domain implicated in recognizing histone H3K4me3 (Pena et al. 2006; Shi et al. 2006; Matthews et al. 2007; Ramon-Maiques et al. 2007). Given that the euchromatin marker H3K4me3 is enriched at the center region of the autosomes in C. elegans, we hypothesize that SLX-1 might be recruited to that region through recognition of H3K4me3 by its PHD/RING finger domain or potentially function as an ubiquitin ligase to activate non-crossover pathways at the center region. It is known that H3K4me2, H3K4me3 and H3K9ac are specifically enriched at the DSB hotspots recognized by the PRDM (PRDI-BF1 (positive regulatory domain I- binding factor 1) and RIZ (retinoblastoma-interacting zinc-finger protein) homology domain)-containing histone H3 methyltransferase Prdm9 in mammals (Hayashi et al. 2005; Buard et al. 2009; Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). After DSBs are introduced, phosphorylation of histone H2AX and hyperacetylation of histone H4 are observed near the hotspots (Mahadevaiah et al. 2001; Buard et al. 2009). H3K4me3 is associated with DSB formation in yeasts and plants (Choi et al. 2013) and the PHD finger of Spp1, a component of the histone H3K4 methyltransferase Set1 complex (also known as COMPASS), reads H3K4me3 to promote DSBs (Acquaviva et al. 2013; Sommermeyer et al. 2013). However, whether H3K4me3 is associated with non-crossover pathways and whether crossover hotspots are marked by H3K9me2/3 remains unknown. One possible explanation for why DSBs are introduced at the arm region is that the DSB machinery could access small euchromatic (H3K4me3) sites dispersed throughout the H3K9me2/3 marked heterochromatin on the arm regions (Ikegami et al. 2010). Combined strategies, described below, allow for direct testing of whether SLX-1 localizes at the center region of chromosomes and the roles of its PHD/RING domain.
INDUCIBLE SINGLE DSB SYSTEM – HOW DOES CROSSOVER POSITION IMPACT CHROMOSOME SEGREGATION IN A METAZOAN?
Although improper crossover distribution has been detected in aneuploid gametes and embryos in humans, the impact of improper CO distribution has not been tested directly in metazoans. C. elegans is a suitable model organism to assess how improper crossover distribution can cause errors in meiotic chromosome segregation given how tightly crossovers are regulated in this organism. We previously showed that starting in late pachytene chromosomes remodel around the off-centered crossover resulting in the characteristic cruciform configuration observed for bivalents in late diakinesis consisting of a long and a short arm intersecting at the chiasma (Nabeshima et al. 2005) (Fig. 5B). Aurora B kinase, AIR-2, localizes at the short arms by the end of diakinesis where it phosphorylates the meiosis-specific cohesin REC-8. This triggers degradation of sister chromatid cohesion along the short arm. LAB-1, a functional Shugoshin analog, antagonizes AIR-2 along the long arms thereby protecting these chromosome subdomains from premature loss of sister chromatid cohesion. Therefore, we hypothesized that achieving proper chromosome remodeling must be critical for an accurate reductional division at meiosis I. To test this directly, we used a single DSB-inducible system in which the Mos1 transposon from Drosophila was introduced into C. elegans along with a transposase under a heat shock promoter and a spo-11 mutation that eliminates endogenous meiotic DSBs (Bessereau et al. 2001). We obtained lines in which a single DSB was produced at either the physical center or at the subtelomeric region in an autosome and compared that to lines in which a single DSB was produced at an off-centered position (Fig. 5A,B). We observed impaired chromosome remodeling based on mislocalization of AIR-2 and LAB-1 as well as subsequent increased non-disjunction when the single DSB was converted into a crossover at the center position. We did not observe a single crossover event at the subtelomeric position although a DSB at such position did get designated into a crossover, suggesting these either cannot be stably maintained or are not effectively processed into a mature crossover at that position (Altendorfer and Colaiacovo, unpublished results). This is supported by the lack of crossovers previously reported at subtelomeric regions by (Rockman and Kruglyak 2009). In contrast, a DSB at an off-centered position proceeded as wild type. We are currently assessing whether bivalent formation is increased if a single DSB is induced at the center region of the chromosomes in slx-1 mutants to confirm our genetic observation that SLX-1 suppresses crossovers at that region. This is being combined with ChIP-seq for epigenetic markers with the goal of unveiling how crossover position impacts chromosome remodeling and proper chromosome segregation and how structure-specific endonucleases are involved in this regulation.
Figure 5. Site-specific analysis of meiotic recombination in C. elegans.
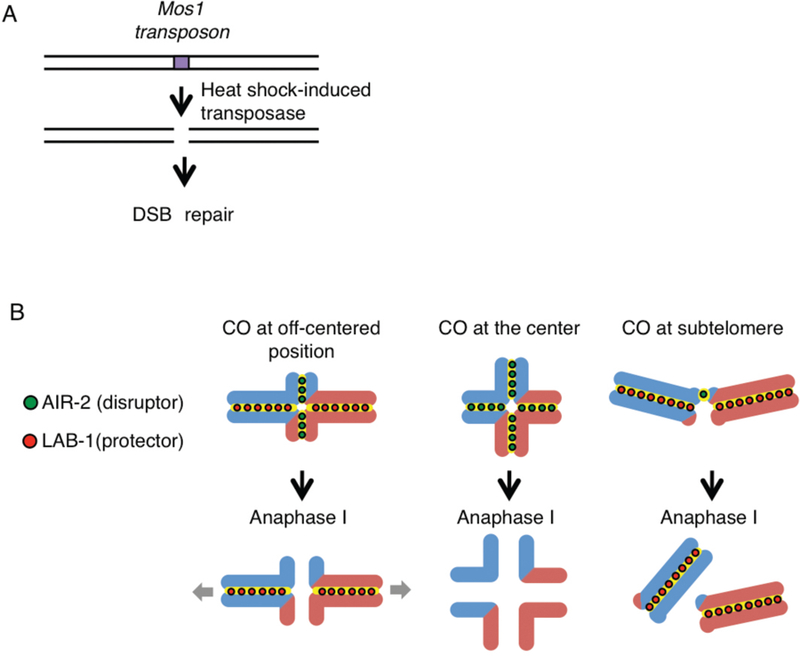
(A) Mos1-based single inducible DSB system. Mos1 transposons and transposases are integrated into chromosomes in a spo-11 mutant background. The Mos1 integrated strain library is available at Nemagentag (http://elegans.imbb.forth.gr/nemagenetag/). Heat shock induces expression of the transposase which excises the Mos1 transposon resulting in a single DSB at a specific genomic position. (B) The single DSB system can be used to investigate the positional effect of crossovers on meiotic chromosome segregation. Blue and red lines are paternal and maternal chromatids, respectively. The system allows us to analyze the outcome of a single crossover forming at specific chromosomal sites. A crossover at the very center region disrupts the asymmetric configuration of the bivalent resulting in premature sister chromatid separation or homolog nondisjunction. Crossovers at subtelomeres result either in potentially fragile connections that are not stably retained at the ends of very short arms or events that fail to mature into crossovers at those positions.
IS THE CLASS II CROSSOVER PATHWAY CONSERVED IN C. ELEGANS?
It has been proposed that only class I crossovers exist in C. elegans because almost no crossovers are detected in worm mutants of the zmm counterparts (syp-1, syp-2, syp-3, syp-4, zhp-3, him-14/msh4, msh-5) (Zalevsky et al. 1999; Kelly et al. 2000; MacQueen et al. 2002; Colaiacovo et al. 2003; Jantsch et al. 2004; Smolikov et al. 2007; Smolikov et al. 2009) and crossover interference is extremely robust given that only a single crossover occurs per bivalent in normal conditions (Brenner 1974; Barnes et al. 1995). While in budding yeast, the structure-specific endonucleases Slx1-Slx4 and Mus81-Mms4 are required for class II crossovers (Zakharyevich et al. 2012), in C. elegans SLX-1-HIM-18/SLX4 and MUS-81 appear to be involved in 20–60% of total crossovers together with XPF-1, another structure-specific endonuclease (Saito et al. 2009; Saito et al. 2012; Agostinho et al. 2013; O’Neil et al. 2013; Saito et al. 2013).
Interestingly, class II crossovers are observed in C. elegans under conditions where there is either an excess of DSBs or a mutation in the antirecombinase RTEL-1 helicase (Youds et al. 2010). DSBs induced artificially by gamma-irradiation (IR) cause a dose-dependent (10 Gy and 75 Gy) increase in the number of crossovers, while the number of crossover designated sites marked by ZHP-3 is kept at normal levels after IR treatment (Youds et al. 2010). Moreover, these excess crossovers are suppressed back to wild-type levels in mus-81 mutants (Youds et al., 2010). Therefore, there must be designation-independent and MUS81-dependent class II crossovers in C. elegans as well. Furthermore, crossovers in rtel-1 mutants are randomly distributed, sugessting that RTEL-1-inhibited crossovers exhibit no interference (Youds et al. 2010). Helicase-dependent inhibition of class II crossovers is conserved in budding yeast where it is exerted by Sgs1 (De Muyt et al. 2012; Zakharyevich et al. 2012), in fission yeast by Fml1 (Lorenz et al. 2012) and in plants by FANCM (Crismani et al. 2012; Knoll et al. 2012). These data and the wide conservation of class II COs in yeast (de los Santos et al. 2003), plants (Berchowitz et al. 2007; Higgins et al. 2008) and mammals (Holloway et al. 2008; Svetlanov et al. 2008) suggest that a “dormant” class II crossover pathway exists in C. elegans.
Mapping of crossover frequency and distribution in either IR-treated combinatorial mutants of structure-specific endonucleases or in mutants for the structure-specific endonucleases combined with an rtel-1 mutation will reveal whether interference-independent class II crossovers are structure-specific endonuclease-dependent as well in C. elegans.
SEXUAL DIMORPHISM IN CROSSOVER REGULATION DURING C. ELEGANS MEIOSIS
The biggest difference regarding crossover control between sexes is observed in Drosophila, where crossovers occur in females but not in males. However, differences are observed in other organisms as well. For example, while the number of crossovers seem similar between female and male meiosis in mice, based on the number of MLH1 foci (females= 23.0 and males=22.7) (Holloway et al. 2008), high resolution sex-specific linkage maps revealed that crossover frequency on autosomes in females (~71 cM/autosome) is higher than in males (~66 cM/autosome) (Liu et al. 2014). Interestingly, the genomic distribution of crossovers between female and male meiosis is also significantly different; crossover distributions are uniform in females, but subtelomerically enhanced and pericentromerically suppressed in males (Liu et al. 2014). In contrast, opposite results regarding crossover frequency were observed in plants, where male crossover frequency (2.23 crossovers/bivalent) is 1.7-fold higher than in females (1.33 crossovers/bivalent) (Giraut et al. 2011), even though the distribution of crossovers follows a similar pattern to that observed in mice. Specifically, crossover frequencies at subtelomeric regions are very high in males and very low in females (Giraut et al. 2011). In human females, the number of MLH1 foci is greater than in males, while the average crossover frequency is similar between the sexes (Wang et al. 2017). This suggests that crossover maturation after designation may be less efficient in females than in males. In C. elegans, crossover frequency is higher in male spermatogenesis than in oogenesis. The ratio of double crossovers is around 4% at chromosomes IV and V in males while almost no double crossovers are detected in oogenesis (Henzel et al. 2011; Gabdank and Fire 2014). Furthermore, him-8 and meDf2 mutants, which lack crossovers on the X chromosome due to impaired X chromosome synapsis, exhibit a higher crossover frequency (including double crossovers) on the autosomes compared with wild type (Carlton et al. 2006). These observations suggest that a crossover homeostasis-like regulation that maintains the crossover number per “nucleus” (referred to as an interchromosomal effect (Sturtevant 1919; Lucchesi and Suzuki 1968) exists in animals which have an XO type sex such as C. elegans males as well. How the “additional” crossovers are formed/allowed is an interesting question. The following aspects should be considered: (1) Crossover interference is stronger in hermaphrodite oogenesis than in male spermatogenesis in C. elegans (Gabdank and Fire 2014); (2) Surveillance mechanisms to check the number and distribution of crossovers (crossover checkpoint) may be weak in XO animals. In fact, while either accumulated DNA damage or aberrant synapsis induce apoptosis resulting in the removal of the affected cell in hermaphrodite gonads, there is no germline apoptosis in C. elegans males due to lack of CED-3 activation in the male germline even though a recombination checkpoint is activated in male gonads (Jaramillo-Lambert et al. 2010). Whether there are also differences in crossover control between spermatogenesis and oogenesis in hermaphrodites remains to be investigated.
CROSSOVER REGULATION IN AUTOSOMES AND SEX CHROMOSOMES
Crossover regulation is different between autosomes and sex chromosomes. X and Y chromosomes in mammals, including humans, pair via short homologous sequences referred to as the pseudoautosomal region (PAR) located at their subtelomeres. C. elegans provides a useful experimental system to understand the evolution of heteromorphic sex chromosomes (Henzel et al. 2011). The karyotype of the C. elegans hermaphrodite consists of 5 autosomes (5A) and XX, and of 5A and XO in males. An end-to-end fusion of chromosomes X and IV generates a new chromosome named mnT12 and males with mnT12 form a neo-sex body (IV = neo-Y and IV-X fusion = neo-X), in which a portion of chromosome IV mimics the mammalian PAR during meiotic prophase (Sigurdson et al. 1986). Meiotic sex chromosome inactivation (MSCI) occurs during meiotic prophase and several factors have been identified as required for mammalian MSCI such as γH2AX, ATR kinase, ubiquitin, SUMO and the BRCA1-A complex (Turner 2007; Lu and Yu 2015). However, the links between MSCI and crossover control remain to be determined. Whether MCSI is conserved in the C. elegans neo-sex body also requires further investigation. The unsynapsed X chromosome region of the neo-sex body in C. elegans males harbors high levels of the heterochromatic marker H3K9me2, similar to the X-Y bodies in mammals (Henzel et al.2011). The PAR must receive DSBs to make an obligate crossover like other autosomes. In mice, chromatin axis length at the PAR is long relative to DNA length and in contrast to autosome axes where DNA content correlates well with axis length (Kauppi et al. 2011). This was proposed to result in shorter chromatin loops on the PAR and contribute to higher DSB levels at this region compared to autosomes. Longer axes are also observed along the paired chromosome IV and mnT12 in C. elegans (Henzel et al. 2011). Because Spo11α-dependent induction of DSBs is observed at the PAR in mice (Kauppi et al. 2011), it would be interesting to investigate whether there are differences between the paired chromosome IV and mnT12 and other normal chromosome pairs regarding the machinery being engaged for DSB formation.
DSBs are also induced at the non-homologous regions of sex bodies in mammals and on the X chromosome in males in C. elegans that lack a homologous partner. The biological function for DSB repair in these cases is unknown, but interestingly, DSBs on the male X chromosome may be partially inhibited by the structure-specific endonuclease XPF-1 in C. elegans (Checchi et al. 2014). Another possible explanation is that XPF-1 functions for repair via single strand annealing at repeat sequences when RAD-51-dependent homologous recombination is not available on male X chromosomes (Checci et al. 2014). Further studies are required to understand how DSBs are induced and repaired in XO animals.
In the C. elegans hermaphrodite, 0.2 % of its progeny are males resulting from X chromosome non-disjunctions during meiosis. Based on the observations that most autosomal aneuploidies are lethal and that nearly 100% of the embryos hatch during hermaphrodite reproduction, X chromosomes in C. elegans are more vulnerable to disjoining during meiosis compared with autosomes. We reported that combinatorial mutants of different structure-specific endonucleases caused higher reductions in crossover frequencies on X chromosomes than on autosomes (Saito et al. 2009; Saito et al 2012; Saito et al. 2013). Understanding the fragility of the X chromosome in the context of crossover control is important. Several known differences were reported between the X and the autosomes. First, the right third of the X chromosome has fewer heterochromatin marks such as H3K9me2/3 and nuclear membrane binding regions compared to the autosomes (Ikegami et al. 2010; Liu et al. 2011). Second, the X chromosome undergoes less DSBs than the autosomes (Gao et al. 2015). Further, both the timing of replication as well as the onset and completion of synapsis are delayed for the X chromosome in C. elegans (Jaramillo-Lambert et al. 2007; Mlynarczyk-Evans and Villeneuve 2017). In yeasts and plants, the regulation of DNA replication and induction of meiotic DSBs are well connected (Murakami and Nurse 2001; Higgins et al. 2012; Murakami and Keeney 2014). Whether the delay in replication is related to DSB induction on the X chromosomes in C. elegans remains to be analyzed. Now we can trace the fate of specific numbers and locations of DSBs by using an inducible single DSB system in C. elegans (Fig. 5A,B). Use of this system, in combination with high-resolution microscopy, SNP-mapping and sequence-based genomics approaches, will allow us to address the remaining questions described above.
CONCLUSIONS
Although crossover control has been a topic of extensive studies, the molecular mechanisms underlying its regulation are largely unknown. Technological advancements now allow for the introduction and analysis of a single (or more) crossovers at specific genomic positions in metazoans. In addition to Mos1 excision, a CRISPR-based DSB induction system will be established shortly. These approaches will unveil the positional effects of crossovers allowing us to understand the origin of aneuploidies. This in turn may have clinical repercussions for treatments of infertility and in finding targets for cancer therapy in humans.
ACKNOWLEDGMENTS
We thank Marina Martinez Garcia for critical reading of this manuscript. This work was supported by National Institutes of Health grant R01GM105853 to MPC. We apologize to authors whose work was not cited due to space constraints.
REFERENCES
- Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, Nicolas A, Geli V 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. [DOI] [PubMed] [Google Scholar]
- Agostinho A, Meier B, Sonneville R, Jagut M, Woglar A, Blow J, Jantsch V, Gartner A. 2013. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes TM, Kohara Y, Coulson A, Hekimi S. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle GW. 1932. A Possible Influence of the Spindle Fibre on Crossing-Over in Drosophila. Proc Natl Acad Sci U S A 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. 2007. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessereau JL, Wright A, Williams DC, Schuske K, Davis MW, Jorgensen EM. 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413: 70–74. [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Barthes P, Grey C, de Massy B. 2009. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J 28: 2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton PM, Farruggio AP, Dernburg AF. 2006. A link between meiotic prophase progression and crossover control. PLoS Genet 2: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checchi PM, Lawrence KS, Van MV, Larson BJ, Engebrecht J. 2014. Pseudosynapsis and decreased stringency of meiotic repair pathway choice on the hemizygous sex chromosome of Caenorhabditis elegans males. Genetics 197: 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. 2008. Global analysis of the meiotic crossover landscape. Dev Cell 15: 401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FC et al. 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. 1998. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359. [DOI] [PubMed] [Google Scholar]
- Chung G, Rose AM, Petalcorin MI, Martin JS, Kessler Z, Sanchez-Pulido L, Ponting CP, Yanowitz JL, Boulton SJ. 2015. REC-1 and HIM-5 distribute meiotic crossovers and function redundantly in meiotic double-strand break formation in Caenorhabditis elegans. Genes Dev 29: 1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14: 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium CeS. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Cooper KW. 1949. The cytogenetics of meiosis in Drosophila; mitotic and meiotic autosomal chiasmata without crossing over in the male. J Morphol 84: 81–121. [DOI] [PubMed] [Google Scholar]
- Cortes DB, McNally KL, Mains PE, McNally FJ. 2015. The asymmetry of female meiosis reduces the frequency of inheritance of unpaired chromosomes. Elife 4: e06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. 2012. FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. 2014. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res 24: 1650–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabdank I, Fire AZ. 2014. Gamete-type dependent crossover interference levels in a defined region of Caenorhabditis elegans chromosome V. G3 (Bethesda) 4: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Kim HM, Elia AE, Elledge SJ, Colaiacovo MP. 2015. NatB domain-containing CRA-1 antagonizes hydrolase ACER-1 linking acetyl-CoA metabolism to the initiation of recombination during C. elegans meiosis. PLoS Genet 11: e1005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB Lu ZJ Van Nostrand EL Cheng C Arshinoff BI Liu T Yip KY Robilotto R Rechtsteiner A Ikegami K et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mezard C. 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7: e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann MA, Sekelsky J. 2017. The absence of crossovers on chromosome 4 in Drosophila melanogaster: Imperfection or interesting exception? Fly (Austin): 1–7. [DOI] [PMC free article] [PubMed]
- Hatkevich T, Sekelsky J. 2017. Bloom syndrome helicase in meiosis: Pro-crossover functions of an anti-crossover protein. Bioessays 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378. [DOI] [PubMed] [Google Scholar]
- Henzel JV, Nabeshima K, Schvarzstein M, Turner BE, Villeneuve AM, Hillers KJ. 2011. An asymmetric chromosome pair undergoes synaptic adjustment and crossover redistribution during Caenorhabditis elegans meiosis: implications for sex chromosome evolution. Genetics 187: 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashide M, Shinohara M. 2016. Budding Yeast SLX4 Contributes to the Appropriate Distribution of Crossovers and Meiotic Double-Strand Break Formation on Bivalents During Meiosis. G3 (Bethesda) 6: 2033–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FC, Jones GH. 2008. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54: 152–162. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, Halpin C, Armstrong SJ, Franklin FC. 2012. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24: 4096–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn KA, Minoda A, Tolstorukov MY, Appert A et al. 2014. Comparative analysis of metazoan chromatin organization. Nature 512: 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. 2008. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4: e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Sonneville R, Agostinho A, Meier B, Wang B, Blow JJ, Gartner A. 2016. The SMC-5/6 Complex and the HIM-6 (BLM) Helicase Synergistically Promote Meiotic Recombination Intermediate Processing and Chromosome Maturation during Caenorhabditis elegans Meiosis. PLoS Genet 12: e1005872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Smith GR. 2010. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD. 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 11: R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagut M, Hamminger P, Woglar A, Millonigg S, Paulin L, Mikl M, Dello Stritto MR, Tang L, Habacher C, Tam A et al. 2016. Separable Roles for a Caenorhabditis elegans RMI1 Homolog in Promoting and Antagonizing Meiotic Crossovers Ensure Faithful Chromosome Inheritance. PLoS Biol 14: e1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. 2004. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol 24: 7998–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J. 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol 308: 206–221. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Harigaya Y, Vitt J, Villeneuve A, Engebrecht J. 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr Biol 20: 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M. 2011. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. 2004. A mechanical basis for chromosome function. Proc Natl Acad Sci U S A 101: 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A, Higgins JD, Seeliger K, Reha SJ, Dangel NJ, Bauknecht M, Schropfer S, Franklin FC, Puchta H. 2012. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24: 1448–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Morgan AP, Chesler EJ, Wang W, Churchill GA, Pardo-Manuel de Villena F. 2014. High-resolution sex-specific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics 197: 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P et al. 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LY, Yu X. 2015. Double-strand break repair on sex chromosomes: challenges during male meiotic prophase. Cell Cycle 14: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J, Suzuki DT. 1968. The interchromosomal control of recombination. Annu Rev Genet 2: 53–86. [Google Scholar]
- Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R. 2008. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18: 1432–1437. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E, Colaiacovo MP. 2009. Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Genet Dev 19: 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC. 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3’−5’ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- Meneely PM, McGovern OL, Heinis FI, Yanowitz JL. 2012. Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S, Villeneuve AM. 2017. Time-Course Analysis of Early Meiotic Prophase Events Informs Mechanisms of Homolog Pairing and Synapsis in Caenorhabditis elegans. Genetics 207: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Keeney S. 2014. DDK links replication and recombination in meiosis. Cell Cycle 13: 3621–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Nurse P. 2001. Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat Genet 28: 290–293. [DOI] [PubMed] [Google Scholar]
- Muscat CC, Torre-Santiago KM, Tran MV, Powers JA, Wignall SM. 2015. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. Elife 4: e06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Villeneuve AM, Colaiacovo MP. 2005. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J Cell Biol 168: 683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H. 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J 18: 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil NJ, Martin JS, Youds JL, Ward JD, Petalcorin MI, Rose AM, Boulton SJ. 2013. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet 9: e1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolini CS, Newnham L, Capalbo A, Natesan SA, Joshi HA, Cimadomo D, Griffin DK, Sage K, Summers MC, Thornhill AR et al. 2015. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet 47: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. 2007. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A 104: 18993–18998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Ferguson K, Kirkpatrick G, Vinning T, Chow V, Ma S. 2016. Altered Crossover Distribution and Frequency in Spermatocytes of Infertile Men with Azoospermia. PLoS One 11: e0156817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet 5: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Libuda DE, Villeneuve AM. 2011. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334: 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Lui DY, Kim HM, Meyer K, Colaiacovo MP. 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet 9: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Mohideen F, Meyer K, Harper JW, Colaiacovo MP. 2012. SLX-1 is required for maintaining genomic integrity and promoting meiotic noncrossovers in the Caenorhabditis elegans germline. PLoS Genet 8: e1002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. 2009. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet 5: e1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears DD, Hegemann JH, Shero JH, Hieter P. 1995. Cis-acting determinants affecting centromere function, sister-chromatid cohesion and reciprocal recombination during meiosis in Saccharomyces cerevisiae. Genetics 139: 1159–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR et al. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A. 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309. [DOI] [PubMed] [Google Scholar]
- Sigurdson DC, Herman RK, Horton CA, Kari CK, Pratt SE. 1986. An X-autosome fusion chromosome of Caenorhabditis elegans. Mol Gen Genet 202: 212–218. [DOI] [PubMed] [Google Scholar]
- Smolikov S, Eizinger A, Hurlburt A, Rogers E, Villeneuve AM, Colaiacovo MP. 2007. Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 176: 2027–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S, Schild-Prufert K, Colaiacovo MP. 2009. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet 5: e1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. 2004. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Sommermeyer V, Beneut C, Chaplais E, Serrentino ME, Borde V. 2013. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. 1915. Castle and Wright on Crossing over in Rats. Science 42: 342. [DOI] [PubMed] [Google Scholar]
- Sturtevant A 1919. Contributions to the genetics of Drosophila melanogaster. III. Inherited linkage variations in the second chromosome. Carnegie Inst Wash Pub 278: 305–341. [Google Scholar]
- Sun L, Wang J, Sang M, Jiang L, Zhao B, Cheng T, Zhang Q, Wu R. 2017. Landscaping Crossover Interference Across a Genome. Trends Plant Sci 22:894–907. [DOI] [PubMed] [Google Scholar]
- Svetlanov A, Baudat F, Cohen PE, de Massy B. 2008. Distinct functions of MLH3 at recombination hot spots in the mouse. Genetics 178: 1937–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Roeder GS. 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. [DOI] [PubMed] [Google Scholar]
- Turner JM. 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831. [DOI] [PubMed] [Google Scholar]
- Varas J, Sanchez-Moran E, Copenhaver GP, Santos JL, Pradillo M. 2015. Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Atfas1–4 Mutant. PLoS Genet 11: e1005301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas E, McNally K, Friedman JA, Cortes DB, Wang DY, Korf IF, McNally FJ. 2017. Autosomal Trisomy and Triploidy Are Corrected During Female Meiosis in Caenorhabditis elegans. Genetics [DOI] [PMC free article] [PubMed]
- Wagner CR, Kuervers L, Baillie DL, Yanowitz JL. 2010. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zickler D, Kleckner N, Zhang L. 2015. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle 14: 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Choi K, Chelysheva L, Macaulay M, de Snoo B, Wijnker E, Miller N, Drouaud J, Grelon M, Copenhaver GP et al. 2012. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet 8: e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ. 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Rose AM. 1995. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics 141: 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. 2014. Topoisomerase II mediates meiotic crossover interference. Nature 511: 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]


