Abstract
Predicting fish acute toxicity of chemicals in vitro is an attractive alternative method to the conventional approach using juvenile and adult fish. The rainbow trout (Oncorhynchus mykiss) cell line assay with RTgill-W1 cells has been designed for this purpose. It quantifies cell viability using fluorescent measurements for metabolic activity, cell- and lysosomal-membrane integrity on the same set of cells. Results from over 70 organic chemicals attest to the high predictive capacity of this test. We here report on the repeatability (intralaboratory variability) and reproducibility (interlaboratory variability) of the RTgill-W1 cell line assay in a round-robin study focusing on 6 test chemicals involving 6 laboratories from the industrial and academic sector. All participating laboratories were able to establish the assay according to preset quality criteria even though, apart from the lead laboratory, none had previously worked with the RTgill-W1 cell line. Concentration-response modeling, based on either nominal or geometric mean-derived measured concentrations, yielded effect concentrations (EC50) that spanned approximately 4 orders of magnitude over the chemical range, covering all fish acute toxicity categories. Coefficients of variation for intralaboratory and interlaboratory variability for the average of the 3 fluorescent cell viability measurements were 15.5% and 30.8%, respectively, which is comparable to other fish-derived, small-scale bioassays. This study therefore underlines the robustness of the RTgill-W1 cell line assay and its accurate performance when carried out by operators in different laboratory settings.
Keywords: in vitro alternatives, round-robin study, validation
The quantification of fish acute toxicity is one of the corner stones of environmental hazard assessment of chemicals. Under the European Chemical Legislation REACH (Registration, Evaluation, Authorization and restriction of CHemicals), for example, any chemical produced or imported ≥10 tons/year needs to be tested on fish according to an accepted test guideline, such as of the Organization for Economic Co-operation and Development (OECD, 1992). Similar requirements for fish acute toxicity testing are put forth in other jurisdiction, such as the US TSCA (Toxic Substances Control Act) and its revisions in the form of the LCSA (Lautenberg Chemical Safety Act). Fish acute toxicity is also important for monitoring of effluents. However, given the high number of animals required and the high degree of severity imposed on the animals to monitor death as an integrative but crude endpoint, alternative test methods are desired as highlighted, for example, under both REACH and US TSCA-LCSA. They should be in line with animal use legislation and meet the societal aspiration to substitute animal tests. Alternative test methods would ideally also be less resource intensive in terms of time, infrastructure maintenance and waste management, and be amenable to high throughput screening.
Complying with the demand for alternative test methods to assess fish acute toxicity, the CEFIC-LRI Eco8 endeavor (see weblink CEFIC-LRI), also referred to as the CEllSens project, was established to explore the conditions under which 2 fish-based model systems might be applicable to predict fish acute toxicity of organic chemicals: a permanent fish cell line from rainbow trout gill (Oncorhynchus mykiss), RTgill-W1 (Bols et al., 1994), and the embryo of zebrafish (Danio rerio, Nagel, 2002). The hypothesis underlying the use of these models is that fish acute toxicity is primarily due to nonspecific modes of action, such as disturbance of cell membrane integrity and function. Therefore, a cell line and embryonal life stages of fish face the same degree of damage as juvenile or adult fish, provided that the bioavailability of the chemical is accounted for in quantifying the effects (Gülden and Seibert, 2005; Knöbel et al., 2012; Tanneberger et al., 2013). Assay conditions for the 2 alternative systems were carefully explored in the CEllSens project, most notably with regard to the type of exposure medium and method of dosing used for the cell line assay (Tanneberger et al., 2010). Toxicity endpoints and quantification of the chemicals were evaluated in multiwell plates for both, the fish gill cell line and the fish embryo. A list of organic chemicals was systematically derived. This list was based on predefined criteria in order to cover chemicals with a wide range of physico-chemical properties, toxicities, and different modes of action (Schirmer et al., 2008). A subset of 35 chemicals from this list was subsequently analyzed for their acute toxicity toward RTgill-W1 cells and zebrafish embryos. Chemical concentrations causing 50% reduction in RTgill-W1 cell viability (EC50) or embryo survival (LC50) were determined based on analytically confirmed exposure concentrations. These data were compared with the acute toxicity reported for the same chemicals in the US EPA fathead minnow database (Russom et al., 1997). Aside from the presence of a few striking outliers, namely 2 specifically acting neurotoxic chemicals (permethrin and lindane) and one chemical requiring metabolic activation (allyl alcohol), both RTgill-W1 cell line and zebrafish embryo toxicity were strongly correlated with the toxicity seen in the traditional acute fish test. In fact, the toxicity data approached the line of unity, meaning that both the cell line and the embryo assay yield responses similar to juvenile or adult fish (Knöbel et al., 2012; Tanneberger et al., 2013).
These and other developments, including the acceptance of the zebrafish embryo test in Germany as an alternative for assessing the acute toxicity of effluents (International Standardization Organization ISO 15088, 2007) and extensive intralaboratory and interlaboratory comparison trials (Belanger et al., 2013; Busquet et al., 2014; Lammer et al., 2009), have laid the foundation for the adoption of the fish embryo toxicity (FET) test as an OECD guideline (OECD, 2013). The excellent performance of the RTgill-W1 cell line assay provided the impetus to start bringing this in vitro method, which could serve as a replacement for fish with an unambiguous nonanimal context, to the same level of international standardization. Therefore, an international round-robin study, involving 6 industrial and academic laboratories from Europe and the United States, was conducted. The overall aim was to test the robustness of the established methodology with particular focus on the transferability of the RTgill-W1 cell line assay from the lead laboratory to the other laboratories, and the assessment of its intralaboratory and interlaboratory variability. The outcome of this round-robin study is presented herein.
MATERIALS AND METHODS
Study Design
The study was divided into 3 rounds of activity. In round I, members of 2 laboratories were trained at the lead laboratory (Eawag), transferred the methodology to their own laboratories, and tested 3, 4-Dichloroaniline (DCA) as positive control. During this process, the detailed standard operation procedure (SOP), established as part of the CEllSens project, was further improved and simplified. Success of training was judged based on quality criteria for cell culture and DCA concentration-response curves for RTgill-W1 cell viability as described in more detail in the respective sections below. Once routine cell culture was established and quality criteria were met with DCA by the 2 laboratories, round II was initiated. This round comprised the training of the remaining laboratories in the same manner at the lead laboratory, using the amended SOP, methodology transfer, and testing of the positive control chemical until quality criteria were met. Finally, in round III, all laboratories tested the set of 6 chemicals with some exceptions as specified below and in the Supplementary information (SI, Section 1.a., Supplementary Table S1). In this last round of testing, samples were taken for subsequent chemical analysis.
Participating Laboratories
Six laboratories (5 from Europe, 1 from the United States) were involved in training, methodology transfer, and assay performance testing based on DCA (laboratories A–F). Thereafter, 1 laboratory (F) had to resign from the round-robin study, so that the remaining set of chemicals was tested by 5 laboratories (laboratories A–E) with one further exception: laboratory D was unable to test pentachlorophenol (PCP) due to facility restrictions in working with chemicals designated as acutely toxic to humans.
Test Chemicals
Six test chemicals (for details see SI Section 1.a, Supplementary Table S1) were selected from the CEllSens list of chemicals (Knöbel et al., 2012; Schirmer et al., 2008; Tanneberger et al., 2013) to meet predefined criteria. Specifically, chemicals were selected to: (1) be characterized by a wide range of logHLC (Henry’s law constant, a measure of air/water distribution and thus of volatility) and logKow (octanol-water partition coefficient, a measure of hydrophobicity); (2) cover a wide range of acute toxicity based on available fish acute toxicity LC50 data; (3) belong to different modes of action according to Russom et al. (1997); and (4) be relatively easily quantifiable based on analytical protocols established during the CEllSens project (Knöbel et al., 2012; Tanneberger et al., 2013).
Taking the above-mentioned criteria into account, the selected chemicals comprised: DCA; 2, 2, 2-trichloroethanol (TCE); 1, 2-dichlorobenzene (DCB); hexachlorophene (HCP); malathion (MAL); and PCP. Their coverage in terms of physico-chemical properties, acute toxicity, and modes of action is depicted in Figure 1. All chemicals were purchased with the same lot number from Sigma-Aldrich (Germany) with ≥99% purity as detailed in SI (Section 1.a, Supplementary Table S1) with chemical analysis being performed as described further below.
Figure 1.
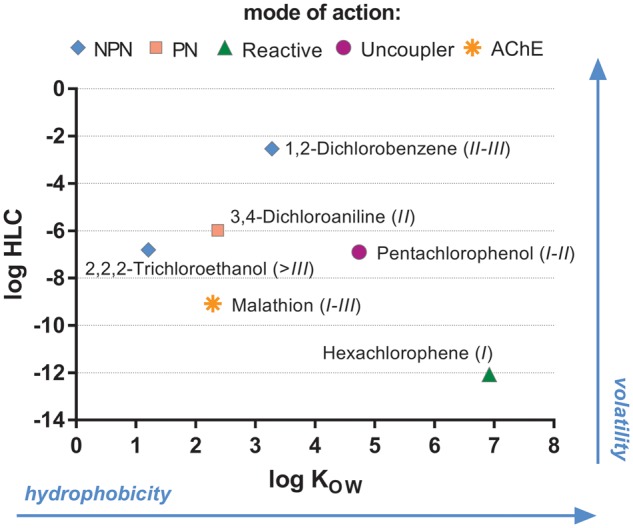
Overview of selected test chemicals, giving reference to their physico-chemical properties (logHLC, logKow), their acute fish toxicity category and their mode of action. The numbers in brackets behind each chemical name indicate the acute fish toxicity category of the chemical (ENV/JM/MONO(2001)8; I = LC50 ≤ 1 mg/L; II = LC50 > 1−≤ 10 mg/L; III = LC50 > 10−≤100 mg/L; >III = LC50 > 100 mg/L; SI Section 4.b). Mode of action according to Russom et al. (1997): NPN, nonpolar narcotic; PN, polar narcotic; AChE, acetyl cholin esterase inhibitor.
The chemical DCA was selected as a positive control primarily due to it being recommended as a positive control in the zebrafish FET test (ISO 15088, 2007; OECD, 2013). Inasmuch as the positive control solely functions to confirm that the RTgill-W1 cell line is responsive under the conditions of the assay, nominal concentrations of DCA are used. Based on the experience of the lead laboratory, EC50 values for the DCA positive control were defined, prior to the round-robin study, to fall within a 2.5 standard deviation (SD) range of the previously determined EC50 values, which were 33.9 mg/L ± 10.1 mg/L, 46.7 mg/L ± 15.0 mg/L, and 46.0 mg/L ± 17.7 mg/L for cell metabolic activity (AlamarBlue, AB), cell membrane integrity (5-CarboxyFluorescein DiAcetate AcetoxyMethyl ester, CFDA-AM) and lysosomal membrane integrity (Neutral Red, NR), respectively (n = 9). The 2.5 SD range was judged fit for purpose when considering the small sample size and the uncertainties for using nominal concentrations; thus, 98.9% of all values would be expected to fall in this range whereas values out of this range should be treated with suspicion. With these considerations, the acceptable EC50 ranges were defined as 8.6–59.1 mg/L for AB, 9.3–84.2 mg/L for CFDA-AM, and 1.7–90.3 mg/L for NR, all based on nominal concentrations. These ranges also cover the historical data published by Tanneberger et al. (2013), though the mean EC50 values for DCA in this initial data set were between 1.6- and 2.4-fold lower than the mean EC50 data established since then.
Routine Cell Culture
The RTgill-W1 cell line is an immortal cell line originally developed from a primary gill cell culture established from pieces of gill filaments of a rainbow trout (Bols et al., 1994). This cell line was chosen based on the consideration that the gill epithelia is the primary uptake site of water-born contaminants into fish and thus a primary target for many toxicants in an acute exposure scenario where epithelial cell membrane integrity, or other vital epithelial cell functions, could be destroyed (Tanneberger et al., 2013). Project partners received the cell line from the nearest possible location, ie, either purchased from ATCC® (CRL-2523TM) or delivered from the lead laboratory, being tested negative for mycoplasma. Overall, passage numbers between 9 and 84 were used for testing; appearance and performance remained unchanged throughout. Routine cell culture of RTgill-W1 was performed exactly as previously described (Tanneberger et al., 2013): cells were cultured at 19°C ± 1°C in 75 cm2 cell culture flasks with L-15 culture medium supplemented with 5% fetal bovine serum (FBS) and 1% gentamicin sulfate solution (10 mg/ml). No restrictions were imposed with regard to supplier or material for cell culture consumables except for the following: cell culture flasks were 75 cm2 with vent screw cap (90075, TPP, Trasadingen, Switzerland) and cell culture medium for routine cell culture was L-15 with glutamine and without phenol red (21083027, Gibco, Thermo Fisher Scientific).
RTgill-W1 Cell Line Assay Performance
The testing procedure closely followed that published by Tanneberger et al. (2013) with a few amendments that resulted from round I of the round-robin study. The resulting overall procedure is described below and depicted in SI (Section 2.a, Supplementary Figure S3).
Preparation of chemical stock solutions
All chemicals were dissolved and serially diluted (1:2.38) in Dimethly sulfoxide (DMSO). The dilution factor of 2.38 was calculated as overall dilution factor based on the dilutions used to test the 35 organic chemicals in the CEllSens project (Tanneberger et al., 2013). The final DMSO concentration was 0.5% vol/vol throughout all tested concentrations to ensure consistent dosing of all chemicals despite their wide range of water solubility. For each experiment, chemical stock solutions and dilution series were freshly prepared.
Exposure of cells to the test chemicals
Exposures were performed on confluent RTgill-W1 cell monolayers in 24-well plates (662160, Greiner Bio-One, Frickenhausen, Germany), where cells were seeded in 22 out of the 24-wells at 350 000 cells per 1 ml of cell culture medium (184 210 cells/cm2; SI Section 2.a, Supplementary Figure S3). With this cell density, confluent monolayers generally form within 24–48 h. Therefore, if monolayer formation was incomplete after 24 h as judged by microscopy, cells were incubated for another 24 h before initiation of chemical exposure.
Exposure of cells to the chemicals was carried out in L-15/ex, a simplified version of the L-15 cell culture medium, containing only the salts, galactose, and pyruvate of the complete L-15 medium (Schirmer et al., 1997). To accomplish dosing, chemical dosing mixtures (DM) were prepared by adding 12 ml L-15/ex and 60 μl of the respective chemical stock solution in DMSO, or DMSO alone for solvent control, into amber glass vials of sufficient size (15–40 ml) followed by vigorous shaking for 10 min. A 500 μL aliquot was collected from each dosing mixture and placed into an autosampler vial for chemical analysis to confirm the intended chemical starting concentration (for details see below). Chemical exposure was initiated by first rinsing the RTgill-W1 cell monolayers once with 1 ml L-15/ex. Afterwards, 2.5 ml of the respective dosing mixture were transferred to the designated wells. Each concentration of chemical was dosed in triplicates. Likewise, triplicates were used for the solvent control (DMSO only). The 2 wells without cells served as control to detect potential background fluorescence by test chemicals. Though apparently rare, such autofluorescence could theoretically interfere with the fluorescence of the cell viability indicator dyes (Schirmer et al., 2000), which would require a separate cell-free control plate with the full chemical concentration range for background values. To be able to detect such cases, one of the cell-free control wells receives L-15/ex exposure medium only whereas the second one receives L-15/ex exposure medium with the highest chemical concentration in DMSO (SI Section 2.a, Supplementary Figure S3).
Immediately after dosing, a 500 μl aliquot was collected from each well and placed in an autosampler vial for chemical analysis at the beginning of the test (C0h). Plates were then sealed with an adhesive foil (polyester film with acrylic adhesive) and placed in the incubator (19°C ± 1°C, normal atmosphere, in the dark) for 24 h. Exposure was terminated by lifting the adhesive foil, removing a 500 μl aliquot for determination of the concentration at the end of exposure (C24h), followed by the cell viability measurement using 3 fluorescent indicator dyes on the same set of cells as previously described (Schirmer et al., 1998; Tanneberger et al., 2013). The fluorescent dyes were Alamar Blue® (AB) or Presto Blue® (PB; DAL1100 or A13262, respectively; Invitrogen, Thermo Fisher Scientific) as a measure of cell metabolic activity, CFDA-AM (C1354, Invitrogen, Thermo Fisher Scientific) as indicator of cell membrane integrity, and NR (N2889, Sigma-Aldrich) as surrogate for lysosomal membrane integrity. Like AB, PB is a commercially available resazurin-based dye preparation, which is used in exactly the same way as AB. Fluorescence was measured with the best-suited excitation/emission wavelength depending on the instrument available in the different laboratories (SI Section 2.b, Supplementary Table S5).
Each experiment was carried out at least 3 times with cells from different passages, starting on different days, and using freshly prepared chemical test solutions, thus yielding a minimum of 3 biological replicates. Within round III of testing, every sixth test plate was a dedicated DCA positive control plate.
Chemical Analysis
Chemical concentrations were measured in the aliquots taken from DM, from wells at the onset of exposure (C0h) and from wells at the end of the exposure (C24h). In case of chemical analysis performed for the samples taken from laboratories A–C and E–F, the respective 500 μl aliquots were transferred into 1.5 ml autosampler vials (548-0029; VWR), which were preloaded with 500 μl cyclohexane (33117; puriss. p.a., ACS reagent, ≥99.5% [GC]; Sigma-Aldrich). The vials were immediately capped (09 04 1220; La-Pha-Pack), vortexed for at least 10 s and stored at −20°C until shipment to analytical laboratory I for chemical quantification. Directly before analysis, the samples were thawed and shaken for 15 min to aid further liquid/liquid extraction before the cyclohexane phase was transferred to a new sampling vial. The chemical concentration in the extracts was determined either by gas chromatography or high-pressure liquid chromatography (SI Section 1.b, Supplementary Table S2). All samples collected by laboratory D were analyzed on site. After transfer of the respective 500 μl aliquots per well into 1.5 ml autosampler vials, samples were immediately processed for analysis. Samples were subjected to liquid dilution with cyclohexane. The chemical concentration in the extracts was determined either by gas chromatography or liquid chromatography (SI Section 1.c, Supplementary Table S3.). For logistical reasons, it was not possible to obtain chemical analytical data from the exact same wells as those used for the cell viability measurements in laboratory A. Instead, average chemical concentrations from 3 separate experimental sets were used and prepared exactly as described above.
Data Analyses
Reporting
Documentation included every step of the assay, starting from a ‘test cover sheet’ (stating, eg, the date, cell passage number, time from plating to exposure) to preparation of chemical stock solutions and raw data sheets containing the measurements for the 3 fluorescent indicator dyes. All forms and data sheet templates were sent to the lead laboratory, where data analysis was performed as previously described (Tanneberger et al., 2013). Concentration-response curves for the 3 measures of cell viability were expressed either based on nominal (ie, intended concentration) or as the geometric mean of the concentrations measured at the beginning (C0h) and at the end (C24h) of exposure (Tanneberger et al., 2013).
Concentration-response modeling
Concentration-dependent effects on cell viability were modeled with the profile likelihood (PL) method (Raue et al., 2009) using the following nonlinear 2-parameter logistic equation in an in-house R script (https://github.com/UtoxEawag/RTgillRoundRobin):
where S is the cell viability in %, EC50 is the log10 transformation of the concentration where cell viability drops to 50%, X is the log10 transformation of the concentration used, and h is the Hill slope that determines the shape of the log10concentration-response curve. The gradient-descent algorithm was used for the optimization, whereas the sum of squared residuals (chi-square) was used as objective function. EC50 values were expressed as the mean of all performed independent replicates (n ≥ 3) per fluorescent cell viability indicator dye.
Confidence intervals were inferred with the PL method (Raue et al., 2009), using an in-house R script (https://github.com/UtoxEawag/RTgillRoundRobin). Starting from the optimum parameter values, determined by fitting the concentration-response curve (see above), the PL confidence intervals for each parameter are determined by fixing the values of this parameter (eg, the EC50), whereas using the other parameter for curve fitting. This is repeated by changing the value of the fixed parameter away from the optimum, until the quality of the fit, as determined by sum of squared residuals, is worse than the 0.05 quantile of the chi-squared distribution (df = 3).
Intralaboratory and interlaboratory variability
The coefficient of variation (CoV = SD/mean) was calculated as a measure of variability: for intralaboratory comparison, the EC50 value of each biological replicate was taken into account; for interlaboratory comparison, the mean EC50 value per laboratory and chemical was used for CoV calculation.
Outlier analyses
As another measure of intralaboratory and interlaboratory variability, we defined outliers at the laboratory level and at the level of individual biological replicates. Laboratory outliers were defined as those for which the determined EC50 values, across all replicates for each chemical and dye combination, fell outside 2 SDs of the calculated mean EC50 between all laboratories. Additionally, we defined biological replicate outliers in the following way: (1) the EC50 value for each biological replicate was normalized to the intralaboratory mean EC50 value; (2) the SD of the normalized value across all biological replicates of all laboratories was calculated; (3) biological replicates that fell outside 2 SDs of the intralaboratory mean were defined as outliers. The outlier analysis was performed using an in-house R script (https://github.com/UtoxEawag/RTgillRoundRobin).
Factors contributing to EC50 value variability
We performed 2 kinds of analyses in order to explore factors potentially contributing to EC50 variability.
Are fluorescent indicator dyes equally reliable?
The CoVs, intralaboratory and interlaboratory, were compared between dyes using the Kruskal–Wallis test with a significance level of α = 0.05.
Which factors contribute most to EC50 variability?
To determine which factors contributed the most to the variability in measured EC50 values, we used a linear mixed effects (LME) model:
where EC50 is the log10 transformation of the 50% effect concentration determined from measurements of chemical i, at laboratory j and with dye k; Ci is the mean (fixed) effect specific to the chemical i; Lj is the (random) effect of laboratory j, assumed to be normally distributed with a mean of 0; Dk is the mean (fixed) effect specific of the dye k; and eijk is the residual error, assumed to be normally distributed with mean 0.
The LME model was fitted with maximum likelihood to model the EC50 as the dependent variable, chemical and dye as fixed effects, and laboratory as random effect. Likelihood-ratio tests were performed to determine statistical significance of the contribution of any random effect variance component to the overall variability of the data.
Correlation with acute fish toxicity data
Fish acute toxicity (LC50) and RTgill-W1 cell viability data (EC50) were plotted assuming an approximately 1:1 relationship. Fish acute toxicity data were, on the one hand, taken from the US EPA fathead minnow database as previously described (Schirmer et al., 2008; Tanneberger et al., 2013). To expand the species coverage beyond fathead minnow, additional fish acute toxicity studies were extracted from the US EPA Ecotox database (SI Section 4.b).
RESULTS
This round-robin study was based on the SOP initially prepared in the CEllSens project; discussing and testing this SOP with the 2 laboratories participating in round I of the study resulted in further improvements of the SOP with the greatest difference being that the entire assay can now be performed in one 24-well plate. Thus, the SOP now describes the entire assay, including sampling for chemical quantification and performance of 3 cell viability tests, to be completed in a single 24-well culture plate per chemical and biological replicate. The complete workflow is schematically depicted in the SI (Section 2.a, Supplementary Figure S3).
Establishment of RTgill-W1 Cell Culture in Participating Laboratories
Except for the lead laboratory, all other laboratories needed to establish the RTgill-W1 cell line culture. Routine cell culture was considered successful when (1) cell morphology corresponded to expectations as documented in the SOP and as judged by light microscopy, and (2) a confluent cell culture, passaged routinely at a ratio of 1:2, reached confluency within 10–12 days. All participating laboratories were able to accomplish this task based on the information provided in the SOP and by reporting back to the lead laboratory. Fetal bovine serum of various sources was used by different laboratories and found suitable for successful routine cultivation of the cells (SI Section 2.b, Supplementary Table S5).
DCA as Positive Control
Each laboratory was asked to establish the workflow from seeding cells in 24-well plates to assessing cell viability upon exposure to DCA as quality control. Three acceptance criteria were applied. The first criterion was that the DMSO versus solvent-free control wells generally did not differ in raw fluorescence values by a more than 10% reduction. Although it is known that 0.5% DMSO does not impair cell viability of RTgill-W1 cells as indicated by the fluorescent indicator dyes, the DMSO control in this case serves to detect potential contamination of this solvent. The second criterion was that the raw fluorescence values in the cell-free control well containing the highest chemical test concentration did not consistently vary by more than 20% for any of the dyes from the cell-free control well containing the L-15/ex exposure medium only. This control is used to confirm that the chemical per se does not possess fluorescence at the respective wavelengths, or otherwise interfere with the indicator dye (see dosing scheme in SI Section 2a, Supplementary Figure S3). The third criterion was that, based on the experience of the lead laboratory, EC50 values were expected to be in the given range (see Materials and Methods section). Because the purpose of this latter quality control is to judge cell handling and cell viability assay performance, these respective EC50 values are based on nominal (ie, intended) DCA concentrations. All laboratories were able to achieve all preset criteria with few exceptions regarding the initially set EC50 range for CFDA-AM (4 replicates out of 27) and NR (3 replicates out of 27). In these cases, the mean EC50 was slightly higher than the upper range limit; yet, all fell into the newly adjusted range that resulted from this round-robin study (see below).
To use the newly gained data from this round-robin study for better estimation of the mean and SDs due to the increasing number of independent observations, the acceptance range for the DCA positive control was adjusted in the final SOP. It is now given as: 43.6 mg/L ± 6.1 mg/L (2.5 SD range: 28.4–58.9 mg/L) for cell metabolic activity (AB/PB), 62.5 mg/L ± 18.9 mg/L (2.5 SD range: 15.2–109.7 mg/L) for cell membrane integrity (CFDA-AM), and 58.6 mg/L ± 18.6 mg/L (2.5 SD range: 12.1–105.0 mg/L) for lysosomal membrane integrity (NR) (n = 27; SI Section 3.a, Supplementary Figure S4).
Testing the 6 Selected Chemicals
All cell-based data sets were sent in a standardized format to the lead laboratory and confirmed to fulfill the overall background criteria for cell viability in the DMSO versus solvent-free control wells and for lack of interference by the chemical with the fluorescence measurements as described for DCA above. Chemical analytical data were also provided to the lead laboratory by the 2 analytical laboratories.
Chemical analysis revealed a high concordance of confirmed chemical concentrations in all samples generated by the same laboratory as well as across the different laboratories based on measurements made by the 2 analytical laboratories (samples from laboratories A–C and E–F = analytical laboratory I and laboratory D = analytical laboratory II; SI Section 1.d, Supplementary Table S4—separate excel file). Exposure concentrations at time 0 (C0h) agreed very well with the concentrations of the respective DM, generally deviating less than 10%, demonstrating that the dosing step itself had no pronounced influence on the achieved exposure concentrations (SI Section 1.e, Supplementary Figure S1). Chemical analytical data were used to convert nominal concentrations to measured values. Exposure concentrations were expressed as the geometric mean values of C0h and C24h in order to account for the time-dependent chemical loss. Indeed, significant losses were measured for DCA (up to 43%) and DCB (up to 92%), the 2 most volatile chemicals of this data set, and for HCP (an average of 60% loss), the least volatile but most hydrophobic chemical. In contrast, chemical exposure concentrations remained stable over the 24 h in the case of MAL, TCE, and PCP (SI Section 1.f, Supplementary Figure S2).
Concentration-response modeling was performed for both nominal as well as geometric mean-derived concentrations. As expected from the stability of the respective chemicals in the exposure medium, EC50 values remained essentially the same when corrected or not for measured concentrations in the case of MAL, TCE, and PCP. Yet, corrected EC50 values were half of those obtained for nominal concentrations in the case of DCA and HCP and about one-fourth in the case of DCB. All subsequent considerations were based on the measured concentration data.
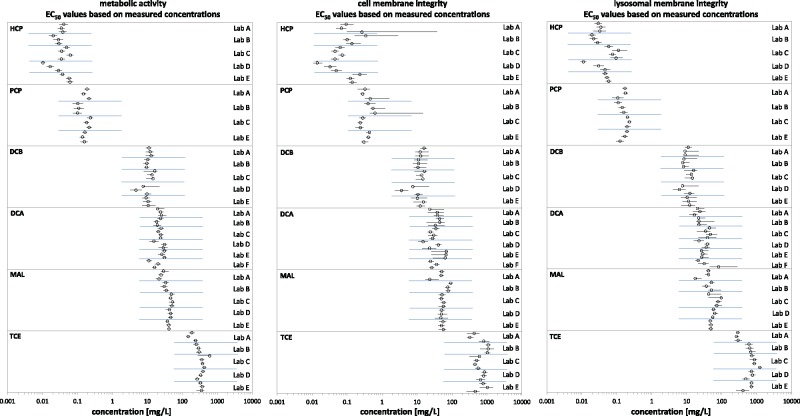
Figure 2 demonstrates that all 3 cell viability indicator dyes yielded EC50 values for each biological replicate with values spanning approximately 4 orders of magnitude over the chemical range, covering all fish acute toxicity categories (see also Figure 1). Thus, EC50 values ranged from about 10 to 100 μg/L for HCP and PCP (category I, most toxic) to >100 mg/L for TCE (category >III, least toxic). All concentration-response curves based on measured concentrations are shown in the SI (Section 3.b, Supplementary Figures S5-1 to S5-18).
Figure 2.
EC50 values (circles) and respective 95% confidence interval (horizontal lines) for each experimental run of the round-robin study obtained for the different cell viability indicators (left: metabolic activity [AB/PB], middle: cell membrane [CFDA-AM] and right: lysosomal membrane [NR] integrity). EC50 values are arranged by test chemical toxicity and laboratory and are based on measured chemical concentrations.
Assessment of Variability
Confidence intervals for EC50 values obtained from individual concentration-response curves using 3 technical replicates (ie, data from 3 culture wells) were overall well below 30% of the EC50 value, with a marked exception being CFDA-AM for the 2 most toxic chemicals, HCP and PCP. EC50 values obtained by each laboratory from biological replicates were comparable as indicated by overlapping confidence intervals. No laboratory outliers were found for any of the chemical/dye combinations (SI Section 3.c, Supplementary Table S6—separate excel file). Yet, in a few cases, one of the biological replicates appeared to be further away from all the others. Out of the 282 concentration-response curves obtained, 13 were classified as outliers in this way (SI Section 3.c Supplementary Table S6—separate excel file). These outliers appeared to be randomly distributed between labs, chemicals, and dyes for the same biological replicate with the exception of one biological replicate for DCB, where all 3 dyes signaled an outlier. For subsequent calculations, these outliers were replaced by the respective average EC50 value calculated from the remaining biological replicates from the same laboratory (=data set without outliers) and compared with the full data set (=including outliers).
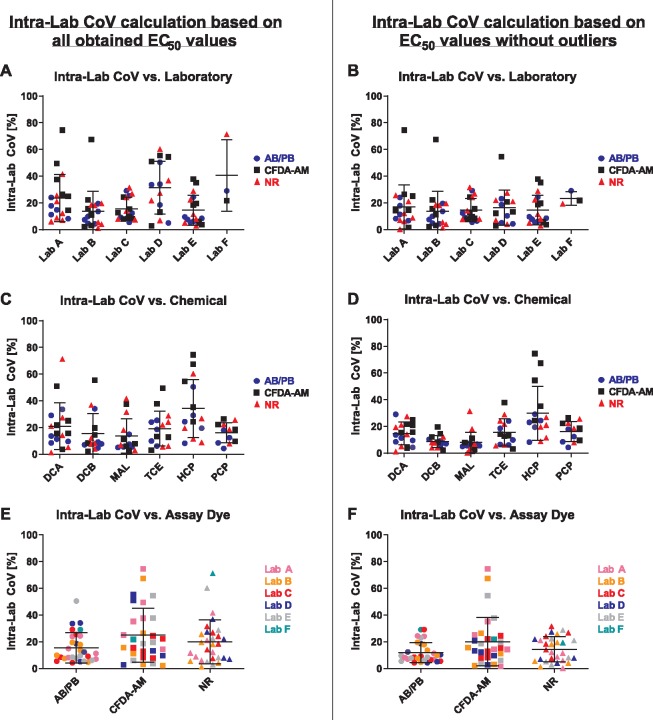
The CoVs were calculated from EC50 values by dividing the SD by the mean for each laboratory (intralaboratory variability or repeatability) and across laboratories (interlaboratory variability or reproducibility) for each dye/chemical/laboratory combination (Table 1; Figures 3 and 4 for intralaboratory and interlaboratory variability, respectively; SI Section 3.c, Supplementary Table S6—separate excel file). Average CoVs for intralaboratory variability across all chemicals and dyes were between 13.8% and 40.7% for the entire data set, with the highest value originating from the laboratory testing only DCA (laboratory F). When the outliers were removed, CoVs were homogeneous, ranging from 13.8% to 17.0% for laboratories A–E, and amounting to 23.4% for laboratory F (Figure 3A and 3B). Average intralaboratory CoVs across all chemicals and laboratories were between 13.6% and 21.0% for all chemicals except for HCP, where the CoV was 34.3%; when the outliers were removed, CoVs were between 7.9% and 16.1% for all chemicals other than HCP, for which the CoV was 29.8% (Figure 3C and 3D). Finally, average intralaboratory CoVs across all chemicals and labs to depict the variation for the cell viability indicator dyes were 15.5%, 24.9%, and 19.9% for metabolic activity (AB/PB), cell membrane integrity (CFDA-AM) and lysosomal membrane integrity (NR), respectively. In the same sequence of dyes, these values were 11.9%, 20.1%, and 14.4% when the outliers were removed (Figure 3E and 3F;Table 1).
Table 1.
Intralaboratory and Interlaboratory CoVs
| CoVs based on all obtained measured concentrations-derived EC50 values |
CoVs based on measured concentrations-derived EC50 values without outliers |
|||||||
|---|---|---|---|---|---|---|---|---|
| Chemical | Lab | AB/PB | CFDA-AM | NR | AB/PB | CFDA-AM | NR | |
| Intralaboratory | HCP | Lab A | 8.2 | 74.4 | 8.8 | 8.2 | 74.4 | 8.8 |
| Lab B | 19.4 | 67.4 | 19.5 | 19.4 | 67.4 | 19.5 | ||
| Lab C | 29.1 | 24.4 | 26.8 | 29.1 | 24.4 | 26.8 | ||
| Lab D | 50.5 | 54.4 | 60.2 | 23.1 | 54.4 | 20.9 | ||
| Lab E | 24.4 | 35.2 | 11.6 | 24.4 | 35.2 | 11.6 | ||
| PCP | Lab A | 17.9 | 26.3 | 25.6 | 17.9 | 26.3 | 25.6 | |
| Lab B | 4.3 | 22.2 | 18.4 | 4.3 | 22.2 | 18.4 | ||
| Lab C | 12.4 | 9.5 | 7.9 | 12.4 | 9.5 | 7.9 | ||
| Lab E | 8.5 | 18.5 | 22.3 | 8.5 | 18.5 | 22.3 | ||
| DCB | Lab A | 7.1 | 14.4 | 12.2 | 7.1 | 14.4 | 12.2 | |
| Lab B | 4.7 | 2.2 | 3.9 | 4.7 | 2.2 | 3.9 | ||
| Lab C | 9.1 | 8.3 | 7.5 | 9.1 | 8.3 | 7.5 | ||
| Lab D | 34.0 | 55.4 | 36.8 | 10.3 | 12.2 | 4.0 | ||
| Lab E | 7.7 | 19.6 | 8.0 | 7.7 | 19.6 | 8.0 | ||
| DCA | Lab A | 11.3 | 25.0 | 21.1 | 11.3 | 25.0 | 21.1 | |
| Lab B | 13.6 | 15.6 | 1.2 | 13.6 | 15.6 | 1.2 | ||
| Lab C | 8.5 | 15.4 | 14.1 | 8.5 | 15.4 | 14.1 | ||
| Lab D | 33.6 | 51.0 | 27.3 | 4.3 | 20.8 | 27.3 | ||
| Lab E | 9.5 | 3.7 | 5.0 | 9.5 | 3.7 | 5.0 | ||
| Lab F | 29.0 | 21.7 | 71.2 | 29.0 | 21.7 | 19.4 | ||
| MAL | Lab A | 14.9 | 37.5 | 41.6 | 6.7 | 1.6 | 0.2 | |
| Lab B | 7.4 | 11.2 | 18.0 | 7.4 | 11.2 | 18.0 | ||
| Lab C | 5.5 | 7.9 | 31.4 | 5.5 | 7.9 | 31.4 | ||
| Lab D | 5.1 | 2.8 | 6.8 | 5.1 | 2.8 | 6.8 | ||
| Lab E | 5.4 | 6.0 | 2.8 | 5.4 | 6.0 | 2.8 | ||
| TCE | Lab A | 24.1 | 49.5 | 6.0 | 24.1 | 14.3 | 6.0 | |
| Lab B | 9.8 | 3.1 | 5.5 | 9.8 | 3.1 | 5.5 | ||
| Lab C | 25.3 | 12.8 | 24.1 | 6.4 | 12.8 | 24.1 | ||
| Lab D | 18.5 | 13.2 | 21.9 | 18.5 | 13.2 | 21.9 | ||
| Lab E | 6.2 | 37.8 | 28.7 | 6.2 | 37.8 | 28.7 | ||
| Average per dye | 15.5 | 24.9 | 19.9 | 11.9 | 20.1 | 14.4 | ||
| SD per dye | 11.3 | 20.1 | 16.4 | 7.6 | 18.1 | 9.3 | ||
| Interlaboratory | HCP | 38.3 | 59.5 | 57.1 | 44.2 | 59.5 | 52.5 | |
| PCP | 28.3 | 29.7 | 18.6 | 28.3 | 29.7 | 18.6 | ||
| DCB | 25.4 | 18.0 | 22.4 | 21.3 | 11.8 | 22.9 | ||
| DCA | 19.6 | 38.8 | 30.7 | 22.4 | 44.0 | 25.8 | ||
| MAL | 25.3 | 27.6 | 28.2 | 27.4 | 23.7 | 23.2 | ||
| TCE | 28.9 | 30.3 | 36.6 | 24.6 | 37.3 | 36.6 | ||
| Average per dye | 27.6 | 34.0 | 32.3 | 28.0 | 34.3 | 30.0 | ||
| SD per dye | 6.2 | 14.2 | 13.7 | 8.4 | 16.6 | 12.6 | ||
Figure 3.
Intralaboratory coefficients of variation (CoV) with respect to different combinations of laboratory (A–F), chemical (DCA, DCB, MAL, TCE, HCP, PCP), or cell viability indicator dye (metabolic activity—AB/PB; cell membrane integrity—CFDA-AM; and lysosomal membrane integrity—NR). For EC50 values underlying the CoV calculations, please refer to Table 1. All data are derived based on measured concentrations.
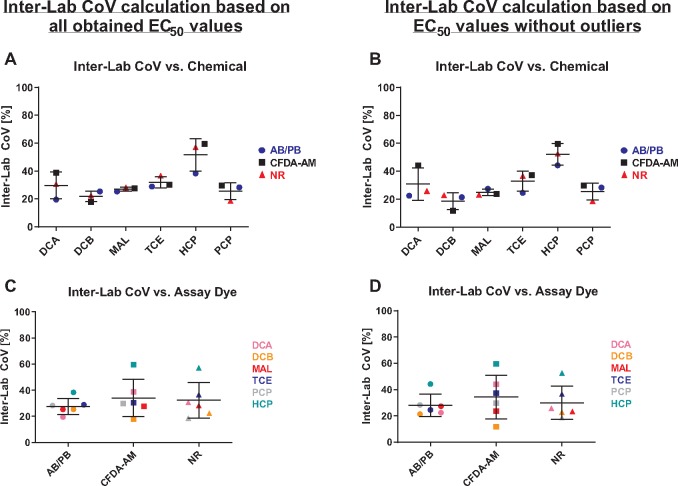
Figure 4.
Interlaboratory coefficients of variation (CoV) with respect to the combinations of chemical (DCA, DCB, MAL, TCE, HCP, PCP) and cell viability indicator dye (metabolic activity—AB/PB; cell membrane integrity—CFDA-AM; and lysosomal membrane integrity—NR). For EC50 values underlying the CoV calculations, please refer to Table 1. All data are derived based on measured concentrations.
Average CoVs for interlaboratory variability across all chemicals were between 21.9% and 31.9% for all chemicals except for HCP, where the interlaboratory CoV was 51.6%. When the outliers were removed, interlaboratory CoVs were between 18.7% and 32.8% for all chemicals other than HCP, for which the interlaboratory CoV was 52.1% (Figure 4A and 4B). Average interlaboratory CoVs across all cell viability indicator dyes were 27.6%, 34.0%, and 32.3% for metabolic activity (AB/PB), cell membrane integrity (CFDA-AM), and lysosomal membrane integrity (NR), respectively. In the same sequence of dyes, these values were 28.0%, 34.3%, and 30.0% when the outliers were removed (Figure 4C and 4D;Table 1). If the highest interlaboratory CoV for HCP was omitted, average interlaboratory CoVs across all cell viability indicator dyes were 25.5%, 28.9%, and 27.3% for, respectively, AB/PB, CFDA-AM, and NR; and 24.8%, 29.3%, and 25.4% if the outliers were removed.
In order to explore if the intralaboratory or interlaboratory variability is different between the dyes, CoVs were compared using a Kruskal–Wallis test. Neither intralaboratory nor interlaboratory CoVs differed significantly between the dyes; as well, for all chemical/dye combinations, the intralaboratory variability was smaller than the interlaboratory variability. This was true for both the full data set as well as for the data set with the outliers removed.
Nonetheless, when the EC50 values were compared, it became apparent that the measurements for cell- (CFDA-AM) and lysosomal (NR) membrane integrity yielded, on average, 1.8- and 1.3-fold higher EC50 values than for metabolic activity (AB/PB; SI Section 3.d, Supplementary Figure S6). Applying the LME model revealed that these differences were significant (p < .0001) for both, the full data set as well as when the outliers were removed. The linear model for the full data set furthermore showed that, as expected, the measured EC50 values depended significantly on the chemical used (F[5] = 2333, p < .0001), and that the chemicals alone explained approximately 98% of the variability in the measured EC50 values (adj R2 = 0.977). The dyes used affected the measured EC50 values to a very small extent, but the contribution was still statistically significant (F[2] = 41.85, p < .0001). Specifically, using dyes and chemicals as explanatory variables together explained 0.5% more of variability in the data set than chemical alone (adj R2 = 0.982 vs. 0.977 for chemical only). After removing individual dyes from the data set, the numbers remained similar, meaning that all 3 dyes contributed equally to the variability. The laboratory performing the experiments (random effect in the linear model) explained 30% of the remaining 1.8% unexplained variability, also significantly contributing to the model (χ2[1] = 16.77, p ≤ .0001). These results were essentially the same when the outliers were removed.
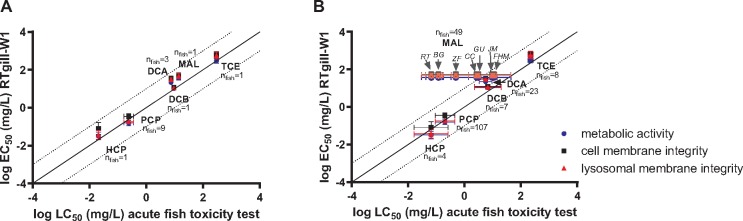
Correlation with Fish Acute Toxicity
The data obtained in this round-robin study fit very well into the previously established relationship between RTgill-W1 derived EC50 values and in vivo derived LC50 values (Figure 5A; SI Section 4.a, Supplementary Figure S7), with all data of this round-robin study falling within the 10-fold range of the line of unity, as expected from the historical data (Tanneberger et al., 2013). The in vivo data used in this earlier study originated from the US EPA fathead minnow database, which contains LC50 values for a large set of chemicals, all tested in a flow-through set-up and confirmed, ie, measured, exposure concentrations (Russom et al., 1997). In contrast, the US EPA Ecotox database contains much more diverse data from different species of fish, mostly from static exposures and nominal (ie, not measured) exposure concentrations. When these additional data were taken into account for the chemicals of this study (SI Section 4.b, Supplementary Figure S8 and Table S7—separate excel file), the excellent relationship between acute fish toxicity and the cell line data remained (Figure 5B). A marked exception is malathion. For this chemical, the database reveals highly variable LC50 values, which could be due to underlying species sensitivity differences or large experimental uncertainty. Rainbow trout and bluegill sunfish appear to be about one order of magnitude more sensitive to this chemical than zebrafish, and approximately 2 orders of magnitude more sensitive than common carp, guppy, medaka, and fathead minnow (SI Section 4.b, Supplementary Figure S8 and Table S7—separate excel file). The RTgill-W1 cell line EC50 data in this case closely match the LC50 values of the species with the lower sensitivity to malathion (Figure 5A and 5B).
Figure 5.
Correspondence of round-robin study-derived average log EC50 values obtained for the 3 different cell viability measurements per test chemical with average logLC50 values from fish acute toxicity testing. A, Correspondence of the cell line derived data with in vivo data obtained from the US EPA fathead minnow database (see Russom et al., 1997 and Tanneberger et al., 2013). B, Correspondence of the cell line derived data with the in vivo data for different fish species (SI Section 4.b). For Malathion (MAL), species for fish acute toxicity data are indicated: RT, rainbow trout (n = 19); BG, bluegill (n = 10); ZF, zebrafish (n = 3); GU, guppy (n = 1); CC, common carp (n = 12); JM, Japanese medaka (n = 1); FHM, fathead minnow (n = 3). All RTgill-W1 data are based on measured concentrations.
DISCUSSION
This study confirms the robustness of the RTgill-W1 cell viability assay: it can be easily established in different laboratories and its intralaboratory and interlaboratory variabilities are very well within the range of those previously reported for other fish bioassay-focused round-robin analyses (Busquet et al., 2014; Nichols et al., 2018). The mean intralaboratory variability of the RTgill-W1 assay in this study was 11.9% ± 7.6%, 20.1% ± 18.1%, and 14.3% ± 9.3% for cell metabolic activity (AB/PB), cell membrane (CFDA-AM), and lysosomal membrane (NR) integrity, respectively. The interlaboratory variability amounted to 28% ± 8.4%, 34.3% ± 16.6%, and 30.0% ± 12.6% for AB/PB, CFDA-AM, and NR, respectively, all with 5–6 chemicals tested in 4–6 laboratories and outliers being removed (Table 1). For comparison, in a round-robin study with the zebrafish embryo acute toxicity test (ZFET), intralaboratory and interlaboratory variability for the 96 h exposure set-up resulted in 13.6% ± 3.6% and 21.3% ± 15.4%, respectively (3–20 chemicals in 3–9 laboratories, outliers removed; Busquet et al., 2014). Furthermore, in a recently published study by Nichols et al. (2018) regarding the determination of intrinsic clearance rates of chemicals using rainbow trout hepatocytes, intralaboratory and interlaboratory variability values were 18.6% ± 3.0% and 32.4% ± 4.1% (5 chemicals and 6 labs). The fact that, apart from the lead laboratory, none of the participants of the RTgill-W1 study had previously worked with this fish cell line additionally supports the vigor of this cell line and this assay.
Exposure concentrations were analytically confirmed in this round-robin study at the beginning (C0h) and end (C24h) of exposure and concentration-response curves expressed on the basis of the geometric mean of these measured concentrations as previously performed (Natsch et al., 2018; Tanneberger et al., 2013). Compared with nominal (ie, intended) concentrations, EC50 values derived from these concentration-response curves differed in accordance to the degree of chemical loss, with the greatest difference being 4-fold for DCB as also observed in Tanneberger et al. (2013). Natsch et al. (2018) also found significant losses in their fragrance chemical data set over the 24 h incubation period owing to the fragrances’ physico-chemical properties. Yet, correlations of in vitro derived EC50 data versus in vivo LC50 values were excellent and very similar whether or not nominal or measured concentrations were taken into account. Natsch et al. (2018) therefore suggested that, in principle, nominal concentrations could be used directly to predict fish acute toxicity. We support this suggestion for certain applications, such as the screening of large numbers of chemicals in a prioritization exercise because omitting chemical analysis lowers the overall effort and cost significantly and also because chemical analysis is a common source of uncertainty. To back such a strategy, physico-chemical properties, along with dose-metric and/or mass balance considerations, can be consulted to estimate the need for chemical quantification (Armitage et al., 2014; Groothuis et al., 2015; Gülden and Seibert, 2005; Stadnicka et al., 2014).
One could argue that losses may also stem from biotransformation of the chemicals by the cells. Indeed, in a study to explore biotransformation for bioaccumulation prediction in fish, RTgill-W1 cells were shown to biotransform benzo(a)pyrene, albeit in the presence of 5% FBS in the exposure medium (Stadnicka-Michalak et al., 2018a). To what extent RTgill-W1 cells are capable of biotransformation within 24 h of exposure in L-15/ex is not yet known, though the relatively short exposure time combined with the simple nature of the medium suggests that biotransformation might, at minimum, be slowed. In any case, measured concentrations add valuable information for data interpretation and for credibility to the results and the chosen 24-well plate format provides sufficient material to accomplish quantification. Exposure quantification is a study requirement for most environmental test guidelines (eg, for the base set of toxicity evaluation: OECD 201 for algae [OECD, 2011]; OECD 202 for Daphnia [OECD, 2004]; OECD 203 for fish [OECD, 1992]). This round-robin study confirms that exposure quantification would not be a barrier for routine use of the assay.
The DCA was established as positive control chemical. A full concentration-response curve, based on nominal test concentrations, should be carried out approximately every sixth plate to prove proper assay functioning and consistency over time. The expected EC50 values and allowable ranges set in this round-robin test will aid in establishing the assay procedure in laboratories new to this assay and evaluate assay performance frequently as part of the intralaboratory quality control. An experienced technician can easily run six plates per batch twice per week (2× 6 plates), meaning that each of these runs would contain 1 plate of DCA and 5 test chemicals. The trend toward positive controls in environmental assays, as also included in the ZFET (OECD, 2013), is a step forward for establishing individual study validity, an aspect that other environmental test guidelines, such as the once mentioned above, could benefit from.
Three types of cell viability indicators were applied on the same set of cells. They ultimately indicate general damage of the cytoplasmic (cell) membrane, whereas 2 of them, AB/PB and NR, also allow the detection of subcellular alterations (Dayeh et al., 2013; Tanneberger et al., 2013). All 3 dyes attested to their reliability in this study by providing full concentration-response curves and CoVs that did not differ statistically within and between laboratories. However, on average, the EC50 values obtained for metabolic activity (AB/PB) were 1.8- and 1.3-fold lower than for cell- and lysosomal membrane integrity (CFDA-AM and NR), independent of the chemical’s mode of action. These results are, in fact, very similar to the findings by Tanneberger et al. (2013) and Natsch et al. (2018), where metabolic activity (AB/PB) overall gave the lowest EC50 values, followed by lysosomal (NR) and cell membrane (CFDA-AM) integrity. On the one hand, it could be reasoned that measuring metabolic activity with a resazurin-based dye is sufficient for cell viability screening (Natsch et al., 2018). In fact, because this assay is noninvasive, it could even be repeatedly applied to the same set of cells after chemical re-exposure or for determining the potential of cells to recover from chemical stress (Schirmer et al., 2000). This assay also showed the lowest intralaboratory and interlaboratory variability in this study, attesting to its particularly high experimental robustness. On the other hand, the additional cell viability measurements are easily added, especially with the dye to measure cell membrane integrity (CFDA-AM) being simply mixed with the resazurin-based dye (Schirmer et al., 1997). Thus, we value the prospect of distinguishing subcellular disruption of metabolic activity and/or lysosomal membrane integrity over the reduction of efforts if only one indicator dye is used.
The EC50 values obtained throughout this study compared very well with the fish LC50 values in accordance with Tanneberger et al. (2013). Obtaining EC50 values for the same test chemicals in different laboratories allowed in vitro–in vivo correlation on a broader set of data, and this comparison was expanded to include additional in vivo data (Figure 5B). Two important points can be deduced from this comparison. First, the variability with the fish cell line assay overall is much less than with the fish data, which speaks for the robustness and simplicity of the cell-based procedure. Second, in the case of malathion, fish data reveal apparent species sensitivity differences over several orders of magnitude, where the RTgill-W1 cell line assay more closely reflects the sensitivity of the less sensitive species of fish (namely common carp, guppy, medaka, and fathead minnow). Malathion is an acetyl choline esterase (AChE) inhibitor, which was somewhat surprisingly detected as a baseline toxic chemical in quantitative structure activity relationships for the RTgill-W1 cell line (in vitro) and for fathead minnow (in vivo; Tanneberger et al., 2013). In this prior study, we hypothesized that malathion is only a weak AChE inhibitor and primarily acts as a baseline toxicant in acute exposures scenarios in fish. It now seems that this hypothesis may not hold for all the species recommended by the OECD for fish acute toxicity testing, especially not for rainbow trout. With regard to the RTgill-W1 cell line, this finding underlines again that specific modes of neurotoxic action cannot be distinguished in the way the assay is currently carried out, as previously emphasized (Tanneberger et al., 2013). Gene editing to explicitly (over)express rainbow trout AChE in the RTgill-W1 cell line, or establishment of brain or muscle cell lines for chemical screening purposes, may be ways to combine the specific mode of action and baseline toxicity analyses for this species of fish. Considering the crudeness and severity of the fish acute toxicity test it becomes very important to integrate understanding of specific modes of action, which may not only result from fish cell-based assays but from nonfish species as well.
This international round-robin study clearly demonstrated the reliability of the RTgill-W1 cell line assay. Combined with its predictive capacity for fish acute toxicity it constitutes an alternative experimental route to conventional toxicity testing with fish. Indeed, the RTgill-W1 cell line assay is currently considered as draft international standard by ISO and has been proposed to be included into the OECD test guideline program. One other alternative is the ZFET, which is standardized under ISO (15088, 2007) and OECD (2013). In fact, a comparative analysis demonstrates that the outcomes of the RTgill-W1 cell line assay and the ZFET are similar (SI Section 4.c, Supplementary Figure S9). The ZFET comprises an organism model (albeit at early developmental stage) which requires the cultivation of fish. The RTgill-W1 cell line is a homogeneous, and commercially available culture which only requires sterile culture techniques. Thus, a decision for 1 or the other of these 2 alternative models maybe guided by the availability of resources and needs. Overall, the RTgill-W1 cell line assay requires even less time and testing material than the ZFET, it is even better suitable for higher throughput and it completely avoids the need to use fish for acute toxicity testing. The assay as such is also done in the absence of any supplements originating from animals. Only the serum component for routine RTgill-W1 culture remains an animal-derived resource in this assay, calling for the development of serum-free culture media for fish cell lines in the future. A rough calculation shows that serum of one calf (FBS) is sufficient to produce RTgill-W1 cells for about 150 assays, which are performed serum-free. As it stands, the RTgill-W1 cell line assay has great potential in taking a unique role not only in predicting fish acute toxicity but as well for defining nontoxic concentrations in fish for downstream applications (Stadnicka-Michalak et al., 2018b). Furthermore, it could be utilized as part of a weight of evidence approach, or in higher level strategies, such as the OECD fish testing framework (ENV/JM/MONO(2012)16, 2012), and Integrated Approaches to Testing and Assessment (IATA) as currently being developed for fish acute toxicity in project 2.54 of the OECD Test Guidelines Program.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DECLARATION OF CONFLICTING INTEREST
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the engagement of all project partners in this round-robin study despite not being able to financially support the work in the respective laboratories. We are grateful to Graham Whale for his unwavering support for this effort and for propagating this work in many different ways. Mike Karb supported the chemical analysis work at P&G. We thank Jenny Maner for collecting the additional fish acute toxicity (LC50) data.
FUNDING
We gratefully acknowledge funding of this round-robin study by CEFIC-LRI and UK NC3Rs (CEllSens Eco8.3). Significant additional resources were provided by the participating laboratories either in-kind or through grants: to RECETOX infrastructure (LM2015051 and CZ.02.1.01/0.0/0.0/16_013/0001761); to NIVA (Research Council of Norway project # 196318, ‘Non-animal [alternative] testing methods for REACH alterREACH’; Hultman, Lillicrap and Tollefsen).
REFERENCES
- Armitage J. M., Wania F., Arnot J. A. (2014). Application of mass balance models and the chemical activity concept to facilitate the use of in vitro toxicity data for risk assessment. Environ. Sci. Technol. 48, 9770–9779. [DOI] [PubMed] [Google Scholar]
- Belanger S. E., Rawlings J. M., Carr G. J. (2013). Use of fish embryo toxicity tests for the prediction of acute fish toxicity to chemicals. Environ. Toxicol. Chem. 32, 1768–1783. [DOI] [PubMed] [Google Scholar]
- Bols N. C., Barlian A., Chirino-Trejo M., Caldwell S. J., Goegan P., Lee L. E. J. (1994). Development of a cell-line from primary cultures of rainbow-trout, Oncorhyrnchus mykiss (Walbaum), gills. J. Fish Dis. 17, 601–611. [Google Scholar]
- Busquet F., Strecker R., Rawlings J. M., Belanger S. E., Braunbeck T., Carr G. J., Cenijn P., Fochtman P., Gourmelon A., Hübler N., et al. (2014). OECD validation study to assess intra- and inter-laboratory reproducibility of the zebrafish embryo toxicity test for acute aquatic toxicity testing. Regul. Toxicol. Pharmacol. 69, 496–511. [DOI] [PubMed] [Google Scholar]
- CEFIC-LRI. Available at: http://cefic-lri.org/projects/eco8-development-of-a-strategy-to-predict-acute-fish-lethality-using-fish-cell-lines-and-fish-embryos/ Accessed March 14, 2019.
- Dayeh V. R., Bols N. C., Tanneberger K., Schirmer K., Lee L. E. J. (2013). The use of fish-derived cell lines for investigation of environmental contaminants: An update following OECD’s fish toxicity testing framework No. 171. Curr. Protoc. Toxicol. 1.5.1–1.5.20. doi:10.1002/0471140856.tx0105s56 [DOI] [PubMed]
- ENV/JM/MONO(2012)16—Fish toxicity testing framework. Series on Testing and Assessent Number 171 (2012).
- ENV/JM/MONO(2001)8—OECD Series On Testing And Assessment Number 27 (2001). Guidance document on the use of the harmonized system for the classification of chemicals which are hazardous for the aquatic environment.
- Groothuis F. A., Heringa M. B., Nicol B., Hermens J. L. M., Blaauboer B. J., Kramer N. I. (2015). Dose metric considerations in in vitro assays to improve quantitative in vitro-in vivo dose extrapolations. Toxicology 332, 30–40. [DOI] [PubMed] [Google Scholar]
- Gülden M., Seibert H. (2005). Impact of bioavailability on the correlation between in vitro cytotoxic and in vivo acute fish toxic concentrations of chemicals. Aquat. Toxicol. 72, 327–337. [DOI] [PubMed] [Google Scholar]
- International Standardization Organization (ISO) (2007). Water quality – Determination of the acute toxicity of waste water to zebrafish eggs (Danio rerio). ISO 15088:2007. (E).
- Knöbel M., Busser F. J. M., Rico-Rico A., Kramer N. I., Hermens J. L. M., Hafner C., Tanneberger K., Schirmer K., Scholz S. (2012). Predicting adult fish acute lethality with the zebrafish embryo: Relevance of test duration, endpoints, compound properties, and exposure concentration analysis. Environ. Sci. Technol. 46, 9690–9700. [DOI] [PubMed] [Google Scholar]
- Lammer E., Carr G. J., Wendler K., Rawlings J. M., Belanger S. E., Braunbeck T. (2009). Is the fish embryo toxicity test (FET) with the zebrafish (Danio rerio) a potential alternative for the fish acute toxicity test? Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 149, 196–209. [DOI] [PubMed] [Google Scholar]
- Nagel R. (2002). DarT: The embryo test with the Zebrafish Danio rerio–A general model in ecotoxicology and toxicology. ALTEX 19(Suppl. 1), 38–48. [PubMed] [Google Scholar]
- Natsch A., Laue H., Haupt T., von Niederhäusern V., Sanders G. (2018). Accurate prediction of acute fish toxicity of fragrance chemicals with the RTgill-W1 cell assay. Environ. Toxicol. Chem. 37, 931–941. [DOI] [PubMed] [Google Scholar]
- Nichols J., Fay K., Bernhard M. J., Bischof I., Davis J., Halder M., Hu J., Johanning K., Laue H., Nabb D., et al. (2018). Reliability of in vitro methods used to measure intrinsic clearance of hydrophobic organic chemicals by rainbow trout: Results of an international ring trial. Toxicol. Sci. 164, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organisation for Economic Co-operation and Development (1992). OECD Guideline for Testing of Chemicals. Test No. 203: Acute fish test. OECD, Paris, France.
- Organisation for Economic Co-operation and Development (2004). OECD Guidelines for Testing of Chemicals. Test No. 202: Daphnia sp. Acute Immobilisation Test. OECD, Paris, France.
- Organisation for Economic Co-operation and Development (2011). OECD Guidelines for Testing of Chemicals. Test No. 201: Freshwater alga and cyanobacteria, growth inhibition test. OECD, Paris, France.
- Organisation for Economic Co-operation and Development (2013). OECD Guidelines for Testing of Chemicals. Test No. 236: Fish embryo acute toxicity (FET) test. OECD, Paris, France.
- Raue A., Kreutz C., Maiwald T., Bachmann J., Schilling M., Klingmüller U., Timmer J. (2009). Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 25, 1923–1929. [DOI] [PubMed] [Google Scholar]
- Russom C. L., Bradbury S. P., Broderius S. J., Hammermeister D. E., Drummond R. A. (1997). Predicting modes of toxic action from chemical structure: Acute toxicity in the fathead minnow (Pimephales promelas). Environ. Toxicol. Chem. 16, 948–967. [DOI] [PubMed] [Google Scholar]
- Schirmer K., Tanneberger K., Kramer N. I., Völker D., Scholz S., Hafner C., Lee L. E. J., Bols N. C., Hermens J. L. M. (2008). Developing a list of reference chemicals for testing alternatives to whole fish toxicity tests. Aquat. Toxicol. 90, 128–137. [DOI] [PubMed] [Google Scholar]
- Schirmer K., Chan A. G. J., Bols N. C. (2000). Transitory metabolic disruption and cytotoxicity elicited by benzo[a]pyrene in two cell lines from rainbow trout liver. J. Biochem. Mol. Toxicol. 14, 262–276. [DOI] [PubMed] [Google Scholar]
- Schirmer K., Dixon D. G., Greenberg B. M., Bols N. C. (1998). Ability of 16 priority PAHs to be directly cytotoxic to a cell line from the rainbow trout gill. Toxicology 127, 129–141. [DOI] [PubMed] [Google Scholar]
- Schirmer K., Chan A. G. J., Greenberg B. M., Dixon D. G., Bols N. C. (1997). Methodology for demonstrating and measuring the photocytotoxicity of fluoranthene to fish cells in culture. Toxicol. in Vitro 11, 107–119. [DOI] [PubMed] [Google Scholar]
- Stadnicka-Michalak J., Weiss F. T., Fischer M., Tanneberger K., Schirmer K. (2018a). Biotransformation of benzo[a]pyrene by three rainbow trout (Onchorhynchus mykiss) cell lines and extrapolation to derive a fish bioconcentration factor. Environ. Sci. Techol. 52, 3091–3100. [DOI] [PubMed] [Google Scholar]
- Stadnicka-Michalak J., Knöbel M., Županič A., Schirmer K. (2018b). A validated algorithm for selecting non-toxic chemical concentrations. ALTEX 35, 37–50. [DOI] [PubMed] [Google Scholar]
- Stadnicka J., Tanneberger K., Schirmer K., Ashauer R. (2014). Measured and modeled toxicokinetics in cultured fish cells and application to in vitro-in vivo toxicity extrapolation. PLoS One 9, e92303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanneberger K., Knoebel M., Busser F. J. M., Sinnige T. L., Hermens J. L. M., Schirmer K. (2013). Predicting fish acute toxicity using a fish gill cell line-based toxicity assay. Environ. Sci. Technol. 47, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Tanneberger K., Rico-Rico A., Kramer N. I., Busser F. J. M., Hermens J. L. M., Schirmer K. (2010). Effects of solvents and dosing procedure on chemical toxicity in cell-based in vitro assays. Environ. Sci. Technol. 44, 4775–4781. [DOI] [PubMed] [Google Scholar]
- Utox-github: https://github.com/UtoxEawag/RTgillRoundRobin. Accessed March 14, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.