Abstract
Esophageal adenocarcinoma (EAC) is a molecularly heterogeneous disease that is rising rapidly in incidence and has poor prognosis. We developed a heterobivalent peptide to target detection of early Barrett’s neoplasia by combining monomer heptapeptides specific for either EGFR or ErbB2 in a heterodimer configuration. The structure of a triethyleneglycol linker was optimized to maximize binding interactions to the surface receptors on cells. The Cy5.5-labeled heterodimer QRH*-KSP*-E3-Cy5.5 demonstrated specific binding to each target and showed 3-fold greater fluorescence intensity and 2-fold higher affinity compared with either monomer alone. Peak uptake in xenograft tumors was observed at 2 hours post-injection with systemic clearance by ~24 hours in vivo. Furthermore, ligand binding was evaluated on human esophageal specimens ex vivo, and 88% sensitivity and 87% specificity were found for detection of either high-grade dysplasia (HGD) or EAC. This peptide heterodimer shows promise for targeted detection of early Barrett’s neoplasia in clinical study.
INTRODUCTION
Esophageal adenocarcinoma (EAC) is a deadly cancer that is rising in incidence rapidly, and is associated with a poor prognosis and low 5-year survival.1 Barrett’s esophagus (BE) represents a metaplastic transformation of squamous (SQ) into specialized columnar epithelium in response to long-standing acid and bile reflux.2 BE is known to transform stepwise through low-grade dysplasia (LGD) and high-grade dysplasia (HGD) before developing into EAC. HGD is a pre-malignant condition that provides a window of opportunity for intervention with either curative resection or ablation therapy.3 Therefore, new methods for accurate detection of HGD and early EAC is needed.4 Conventional surveillance using white light illumination with random four-quadrant biopsy has been found to have limited effectiveness for localizing dysplasia that is flat in appearance, focal in extent, and patchy in distribution.5 Chemoprevention with nonsteroidal anti-inflammatory drugs, such as COX-2 inhibitors, has been limited by unacceptable side effects.6
The ability to identify expression of molecular targets can be used to diagnose, stage, and classify tumors, and to monitor response to therapy. Gene expression of EAC is heterogeneous,7 and strategies that focus on a single target are limited. Epidermal growth factor receptor (EGFR) and ErbB2 have been found to be high-frequency gene amplified and overexpressed in HGD and early EAC.8,9 These receptor tyrosine kinases have been validated as cancer biomarkers and function to stimulate epithelial cell growth, proliferation, and differentiation.10 Emerging evidence supports early expression in progression of BE to EAC when intervention can improve patient outcome.11 In addition, targets located on the cell surface are well suited for development of either diagnosis or therapy. Minimal overlap in expression has been found in studies of surgically resected HGD and EAC specimens.12 These observations motivate a need for a combined approach that detects multiple targets concurrently.
Multivalent ligands generate synergistic effects that can increase their affinity, avidity, selectivity, and potency by binding multiple targets concurrently.13–17 These properties can enhance in vivo diagnostic imaging performance by improving target-to-background (T/B) ratio and detecting targets at lower levels of expression.18 Also, this strategy can be effective for therapy by connecting different cell signaling pathways.19 The ability to interact with multiple targets simultaneously may reduce acquired resistance that arises with use of a single target.20 This heterobivalent targeting strategy was first tested as a proof-of-principle study on receptors of human melanocortin-4 and δ-opioid in vitro21 and other G protein-coupled receptors (GPCR).22–24 Peptides specific for αvβ3 integrin and gastrin-releasing peptide receptor (GRPR) have been arranged in a heterodimeric configuration and radiolabeled with 18F for PET imaging.25–27 Recently, the heterodimeric peptide 68Ga-BBN-RGD was evaluated for prostate cancer diagnosis and clinically translated.28 A number of bispecific antibodies have been developed for multivalent targeting,29 however effectiveness may be limited by poor tumor uptake, immunogenicity, and high manufacture costs.30–31 We have previously identified and validated heptapeptide monomers specific for EGFR and ErbB2.32,33 In this study, we arrange heptapeptide monomers in a heterodimer configuration, and present the first heterodimeric peptide that targets EGFR and ErbB2. This bivalent probe was screened using variable length linkers to optimize binding efficiency and was labeled with a near-infrared dye. Further validation was conducted in an OE33 tumor-bearing mouse model in vivo, and with antibodies for binding to HGD and EAC found in human esophageal specimens.
RESULTS
Heterodimeric peptide configuration
The heptapeptide sequences QRHKPRE (green) and KSPNPRF (purple) were synthesized to bind specifically to EGFR and ErbB2, respectively.32,33 The fluorophore Cy5.5 (red) was attached to the C-termini via a GGGSK linker (blue), hereafter QRH*-Cy5.5 and KSP*-Cy5.5, respectively, Figure 1. Cy5.5 was chosen because of good photostability and high quantum yield in the near-infrared (NIR) spectrum.34 The linker separates the peptide and fluorophore to prevent steric hindrance. Purity >95% by analytical HPLC was achieved for both monomers, and an experimental mass-to-charge (m/z) ratio of 1794.98 and 1900.04 was measured by mass spectrometry that agrees with expected values, Figure S1A,B. The monomers were combined in a heterodimer configuration by introducing a linker with either a variable PEG spacer, ranging in size from 17–74 atoms, or a hydrophobic 6-aminohexonic acid linker in the α-, ξ- amino groups of the second lysine attached to Cy5.5 (red), Figure 1. The linker length was varied to determine the optimal spacing between monomers to maximize binding interactions to EGFR and ErbB2.
Figure 1. Design of peptide heterodimer.
Chemical structures for heptapeptide monomers QRHKPRE (QRH*) and KSPNPRF (KSP*) are shown along with arrangement of heterodimer configuration with variable length linkers.
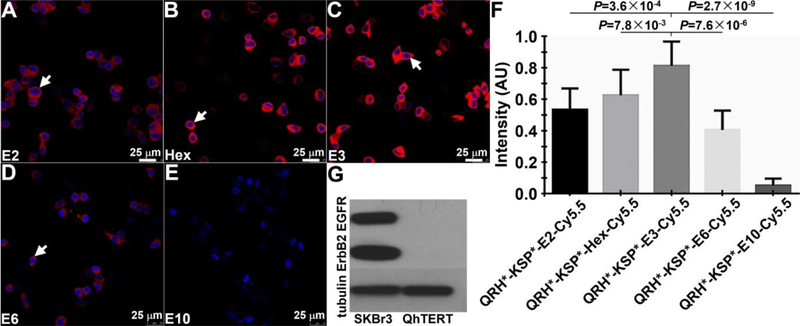
The fluorescence spectra of the Cy5.5-labeled peptide monomers and optimized heterodimer revealed peak fluorescence absorbance and emission at λ = 680 and 708 nm, respectively, Figure S2A,B. On confocal microscopy, the strength of binding of each candidate heterodimer with different length linkers to the surface (arrow) of SKBr3 cells is shown, Figure 2A–E. These cells are well-known to express EGFR and ErbB2.35 The mean intensity for each heterodimer was quantified, Figure 2F. The heterodimer QRH*-KSP*-E3-Cy5.5 with the triethyleneglycol linker (E3) showed highest signal, and was found to have good hydrophilicity, as well. The signal decreased dramatically with longer linkers because of either increased flexibility or internal entropy. Aminohexonic acid linkers can also be used to provide comparable binding affinities, however their decreased hydrophilicity as compared with PEG linkers may be more challenging for clinical translation. A Western blot of the cells is shown, Figure 2G.
Figure 2. Optimization of peptide heterodimer.
On confocal microscopy, fluorescence from binding of candidate heterodimer with linkers A) E2, B) Hex, C) E3, D) E6, and E) E10 to the surface (arrow) of SKBr3 cells can be seen. F) Quantified results show that the E3 linker provides the highest mean fluorescence intensity. P-values were determined using unpaired t-test. Measurements are an average of 10 randomly chosen cells from 4 images collected independently. G) Western blot shows EGFR and ErbB2 expression in SKBr3 and QhTERT cells.
We also evaluated heterodimer binding using structural models for EGFR (1IVO) and ErbB2 (2A91), and found that the triethyleneglycol linker (E3) provides spacing for the heterodimer QRH*-KSP*-E3-Cy5.5 to bind to domain 2 of EGFR and domain 3 of ErbB2, Figure S3A–C. The model suggests that a linker length of ∼60 Å is needed for the heterodimer to conform to the spacing between these binding domains. We then synthesized QRH*-KSP*-E3-Cy5.5 with >95% purity by analytical HPLC, and measured an experimental mass-to-charge (m/z) ratio by mass spectrometry of 2974.69 that agreed with the expected value, Figure S1C. The spectra revealed no change in peak values after arranging the peptide monomers in this heterodimeric configuration.
Stability
The stability of the peptide heterodimer in PBS containing 0.1% BSA at pH 7.4 and 37°C was monitored by HPLC at 0.5, 2, 6 and 12 hours, Figure S4A. No noticeable degradation was observed. We also monitored enzymatic stability by incubating the heterodimer at a concentration of 30 μM in mouse serum at 37°C at 0.5, 1.0, 1.5 and 2 hours of incubation, Figure S4B. We observed a new peak formed that may arise from degradation and at 2 hour the area of new formed peak is about 20%.
Validation with confocal microscopy
On confocal microscopy, strong binding by the peptide heterodimer (red) to the surface (arrow) of OE33 human esophageal adenocarcinoma cells was observed, Figure S5A. By comparison, we visualized much less fluorescence intensity with either monomer, Figure SB,C. Minimal binding was observed for all peptides to QhTERT human non-dysplastic Barrett’s esophagus cells (control) that do not express either EGFR or ErbB2, Figure S5D–F. Quantified results show a greater signal for the heterodimer compared with that of the individual monomers combined, Figure S5G. Western blot is shown, Figure S5H. These results support effective use of peptide heterodimers at much lower concentrations than that of monomers.
siRNA knockdown
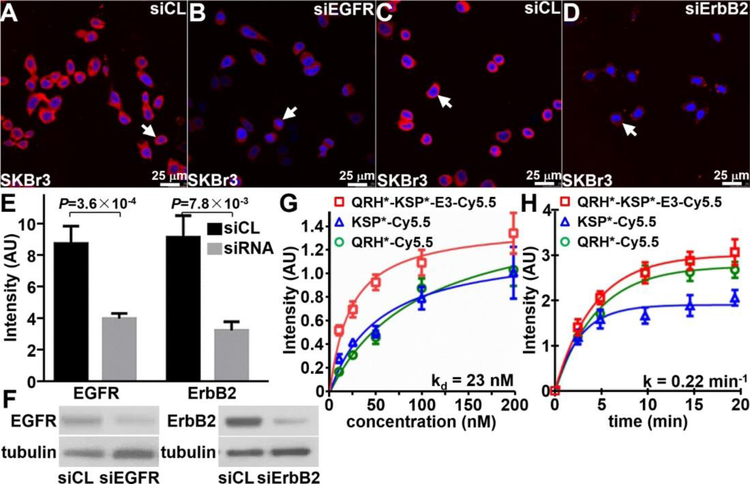
The siRNA knockdown experiments were performed to validate specific binding of the heterodimer to either EGFR or ErbB2 using SKBr3 cells. On confocal microscopy, QRH*-KSP*-E3-Cy5.5 (red) binds strongly to the surface (arrow) of cells transfected with siCL non-targeting siRNA (control), Figure 3A. Reduced fluorescence intensity was seen with knockdown cells transfected with siEGFR targeting siRNA, Figure 3B. Similar results were found with the heterodimer and monomer for ErbB2, Figure 3C,D. The fluorescence intensities were quantified, and the differences were found to be significant, Figure 3E. Western blot is shown for control and knockdown SKBr3 cells, Figure 3F.
Figure 3. Characterization of peptide heterodimer.
In siRNA knockdown experiments, QRH*-KSP*-E3-Cy5.5 (red) shows significantly greater binding to the surface (arrows) of A) siCL (control) SkBr3 cells compared with that for B) siEGFR (knockdown) cells. Similar results were found for C) siCL and D) siErbB2 (knockdown) cells. E) Quantified results show significantly greater intensity for siCL versus siEGFR and siCL versus siErbB2, P=3.6×10−4 and P=7.8×10−3, respectively, by unpaired t-test. The mean value was calculated from 5 cells chosen randomly from 3 images collected independently. F) Western blot shows EGFR and ErbB2 expression in control and knockdown cells. G) The apparent dissociation constant (binding affinity) for QRH*-KSP*-E3-Cy5.5 was found to be kd = 23 versus 98 and 54 nM for QRH*-Cy5.5 and KSP*-Cy5.5. H) The apparent association time constant for QRH*-KSP*-E3-Cy5.5 was found to be k = 0.22 min−1 (4.5 min) versus 0.21 min−1 (4.8 min) and 0.35 min−1 (2.9 min) for QRH*-Cy5.5 and KSP*-Cy5.5. Results for each measurement are representative of 3 independent experiments.
Binding properties
The binding parameters of the peptide heterodimer were measured using flow cytometry with OE33 cells, and an apparent dissociation constant of kd = 23 nM was measured, Figure 3G. This result is much improved compared with that of kd = 98 or 54 nM for peptide monomers QRH*-Cy5.5 or KSP*-Cy5.5, respectively. The apparent association time constant of k = 0.22 min−1 (4.5 min) was measured for the heterodimer compared with k = 0.21 min−1 (4.7 min) and 0.35 min−1 (2.8 min) for the monomers QRH*-Cy5.5 and KSP*-Cy5.5, respectively, Figure 3H.
Co-localization
Co-localization experiments were performed to further validate specific binding of the peptide heterodimer to either EGFR or ErbB2. On confocal microscopy, QRH*-KSP*-E3-Cy5.5 (red) binds strongly to the surface (arrow) of SKBr3 cells, Figure S6A. Binding by AF568-labeled anti-EGFR (yellow) and AF488-labeled anti-ErbB2 (green) to the surface (arrows) of the same cells are shown for comparison, Figure S6B,C. We measured a Pearson’s correlation coefficient of ρ = 0.77 and 0.65 between the heterodimer and the anti-EGFR and anti-ErbB2 antibodies, respectively, Figure S6D,E. The DAPI image shows the locations of the cell nuclei, Figure S6F.
Internalization
We collected confocal images over time to demonstrate heterodimer internalization. From 1–4 min, QRH*-KSP*-E3-Cy5.5 accumulates on the surface of SKBr3 cells, Figure S7A–C. At 8 min, internalization begins, and reaches near completion at 40 min, Figure S7D–G. DAPI images show cell nuclei location, Figure S7H–N. The spatial distribution of the ligand can be appreciated on merged images, Figure S7O–U. Ligand-mediated endocytosis can result in signal amplification, and supports use for mediating targeted therapy.
Effect on cell signaling
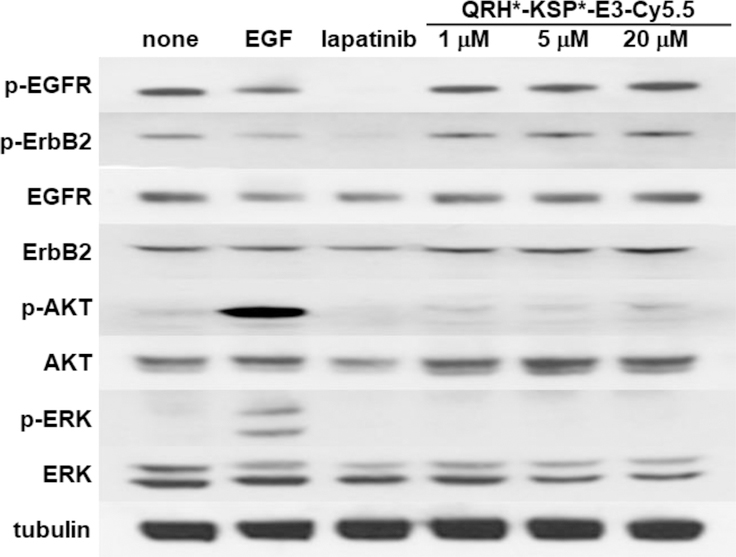
The effect of heterodimer binding on downstream signaling was evaluated in SKBr3 cells. Western blot shows no change in phosphorylation of either EGFR (p-EGFR) or ErbB2 (p-ErbB2) with addition of up to 20 μM of QRH*-KSP*-E3-Cy5.5, Figure 4. Similarly, no downstream change was observed in AKT (p-AKT) and ERK (p-ERK). By comparison, strong phosphorylation activity of AKT (p-AKT) and ERK (p-ERK) was observed with addition of EGF used as a positive control. Also, the addition of lapatinib, a tyrosine kinase inhibitor known to disrupt ErbB2 signaling in solid tumors, showed reduced expression of p-ErbB2, p-AKT, and p-ERK.
Figure 4. No effect of peptide heterodimer on cell signaling.
We evaluated the effect of the QRH*-KSP*-E3-Cy5.5 on downstream cell signaling after binding to SKBr3 cells. On Western blot, we observed no change in phosphorylation of EGFR (p-EGFR), ErbB2 (p-ErbB2) or of downstream AKT (p-AKT) and ERK (p-ERK) with incubation of heterodimer at 1, 5, and 20 μM. By comparison, the addition of EGF, an endogenous ligand for EGFR, showed increased expression of p-AKT and p-ERK. The addition of 100 nM of lapatinib, a tyrosine kinase inhibitor known to interrupt EGFR/ErbB2 signaling in solid tumors, showed reduced expression of p-EGFR, p-ErbB2 and p-AKT. Cells treated with 1% DMSO and untreated cells showed no suppression of EGFR and ErbB2 mediated signaling. β-tubulin is used as loading control.
Pharmacokinetics
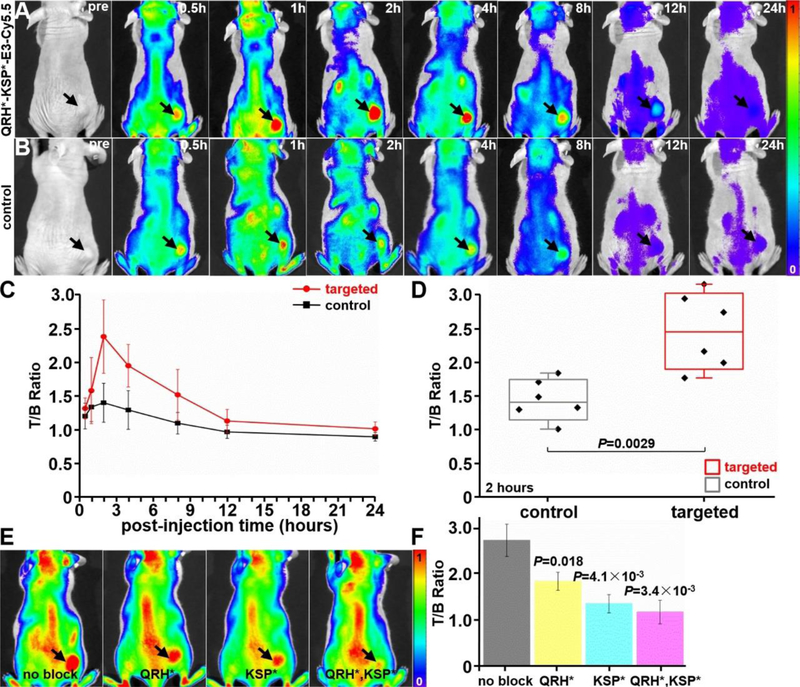
The heterodimer (300 μM, 150 μL) was injected via tail vein to evaluate in vivo uptake by OE33 xenograft tumors. Fluorescence images were collected before peptide injection to assess baseline, and then at various time points up to 24 hours when the probe cleared, Figure 6A. A set of images was also collected with a Cy5.5-labeled control peptide, Figure 6B. Peak intensity was observed at 2 hours post-injection, and the mean result for the targeted peptide was significantly greater than that for control, Figure 6C,D. We also performed blocking studies by injecting unlabeled QRH*, KSP*, and both monomers together prior to measuring the fluorescence intensity from the tumor (arrow) at 2 hours post-injection, Figure 6E. A significant reduction in signal was observed for each condition, Figure 6F.
Figure 6. Biodistribution of peptide heterodimer.
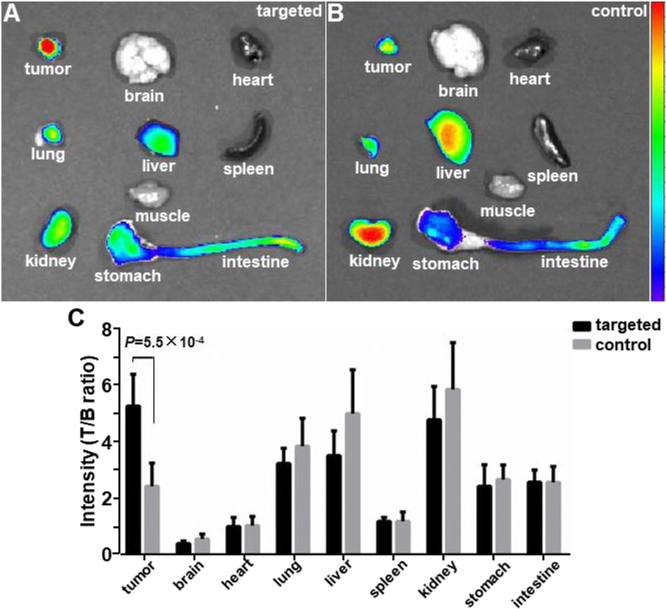
Representative fluorescence images are shown from excised mouse organs at ~2 hours post-injection of A) targeted peptide heterodimer QRH*-KSP*-E3-Cy5.5 and B) control peptide (GGGAGGG)2KK-Cy5.5. C) Fluorescent signals were quantified from individual organs (n = 6) for each peptide. The mean fluorescence intensity for targeted was significantly greater than that for control, 5.28±1.14 versus 2.24±0.82, P=5.5×10−4 by unpaired t-test.
Biodistribution
Heterodimer biodistribution was evaluated in nude mice bearing OE33 xenograft tumors following systemic injection (300 μM, 150 μL PBS) of either targeted peptide QRH*-KSP*-E3-Cy5.5 (n=6) or control (GGGAGGG)2KK-Cy5.5 (n=6). The mice were sacrificed 2 hours later, and heterodimer uptake was found to be significantly higher in tumors than in other organs, including 3-fold higher than in muscle, Figure 7A. High uptake was also visualized in kidney to support renal clearance. Significantly less intensity was observed in tumor using the control peptide, Figure 7B. Quantified results show a significantly greater mean value in tumor for the heterodimer versus control, Figure 7C. Overexpression of EGFR and ErbB2 was validated in sections of OE33 xenograft tumors ex vivo with immunohistochemistry (IHC), Figure S8, and no evidence of acute toxicity was found on histology (H&E), Figure S9.
Figure 7. Specific binding of peptide heterodimer to Barrett’s neoplasia.
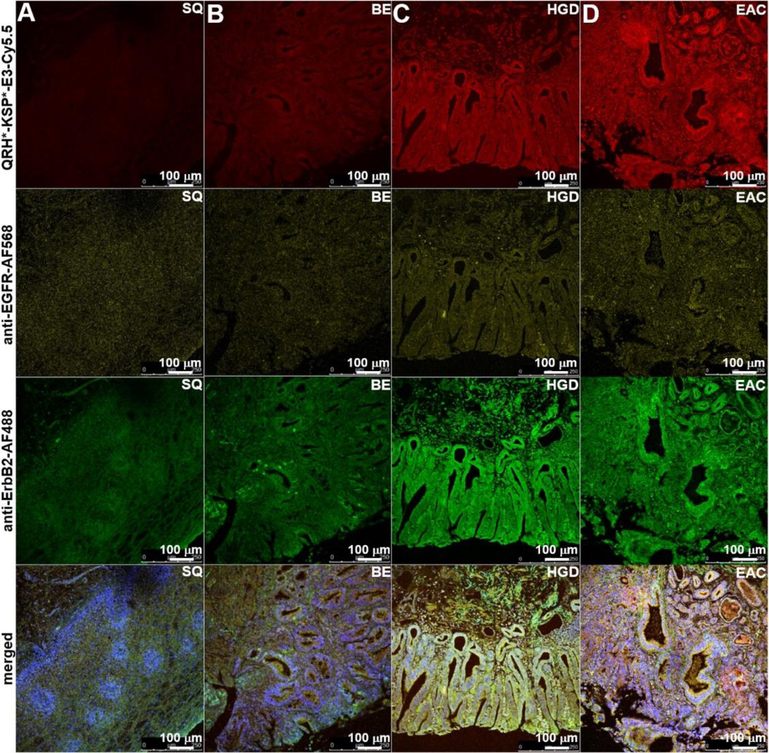
On representative confocal microscopy images of human esophageal specimens ex vivo, QRH*-KSP*-E3-Cy5.5 (red) shows minimal staining to A) squamous (SQ) and B) Barrett’s esophagus (BE), and increased intensity with C) high-grade dysplasia (HGD) and D) esophageal adenocarcinoma (EAC). Similar results were found with AF568-labeled anti-EGFR antibody (yellow) and for AF488-labeled anti-ErbB2 antibody (green). Merged images show co-localization of peptide and antibody binding.
Validation with human esophageal specimens
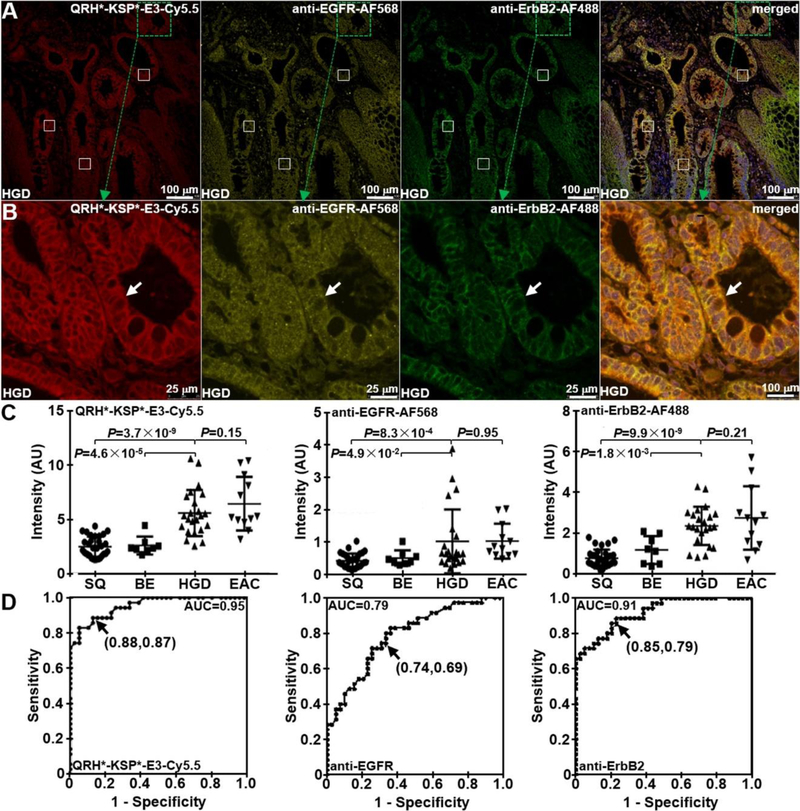
On confocal microscopy, specific heterodimer binding was validated to human specimens of Barrett’s neoplasia ex vivo. We observed minimal fluorescence intensity with SQ and BE, and increased signal with HGD and EAC, Figure 7A–D. The same set of specimens was also stained with AF568-labeled anti-EGFR antibody (yellow), and AF488-labeled anti-ErbB2 antibody (green). Co-localization of heterodimer and antibody binding can be appreciated on the merged images. Fluorescence intensities were measured from a set of 3 boxes with dimensions of 20×20 μm2 placed within random crypts of heterodimer and antibody images, Figure 8A. At higher magnification, cell surface binding (arrows) can be appreciated, Figure 8B. Co-localization of binding was evaluated between the heterodimer QRH*-KSP*-E3-Cy5.5 (red) and the anti-EGFR-AF568 (yellow) and anti-ErbB2-AF488 (green) antibodies, and a Pearson’s correlation coefficient of ρ = 0.60 and 0.75, respectively, was measured.
Figure 8. Co-localization of peptide heterodimer and antibody binding to Barrett’s neoplasia.
A) On confocal microscopy, serial sections of HGD in human esophageal specimens are shown following staining with QRH*-KSP*-E3-Cy5.5 (red), anti-EGFR antibody labeled with AF568 (yellow) and anti-ErbB2 antibody labeled with AF488 (green). Fluorescence intensities were quantified from the mean of a set of 3 boxes with dimensions of 20×20 μm2 placed over random crypts. Co-localization of binding can be appreciated on the merged image. B) High-magnification images are shown from dashed boxes. On the merged image, Pearson’s correlation coefficient of ρ = 0.60 and 0.75 was measured for EGFR and ErbB2, respectively. C) From n = 31, 8, 23, and 12 specimens of SQ, BE, HGD, and EAC, respectively, we found significantly greater mean fluorescence intensity from HGD and EAC compared with that for BE and SQ with QRH*-KSP*-E3-Cy5.5, the P-value for difference are calculated by Tukey’s multiple comparisons. A similar result was found for anti-EGFR-AF568 and anti-ErbB2-AF488. D) ROC curve shows 88% sensitivity, 87% specificity and 0.95 AUC with QRH*-KSP*-E3-Cy5.5; 74% sensitivity, 69% specificity, and 0.79 AUC with QRH*-Cy5.5, and 85% sensitivity, 79% specificity, and 0.91 AUC with KSP*-Cy5.5.
We measured the mean (±SD) fluorescence intensity for the heterodimer from n = 31, 8, 23, and 12 human specimens of SQ, BE, HGD, EAC, respectively, Figure 8C. The result for HGD was significantly higher than that for BE and SQ. Similar findings were found with the anti-EGFR and anti-ErbB2 antibodies. From receiver-operator characteristic (ROC) curves, we found 88% sensitivity and 87% specificity for the heterodimer to detect Barrett’s neoplasia (HGD and EAC) at a T/B ratio of 5.9 when compared with pathology, Figure 8D. The sensitivity and specificity for the heterodimer were higher than that for either antibody.
DISCUSSION AND CONCLUSIONS
Progression of BE to EAC is characterized by high molecular heterogeneity that has limited the usefulness of conventional ligands that recognize a single target. Here, we demonstrate a peptide heterodimer that binds either EGFR or ErbB2 concurrently for improved performance by matching the linker length in the heterodimer with the mean distance between the extracellular binding domains of either target. We found that QRH*-KSP*-E3-Cy5.5 with triethyleneglycol (PEG3) consisting of 30 atoms as the linker produced 3-fold greater fluorescence intensity from binding to OE33 cells than either monomer alone. On immunofluorescence, this heterobivalent probe provided higher sensitivity and specificity for detection of Barrett’s neoplasia in human esophageal specimens by comparison with that for known monoclonal antibodies to either target. The in vivo imaging results in OE33 xenograft tumors showed peak uptake in ~2 hours and systemic clearance within ~24 hours to provide pharmacokinetic parameters that are compatible with diagnostic imaging. No evidence of acute toxicity was found on pathology. This combination strategy for target binding is promising for developing novel diagnostic and therapeutic agents for clinical use to manage this heterogeneous disease.
Bi-specific ligands are promising for their potential to improve in vivo binding performance. By covalently linking two unique peptide sequences, multivalent ligand-target interactions were generated to improve binding affinity by >2-fold. This synergy is reflected by the mean fluorescence intensity for heterodimer binding to OE33 cells being greater than the sum of that for the two individual monomers, allowing for targets to be detected at lower levels of expression. With the heterodimer, we achieved 88% sensitivity and 87% specificity for detection of either HGD or EAC, a result that was better than that for the monoclonal antibody to either target. Higher sensitivity results from detection of two independent targets, and greater specificity arises from dual binding to a larger combined target epitope by comparison to the monomer.36 High T/B ratio was achieved using Cy5.5 to emit NIR fluorescence, which is least affected by hemoglobin absorption, tissue scattering, and autofluorescence background,37 and provides maximum tumor imaging depth.
To our knowledge, this is the first bivalent heterodimer demonstrated for targeting of Barrett’s neoplasia. Peptides offer a number of advantages over bulky antibodies for in vivo targeting.38 Their small size and low molecular weight facilitate greater vascular permeability, enhanced tissue diffusion, and deeper tumor penetration. Peptides have less potential for immunogenicity than antibodies, allowing for repetitive use.39 In the future, this dual targeting agent will undergo rigorous pharmacology/toxicology studies and will be synthesized under Good Manufacturing Practice (GMP) conditions for FDA approval to be used in human clinical studies with fluorescence endoscopy. This combination approach greatly simplifies the process for FDA review by comparison with that for two ligands separately. Development of bivalent peptides can be performed at a substantially lower cost than that for other protein based agents separately to be used in combination therapy. Established peptide synthesis processes are easy to scale up for mass manufacture with reproducible results.40 By recognizing two different cell surface targets that are validated EAC biomarkers, enhanced diagnostic and therapeutic effects can be achieved as compared with ligands that bind only one target.
EXPERIMENTAL SECTION
Cells, media and chemicals
All cells were maintained at 37°C and 5% CO2 and were supplemented with 10% FBS and 1% penicillin/streptomycin. Penicillin/streptomycin was omitted for the siRNA knockdown studies, and FBS was omitted in keratinocyte-SFM media. Human SKBr3 breast cancer cells were obtained from the American Type Culture Collection (ATCC) and cultured in McCoy’s 5A media. Human OE33 and QhTERT esophageal cells were cultured with Roswell Park Memorial Institute (RPMI) 1640 media and keratinocyte-SFM media (Gibco), respectively. Cells were passaged using 0.25% EDTA containing trypsin (Mediatech), and a hemocytometer was used to count cell number. Peptide synthesis reagents (Nα-Fmoc protected amino acids, HBTU and HOBt) were purchased from either Anaspec or AAPPTec. The side-chain protecting groups used the following amino acids: Arg(Nω-Pbf), Asn(Nγ-Trt), Glu(O-tBu), His(Nim-Trt), Ser(tBu), Lys(Nε-Alloc), Lys(Nε-Ivdde). Fmoc-AEA, Fmoc-AEEA, Fmoc-AE3A, Fmoc-AE6A Fmoc-AE10A and 6-aminohexanoic acid were purchased from AAPPTec. These reagents are analytical grade with >99% purity, and were used without further purification. Rink amide MBHA resin with initial loading of 0.2 mmol/g was acquired from Protein Technologies, Inc. Cy5.5-NHS ester was acquired from Lumiprobe. Analytical grade solvents for peptide synthesis and HPLC were purchased from Fisher Scientific, and were used without further purification unless otherwise stated.
Peptide monomers and heterodimer
Peptide monomers and heterodimers were synthesized and labeled with Cy5.5 using standard Fmoc solid-phase chemical synthesis with rink amide MBHA resin in a PS3 automatic synthesizer (Protein Technologies Inc). Fmoc protected L-amino acids were applied with standard HBTU/HOBt activation. The C-terminus lysine was incorporated as Fmoc-Lys(Alloc)-OH for fluorophore labeling. Upon completion of peptide assembly, the resin was transferred to a reaction vessel for manual labeling with dye. The Alloc side chain protecting group was removed with a palladium catalyst. The resin was washed with dimethylformamide (DMF) and dichloromethane (DCM) for 1 min 3X. The protected resin-bound peptide was reacted with Cy5.5-NHS ester and (N,N-Diisopropylethylamine) DIPEA for 12 hours. The completion of the reaction was monitored with a qualitative ninhydrin test. A cleavage cocktail reagent TFA: TIS: H2O (95:2.5:2.5 v/v/v) was mixed with the resin, and stirred for 2 hours in dark conditions at 25°C. The crude peptides were isolated from the resin by filtration and evaporated with N2 gas followed by precipitation with chilled diethyl ether and stored at −20°C for 12 hours. The precipitated peptides were centrifuged and washed 2X with ether, dried, dissolved in water, and lyophilized to produce a dark-green powder. The crude peptides were purified by prep-HPLC with a C18 column (Waters Inc) using a water (0.1% TFA)-acetonitrile (0.1% TFA) gradient. Peptide purity was tested using an analytical C18-column. Further characterization was confirmed by either ESI (Waters Inc.) or Q-TOF (Agilent Technologies) mass spectrometry.
The absorbance spectra of the Cy5.5-labeled peptides were characterized with a spectrophotometer (NanoDrop 2000, Thermo Scientific). Fluorescence excitation and emission from a 1 μM peptide solution diluted in PBS was collected with a fiber coupled spectrophotometer (Ocean Optics) using a diode-pumped solid-state laser (Technica Laser Inc) with λex = 671nm. The spectra were plotted with Origin 8.5 software (OriginLab Corp).
Peptide heterodimer validation with confocal microscopy
Cells were cultured to ~70% confluence on glass coverslips, washed with PBS, and incubated with 2% BSA in PBS for 30 min to block non-specific binding. 1 μM of Cy5.5-labeled peptides were added and incubated for 30 min at 4°C. The cells were then washed 3X with PBS, fixed with ice cold 4% paraformaldehyde (PFA) for 10 min, washed with PBS 1X, and then mounted on glass slides with ProLong Gold reagent containing DAPI (Invitrogen). For antibody staining, cells were incubated with either anti-EGFR (1:500, #2232S) or anti-ErbB2 (1:500, #29D8) primary antibody from Cell Signaling Inc overnight at 4°C after fixation, and then washed with PBS 3X and processed with secondary antibody staining. Either goat anti-rabbit IgG (H+L) labeled with Alexa-Fluor 488 (1:1000, #A-11008, Invitrogen) or goat anti-mouse IgG (H&L) labeled with Alexa-Fluor 568 (1:1000, #ab175473, Abcam) was added and incubated for 1 hour at room temperature (RT). The cells were washed with PBS 3X, and mounted onto glass coverslips. Confocal fluorescence images were collected using DAPI, AF488, AF568 and Cy5.5 filter sets. Fluorescence intensities from 3 independent images were quantified using custom Matlab (Mathworks) software.
Western blot
Western blot was performed by lysing cells in Pierce® RIPA lysis buffer containing HaltTM protease inhibitor cocktail (#87786, Thermo Scientific) for 30 min on ice. The lysates were centrifuged at 10,000 rpm for 10 min at 4oC. A BSA protein assay kit (#23227, Thermo Scientific) was used to quantify protein concentration in the supernatant. Aliquots of protein (10 μL) were used for electrophoresis on NuPAGETM 4–12% Bis-Tris gels (#17031471, Invitrogen) followed by electrophoretic transfer onto membranes (#ISEQ00010, Merck Millipore Ltd). The membranes were incubated with primary rabbit anti-EGFR monoclonal antibody (1:1000, #2232S) and rabbit anti-ErbB2 monoclonal antibody (1:1000, #2165S) from Cell Signaling Inc. Mouse anti-β-tubulin monoclonal antibody (1:500, #32–2600, Invitrogen) was used for loading control. For visualization, horseradish peroxiase (HRP)-conjugated secondary antibodies consisting of goat anti-rabbit IgG (H+L) HRP (1:5000, #65–6120) and goat anti-mouse IgG (H+L) HRP (1:5000, #62–6520) from Thermo Scientific were used, and followed by the ECL kit (#RPN2106, GE Healthcare) per manufacturer instructions.
Heterodimer stability
The stability of the heterodimer was evaluated in 0.01 mol/L PBS containing 0.1% bovine serum albumin (BSA, pH 7.4) at 37°C. Degradation was monitored by HPLC at 0.5, 2, 6 and 12 hours. Enzymatic stability was also evaluated by incubating 30 μM of the heterodimer in mouse serum at 37°C. The samples were centrifuged at 14,000 rpm, and 20 μL aliquots of the supernatant were analyzed by HPLC at 0.5, 1.0, 1.5 and 2 hours of incubation.
siRNA knockdown
We performed separate knockdown of either EGFR or ErbB2 expression in SKBr3 cells to validate specific peptide binding. ON-TARGETplus human EGFR siRNA (#L-003114–00-0005), ONTARGETplus human ErbB2 siRNA (#L-003126–00-0005) and ON-TARGET plus non-targeting pool (#D-001810–10-05) were used. siRNA at 5 μΜ concentration in 5 μL was transfected into SKBr3 cells using DharmaFECT transfection reagents (Thermo Scientific). Briefly, cells were seeded in 6-well culture plates at 30% confluence in McCoy’s 5A media supplemented with 10% FBS without antibiotics. The cells were transfected with siRNA at a final concentration of 5 μM using oligofectamine (Thermo Scientific). Knockdown of either EGFR or ErbB2 was confirmed by Western blot.
Binding properties
We measured the apparent dissociation constant kd of the peptide heterodimer to OE33 cells to assess binding affinity. The heterodimer was serially diluted in PBS at concentrations of 0, 10, 25, 50, 100, and 200 nM with ~105 OE33 cells at 4°C for 1 hour and washed with cold PBS. The mean fluorescence intensities were measured with flow cytometry (BD LSRII, BD Biosciences). The equilibrium dissociation constant kd = 1/ka was calculated with a least-squares fit of the data to the nonlinear equation I = (I0 + Imaxka[X])/(I0 + ka[X]). I0 and Imax are the initial and maximum fluorescence intensities, corresponding to no peptide and at saturation, respectively, and [X] represents the concentration of bound peptide. Prism 5.0 software (Graphpad Inc) was used to calculate kd and ka.
We measured the apparent association time constant k to OE33 cells to assess binding kinetics. OE33 cells were grown to ~80% confluence in 10 cm dishes, and detached with PBS-based cell dissociation buffer (Invitrogen). ~105 cells were incubated with 1 μM of the heterodimer at 4°C for different time periods ranging from 0–60 min. The cells were centrifuged, washed with cold PBS, and fixed with 4% PFA. Flow cytometry was performed, and the median fluorescence intensity (y) was ratioed with that of OE33 cells without addition of peptide at various time points (t) using Flowjo software. The rate constant k was calculated by fitting the data to the first-order kinetics model, y(t) = Imax[1-exp(−kt)], where Imax = maximum value, using Prism 5.0 software.
Co-localization
We evaluated co-localization of binding by the peptide heterodimer, anti-EGFR antibody and anti-ErbB2 antibody to SKBr3 cells. The heterodimer at 1 μM concentration was incubated with cells for 1 hour at 4°C. The cells were washed and fixed with 4% PFA for 10 min, and then incubated with either primary mouse anti-EGFR and secondary AF568-labeled antibody or primary rabbit anti-ErbB2 and secondary AF488-labeled antibody.
Effect on cell signaling
SKBr3 cells were seeded in 6-well flat-bottom plates. For EGFR activation, serum free media was used to culture the cells for 16 hours. EGF (#E9644, Sigma) was reconstituted to 1 mg/mL using 10 mM acetic acid, diluted with 0.1% BSA, and added to SKBr3 cells at a concentration of 100 ng/mL. Lapatinib (CDS022971, Sigma) was diluted to 100 nM in DMSO and PBS from 1 mg/mL stock solution, and was added to the cells incubated with the heterodimer at concentrations of 1, 5, 20 μM in separate wells. The cells were washed and harvested by RIPA buffer containing protease inhibitors (#11836170001, Roche), and evaluated by Western blot. Anti-EGFR antibody (#2232S), anti-HER2 antibody (#2165), anti-phospho-EGFR sampler kit (#9922s), anti-phospho-HER2/ErbB2 (Tyr1248) antibody (#2247), anti-AKT (#4691P), anti-ERK1/2 (#4695P), anti-phospho-AKT (pS473; #4060P), anti-phospho-ERK1/2 (#4370P) were used per manufacturer’s instructions (Cell Signaling, Inc).
Pharmacokinetics
All experimental procedures were performed in accordance with relevant guidelines and regulations of the University of Michigan, and all animal studies were conducted with approval by the University Committee on the Use and Care of Animals (UCUCA). Animals were housed per guidelines of the Unit for Laboratory Animal Medicine (ULAM).
Female athymic nude mice (002019 Foxn1, Jackson Laboratories) were obtained at 4–5 weeks of age, housed with 4 animals per cage, and fed with sterilized pellet chow and water. Mice were anesthetized in an isoflurane chamber, and OE33 xenograft tumors were generated by subcutaneous injection of ~5×107 tumor cells suspended in 100 μL of sterile PBS in the hind limb flank. Mice were studied at ~3–4 weeks post-inoculation when the tumor volume reached ~0.5–0.8 cm in size.
We collected fluorescence images in vivo using the Xenogen IVIS™ 200 small animal system. The fluorescence intensity was measured and normalized. Mice were anaesthetized using isoflurane, administered 300 μM of either targeted or control peptide diluted in 150 μL of PBS via tail vein. Images were collected prior to peptide injection to assess baseline autofluorescence and at various time points post-injection. Fluorescence images were excited at λex = 675 nm, and emission was collected at λem = 740 nm with 5 sec exposure (f/stop = 8).
Competition
In vivo heterodimer specificity was assessed by competition with 3000 μM of unlabeled peptide monomer or mixture of two monomers diluted in 50 μL of PBS injected 20 min beforehand in tumor-bearing mice (n = 3 per group). Images were collected at two hours after heterodimer injection of when peak uptake was expected. The ratio of fluorescence intensity in the tumor was calculated using regions of interest.
Biodistribution
We evaluated peptide biodistribution in nude mice bearing OE33 xenograft tumors. Mice (n = 6 per group) were injected with either the heterodimer or control at a concentration of 300 μM, 150 μL in PBS, and were euthanized 2 hours later. Organs were harvested, separated, and imaged ex vivo using the Xenogen IVISTM 200 small animal imaging system. Fluorescence intensity from each organ was quantified using regions of interest (ROI).
Validation with human esophageal specimens
Formalin-fixed, paraffin-embedded (FFPE) specimens of human esophagus were obtained from the archived tissue bank in the Department of Pathology. The tissues were cut into 10 μm sections, deparaffinized, and rehydrated using standard conditions. Two-step acidic and protease-based antigen retrieval was performed. The sections were blocked with protein serum for 10 min at RT followed by rinsing with PBS, and then incubated with 0.5 μM of the heterodimer for 10 min in dark conditions at RT. The sections were then washed for 3 min with PBS 3X and further incubated sequentially with 1:200 dilution of EGFR monoclonal antibody (H11, #MA5–13070, Invitrogen) and 1:500 goat anti-mouse IgG (H+L) AF-568-labeled secondary antibody (#A-11004, Thermo Fisher Scientific), 1:500 dilution of ErbB2 primary antibody (#29D8, rabbit monoclonal antibody #2165S, Cell Signaling Inc) and 1:1000 goat anti-rabbit IgG (H+L) AF-488-labeled secondary antibody (#A11008, Life Technologies Corp). Each antibody was incubated for 1 hour at RT. Sections were washed with TBST 3X and mounted with Prolong Gold reagent containing DAPI (Invitrogen) using #1 cover glass (1.5 μm thickness). Confocal fluorescence microscopy was performed using DAPI, AF488, AF568 and Cy5.5 filter sets. Image quantification was performed by placing 3 boxes with dimensions of 20×20 μm2 completely within cells found in the epithelium. Mean fluorescence intensities were measured using custom Matlab software. Regions of saturated intensities were avoided.
Statistical Analysis
We fit the fluorescence intensities from the human esophageal specimens according to 4 histological classifications, including squamous (SQ), Barrett’s esophagus (BE), high-grade dysplasia (HGD), and esophageal adenocarcinoma (EAC), and used a unpaired t test and one-way ANOVA. Co-localization of peptide and antibody binding was evaluated using Pearson’s correlation coefficient.
Supplementary Material
Figure 5. Pharmacokinetics of peptide heterodimer.
A) Representative whole-body NIR fluorescence images show uptake of targeted peptide heterodimer QRH*-KSP*-E3-Cy5.5 in xenograft tumor (arrow) over time. B) (GGGAGGG)2KK-Cy5.5 was used as control. C) Quantitative analysis shows peak mean signal from (n = 6) tumors at 2 hours following iv injection. A mean (±SD) T/B ratio of 2.39±0.52 and 1.43±0.28 was measured at the tumor for the targeted (n = 6) and control (n = 6) peptides, respectively. Signal returns to baseline by ~24 hours. D) The mean fluorescence intensity from individual tumors at 2 hours is significantly greater for the targeted versus control peptide, P=2.9×10−3 by unpaired t-test. E) Unlabeled QRH*, KSP*, and both (QRH*, KSP*) monomer peptides were injected iv prior to heterodimer for blocking. F) Quantified results show significant reduction in mean intensity from tumor (n = 3).
ACKNOWLEDGMENTS
Funding was provided in part by NIH U54 CA163059 and U01 CA189291 (TDW) and P30 CA046592 (RK).
ABBREVIATIONS
- AFI
autofluorescence imaging
- BE
Barrett’s esophagus
- EAC
esophageal adenocarcinoma
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EMR
endoscopic mucosal resection
- ErbB2
epidermal growth factor receptor 2
- FFPE
formalin-fixed paraffin-embedded
- HGD
high-grade dysplasia
- NBI
narrow band imaging
- NIR
near infrared
- RFA
radio-frequency ablation
- ROI
regions of interest
- RT
room temperature
- T/B
target-to-background
Footnotes
Conflict of interest
Authors (JC, JZ, BPJ, TDW) are inventors on patents filed by the University of Michigan on the peptides used in this study. The other authors disclose no conflicts.
REFERENCES
- 1.Torre LA; Siegel RL; Ward EM; Jemal A Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol. Biomarkers Prev 2016, 25, 16–27. [DOI] [PubMed] [Google Scholar]
- 2.Spechler SJ; Souza RF Barrett’s esophagus. N. Engl. J. Med 2014, 371, 836–845. [DOI] [PubMed] [Google Scholar]
- 3.Levine DS; Haggitt RC; Blount PL; Rabinovitch PS; Rusch VW; Reid BJ An endoscopic biopsy protocol can differentiate high-grade dysplasia from early adenocarcinoma in barrett’s esophagus. Gastroenterology 1993, 105, 40–50. [DOI] [PubMed] [Google Scholar]
- 4.Lekakos L; Karidis NP; Dimitroulis D; Tsigris C; Kouraklis G; Nikiteas N Barrett’s esophagus with high-grade dysplasia: focus on current treatment options. World J.Gastroenterol 2011, 17, 4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaheen NJ; Falk GW; Iyer PG; Gerson LB ACG clinical guideline: diagnosis and management of barrett’s esophagus. Am. J. Gastroenterol 2016, 111, 30–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrams JA Chemoprevention of esophageal adenocarcinoma. Therap. Adv. Gastroenterol 2008, 1, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulak AM; Schumacher SE; van Lieshout J; Imamura Y; Fox C; Shim B; Ramos AH; Saksena G; Baca SC; Baselga J Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012, 72, 4383–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cronin J; McAdam E; Danikas A; Tselepis C; Griffiths P; Baxter J; Thomas L; Manson J; Jenkins G Epidermal growth factor receptor (EGFR) is overexpressed in high-grade dysplasia and adenocarcinoma of the esophagus and may represent a biomarker of histological progression in barrett’s esophagus (BE). Am. J. Gastroenterol 2011, 106, 46–56. [DOI] [PubMed] [Google Scholar]
- 9.Dahlberg PS; Jacobson BA; Dahal G; Fink JM; Kratzke RA; Maddaus MA; Ferrin LJ ERBB2 amplifications in esophageal adenocarcinoma. Ann. Thorac. Surg 2004, 78, 1790–1800. [DOI] [PubMed] [Google Scholar]
- 10.Citri A; Yarden Y EGF–ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol 2006, 7, 505–516. [DOI] [PubMed] [Google Scholar]
- 11.Paterson AL; O’donovan M; Provenzano E; Murray LJ; Coleman HG; Johnson BT; McManus DT; Novelli M; Lovat LB; Fitzgerald RC Characterization of the timing and prevalence of receptor tyrosine kinase expression changes in oesophageal carcinogenesis. J. Pathol 2013, 230, 118–128. [DOI] [PubMed] [Google Scholar]
- 12.Miller CT; Moy JR; Lin L; Schipper M; Normolle D; Brenner DE; Iannettoni MD; Orringer MB; Beer DG Gene amplification in esophageal adenocarcinomas and barrett’s with high-grade dysplasia. Clin. Cancer Res 2003, 9, 4819–4825. [PubMed] [Google Scholar]
- 13.Rao J; Lahiri J; Isaacs L; Weis RM; Whitesides GM A trivalent system from vancomycin d-Ala-d-Ala with higher affinity than avidin biotin. Science 1998, 280, 708–711. [DOI] [PubMed] [Google Scholar]
- 14.Handl HL; Vagner J; Han H; Mash E; Hruby VJ; Gillies RJ Hitting multiple targets with multimeric ligands. Expert Opin.Ther. Targets 2004, 8, 565–586. [DOI] [PubMed] [Google Scholar]
- 15.Landers KA; Burger MJ; Tebay MA; Purdie DM; Scells B; Samaratunga H; Lavin MF; Gardiner RA Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int. J. Cancer 2005, 114, 950–956. [DOI] [PubMed] [Google Scholar]
- 16.Shallal HM; Minn I; Banerjee SR; Lisok A; Mease RC; Pomper MG Heterobivalent agents targeting PSMA and integrin-αvβ3. Bioconjug. Chem 2014, 25, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eder M; Schäfer M; Bauder‐Wüst U; Haberkorn U; Eisenhut M; Kopka K Preclinical evaluation of a bispecific low‐molecular heterodimer targeting both PSMA and GRPR for improved PET imaging and therapy of prostate cancer. Prostate 2014, 74, 659–668. [DOI] [PubMed] [Google Scholar]
- 18.Luo H; Hong H; Yang SP; Cai W Design and applications of bispecific heterodimers: molecular imaging and beyond. Mol. Pharm 2014, 11, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochran JR Engineered proteins pull double duty. Sci. Transl. Med 2010, 2, 17ps5. [DOI] [PubMed] [Google Scholar]
- 20.Rosenzweig SA Acquired resistance to drugs targeting receptor tyrosine kinases. Biochem. Pharmacol 2012, 83, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vagner J; Xu L; Handl HL; Josan JS; Morse DL; Mash EA; Gillies RJ; Hruby VJ Heterobivalent Ligands Crosslink Multiple Cell‐Surface Receptors: The human melanocortin‐4 and δ‐opioid receptors. Angew. Chem. Int. Ed 2008, 47, 1685–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L; Vagner J; Josan J; Lynch RM; Morse DL; Baggett B; Han H; Mash EA; Hruby VJ; Gillies RJ Enhanced targeting with heterobivalent ligands. Mol. Cancer Ther 2009, 8, 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Josan JS; Handl HL; Sankaranarayanan R; Xu L; Lynch RM; Vagner J; Mash EA; Hruby VJ; Gillies RJ Cell-specific targeting by heterobivalent ligands. Bioconjug. Chem 2011, 22, 1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L; Josan JS; Vagner J; Caplan MR; Hruby VJ; Mash EA; Lynch RM; Morse DL; Gillies RJ Heterobivalent ligands target cell-surface receptor combinations in vivo. Proc. Natl. Acad. Sci. U.S.A 2012, 109, 21295–21300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z; Yan Y; Chin FT; Wang F; Chen X Dual integrin and gastrin-releasing peptide receptor targeted tumor imaging using 18F-labeled PEGylated RGD-bombesin heterodimer 18F-FB-PEG3-Glu-RGD-BBN. J. Med. Chem 2009, 52, 425–432. [DOI] [PubMed] [Google Scholar]
- 26.Li Z-B; Wu Z; Chen K; Ryu EK; Chen X 18F-labeled BBN-RGD heterodimer for prostate cancer imaging. J. Nucl. Med 2008, 49, 453–461. [DOI] [PubMed] [Google Scholar]
- 27.Yan Y; Chen K; Yang M; Sun X; Liu S; Chen X A new 18F-labeled BBN-RGD peptide heterodimer with a symmetric linker for prostate cancer imaging. Amino Acids 2011, 41, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J; Niu G; Lang L; Li F; Fan X; Yan X; Yao S; Yan W; Huo L; Chen L; Li Z; Zhu Z; Chen X Clinical translation of a dual integrin αvβ3– and gastrin-releasing peptide receptor–targeting PET radiotracer, 68Ga-BBN-RGD. J. Nucl. Med 2017, 58, 228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razumienko E; Dryden L; Scollard D; Reilly RM MicroSPECT/CT imaging of co-expressed HER2 and EGFR on subcutaneous human tumor xenografts in athymic mice using 111In-labeled bispecific radioimmunoconjugates. Breast Cancer Res. Treat 2013, 138, 709–718. [DOI] [PubMed] [Google Scholar]
- 30.Wu AM; Senter PD Arming antibodies: prospects and challenges for immunoconjugates. Nat. Biotechnol 2005, 23, 1137–1146. [DOI] [PubMed] [Google Scholar]
- 31.Thakur A; Lum LG Cancer therapy with bispecific antibodies: clinical experience. Curr. Opin. Mol. Ther 2010, 12, 340. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J; Joshi BP; Duan X; Pant A; Qiu Z; Kuick R; Owens SR; Wang TD EGFR overexpressed in colonic neoplasia can be detected on wide-field endoscopic imaging. Clin. Transl. Gastroenterol 2015, 6, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi BP; Zhou J; Pant A; Duan X; Zhou Q; Kuick R; Owens SR; Appelman H; Wang TD Design and synthesis of near-infrared peptide for in vivo molecular imaging of HER2. Bioconjug. Chem 2015, 27, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo S; Zhang E; Su Y; Cheng T; Shi C A review of NIR dyes in cancer targeting and imaging. Biomaterials 2011, 32, 7127–7138. [DOI] [PubMed] [Google Scholar]
- 35.Björkelund H; Gedda L; Barta P; Malmqvist M; Andersson K Gefitinib induces epidermal growth factor receptor dimers which alters the interaction characteristics with 125I-EGF. PLoS One 2011, 6, e24739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Y; Chen X Peptide heterodimers for molecular imaging. Amino Acids 2011, 41, 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker A; Hessenius C; Licha K; Ebert B; Sukowski U; Semmler W; Wiedenmann B; Grötzinger C Receptor-targeted optical imaging of tumors with near-infrared fluorescent ligands. Nat. Biotechnol 2001, 19, 327–331. [DOI] [PubMed] [Google Scholar]
- 38.Lee S; Xie J; Chen X Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem. Rev 2010, 110, 3087–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tangri S; LiCalsi C; Sidney J; Sette A Rationally engineered proteins or antibodies with absent or reduced immunogenicity. Curr. Med. Chem 2002, 9, 2191–2199. [DOI] [PubMed] [Google Scholar]
- 40.Bray BL Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat. Rev. Drug Discov 2003, 2, 587–593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.