Abstract
Rationale
Sputum and bronchoalveolar lavage fluid (BALF) are often obtained to elucidate the lower airway microbiota in adults. Acquiring sputum samples from children is difficult and obtaining samples via bronchoscopy in children proves challenging due to the need for anesthesia and specialized procedural expertise; therefore nasopharyngeal (NP) swabs are often used as surrogates when investigating the pediatric airway microbiota. In adults, the airway microbiota differs significantly between NP and BALF samples however, minimal data exist in children.
Objectives
To compare NP and BALF samples in children undergoing clinically indicated bronchoscopy.
Methods
NP and BALF samples were collected during clinically indicated bronchoscopy. Bacterial DNA was extracted from 72 samples (36 NP/BALF pairs); the bacterial V1-V3 region of the 16S rRNA gene was amplified and sequenced on the Illumina Miseq platform. Analysis was performed using mothur software.
Results
Compared to NP samples, BALF had increased richness and diversity. Similarity between paired NP and BALF (intra-subject) samples was greater than inter-subject samples (p=0.0006). NP samples contained more Actinobacteria (2.2% vs. 21%; adjusted p = 1.4 × 10−6), while BALF contained more Bacteroidetes (29.5% vs. 3.2%; adjusted p = 1.2 × 10−9). At the genus level several differences existed, however Streptococcus abundance was similar in both sample types (NP 37.3% vs. BAL 36.1%; adjusted p = 0.8).
Conclusion
Our results provide evidence that NP samples can be used to distinguish differences between children, but the relative abundance of organisms may differ between the nasopharynx and lower airway in pediatric patients. Studies utilizing NP samples as surrogates for the lower airway should be interpreted with caution.
Keywords: microbiota, pediatric, bronchoalveolar lavage fluid, nasopharyngeal swabs
INTRODUCTION
Bronchoscopy with bronchoalveolar lavage fluid (BALF) samples is a common method of evaluating the lower airway in children who cannot expectorate. However, performing bronchoscopy proves challenging due to the need for anesthesia and specialized procedural expertise. Due to ethical concerns, BALF can only be collected from pediatric patients if the procedure is clinically indicated; thereby, prohibiting the collection of lower airway samples in both healthy children and those with lower respiratory disease unless ordered for clinical purposes. Given this, upper airway samples often serve as a surrogate of the lower airway.
In healthy adults, differences exist between the upper and lower airway microbiota1,2. Overlap exists between the oral and lung microbiota while the nasal microbiota does not share bacterial communities with either location in healthy adults2. Differences between sampling location also exist in children and adults with cystic fibrosis3,4. However, there are limited studies comparing the upper and lower airway in children without cystic fibrosis.
It was recently reported that upper airway sampling provided a reliable representation of BALF microbiota in children with protracted bronchial bronchitis and chronic suppurative lung disease with a combination of nasopharyngeal (NP) and oropharyngeal (OP) samples offering the best representation of the lower airway5. Because many studies examining upper airway diseases including early wheeze have focused on the NP microbiota, we focused on comparing the NP microbiota and BALF microbiota in all children regardless of the reason for their bronchoscopy. We hypothesized that the NP microbiota and BALF microbiota would differ in children regardless of their diagnosis. To address this issue, we compared NP swabs and BALF samples from all children undergoing clinically indicated bronchoscopy to determine if NP swabs reflect the BALF microbiota.
METHODS
Participants
Inclusion criteria were children undergoing a clinically indicated bronchoscopy in the Riley Children’s Hospital outpatient center (Indianapolis, IN). Children were excluded if parents and/or children refused to give informed consent. This study was approved by the Indiana University Human Subjects Committee. If the participant was <18 years of age, written informed consent was obtained from the parents and assent was obtained from children ≥11 years of age. If the patient was ≥18 years of age, full consent was obtained from the patient. After consent was obtained, a questionnaire was administered asking basic demographic questions (gender, race, and ethnicity).
NP swabs were obtained prior to bronchoscopy using Copan eSwab 482C in Amies Transport Medium (Copan Diagnostics, Murrieta, CA). Samples were obtained by rubbing the swab on the mid-inferior turbinate for 10 seconds, then placed in transport media (Amies fluid). Samples were then placed on ice while awaiting BALF collection. Bronchoscopy was performed under general anesthesia following current American Thoracic Society (ATS) guidelines6. The bronchoscope was introduced via laryngeal mask airway (LMA) or trans-nasally. Passage of the bronchoscope nasally was utilized when the clinician elected to evaluate the upper airway. If bronchoalveolar lavage was performed, any extra BALF available that exceeded the 10 mL of BALF necessary for clinically indicated studies was collected for this study. BALF was placed in a sterile container, placed on ice with the corresponding NP swab, and taken to the laboratory (within the same building) where it was aliquoted and stored at −80° C until DNA extraction occurred. An average of 13 mL of BALF was collected during the procedure. After removing 10mL for clinically indicated laboratory studies, there was an average of 3 mL remaining for this study.
DNA Extraction
Prior to extraction, 1mL of BALF was centrifuged at 10,000 x g for 10 minutes. The pellet was re-suspended in 200μL of phosphate buffered saline (PBS, GE HyClone). DNA was extracted from NP swabs and BALF using the Qiagen DNeasy Blood and Tissue Kit. A DNA library was constructed using primers that amplify the V1–V3 region of the 16S rRNA gene. 7,8,7,8,7,8,7,8,7,8DNA was amplified using Phusion® High-Fidelity DNA Polymerase mixture (Thermo Scientific). The PCR protocol was 1 cycle of 30 seconds at 98° C followed by 34 cycles of 98° C for 10 seconds, 62° C for 30 seconds, 72° C for 30 seconds and a final elongation at 72° C for 5 minutes. The resulting amplicons were purified with QIAquick® PCR Purification Kit (Qiagen).
Sequencing was performed on an Illumina MiSeq8. The resulting sequence reads were de-multiplexed using CASAVA software installed on the MiSeq Illumina sequencer producing 6,042,668 sequencing reads. Separate pairs of fastq files were generated for each specimen. The splicing of forward and reverse fastq files produced an average of 100,710 ± 48,567 reads per specimen.
Sequence Quality Analysis
16S rRNA sequence processing was performed utilizing mothur (v.1.33.3) software9,10 and adapted from their 16S MiSeq Protocol. Raw paired-end fastq sequences were combined into contigs using make.contigs. Contigs were then aligned against release 123 of the SILVA 16S reference alignment using align.seqs, and contigs that did not match the V1-V3 regions were discarded. Sequencing error correction was performed with pre.cluster, allowing up to three bases to be corrected per read. We then used the UCHIME algorithm11 from within mothur to detect and remove chimeric sequences. The classify.seqs command and RDP training set were used to filter sequences identified as non-bacteria12.
Operational Taxonomic Unit (OTU) clustering and classification
Remaining sequences were clustered using swarm with the fastidious parameter13,14. Swarm is a single-linkage clustering tool that uses a local (rather than global) clustering threshold. Sequence abundance is used to delineate OTUs with high precision. The representative sequences for each OTU was classified using BLAST 15 against release 123 of the SILVA SSU database16. We applied an abundance filter, removing all OTUs that did not compose 2% or more of the read population of at least one sample. Due to concern for contamination, we extracted DNA from sterile water passed through the bronchoscope prior to utilization; the reagents; blank swabs; and sterile water. Sequences detected in control samples clustered into 15 OTUs that constituted >80% of total sequence reads detected in negative controls (Supplemental Table 1). These OTUs were excluded from all subsequent analysis, and represented less than 1.5% abundance in participant samples.
Sequence Analysis
Rarefaction curves describing the number and proportionality of OTUs observed as a function of sampling effort were generated using alpha_rarefaction.py in QIIME (1.9.1)17. Random sub-sampling to the smallest sample size (3935) was performed to address concerns of different sequencing depths across samples affecting the rarefaction curves. Shannon diversity and Chao 1 richness indices were calculated from the sub-sampled OTU abundance data using QIIME.
The align_seqs.py and make_phylogeny.py commands in QIIME were used to align OTU consensus sequences18 and construct a taxonomic tree19. The weighted UniFrac and Bray-Curtis distances were calculated between participants and sample types for variation analysis using the beta diversity.py command in QIIME. Both methods are qualitative measures of community dissimilarity, however weighted UniFrac incorporates phylogenetic relatedness between OTUs. Principal coordinate analysis (PCoA), which employs an eigenvector-based approach, was performed with the QIIME suite to represent the multidimensional data of OTU abundance in three dimensions. OTU frequencies were compared across sample types and other groups at multiple taxonomic levels using the group_significance.py command from QIIME using Student’s t-test. The conservative Bonferroni method of multiple testing correction was followed, with each p-value being multiplied by the number of OTU/taxa tested.
RESULTS
Thirty-six participants were approached and consented to participate in this study between January 2015 and March 2016. One participant was also enrolled in the NIH funded observational Early Cystic Fibrosis study. Because the Early Cystic Fibrosis study is an observational study, we determined that participating would not influence our results. Sequences were successfully measured from all 72 samples.
The 36 participants included in the study consisted of 18 males (50%); had a median age of 3.3 years (interquartile range 1.3–5.2 years) (Table 1). The most common reason for bronchoscopy was chronic cough (61% of participants) followed by noisy breathing (14%) (Table 2). The most common underlying diagnosis was recurrent bronchitis/pneumonia (67%) followed by asthma (61%) and gastroesophageal reflux (22%) (Table 3). Of the 36 samples, 24 contained pooled BALF from 2–3 different lobes, while 12 samples were obtained from a single lobe. These 12 samples were obtained mainly from the middle (n=6) and lower lung lobes (n=5). One sample was obtained from the left upper lobe; none were obtained from the right upper lobe. All pooled samples contained fluid from the middle and/or lower lobes. Four pooled samples contained fluid from the right upper lobe as well as the middle or lower lobe. Four participants were on antibiotics at the time of bronchoscopy. Three of the four participants had positive BALF cultures despite this antibiotic use. The other participants denied antibiotic use for in the two weeks prior to the bronchoscopy. Rarefaction plots (supplemental Figure 1) revealed that Shannon diversity for all samples had stabilized by our chosen cutoff of 3935 reads, indicating all samples had sufficient sequencing depth to capture the diversity of the samples.
Table 1.
Demographics
| Gender | 50% Male (n=18) |
|---|---|
| Median Age (IQR) | 3.3 years (3 months – 18.3 years) |
| Race | Black/African American: 1 White/Caucasian: 29 Other: 3 No response: 3 |
| Ethnicity | Hispanic/Latino: 3 Non-Hispanic/Latino: 24 No response: 9 |
IQR: Interquartile range
Table 2.
Reason for Bronchoscopy.
| Reason for Bronchoscopy | Number of Participants with Diagnosis (n=36) |
|---|---|
| Chronic cough | 22 (61%) |
| Noisy breathing | 5 (14%) |
| Recurrent Wheeze | 4 (11%) |
| Recurrent Pneumonia | 4 (11%) |
| Chronic shortness of breath with recent diagnosis of systemic scleroderma | 1 (3%) |
Table 3.
Underlying Diagnoses prior to Bronchoscopy. Because some participants had more than one diagnosis, there are more diagnoses than participants.
| Underlying Diagnosis | Number of Participants with Diagnosis (n=36) |
|---|---|
| Recurrent Bronchitis/Pneumonia | 24 (67%) |
| Asthma | 22 (61%) |
| Gastroesophageal Reflux | 8 (22%) |
| Cystic Fibrosis | 3 (8%) |
| CFTR related metabolic disorder | 1 (3%) |
| Bronchopulmonary Dysplasia | 1 (3%) |
| Obstructive Sleep Apnea | 1 (3%) |
| Seizures | 1 (3%) |
| Scleroderma | 1 (3%) |
CFTR = Cystic Fibrosis Transmembrane Regulator protein.
Differences between NP swabs and BALF
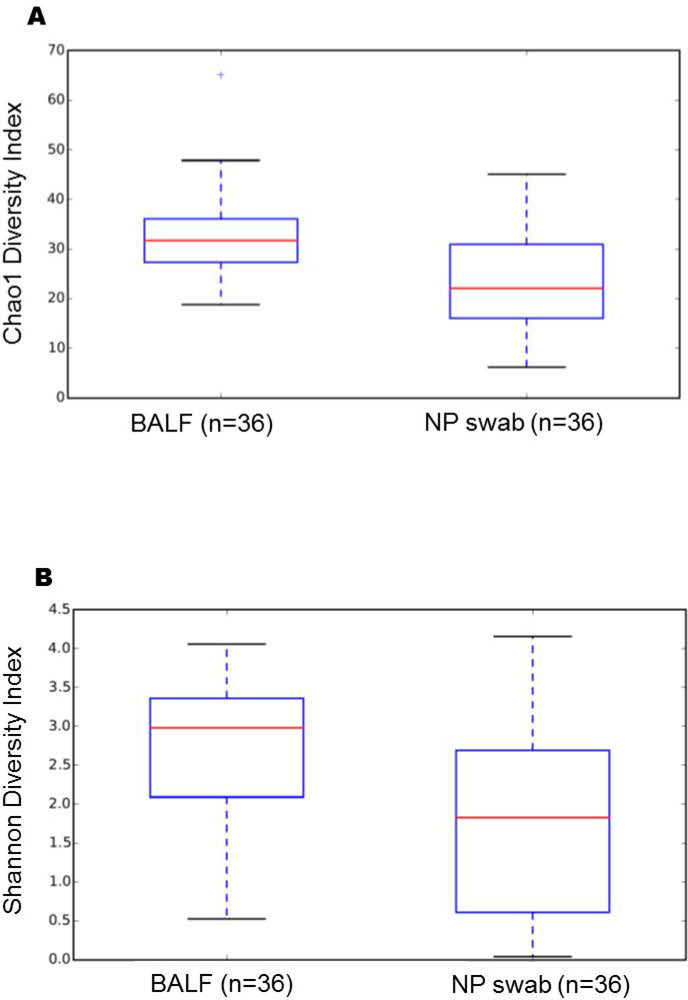
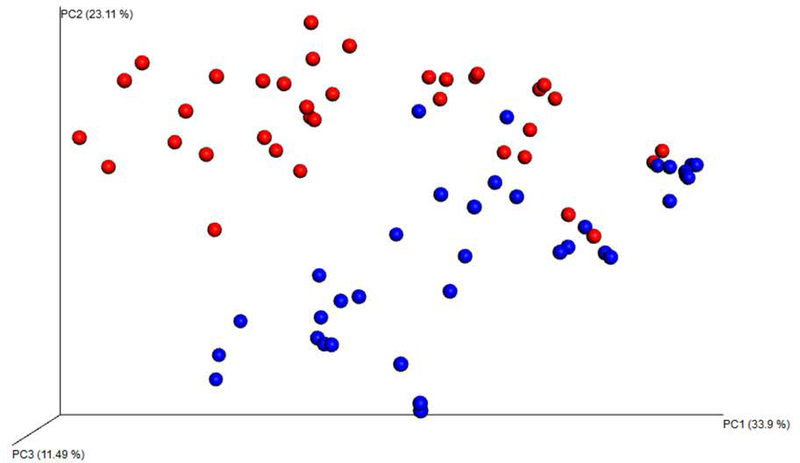
Chao1 richness (a qualitative lower-bounds of OTU richness) and Shannon diversity indices (a quantitative measure of OTU abundance and evenness) revealed that BALF samples are significantly richer (p = 0.001; nonparametric t-test) and more diverse (p = 0.001; nonparametric t-test) than NP swabs (Figure 1). Using principal coordinate analysis (PCoA) of the weighted Unifrac, distinct clustering patterns were present between the two sampling methods (Figure 2) with the top 3 axes accounting for 68.5% of the variation in samples. Monte Carlo permutation testing (n = 999) confirmed that the distance between BALF and NP samples was larger than the distance between BALF samples (adjusted p = 0.003) and the distance between NP samples (adjusted p = 0.003).
Figure 1.
Mean diversity between BALF and NP samples utilizing Chao1 Richness index p=0.001 (A), and Shannon Diversity p=0.001 (B).
Figure 2:
Weighted Unifrac Plot presents the multidimensional data of OTU abundance in three dimensions. It visually demonstrates that distinct clustering patterns were present between the two sampling methods. Red dots = BALF. Blue dots=NP swabs.
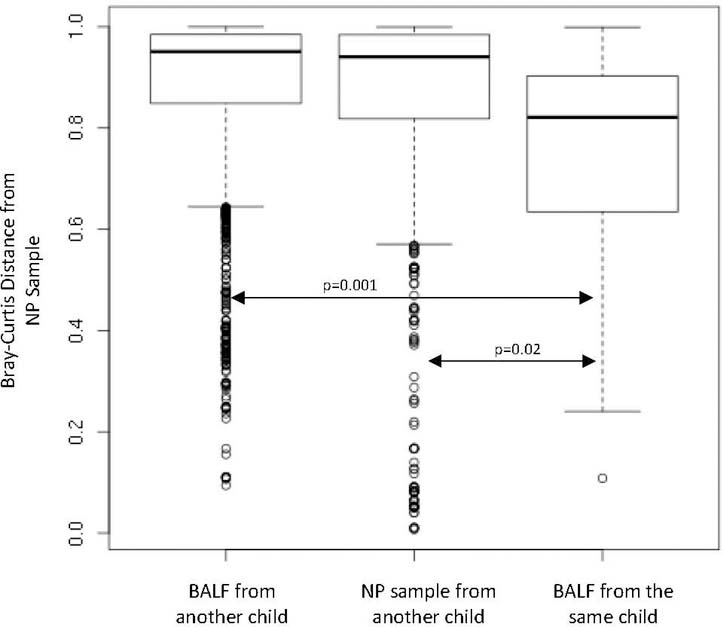
While overall differences were observed between NP and BALF samples, our primary goal was to determine if the NP and BALF samples were similar between each individual. During analysis, it appeared that within each individual, several OTUs were conserved across BALF and NP samples. To formally study this effect, we compared the dissimilarity between the microbial communities within individual subjects (intrasubject) to the dissimilarity across subjects (intersubject). For this test, we chose the Bray-Curtis distance metric to study the conservation of specific OTUs. Compared to NP (p = 0.02; t-test) and BALF samples (p = 0.001; t-test) obtained from other individuals, an individual’s NP sample more closely resembled their own BALF sample (Figure 3). The Bray-Curtis distance metric (utilized above) factors in the relative abundance of bacteria present in a sample. To determine if the presence or absence of bacteria differed between NP and BALF samples, we utilized Sorensen similarity calculations. Compared to unpaired NP/BALF samples, paired NP/BALF samples are more similar (p = 0.0006).
Figure 3:
Comparison of similarity between sample types. Microbiota detected in NP samples were more similar to their corresponding BALF (intra-subject comparison) compared to microbiota in BALF from other children (inter-subject comparison) (p=0.001). Intra-subject NP and BALF microbiota was also more similar than NP samples between children (p=0.02). Bray-Curtis dissimilarity is bounded between 0 and 1, so two samples with a distance of 1 would share no OTUs, where at 0 the samples would contain the same OTUs in the same proportionality.
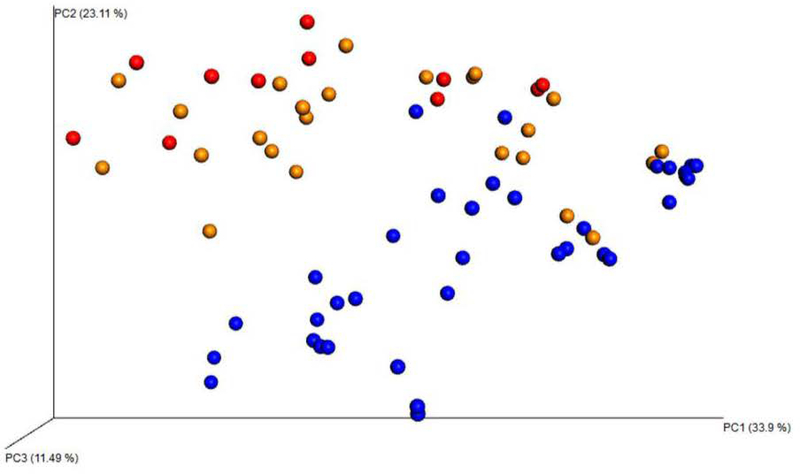
When adjusting for possible confounders such as age and method of scope introduction, no difference was found between the two sample types based on age or method of scope introduction (Figure 4).
Figure 4:
Weighted Unifrac Plot visually demonstrates that distinct clustering patterns were present between the three methods of bronchoscopy. Orange dots = nasal bronchoscopy. Red dots = bronchoscopy via laryngeal mask airway (LMA). Blue dots= NP swabs.
Relative Abundance of NP and BALF
Within the 36 NP samples, the dominant phyla detected were Firmicutes (54.7%); Actinobacteria (21%); Proteobacteria (20.4%); Bacteroidetes (3.2%); Fusobacteria (0.3%); and unclassified (0.4%). The 36 BALF samples were dominated by Firmicutes (48.6%); Bacteroidetes (29.5%); Proteobacteria (14.8%); Fusobacteria (2.9%); Actinobacteria (2.2%); Tenericutes (1.4%); and unclassified (0.6%). Sequences were annotated to the lowest available taxonomic classification.
The most abundant genera in the NP microbiota were Streptococcus (37.3%); Staphylococcus (14.6%); Haemophilus (10.1%); Corynebacteriaceae unknown genus (11.5%); Moraxella (7.7%); Corynebacterium (5.3%); Propionibacterium (3.9%); Porphyromonas (1.4%); Neisseria (1.4%); Veillonella (1.3%); and Neisseriaceae unknown genus (1%). In the BALF, Streptococcus was also the most abundant genera (36.1%); followed by Neisseria (10.3%); Prevotella 7 (9.4%); Alloprevotella (8.5%); Porphyromonas (7.3%); Staphylococcus (4%); Veillonella (3.8%); Prevotella (2.9%); Fusobacterium (2.5%); Haemophilus (1.7%); Propionibacterium (1.5%); Granulicatella (1.4%); Mycoplasma (1.4%); and Gemella (1%). All other classified OTUs belong to genera compromising less than 1% of the total abundance.
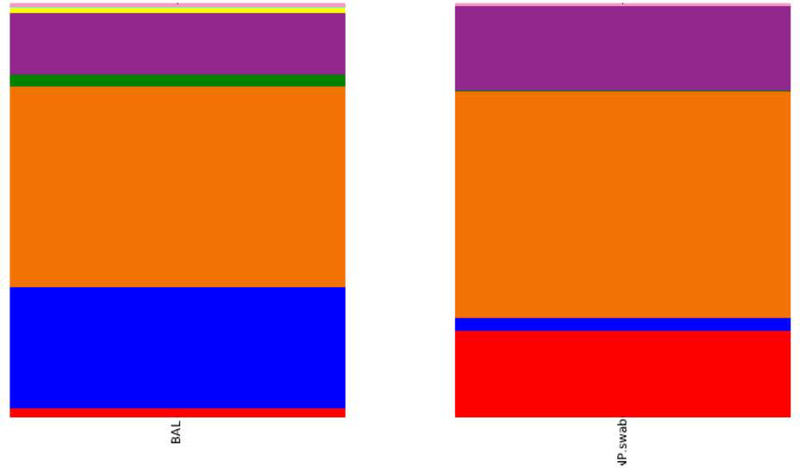
To further explore the differences between upper and lower airway microbiota, we compared the relative abundance of BALF and NP samples using the Kruskal-Wallis nonparametric test and found significant differences at the phyla level. While BALF samples had more Bacteroidetes (29.5% vs. 3.2%; adjusted p = 1.2 × 10−9), NP samples had more Actinobacteria (2.2% vs. 21%; adjusted p = 1.4 × 10−6) (Figure 5).
Figure 5:
BALF versus NP samples reported at phyla level. Purple= Proteobacteria; Orange-Firmicutes; Blue=Bacteroidetes; and red= Actinobacteria.
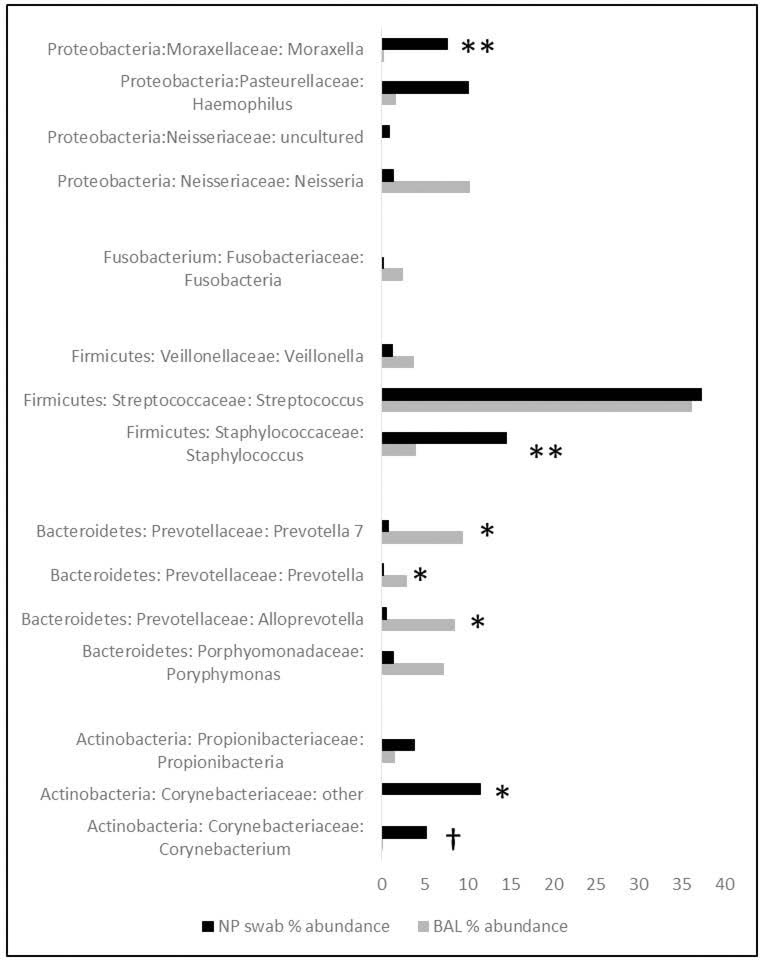
Further classification of the NP samples revealed higher abundance of bacteria within two genera of the Family Corynebacteriaceae. These include an unclassified genera (NP 11.5% vs. 0.04%; adjusted p = 8.9 × 10−5) (Figure 6) and Corynebacterium (NP 5.3% vs. 0.06%; adjusted p = 0.039). In addition, NP samples had a higher abundance of bacteria from the genus Staphylococcus (NP 14.6% vs. 4%; adjusted p = 0.002). In contrast, NP samples contained lower abundance of three genera within Family Prevotellaceae including: Prevotella 7 (NP 0.8% vs. 9.4%; adjusted p = 1.2 × 10−5), Prevotella (NP 0.2% vs. 2.9%; adjusted p = 5.6 × 10−7), and Alloprevotella (NP 0.6% vs. 8.5%; adjusted p = 3.4 × 10−6). At the genera level, Moraxella (BAL 0.2% vs. NP 7.7%, adjusted p = 0.003) and Haemophilus (BAL 1.7% vs. NP 10.1%, adjusted p = 1) abundance were higher in NP samples, while Streptococcus abundance was similar in both BALF and NP samples (NP 37.3% vs. 36.1%; adjusted p = 0.8).
Figure 6:
Bacterial Abundance of 15 most common OTUs. *adjusted p<0.001; **adjusted p<0.01; †adjusted p<0.05.
To determine if organisms detected in clinically obtained BALF cultures were also present in the BALF microbiota, we compared culture results with each participant’s corresponding BALF 16S rRNA sequencing results. BALF cultures were clinically performed in 34 of 36 samples. 22 of 34 samples (65%) were culture positive to at least one organism. In all 22 culture positive participants, the organisms present in their BALF culture were also present in the participant’s corresponding BALF microbiota. A correlation between the presence of bacteria in culture and the abundance of bacteria in the BALF microbiota was not detected.
DISCUSSION
When comparing the microbiota between BALF and NP samples in children undergoing clinically indicated bronchoscopy, NP swabs reflected BALF within an individual. However, when samples were grouped based on type (NP vs. BALF), differences were noted. These differences were likely due to averaging the relative abundance of multiple samples and suggests that caution should be made when making conclusions utilizing this method. Because intra-sample similarity was greater than inter-sample similarity, it is likely that differences will be detected between individuals regardless of the type of sampling method utilized.
Describing a ‘normal’ lung microbiota in children is difficult due to our inability to perform bronchoscopy on healthy children. Previous studies have utilized upper airway samples as a surrogate of lower airway samples. However, adult studies comparing upper and lower airway microbiota demonstrate differences between the two1,2,20, suggesting that upper airway samples do not represent the lower airway microbiota. Due to ethical concerns surrounding the collection of lower airway samples in healthy ‘normal’ children, samples were collected from children undergoing clinically indicated outpatient bronchoscopy (Table 2). While similar bacteria were found in both the upper and lower airway, other bacteria were only reported in one location. One such bacteria, Prevotella, was significantly more abundant in BALF than in NP samples. Similar findings have been reported in adult microbiota studies that examined oral swabs, BALF and gastric fluid2, suggesting that other locations (mouth, GI tract) may contribute to the microbial make-up of the lower airway. Furthermore, in children with cystic fibrosis, the relative abundance of Pseudomonas and Staphylococcus was higher in the lower airway compared to the upper airway, further suggesting that NP samples are not always good surrogates for the lower airway microbiota3,21. It was recently demonstrated that within young children with airway disease, combining OP and NP samples reflected BALF samples better than utilizing only OP or NP samples alone.5 It may be possible that if we had analyzed OP swabs as well, we would have seen Prevotella in higher quantities in the upper airway. Future studies have been planned to utilize both methods of upper airway sampling in our pediatric studies.
In our pediatric participants the most common genera in both BALF and NP samples was Streptococcus. Interestingly, the relative abundance of Streptococcus was similar in both locations. Bronchoscope contamination or carryover from the upper airway can occur and influence BALF samples. While Streptococcus abundance was similar between the two types of samples, there was a lack of similar abundance of other OTUs. Therefore, we believe upper airway contamination of lower airway samples did not occur in our population. To determine if the route of bronchoscope entry influenced the BALF results, we compared the two routes of performing bronchoscopy (through a laryngeal mask airway and trans-nasally) finding no difference in OTU richness (p = 0.41; nonparametric t-test) or community diversity (p = 0.17; non-parametric t-test) between the routes of bronchoscope introduction. Principal coordinate analysis of weighted Unifrac did not show distinct clusters for route of entry, and Monte Carlo permutation testing did not find a significant difference in the distance between laryngeal mask BALF and trans-nasal BALF samples as compared to the distance within laryngeal mask samples (adjusted p = 0.52 ) or trans-nasal samples (adjusted p = 0.21).
A limitation of this study may be an insufficient number of samples to measure significant differences in community structure, and differences based on age. Because power calculations for microbiota analysis have not been standardized, determining the number needed to reach significance can be challenging.. We divided samples into quartiles based on age in an attempt to examine if age impacted richness, diversity and/or changes in relative abundance but we did not find significant differences. A larger sample size would help further define and verify individual communities of bacteria across the various age groups thereby determining if borderline significant data is actually significant. While we attempted to standardize our sampling methods as much as possible, the procedures were clinically indicated; therefore, the route used to introduce the bronchoscope (laryngeal mask airway (LMA) or nasal) and which lung lobes to sample were selected by the performing physician. However, we collected the NP sample prior to bronchoscopy to limit possible contamination of lower airway bacteria on the upper airway during scope removal. Also, during analysis we compared BALF samples obtained via laryngeal mask airway (LMA) versus nasal entry and did not find a difference, suggesting that the route of entry did not impact sample analysis. Because we could not control which lobe(s) were sampled, BALF may only represent the area(s) of the lung sampled and not the entire lung. Our finding that diversity is greater in BALF than NP samples was previously reported in children with cystic fibrosis3. It is possible that the bacterial load differs between these two sampling methods thereby resulting in differences in diversity. Due to low sample volume, we were unable to perform qPCR on our samples to determine overall bacterial load. Future studies will focus on measuring bacterial load. Contaminating DNA can be a challenge in samples with low microbial biomass22 therefore we extracted DNA from controls and excluded OTUs detected in our negative controls (supplemental Table 1) to prevent reporting bacterial contaminants.
In summary, we have performed a comparison of NP swabs and BALF in a pediatric population undergoing clinically indicated bronchoscopies. We report that the pediatric NP airway microbiota is similar to BALF samples for an individual but often have significantly different relative abundance of bacteria. Studies utilizing NP samples as surrogates for the lower airway, should be interpreted with caution.
Supplementary Material
Supplemental Table 1: OTUs removed because suspected they were contaminants due to presence in negative controls.
Supplemental Figure 1. Rarefaction plots reveal that Shannon diversity for all samples stabilized by our chosen cutoff of 3935 reads, indicating all samples had sufficient sequencing depth to capture the diversity of the samples. It also ensures high reproducibility and repeatability which is mostly attained through robust amplification.
Acknowledgments
We greatly appreciate the efforts put forth by our bronchoscopy coordinator, Kristine Denny, to ensure we were able to obtain the BALF used in these analyses. We would like to acknowledge the cooperation of the pediatric pulmonologists and anesthesiologists within our institution for allowing us to procure samples before and during the procedure. Finally, we thank the patients for participating in this study.
The following grants supported this research:
K-12 Indiana University School of Medicine (IUSM) Indiana Pediatric Scientist Award (IPSA) program through the Child Health Research Career Development Award (CHRCDA). 1K12HD068371–01A1
Children’s Clinical Research Center, a member of the Indiana Clinical and Translational Sciences Institute supported by grant UL1TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Lilly Physician Scientist Initiative Award
Footnotes
Author Disclaimers: none
References
- 1.Dickson RP, et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Annals of the American Thoracic Society 12, 821–830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassis CM, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 6, e00037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prevaes SM, et al. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. The European respiratory journal 49(2017). [DOI] [PubMed] [Google Scholar]
- 4.Rogers GB, et al. Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. Journal of clinical microbiology 44, 2601–2604 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh RL, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 4, 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faro A, et al. Official American Thoracic Society technical standards: flexible airway endoscopy in children. American journal of respiratory and critical care medicine 191, 1066–1080 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Lozupone C, Hamady M, Bushman FD & Knight R Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic acids research 35, e120 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fierer N, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proceedings of the National Academy of Sciences of the United States of America 109, 21390–21395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC, Haas BJ, Clemente JC, Quince C & Knight R UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Garrity GM, Tiedje JM & Cole JR Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahe F, Rognes T, Quince C, de Vargas C & Dunthorn M Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2, e593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahe F, Rognes T, Quince C, de Vargas C & Dunthorn M Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ 3, e1420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. Journal of molecular biology 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic acids research 35, 7188–7196 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price MN, Dehal PS & Arkin AP FastTree 2--approximately maximum-likelihood trees for large alignments. PloS one 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris A, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. American journal of respiratory and critical care medicine 187, 1067–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zemanick ET, et al. Assessment of airway microbiota and inflammation in cystic fibrosis using multiple sampling methods. Annals of the American Thoracic Society 12, 221–229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC biology 12, 87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: OTUs removed because suspected they were contaminants due to presence in negative controls.
Supplemental Figure 1. Rarefaction plots reveal that Shannon diversity for all samples stabilized by our chosen cutoff of 3935 reads, indicating all samples had sufficient sequencing depth to capture the diversity of the samples. It also ensures high reproducibility and repeatability which is mostly attained through robust amplification.