Abstract
Mammalian cardiac myocytes (CMs) stop proliferating soon after birth and subsequent heart growth comes from hypertrophy, limiting the adult heart’s regenerative potential after injury. The molecular events that mediate CM cell cycle exit are poorly understood. To determine the epigenetic mechanisms limiting CM cycling in adult CMs (ACMs) and whether trimethylation of lysine 9 of histone H3 (H3K9me3), a histone modification associated with repressed chromatin, is required for the silencing of cell cycle genes, we developed a transgenic mouse model where H3K9me3 is specifically removed in CMs by overexpression of histone demethylase, KDM4D. Although H3K9me3 is found across the genome, its loss in CMs preferentially disrupts cell cycle gene silencing. KDM4D binds directly to cell cycle genes and reduces H3K9me3 levels at these promotors. Loss of H3K9me3 preferentially leads to increased cell cycle gene expression resulting in enhanced CM cycling. Heart mass was increased in KDM4D overexpressing mice by postnatal day 14 (P14) and continued to increase until 9-weeks of age. ACM number, but not size, was significantly increased in KDM4D expressing hearts, suggesting CM hyperplasia accounts for the increased heart mass. Inducing KDM4D after normal development specifically in ACMs resulted in increased cell cycle gene expression and cycling. We demonstrated that H3K9me3 is required for CM cell cycle exit and terminal differentiation in ACMs. Depletion of H3K9me3 in adult hearts prevents and reverses permanent cell cycle exit and allows hyperplastic growth in adult hearts in vivo.
Keywords: Epigenetics, Cell cycle gene expression, Cardiac myocyte cell cycle, Cardiac development
Introduction
Cardiac myocyte cell cycle activity displays distinct developmentally dependent patterns. Fetal and neonatal CMs proliferate robustly and neonatal mice can replace lost myocytes even after their hearts have been amputated [1]. However, this regenerative capacity is soon lost and mature CMs lose the capacity to proliferate, a process often called terminal differentiation [2, 3]. Although there is limited DNA-synthesis in adult CM (ACMs) after injury, it primarily results in polyploidization and multi-nucleation, rather than CM division [4]. In fact, ACM division in vivo is an extremely rare event [1, 2, 5]. The basis for the essentially irreversible cell cycle exit is poorly understood but cell cycle genes are dramatically downregulated in ACMs compared with embryonic CMs [6]. Further, this downregulation appears to be associated with permanent silencing of these genes. Although, subjecting adult hearts to a growth stimulus leads to upregulation of early G1/S-phase genes, genes that promote later cell cycle phases, mitosis and cytokinesis, remain silenced [6]. As G1/S-phase genes are required for CM hypertrophy [7, 8] their reexpression is consistent with the hypertrophy-restricted growth and the increased DNA-content displayed in ACMs after trans-aortic-constriction (TAC) or myocardial infarction (MI) [4, 9]. Interestingly, the switch to hypertrophic growth in CMs coincides developmentally with the postnatal loss of regeneration capacity [1, 2, 9]. Thus, ACMs have cell size growth that is uncoupled from cell division [10], which can be attributed to selective silencing of mitosis- and cytokinesis-progression genes [6].
The stable silencing of G2/M and cytokinesis genes is a major component of the transcriptome change that occurs when CMs undergo terminal differentiation [9]. Epigenetic mechanisms, such as post-translational modifications of histone proteins, DNA methylation, and non-coding RNAs, have been implicated in directing changes in gene expression that occur during cardiac development and disease but little is known about their role in mediating terminal differentiation [2, 11–13]. Simplistically, there are two types of epigenetically-defined chromatin structure and function: actively transcribed euchromatin and transcriptionally silenced heterochromatin [14, 15]. Each chromatin type is associated with distinct sets of histone modifications and chromatin-associated proteins [15–17]. Histone modifications are thought to establish different states of chromatin by physically altering its structure [18–20], as well as recruiting other adapter and effector proteins which possess modification-specific-binding domains [14, 15]. In general, euchromatin is enriched with histone acetylations, H3K4me3, and H3K36me3, which recruit transcriptional activators [15]. In contrast, heterochromatin is enriched with H3K9me3, H3K27me3, and DNA methylation: repressive methylations that recruit heterochromatin-protein-1 (HP1) family members, polycomb proteins, and other repressive factors [14, 15]. However, it is unclear how these epigenetic mechanisms regulate chromatin accessibility and cell cycle gene expression in ACMs.
The stable cell cycle exit of terminally differentiated cells is shared with senescent cells, which do not proliferate in response to mitogenic stimuli [21, 22]. This permanent cell cycle exit in senescent cells has been proposed to be mediated by Retinoblastoma (Rb)-dependent recruitment of DNA methyltransferases, histone deacetylase HDAC1, and H3K9me3- and H3K27me3-associated proteins to E2F-dependent gene promoters [6, 23–25], as well as through recruitment of repressive DREAM (Rb-E2F-MuvB) complex members that prevent MuvB-Myb-FoxM1 activation of cell cycle genes [26]. ACM-specific knockout of Rb combined with deletion of related Rb-family member p130 resulted in disruption of heterochromatin and cell cycle re-entry [6]. However, how each component of heterochromatin was affected and which were critical for cell cycle activation was unclear. Surprisingly, CM-specific knockout of H3K27me3 methyltransferase Ezh2 resulted in reduced CM proliferation due to increases in expression of cell cycle repressors [27], and CM-specific combined knockout of DNA methyltransferases Dnmt3A/B did not alter cell cycle gene expression in transgenic mouse studies [28] suggesting other epigenetics marks were the critical mediators in ACMs. Interestingly the role of the third heterochromatin marker, H3K9me3, has not been studied in vivo.
To test the significance of H3K9me3 in regulating ACM cell cycle gene silencing we generated a transgenic mouse model where H3K9me3-demethylase KDM4D was overexpressed specifically in CMs. This depleted H3K9me3 and resulted in increased cardiac myocyte proliferation and cell cycle gene expression into adulthood.
Methods
Mouse Studies
All animal studies were performed in accordance with an approved Institutional Animal Care and Use Committee (IACUC protocol #4290-01), the University of Washington institutional guidelines, and the National Institute of Health Guide for the Care and Use of Laboratory Animals. The αMHC-tTA mice used to control transgene expression was generated by the Robbins lab [29]. We used the previously published responder construct, which possesses a tetracycline responsive element upstream of an attenuated αMHC promoter to drive KDM4D expression [29]. Plasmid containing FLAG- and MYC-tagged human KDM4D cDNA (Origene RC212600) had a NotI restriction site present in the cDNA sequence, which we destroyed by inducing a silent mutation (Agilent 200521). The resulting cDNA was subcloned into the responder construct, then freed of vector backbone, purified, and injected into mouse pronuclei (University of Washington transgenic core facility). The resulting tet transgenic was bred to the αMHC-tTA line to generate the CM-specific KDM4D induction model (n=48). Littermate controls were used for all experiments involving transgenic mice (n=93) from breeders backcrossed 8 generations to the C57/B6 strain.
RNA and DHS sequencing
Libraries were constructed for control and BiTg ACMs (Illumina Tru-Seq) and paired-end sequencing (ABI3730XL) was performed by the University of Washington core facility. Read alignment was performed using Bowtie/Cufflinks package. DHS peaks were called using MACS. Partek Genomic Suites was used for mRNA quantification, PCA, differential expression analysis (0.1FDR adjusted p<0.05), and gene ontology using Fisher’s Exact Test and only considering GO categories with at least 3 genes. Sequencing data is available through the NIH’s SRA repository (Bioproject Accession number PRJNA309702). Detailed analysis of all gene expression in control and BiTg ACMs, all significantly upregulated genes, and all GO term enrichment scores are provided (Supplemental Tables 1 through 3).
Imaging of thick sections
For unambiguous determination of cell type in our phospho-H3 staining assays we developed a method for generating, staining, and imaging 100μm-thick heart sections, which will be described in detail in a methodologies article. Briefly, hearts were arrested in KB buffer, perfused with KB, then perfusion fixed with methanol cooled to −20°C. The hearts were rehydrated in Methanol:PBS gradients (100:0, 80:20, 60:40), then washed with PBS and mounted in 5% low-melt agarose. 100μm-thick sections were cut from a Leica 1200s vibratome and were stained in suspension with the following reagents: phospho-H3 (Abcam ab5176), Hoechst (Life Technologies H3570), Phalloidin (ThermoFisher Scientific A22287), and Wheat Germ Agglutinin (WGA, Life Technologies W6748). The stained sections were mounted to glass coverslips coated with 0.01% poly-L-lysine. To increase the transparency of the sections, which is needed to view the interior of the thick sections, they were cleared: sections were incubated in an isopropanol series (70%, 85%, 95%, 100%) followed by incubations in a 1:2 solution of benzyl alcohol and benzyl benzoate. The samples were prepared, imaged with confocal microscopy, and analyzed by a single operator blinded to the genotypes.
2-D Echocardiography
Under 0.5% isoflurane, mice EKG and heart function was assessed using Visual Sonics Vevo 2100. Parasternal short axis images at the plane of the papillary muscle were collected in B- and M-Modes. Images were collected with heart rates ranging from 400–500 BPMs. Imaging and analysis was performed by a single operator who was blinded to the genotypes. Quantification of images was performed using Vevo Labs 1.7.0, according to the manufacturer’s guidelines.
Results
Epigenetic regulation of cell cycle genes in CMs
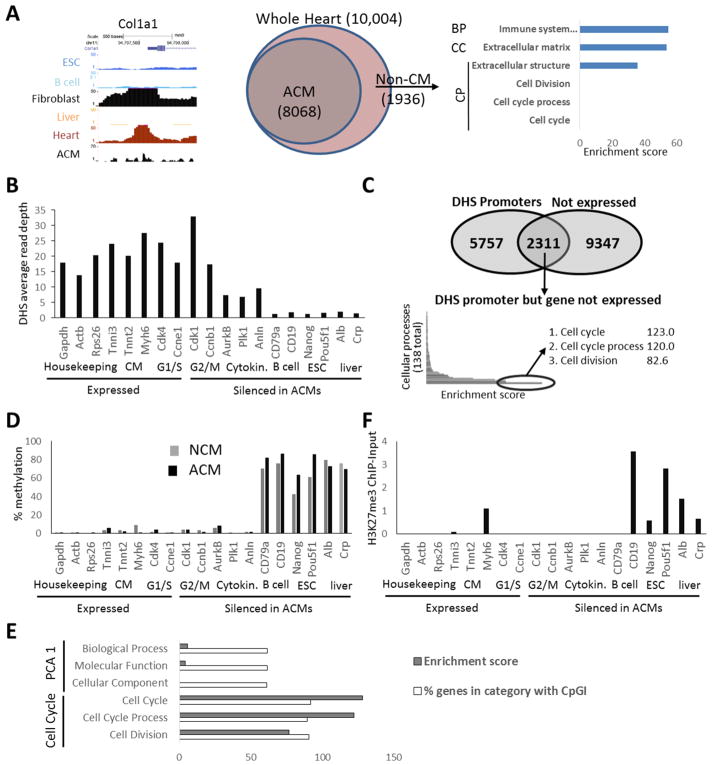
We hypothesized that cell cycle genes are silenced in ACMs through incorporation into heterochromatin or at least tightly packed chromatin that is inaccessible to transcriptional machinery. To investigate this we analyzed the mouse ENCODE DnaseI-hypersensitive (DHS) sequencing datasets[30]. Accessibility of genes to transcription factors and other proteins can be measured by mapping areas of the chromatin sensitive to DnaseI cleavage. As expected DHS signal was observed at the promoters of active genes, with ubiquitously expressed promoters having DHS peaks in all tissues and tissue specific promoters having DHS peaks only in their respective tissues (Supplemental Figure 1). Surprisingly, in adult heart samples, cell cycle gene promoters, including silenced mitosis and cytokinesis genes, had prominent DHS peaks suggesting the promoters were accessible (Supplemental Figure 1). Since these assays were performed on whole heart tissue, which includes non-CM cells that can proliferate in response to stimuli, we reasoned they might be the contrib utor of this DHS signal in the whole heart samples. Therefore, we generated DHS sequencing data sets from isolated ACMs and found the same peaks were present at all cell cycle genes examined (Supplemental Figure 1). Genome-wide analysis confirmed the whole heart samples had additional DHS gene promoters not present in the ACMs consistent with fibroblast and blood cell genes, such as Col1A1, and that the whole heart samples displayed no enrichment for DHS cell cycle gene promoters compared to ACM samples (Figure 1A). These results suggested stably silenced cell cycle genes have accessible promoters in ACMs (Figure 1B and 1C).
Figure 1. Cell cycle gene promoters are accessible and spared from heterochromatin markers in ACMs.
(A) DHS at Col1a1 promoter and GO analysis of DHS+ gene promoters derived from non-CM (1936 genes) in whole heart preparation showing the most enriched term within PCA-level-1 Biological process (BP), Cellular Component (CC), and the most enriched term and cell cycle related terms within Cellular Processes (CP). (B) DHS at expressed and silenced (RPKM<1) gene promoters (TSS +/− 500bp) in ACMs. (C) GO was performed on a list of genes that are not expressed in ACMs (RPKM<1) but display a DHS peak in their promoter. The 3 most enriched terms and enrichment scores (-log(enrichment p-value)) within the cellular processes GO categories are listed. (D) Average % methylation of all CpG dinucleotides within promoter regions of indicated genes in neonatal CMs and ACMs. (E) A list of all genes containing CpG Islands in the promoter region was analyzed by GO, the % of genes with a CpGI-containing promoter compared to all genes within the categories are shown for PCA level 1 and cell cycle related terms. (F) H3K27me3 ChIP normalized to input at promoter regions in ACMs. Sample Number: (A-E) N=1 (ENCODE/publicly available), ACM RNA-seq (expressed vs not expressed) N=2.
To determine whether other epigenetic factors implicated in heterochromatin formation were controlling the silencing, we analyzed DNA methylation patterns using whole-genome bisulfide sequencing datasets [31] from purified ACMs (Figure 1D). Intergenic regions and the promoters and gene bodies of silenced, non-CM-lineage genes were hypermethylated. In contrast, housekeeping- and CM-gene promoters were hypomethylated. Cell cycle gene promoters were hypomethylated (Figure 1D). Moreover, this pattern does not appear to change during postnatal development when these genes are dramatically downregulated as neonatal CMs displayed a similar profile (Figure 1D). Whole-genome analysis of CpG Islands, several hundred base-pair regions in the genome with a high density of CpG dinucleotides that are known to be hypomethylated, showed cell cycle genes were particularly enriched with CpG island-containing promoters, with about 90% of cell cycle-related genes having CpG Islands in their promoters (Figure 1E). We also examined H3K27me3 in ACMs[31], which has been implicated in facultative heterochromatin but did not find enrichment at cell cycle gene promoters (Figure 1F).
Generation of a transgenic mouse model to deplete H3K9me3 specifically in CM
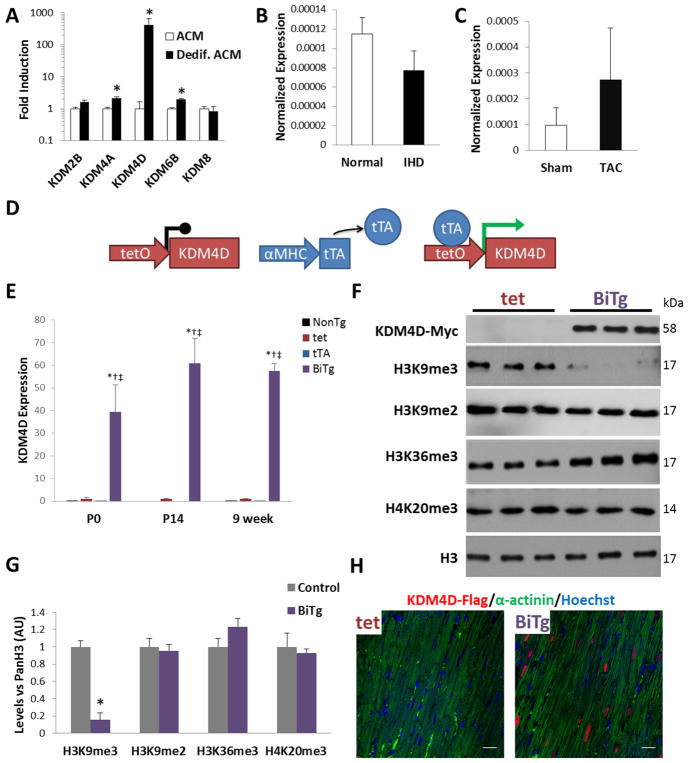
Since DNA methylation and H3K27me3 did not appear to silence cell cycle genes in ACMs, we wished to determine the role of H3K9me3, the classic histone modification associated with heterochromatin, since it is found on cell cycle gene promoters in ACMs (Figure 4C) and is enriched compared to fetal CMs [6]. To deplete H3K9me3 we examined members of the KDM4 family of H3K9 demethylases. The KDM4 family have divergent demethylase activity but KDM4D is specific to H3K9me3 [32–34]. KDM4D is expressed in proliferative fetal CMs that highly express cell cycle genes, downregulated in ACMs and is elevated in ACMs dedifferentiated in vitro that regain cell cycle activity (Supplemental Figure 2 and Figure 2A). It is not re-expressed in either mouse or human cardiomyopathy samples where hypertrophic growth predominates (Figure 2B and 2C).
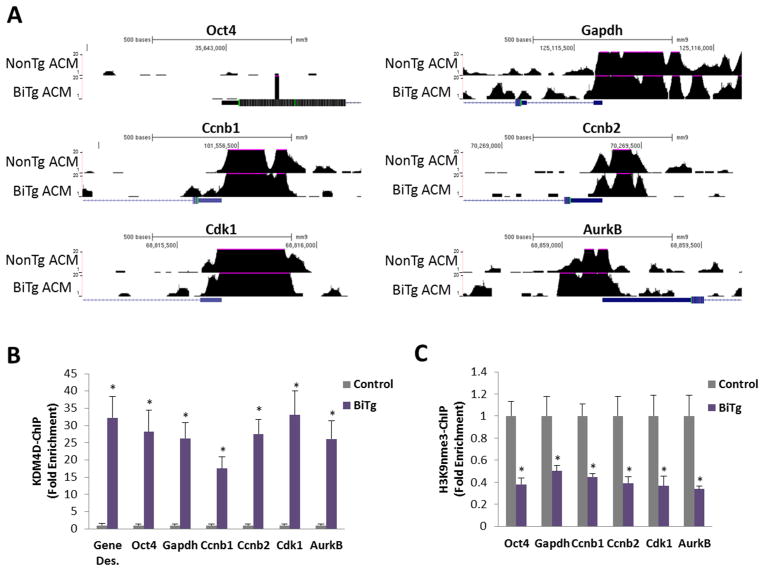
Figure 4. KDM4D binding and H3K9me3 status, but not accessibility, are altered in gene promoters in BiTg ACMs.
(A) Genome browser views of DnaseI-hypersensitivity sequencing at promoter regions (+/− 500bp from transcription start site) of stem cell (silenced), housekeeping (active), and cell cycle genes. (B) KDM4D (Myc-tag) ChIP-PCR in adult ventricle samples analysis of indicated gene promoters. (C) H3K9me3 ChIP-PCR in isolated ACMs. Sample Number: (B–C) Control=3, BiTg=3. Statistics: (B–C) Two-tailed T-test, control vs. BiTg, * P<0.05.
Figure 2. Generation of cardiac myocyte-specific KDM4D model.
(A) Gene expression of HDMs in dedifferentiated mouse ACMs relative to ACMs. (B) KDM4D expression in human ischemic cardiomyopathy samples (IHD) and in (C) ACMs 4-weeks post sham or TAC surgery, expression normalized to GAPDH. (D) Schematic of BiTg model for CM-specific KDM4D induction. (E) KDM4D transgene RNA expression is robustly induced in BiTg P0 and P14 hearts and ACMs, fold induction vs. tet control. (F) KDM4D protein induction and global levels of specific histone methylations in 9-week ACMs. (G) Densitometry quantification of Western Blot. (H) BiTg mice display nuclear KDM4D (FLAG-tag) localization specifically in CMs, bar=20μm. Sample Number: (A) ACM=3, Dedif. ACM=3. (B) Normal=2, IHD=3. (C) Sham=3, TAC=3. (E–G) 3 animals per group. Statistics: (A–C) Two-tailed T-test, * P<0.05. (E) One-way ANOVA/Tukey’s test, * P<0.05 vs NonTg, † P<0.05 vs tet, ‡ P<0.05 vs tTA. (G) Two-tailed T-test, control vs. BiTg, * P<0.05.
We generated a CM-specific H3K9me3-depletion transgenic mouse model containing a MYC- and FLAG-tagged KDM4D cDNA downstream of a tetracycline responsive promoter [29]. Breeding tTA mice with tet-responsive KDM4D (tet) mice yields bi-transgenic (BiTg) mice that constitutively express KDM4D specifically in CMs (Figure 2D) as well as single-transgenic (tet or tTA) and non-transgenic (NonTg) controls. We confirmed that KDM4D expression was robustly induced in BiTg hearts at P0, P14, and 9 weeks (Figure 2E). The KDM4D transgene expression was not detectable in other organs in BiTg mice or non-CM cardiac cells (Supplemental Figure 3), with the exception that low levels could be detected in BiTg lungs, consistent with previous reports using the αMHC promoter[29]. Western blot analysis confirmed KDM4D protein expression and showed global H3K9me3 levels were depleted 84.5% in BiTg ACMs compared to controls (P<0.001, Figure 2F and 2G). Immunofluorescence imaging in heart sections confirmed exogenous KDM4D protein was specifically expressed and localized in the nuclei of BiTg CMs (Figure 2H). In contrast to other KDM4 family members (Supplemental Figure 4), KDM4D demethylase activity is specific to H3K9me3 [32–34] and did not demethylate H3K9me2 or H3K36me3 in ACMs (Figure 2F and 2G).
H3K9me3 is required for CM cell cycle gene silencing
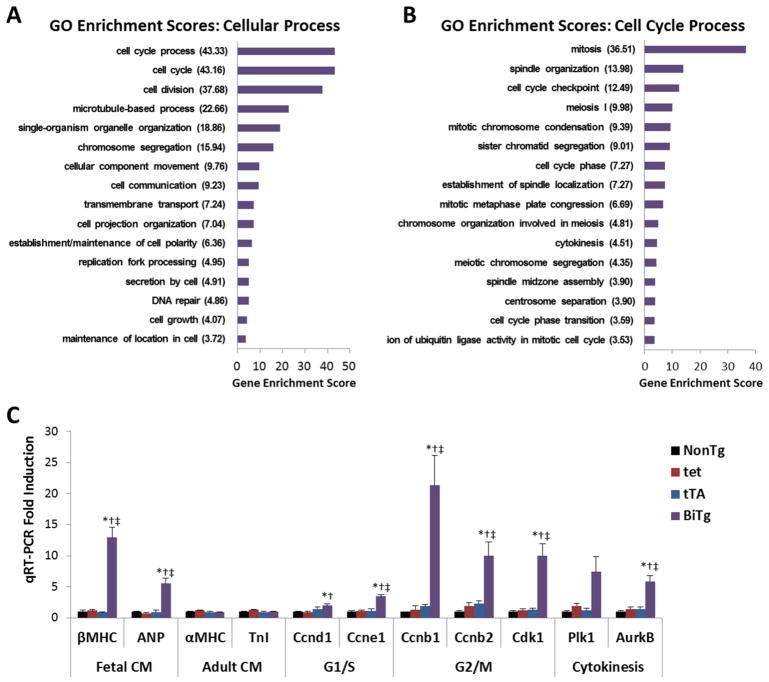
To assess the role of H3K9me3 depletion on cardiac gene expression in vivo we performed RNA-sequencing on ACMs isolated from control and transgenic 9-week-old hearts. Control ACM samples were grouped since NonTg and single transgenic mice showed no differences in gene expression, with the unconstrained slope correlation test showing R2=0.9764 when comparing the whole-genome transcriptome, and principle component analysis (PCA) revealed biological replicates within each group clustered (Supplemental Figure 5). BiTg ACMs had increased expression of genes involved in 16 of 138 cellular processes (Figure 3A). Of these identified cell processes, those involved in later phases of cell cycle were increased preferentially, particularly mitosis (Figure 3B). Of the 7082 GO functional groups, genes in the cell cycle processes were the most overrepresented among genes with increased expression in BiTg ACMs. We confirmed increases in G2/M and cytokinesis genes in BiTg ACMs compared to controls by qPCR (ranging from 5.8- to 21.4-fold, P<0.01; Figure 3C and Supplemental Figure 6). Fetal CM-specific genes were also increased (Figure 3C). Although the expression of fetal CM genes is frequently associated with a pathologic state, it should be noted that expression of less mature CM-specific genes could also be consistent with proliferation-competent CMs in fetal and neonatal hearts [6, 35] (Supplemental Figure 2).
Figure 3. Gene expression in KDM4D-overexpressing ACMs.
(A) Gene Ontology Enrichment scores for “Cellular Process”, and (B) “Cell Cycle Process”. GO enrichment scores, the log(Enrichment p-values), were generated from lists containing genes with >3 fold increase in BiTg ACMs at 9 weeks compared controls. (C) Expression of CM and cell cycle genes in 9-week ACMs measured by qRT-PCR, fold induction vs. NonTg. Sample Number: (A–B) Control=2, BiTg=2. (C) NonTg=3, tet=6, tTA=3, BiTg=5. Statistics: (A–B) One-way ANOVA was used to identify genes with significantly altered expression (FDR corrected P<0.05), Fisher’s exact test was used to identify GO terms with significant enrichment scores (P<0.05). (C) One-way ANOVA/Tukey’s test, * P<0.05 vs NonTg, † P<0.05 vs tet, ‡ P<0.05 vs tTA.
To assess the mechanism by which KDM4D increases cell cycle gene expression in ACMs we performed DHS sequencing in control and BiTg ACMs. KDM4D expression did not alter chromatin conformation of stem cell or housekeeping gene promoters, nor cell cycle gene promoters (Figure 4A), which were already accessible in control ACMs (Figure 1 and Supplemental Figure 1). To understand whether KDM4D-mediated cell cycle gene activation was a direct or indirect mechanism we performed chromatin immunoprecipitation for MYC-tagged-KDM4D and H3K9me3 in control or BiTg ACMs. KDM4D binding was enriched on G2/M and cytokinesis gene promoters (Figure 4B) and this was accompanied with reduced H3K9me3 (Figure 4C). However, this pattern was also observed at gene deserts, active gene promoters, and non-lineage gene promoters that did not have increased mRNA expression in BiTg ACMs (Figure 4B and 4C).
H3K9me3 depletion delays CM cell cycle exit
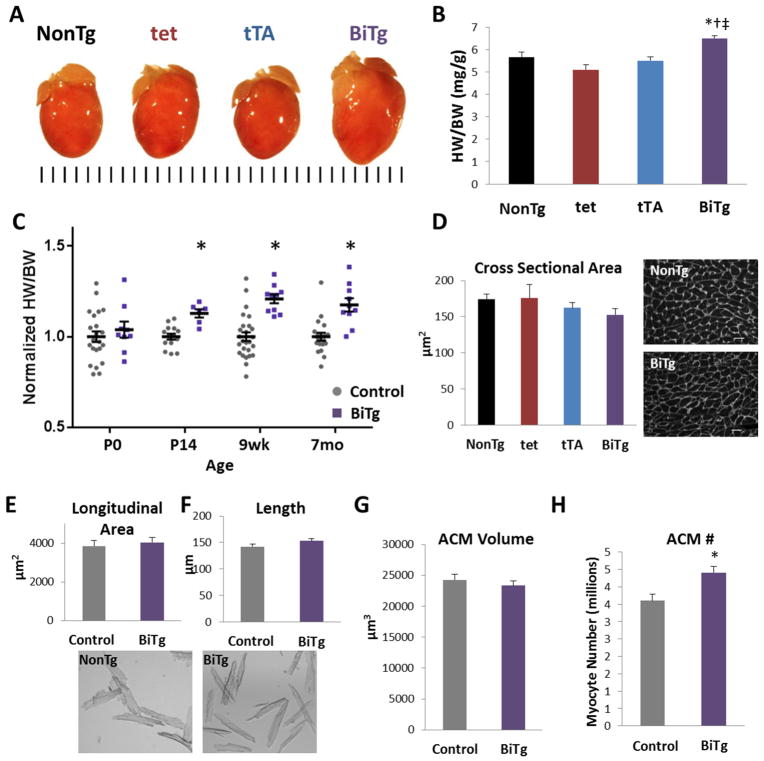
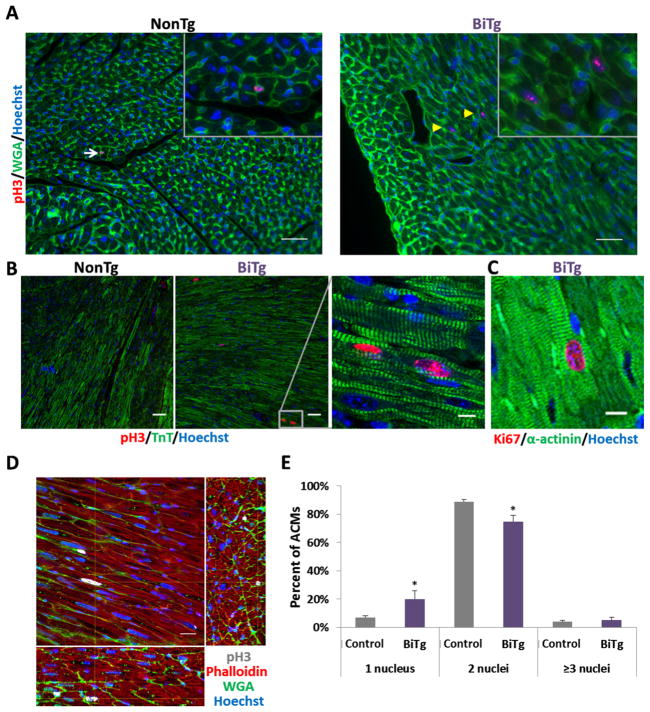
BiTg mice had visibly larger hearts (Figure 5A) with a 20.8% increase in heart weight to body weight ratio (HW/BW) at 9 weeks (P<0.0001; Figure 5B) with no change in body weight (Supplemental Figure 7). This increase in HW/BW first became apparent at P14 (12.9% increase, P<0.001; Figure 5C); suggesting KDM4D overexpression specifically promoted postnatal cardiac growth. This cardiac enlargement was not associated with sarcomere disarray, fibrosis or alteration of vasculature (Figure 2H and Supplemental Figure 8) and there was no increase in extracellular matrix[36] or apoptosis (Supplemental Figure 8B and 8C). To differentiate between cardiac hypertrophy or hyperplasia as the cause of the increased heart mass, we quantified ACM size. Cross sectional area (Figure 5D) and direct measurements of longitudinal area and length did not differ between ACMs isolated from BiTg or controls (Figure 5E and 5F). ACM volume was also unchanged (Figure 5G). Calculated myocyte number suggested BiTg hearts had 22% more ACMs compared to controls (P<0.03; Figure 5H). Consistent with this finding, CM cell cycle activity was increased in BiTg hearts. We immunostained P14 and 9 week myocardial sections for phosphorylated histone H3 serine-10 (pH3), a marker of mitosis. We observed pH3+ CMs only in BiTg hearts at both time points (Figure 6A and 6B). Similar trends were seen for the general cell cycle activity marker Ki67 in 9-week hearts (Figure 6C). We confirmed mitotic activity in BiTg ACMs in thick myocardial sections that allow 3D examination of pH3+ nuclei (Figure 6D). Quantification of the number of nuclei per ACM revealed there was an increase of mononucleated and a decrease in binucleated ACMs in BiTg hearts (Figure 6E). These findings are consistent with a model where the increased heart mass in BiTg mice is secondary to CM hyperplasia. Long-term H3K9me3-depletion had no significant effect on cardiac function or morphology (Table 1).
Figure 5. Heart mass and ACM number are increased in KDM4D induced mice.
(A) Representative image showing PFA-fixed BiTg and control hearts at 9 weeks, tick marks=1mm. (B) Quantification of HW/BW at 9 weeks showing cardiac growth phenotype is specific to BiTg mice. (C) Quantification of HW/BW in different ages of mice, normalized to controls for each time point. (D) Left, quantification of ACM cross sectional area. Right, WGA staining in 9-week NonTg and BiTg PFA-fix hearts, bar=20μm. (E) Quantification of longitudinal area and (F) length measured in dispersed, isolated 9-wk CMs, with representative images below. (G) Calculated ACM volume and (H) ACM number at 9-wks of age. Sample Number: (A–B) NonTg=6, tet=11, tTA=9, BiTg=10. (C) P0, Control=22, BiTg=9; P14, Control=14, BiTg=6; 9wk Control=26, BiTg=10; 7mo Control=19, BiTg=10. (D–H) Each assay had 3 animals per group. Statistics: (B,D) One-way ANOVA/Tukey’s test, * P<0.05 vs NonTg, † P<0.05 vs tet, ‡ P<0.05 vs tTA. (C, E, F) Two-tailed T-test, control vs. BiTg, * P<0.05. (G–H) The Bootstrap method was used to compute standard error and Permutation test was used to compute p-value, * P<0.05.
Figure 6. Persistent low level cardiac myocyte cell cycle activity in adult BiTg hearts.
Mitotic marker phospho-H3 (pH3) staining in NonTg and BiTg heart sections (A) at P14 and (B) 9 weeks, bar=40μm. (A) White arrows point to pH3+ non-CM nuclei, yellow arrowheads point to pH3+ CM nuclei. (B) Right, high magnification of boxed region, bar=10μm. (C) Cell cycling marker Ki67 in 9-week BiTg hearts, bar=10μm. (D) Thick section imaging of adult BiTg heart showing XY plane and reconstructed 75μm depth of XZ and YZ orthogonal planes. Yellow crosshairs indicate position within all 3 planes, bar=20μm. (E) Quantification of nuclei number in 7 month old ACMs. Sample Number: (A–E) Each assay had 3 animals per group. Statistics: (E) Two-tailed T-test, control vs. BiTg, * P<0.05.
Table 1. Normal cardiac function and morphology in BiTg mice at 7 months.
| NonTg | tet | tTA | BiTg | |
|---|---|---|---|---|
| HR (BPM) | 450 ± 9 | 452 ± 11 | 441 ± 11 | 422 ± 10 |
| EF (%) | 74.1 ± 2.3 | 74 ± 4.4 | 79.2 ± 3.1 | 73.8 ± 4.3 |
| FS (%) | 42.1 ± 2.3 | 43 ± 3.8 | 47.6 ± 3.2 | 43 ± 3.8 |
| CO (mL/min) | 15.1 ± 2 | 19.8 ± 1 | 16.4 ± 1.1 | 18.8 ± 1.6 |
| LVEDD(mm) | 3.33 ± 0.13 | 3.73 ± 0.02 | 3.38 ± 0.09 | 3.74 ± 0.1 |
Echocardiography results in 7 month old mice. HR: Heart Rate, EF: Ejection Fraction, CO: Cardiac Output, LVEDD: Left Ventricular End-Diastolic Dimension. Mean and SEM values are shown. Sample Number: N 5 for each genotype. Statistics: One-way ANOVA/Tukey’s test,
P<0.05 vs NonTg,
P<0.05 vs tet,
P<0.05 vs tTA.
H3K9me3 is required for establishment and maintenance of the ACM terminally differentiated state
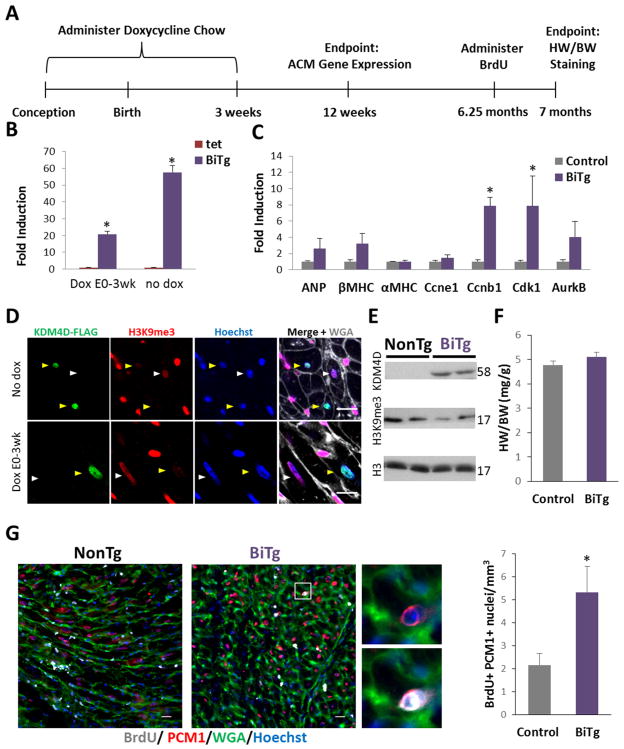
To confirm that the phenotype we observed in BiTg animals was dependent on KDM4D expression in ACMs, we utilized doxycycline to temporally control CM-specific KDM4D expression (Supplemental Figure 9A and 9B). At 2 weeks of age mice were given doxycycline, and by 3 weeks of age no detectable transgene expression was observed (Supplemental Figure 9C). We maintained control and BiTg mice on doxycycline through the endpoint at 9 weeks and did not see significant differences in HW/BW (Supplemental Figure 9D) or expression of cell cycle genes (Supplemental Figure 9E) indicating continued KDM4D expression in ACMs was required to prevent terminal differentiation and allow the hyperplasia phenotype.
To address whether H3K9me3-depletion could reverse terminal differentiation, we suppressed KDM4D expression from conception through 3 weeks of age (Figure 7A). Transgene expression was not detected 3 weeks after removing doxycycline (data not shown); however, 9 weeks after doxycycline withdrawal, we saw induced KDM4D mRNA (Figure 7B), though at lower levels compared to the constitutive model. In this model, cell cycle genes were upregulated (Figure 7C). We examine KDM4D and H3K9me3 levels 6 months post doxycycline withdrawal and confirmed induction of exogenous KDM4D protein and reduced H3K9me3 levels (Figure 7D and 7E). While these mice did not display significantly increased heart mass (Figure 7F), we observed a 2.5-fold increase in cycling PCM1+ ACMs (P<0.03) with ACM-specific KDM4D induction (Figure 7G).
Figure 7. ACM-specific KDM4D overexpression induces ACM cell cycling.
(A) Timeline showing protocol for ACM-specific KDM4D expression and endpoints. (B) qRT-PCR of KDM4D expression in BiTg ACMs with or without Dox treatment, showing KDM4D expression can be partially recovered after 9 weeks after removing dox from chow. (C) qRT-PCR analysis of control and BiTg in 12 week CMs, 9 weeks post dox withdrawal. (D) H3K9me3 and KDM4D-FLAG co-staining in cardiac sections of adult BiTg mice that did not receive dox treatment (top) or were treated with dox (bottom), 6 months post dox withdrawal. Yellow arrows point to KDM4D_FLAG+ ACM nuclei, white arrows point to KDM4D_FLAG- ACM nuclei. (E) Immunoblots of heart lysates from 7 month mice that received dox from E0–3weeks. (F) HW/BW in 7 month mice that received dox from E0–3 weeks. (G) BrdU staining, bar=20μm, and quantitation in cardiac sections in the inducible model. Sample Number: No dox, 9 wk endpoint: tet=6, BiTg=5. DoxE0-P21, 12wk endpoint: control=9 BiTg=3. DoxE0-P21, 7 month endpoint: control= 8, BiTg=7. Statistics: (B, C, E, F) Two-tailed T-test, * P<0.05.
Discussion
The discovery that neonatal mammalian hearts can regenerate by CM proliferation [1, 2] and that ACMs retain some, though very limited, capacity to divide [4], has reignited interest in the regulation of CM cell cycle [3, 37–39]. Recently, epicardial paracrine factor FSTL1 [38], miR-15 inhibition [2], and the NRG1 co-receptor Erbb2 [40] were suggested to promote ACM proliferation, though the molecular mechanisms and relevant targets in ACMs are unknown. We previously found that the heterochromatin marker H3K9me3 was enriched on G2/M and cytokinesis genes in ACMs compared to fetal CMs implicating this mark as a barrier to ACM proliferation [6]. In this study we screened HDM expression levels in CM development, ACM dedifferentiation cell cycle re-entry, and human and mouse cardiomyopathies and found KDM4D expression levels correlated with CM hyperplasia but not hypertrophy models. We overexpressed KDM4D in CMs to explore the role of H3K9me3 in regulating cardiac cell cycling and documented a critical role in silencing cell cycle genes and establishing and maintaining terminal differentiation. Although recent studies have also suggested that non-histone proteins may be substrates of KDM4 demethylases; this has not been shown for KDM4D, which lacks the PHD and Tudor protein-binding domains found in KDM4A-C [41]. Unlike KDM4A (Supplemental Figure 4), we observed KDM4D does not demethylate H3K36me3 in ACMs (Figure 2F and 2G) and our data suggest that only H3K9me3 levels were affected. Thus, we believe the effects of KDM4D are primarily related to a reduction in H3K9me3 levels in CMs.
Surprisingly, our analysis revealed that the mitosis and cytokinesis genes that are silenced in ACMs have DnaseI-sensitive promoters suggesting they are not incorporated into inaccessible heterochromatin, unlike silenced non-lineage genes (Figure 1B and Supplemental Figure 1). Interestingly, the promoters of these genes are spared from repressive DNA methylation in both neonatal CMs that highly express cell cycle genes as well as in ACMs (Figure 1D and Supplemental Figure 2B). Cell cycle genes appear to be intrinsically protected from DNA methylation-mediated silencing as 90% of cell cycle gene promoters contain CpG Island sequences that evade DNA methylation (Figure 1E). Likewise, H3K27me3 was not enriched on cell cycle genes in ACMs and does not appear to mediate their silencing (Figure 1F). These findings are consistent with the absence of cell cycle activation in CM-specific knockout of H3K27me3- or DNA-methyltransferases and suggest other mechanisms are responsible for the silencing of cell cycle genes in ACMs [27, 28]. Our results suggest H3K9me3 depletion preferentially increases the expression of late cell cycle genes (Figure 3) which are enriched with H3K9me3 in terminally differentiated NonTg ACMs [6].
The finding that KDM4D binds to and depletes H3K9me3 at cell cycle gene promoters (Figure 4B and 4C) suggests a direct mechanism for the increases in cell cycle gene expression seen in BiTg ACMs. However, how cell cycle gene expression was preferentially increased is still unclear as we found KDM4D bound to diverse genomic loci, consistent with the observed global reduction of H3K9me3 (Figure 2F and 2G). Non-lineage genes are enriched with both DNA methylation and H3K27me3 (Figure 1D and 1F) which may explain why this class of genes was not expressed in H3K9me3-depleted ACMs. Our analysis suggests that cell cycle gene Dnase-I sensitivity is not altered by H3K9me3-depletion (Figure 4A), as these promoters are accessible even in NonTg control ACMs (Figure 1B and Supplemental Figure 1). One limitation of DHS-seq analysis is that it does not distinguish which regulatory complexes are binding to the accessible promoter. This is particularly important in cell cycle gene regulation as E2F family members with conserved DNA binding motifs have the capacity to bind cell cycle genes, but E2Fs 1–3 lead to gene activation while E2Fs 4–6 recruit repressive Rb/p130-contaning DREAM complexes that inhibit cell cycle gene expression [26, 42, 43]. Interestingly, Rb co-immunoprecipitates with H3K9me3-binding HP1 adapter proteins [25, 44]. Thus, we speculate that depleting H3K9me3 inhibits the recruitment of repressive cell cycle transcriptional complexes. Consistent with this notion, we previously found that ACM-specific Rb deletion combined with p130 knockout led to increased cell cycle gene expression, but H3K9me3 was maintained [6]. That study suggested that Rb recruitment to cell cycle genes was required for their stable repression, and that Rb targeting was mediated by H3K9me3-binding adapter proteins. Therefore, it is possible that H3K9me3-depletion relieves cell cycle gene silencing by inhibiting DREAM complex recruitment to these genes and the observed specificity of cell gene expression increases in the BiTg ACMs is endowed through cell cycle transcription factors with specific DNA binding motifs. In agreement, in BiTg ACMs we found that the E2F/Rb-family members most increased by H3K9me3-depletion were E2F1 and p107 (Supplemental Figure 6), which are the same E2F/Rb-family members that are expressed in proliferative embryonic and neonatal CMs (Supplemental Figure 2C) [6]. We found the inhibitory E2Fs, Rb/p130, and DREAM complex members had normal expression after H3K9me3-depletion, but the activating Myb and FoxM1 were increased (Supplemental Figure 6). The idea that activating and repressive transcription factors can compete for occupancy of cell cycle gene promoters is supported by the finding that these promoters have similar accessibility and DNA methylation in proliferation-competent cells and senescent CMs (Supplemental Figure 1 and Figure 1D) despite the large differences in mRNA expression. These DHS accessible regions in ACMs are bound by repressive E2F4 and activating Myc transcription factors in differentiated myotubes and cancer cell lines, respectively (ENCODE data). Studies in human samples show these cell cycle transcription factors are also present within the highly-conserved cell cycle gene promotors suggesting these mechanisms may be shared in mice and humans (ENCODE data). Therefore, even if KDM4D-binding and H3K9me3 itself is not specific to cell cycle genes, H3K9me3 depletion could specifically increase cell cycle gene expression due to their unique configuration of being transcriptionally silenced but accessible in ACMs (Figure 1C), selective increases in E2F/Rb-family and Myb/FoxM1 expression levels (Supplemental Figure 6), and disruption of cell cycle inhibition complexes that bind H3K9me3 and H3K9me3-adapter proteins.
We demonstrated that ACMs can tolerate increased levels of G2/M and cytokinesis gene expression (Figure 3 and Supplemental Figure 6) and moderate cycling (Figure 6) without deleterious effects on heart function (Table 1). This is similar to other models of limited ACM cycling [45], but contrasts with our previous findings where disrupting heterochromatin formation and inducing cell cycle reentry in ACMs was associated with decreased heart function at baseline [6]. A fundamental difference between the KDM4D mice and that model is that KDM4D overexpression specifically targets one methylation pathway, whereas Rb/p130 KO likely disrupted multiple epigenetic modifications (H3K9me3, H3K27me3, and H4K20me3) [6, 42, 46]. Consistent with this notion, H3K9me3-depleted chromatin in KDM4D mice maintained its global structure including heterochromatin unlike the Rb/p130 model with the exception of cycling pH3+ ACMs (Supplemental Figure 10). Thus, repressive methylations have overlapping roles in maintaining global chromatin structure in ACMs consistent with reports of H3K9me3-depletion in other cell types [47–49].
The vast majority of BiTg ACMs appeared to exit the cell cycle (Figure 6) and HW/BW did not increase after 9 weeks (Figure 5C) even though cell cycle gene expression remained elevated in H3K9me3-depleted CMs (Figure 3). Although highly upregulated compared to controls, late cell cycle gene expression in BiTg ACMs is much less than in wildtype embryonic and postnatal CMs (compare Figure 3C with Supplemental Figure 2B). It is possible that additional stimulation of mitogenic pathways, in addition to removal of repressive H3K9me3, is required for more robust ACM proliferation. Our results are similar to findings in studies of another major cell proliferation regulation signaling pathway, the organ-size-controlling Hippo/Yap pathway. This pathway has been intensely studied with several CM-specific loss of function and gain of function mouse models through CM development and adulthood [39, 50]. Though Yap1 gain of function in adults increased ACM proliferation, the levels were 20-fold less than NonTg neonatal CMs [45]. The authors postulated Yap activation alone is insufficient to overcome the multiple barriers blocking ACM proliferation. In several other systems Yap signaling fails to drive proliferation in the absence of E2F signaling [51–53]. Interestingly, informatics and chromatin immunoprecipitation sequencing approaches found E2F- and Yap- binding sites neighbor each other on many cell cycle gene promoters [52–54], which suggests E2F and Yap might be parallel pathways. Indeed, in liver regeneration models enhanced E2F activation by triple knockout of the Rb-family members resulted in cell proliferation; however, the increased proliferation declined over time due to dampening of Yap signaling [54]. Forced Yap1 activation or reducing liver size by partial hepatectomy allowed the E2F-mediated increases in proliferation to persist. This suggests that the intrinsic Hippo/Yap pathway has a remarkable ability to sense and regulate normal organ size and that E2F-mediated increases in proliferation can be augmented by growth stimulation or Yap signaling. The fact that the E2F pathway, but not the Yap pathway (Supplemental Figure 6C), is activated in BiTg ACMs suggests that additional mitogenic stimulation (ie Yap activation, injury) may promote more robust ACM proliferation in H3K9me3-depleted CMs.
In conclusion, CM-specific KDM4D induction and the subsequent H3K9me3-depletion is sufficient to maintain proliferation competence in ACMs. Depleting H3K9me3 in ACMs after terminal differentiation led to re-expression of cell cycle genes, and CM cell cycle reentry. This suggests H3K9me3 is required for both establishment and maintenance of the terminally differentiated phenotype in ACMs. These results further our understanding of how cardiac growth is regulated and identify a new role for H3K9me3 and common effector pathway for regulation of CM terminal differentiation. KDM4D overexpression did not affect normal heart function at baseline but allowed CM hyperplasia. Additional studies will be required to determine if KDM4D expression can extend the regenerative window in neonates and reverse the permanent CM cell cycle exit after injury and promote a regenerative response in adults. If true, this strategy would be very amenable to gene therapies with localized and temporally controlled KDM4D expression, which may bring us closer to a clinically relevant cardiac therapy.
Supplementary Material
Highlights.
Cell cycle genes are accessible but silenced in adult cardiac myocytes (ACMs).
Depleting H3K9me3 prevents cell cycle gene silencing in ACMs in vivo.
H3K9me3-depleted ACMs have increased cell cycling with normal heart function.
Depleting H3K9me3 in ACMs after differentiation reverses cell cycle silencing.
Acknowledgments
We would like to acknowledge James Gulick and the Robbins lab for providing the tTA mice and the tet-responsive attenuated αMHC promoter construct; Lutz Hein and Ralf Gilsbach for providing DNA methylation datasets; the University of Washington’s high throughput sequencing, microscopy, and transgenic mouse core facilities; April Stempien-Otero for providing human tissue samples; Hidemi Kajimoto for proofreading the manuscript and the Institute of Translation Health Sciences for assistance with statistical analysis.
Sources of Funding
NIH, R01 HL70748 to W.R.M.
American Heart Association, 14PRE20140025 to D.E.
Footnotes
Sequencing Data Accession Number
Bioproject Accession Numbers:
PRJNA309702 (RNA seq)
PRJNA449942 (DnaseI-hypersensitivity Seq)
Disclosures
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2013;110:187–92. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–44. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–6. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sdek P, Zhao P, Wang Y, Huang CJ, Ko CY, Butler PC, et al. Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J Cell Biol. 2011;194:407–23. doi: 10.1083/jcb.201012049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelis E, Garcia A, Chan SS, Schenke-Layland K, Ren S, Goodfellow SJ, et al. A cyclin D2-Rb pathway regulates cardiac myocyte size and RNA polymerase III after biomechanical stress in adult myocardium. Circ Res. 2008;102:1222–9. doi: 10.1161/CIRCRESAHA.107.163550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong W, Mao S, Tobis S, Angelis E, Jordan MC, Roos KP, et al. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25:3869–79. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–44. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neufeld TP, Edgar BA. Connections between growth and the cell cycle. Curr Opin Cell Biol. 1998;10:784–90. doi: 10.1016/s0955-0674(98)80122-1. [DOI] [PubMed] [Google Scholar]
- 11.Oyama K, El-Nachef D, Zhang Y, Sdek P, MacLellan WR. Epigenetic regulation of cardiac myocyte differentiation. Front Genet. 2014;5:375. doi: 10.3389/fgene.2014.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohl M, Wagner M, Reil JC, Muller SA, Tauchnitz M, Zimmer AM, et al. HDAC4 controls histone methylation in response to elevated cardiac load. J Clin Invest. 2013;123:1359–70. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–22. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–35. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 15.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–27. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–18. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–8. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 20.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, et al. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A. 2011;108:12711–6. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007;179:1399–412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao G, Mao S, Baumgarten G, Serrano J, Jordan MC, Roos KP, et al. Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ Res. 2001;89:1122–9. doi: 10.1161/hh2401.100742. [DOI] [PubMed] [Google Scholar]
- 23.Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–62. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahiya A, Wong S, Gonzalo S, Gavin M, Dean DC. Linking the Rb and polycomb pathways. Mol Cell. 2001;8:557–69. doi: 10.1016/s1097-2765(01)00346-x. [DOI] [PubMed] [Google Scholar]
- 25.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 26.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13:585–95. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He A, Ma Q, Cao J, von GA, Zhou P, Xie H, et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res. 2012;110:406–15. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nuhrenberg TG, Hammann N, Schnick T, Preissl S, Witten A, Stoll M, et al. Cardiac Myocyte De Novo DNA Methyltransferases 3a/3b Are Dispensable for Cardiac Function and Remodeling after Chronic Pressure Overload in Mice. PLoS One. 2015;10:e0131019. doi: 10.1371/journal.pone.0131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92:609–16. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 30.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilsbach R, Preissl S, Gruning BA, Schnick T, Burger L, Benes V, et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun. 2014;5:5288. doi: 10.1038/ncomms6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, et al. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J Biol Chem. 2011;286:41616–25. doi: 10.1074/jbc.M111.283689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnan S, Trievel RC. Structural and functional analysis of JMJD2D reveals molecular basis for site-specific demethylation among JMJD2 demethylases. Structure. 2013;21:98–108. doi: 10.1016/j.str.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353:973–7. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 35.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emde B, Heinen A, Godecke A, Bottermann K. Wheat germ agglutinin staining as a suitable method for detection and quantification of fibrosis in cardiac tissue after myocardial infarction. Eur J Histochem. 2014;58:2448. doi: 10.4081/ejh.2014.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–53. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei K, Serpooshan V, Hurtado C, Diez-Cunado M, Zhao M, Maruyama S, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–85. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, Li L, Zhao B, Guan KL. The hippo pathway in heart development, regeneration, and diseases. Circ Res. 2015;116:1431–47. doi: 10.1161/CIRCRESAHA.116.303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Uva G, Aharonov A, Lauriola M, Kain D, Yahalom-Ronen Y, Carvalho S, et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat Cell Biol. 2015;17:627–38. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 41.Ponnaluri VK, Vavilala DT, Putty S, Gutheil WG, Mukherji M. Identification of non-histone substrates for JMJD2A-C histone demethylases. Biochem Biophys Res Commun. 2009;390:280–4. doi: 10.1016/j.bbrc.2009.09.107. [DOI] [PubMed] [Google Scholar]
- 42.Macaluso M, Montanari M, Giordano A. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene. 2006;25:5263–7. doi: 10.1038/sj.onc.1209680. [DOI] [PubMed] [Google Scholar]
- 43.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–5. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 45.Lin Z, von GA, Zhou P, Gu F, Ma Q, Jiang J, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 2014;115:354–63. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–8. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 47.Chandra T, Kirschner K, Thuret JY, Pope BD, Ryba T, Newman S, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell. 2012;47:203–14. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muramatsu D, Singh PB, Kimura H, Tachibana M, Shinkai Y. Pericentric heterochromatin generated by HP1 protein interaction-defective histone methyltransferase Suv39h1. J Biol Chem. 2013;288:25285–96. doi: 10.1074/jbc.M113.470724. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 50.Lin Z, Pu WT. Harnessing Hippo in the heart: Hippo/Yap signaling and applications to heart regeneration and rejuvenation. Stem Cell Res. 2014;13:571–81. doi: 10.1016/j.scr.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dick FA, Mymryk JS. Sweet DREAMs for Hippo. Genes Dev. 2011;25:889–94. doi: 10.1101/gad.2050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehmer U, Sage J. Control of Proliferation and Cancer Growth by the Hippo Signaling Pathway. Mol Cancer Res. 2016;14:127–40. doi: 10.1158/1541-7786.MCR-15-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–35. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehmer U, Zmoos AF, Auerbach RK, Vaka D, Butte AJ, Kay MA, et al. Organ size control is dominant over Rb family inactivation to restrict proliferation in vivo. Cell Rep. 2014;8:371–81. doi: 10.1016/j.celrep.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.