Abstract
Cell-mediated remodeling of extracellular matrix (ECM) plays important roles for cell functions, but it is challenging to develop synthetic materials for mimicking such a dynamic aspect of proteins in ECM. Here we show that intercellular morphological transition of peptide assemblies mimic the unfolding of fibronectin, thus enabling cell to form spheroids from a monolayer of cells. Specifically, the phosphopeptide self-assembles to form nanoparticles, which turns into nanofibers upon partial dephosphorylation catalyzed by enzymes (e.g., phosphatases) at intercellular space. Occurring between HS-5 cells, such an enzyme instructed self-assembly enables a sheet of the HS-5 cells to form cell spheroids. Structure-activity investigation reveals that proteolytic stability, dephosphorylation, and biotin conjugation of the peptides are indispensable for forming the cell spheroids. Further mechanism study indicates that the intercellular assemblies interact with multiple ECM components (e.g., laminin, collagens III and IV) to drive the formation of the cell spheroids. As the first example of intercellular instructed-assembly from homotypic precursors, this work illustrates a new approach that uses cell-responsive peptide assemblies to mimic protein dynamics for control cell behaviors.
Graphical Abstract
Extracellular matrix (ECM) plays multifarious roles in cellular processes, such as proliferation,1 differentiation,2 migration,3 and communication.4 In the development of biomaterials for regenerative medicine and tissue engineering, considerable efforts have centered on designing biomaterials to mimic ECM for guiding the behavior of cells, such as using simple biochemical cues (e.g., cell adhesion sequences) for controlling cell adhesion,5 modulating elasticity of matrix for directing cell differentiation,2,6–7 polyvalent carbohydrates for inhibiting agglutination8 or promoting cell aggregation,9 and applying photochemistry to switch synthetic networks for guiding cell migration and proliferation.10 Despite that significant progresses have achieved by these approaches, most synthetic ECM materials lack the dynamic features exhibited by the proteins that constitute ECM. For example, ECM undergoes cell-mediated remodeling, which play essential physiological roles in regulating tissue architecture, morphogenesis, and homoeostasis.11 Thus, it would be advantageous and necessary to develop a facile approach to generate materials that, like endogenous ECM, undergo cell-mediated remodeling.
A well-studied example of ECM remodeling is the unfolding of fibronectin (FN), a major ECM glycoprotein that plays central role for regulating a variety of cell activities.12 Initiated by integrin receptor on cell surface, the unfolding of FN assembly is a cell-mediated process: the dimeric globular FN bind with integrin, integrin receptors at focal adhesion sites transmit cellular contraction forces, and this local mechanical change of the cell stretches FN molecules to active conformations, and thus promoting FN-FN interactions to form fibrils. The increase of noncovalent interactions between the fibrils further stabilizes the fibrillar networks to enhance fibrillar adhesions.13 Occurring at the intercellular space, such a fibrillar adhesion provides cell-cell adhesive forces that are critical for 2D to 3D cell morphogenesis in vitro.14 So the essence of FN unfolding is to generate cell adhesion force by forming FN fibrils from globular FN. Since the determinant for cell morphogenesis is biophysical in nature (i.e., cell-cell adhesive forces),14 it should be feasible to use molecules other than FN to modulate cell adhesion forces for generating cell spheroids. Stimulated by this insight,14 we intended to build synthetic fibrillar matrix for modulating cell adhesion forces and inducing cell spheroids.
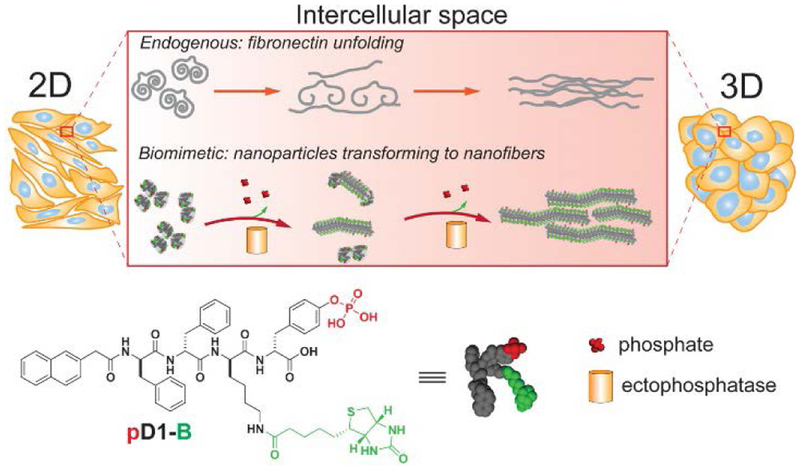
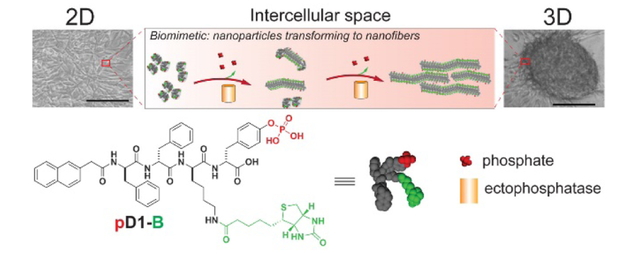
To mimic the essence of FN unfolding and to ensure the mediation by the cells, we decided to use instructed-assembly15–18 to form supramolecular nanofibers at intercellular space as the synthetic fibrillar matrix (Scheme 1). To maintain the adhesion of the nanofibers with the cells and to distinct from the endogenous cell adhesion motifs (e.g., RGD, IKVAV, YIGSR, and carbohydrates),19–22 we used biotin as the proteolytic resistant cell adhesion motif because various cell lines express high level biotin receptor.23 So we designed a biotinylated D-phosphotetrapeptide (pD1-B) as the substrate for the phosphatase instructed assembly. Our studies show that, being partially dephosphorylated by phosphatases at intercellular space, globular assemblies (i.e., nanoparticles) of pD1-B turns into nanofibers (consisting of the mixture of pD1-B and D1-B) at the intercellular space, which transforms a 2D cell sheet to 3D cell spheroids. Dephosphorylation, cell surface binding motif, and proteolytic resistance of the precursor, are essential for the observed cell spheroids. The formed intercellular assemblies interact with multiple ECM components (e.g., laminin, collagens III and IV) within the cell spheroids. As a demonstration of morphological transition of supramolecular assemblies to modulate intercellular mechanical force, this work illustrates a versatile approach that uses enzymatic noncovalent synthesis to mimic protein dynamics and to control biophysical determinants locally for cellular signal transductions.
Scheme 1.
Intercellular instructed-assembly to mimic the essence of the dynamic of an ECM protein (e.g., fibronectin unfolding).
As shown in Scheme 1, pD1-B consists of three key components: a well-studied self-assembling backbone (i.e., a diphenylalanine capped with naphthalene (Nap-ff)24), an enzymatic trigger (i.e., tyrosine phosphate (py) as a substrate of phosphatase25–26), and a cell surface targeting motif (i.e., biotin). We utilized biotin because biotin is a ligand of cell surface receptor and is a biocompatible vitamin.23,27 After synthesizing Fmoc (9-fluorenylmethoxycarbonyl) protected phosphorylated tyrosine via two steps, we synthesized Nap-ffkpy by standard solid-phase (Fmoc) peptide chemistry, and subsequently using N-hydroxysuccinimide (NHS) activated biotin to react with Nap-ffkpy via ε-amino group of lysine to form stable amide bonds to result in pD1-B. After purifying the precursor, we used 1H-NMR and LC-MS to confirm their purity and identity. This facile synthetic route (Scheme S1) produces pD1-B in a good yield (76%).
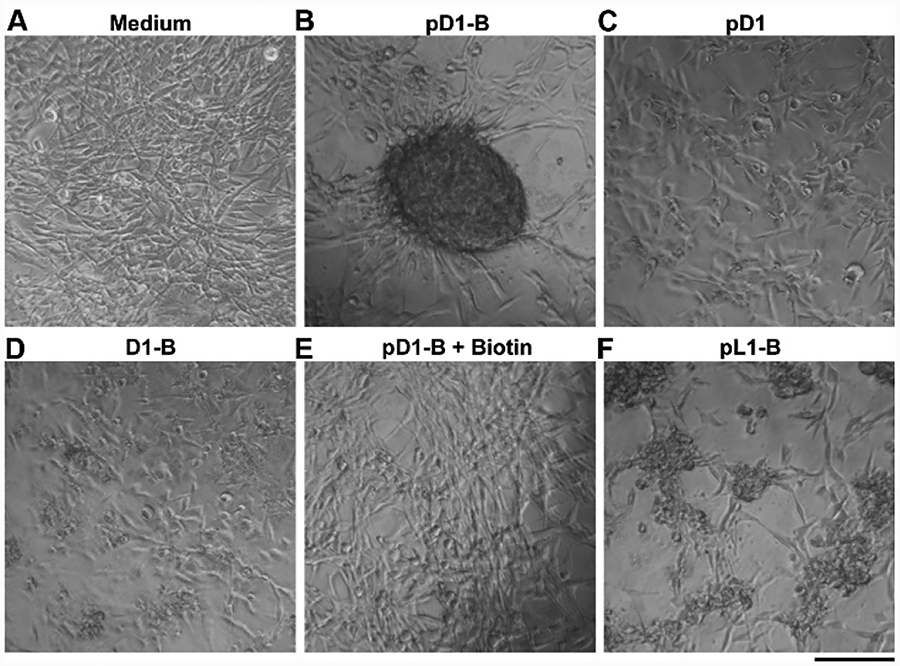
After demonstrating pD1-B is innocuous to HS-5 cells (Figure S1), we investigated the 3D cell spheroid formation of HS-5 cells upon the treatment of pD1-B. We chose HS-5 cells because these fibroblast cells express relatively low level of phosphatases, synthesize 3D ECM slowly (several days at higher density),28 and serve as a stromal cell line for co-culturing with other disease-related cells. After being treated with pD1-B for 24 h, HS-5 cells form 3D cell spheroids (Figure 1B, S2 and S3) from a 2D cell sheet, while the cells remain a 2D sheet without pD1-B in the culture medium. Adding pD1 or D1-B (Scheme S2), which is the analogue of pD1-B without either the biotin motif or the phosphate group, HS-5 cells remain as a 2D cell sheet. These results indicates that both the binding to cell surface and the dynamic change of pD1-B are indispensable for inducing the 3D cell spheroids. Importantly, using biotin molecules to compete with pD1-B to bind the receptors on cell surface blocks the formation of cell spheroids in a concentration dependent manner (Figure 1E and S5), further confirming the crucial role of biotin, as a ligand, for attaching the assemblies of pD1-B on the cell surface to undergo instructed-assembly at the intercellular space. Using pL1-B, the L-enantiomer of pD1-B, results in smaller clusters of cells (Figure 1F and S6), indicating the proteolytic stability (Figure S7 and S8) of the assemblies of pD1-B also play important roles for forming the cell spheroids.
Figure 1.
Optical images of HS-5 cells (A) in culture medium and co-incubation with (B) pD1-B; (C) pD1; (D) D1-B; (E) pD1-B and Biotin (500 μM), and (F) pL1-B for 24h. The concentration of all the molecules is 200 μM. Scale bar is 150 μm.
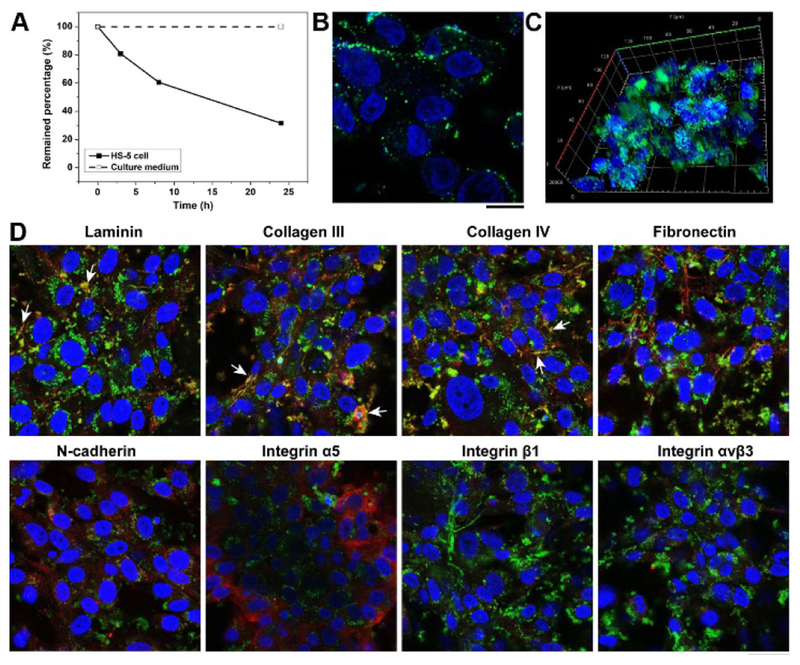
To verify that the enzymatic dephosphorylation and self-assembly of pD1-B on the cells are a dynamic process, we quantified the conversion of pD1-B after incubating it with HS-5 cells at different time points. The results (Figure 2A) show that endogenous phosphatases convert 19.2%, 39.5%, and 68.5% of pD1-B to D1-B at 3, 8, and 24 h, respectively. The results indicate that the process of instructed-assembly15 involves two build blocks, pD1-B and D1-B, which co-assemble to form a continuum phosphorylated nanostructures at intercellular space. We next used FITC-avidin to stain the assemblies of pD1-B and D1-B. Figure 2B (and Figure S9) indicates that the fluorescence of FITC-avidin distribute on cell surface as puncta with slightly different sizes, suggesting that the assemblies anchor on cell surface and form intercellular assemblies. In contrast, HS-5 cells treated with pL1-B exhibit very weak fluorescence in cytoplasm, indicating the stability of molecule is important for its anchoring on cell surface. To directly visualize the distribution of these intercellular assemblies, we introduced a nitrobenzofurazan (NBD)-capped phosphopeptide (NBD1P-B, scheme S3) since NBD fluoresces intensely when forming assemblies.29 3D construction of CLSM images (Figure 2C and Video clip S1) reveals that most fluorescent puncta locate around cells, confirming that the intercellular dynamic assemblies of NBD1P-B drive the formation of the cell spheroids.
Figure 2.
(A) HS-5 cells catalyzed conversion of pD1-B within 24 h. (B) HS-5 cells treated with pD1-B (200 μM) for 24h and then stained with FITC-avidin for 2h. Scale bar is 10 μm. (C) 3D construction of HS-5 cells treated with NBD1P-B (200 μM) for 24 h. (D) 3D construction (see videos S2 and S6) of immunofluorescence staining of HS-5 cells treated with NBD1P-B (200 μM) for 24 h: the cell spheroids stained with antibodies of ECM molecules (i.e., laminin, collagens III and IV, fibronectin) and adhesive molecules (i.e., N-cadherin; integrins α5, β1, and β3). Green indicates the assemblies, red the antibodies, and blue the nuclei. Arrows point to the co-localization of the assemblies and the antibodies.
We used immunofluorescent staining to detect the co-localization of the assemblies of NBD1P-B and NBD1-B and major components of ECM or cell adhesion molecules. The results reveal that the assemblies partially co-localize with laminin, collagen III, and collagen IV (Figure 2D, video clips S2–S4). The assemblies hardly co-localize with fibronectin. These results imply that intercellular instructed-assembly likely acts as a dynamic adhesive force for generating 3D cell spheroids. Further mechanistic studies (Figure 2D) exclude the direct interaction between the assemblies and cell adhesion molecules, such as N-cadherin and integrins. More importantly, antibodies against α5 (fibronectin receptor) show brighter fluorescence in spheroid (Figure 2D) than in the corresponding 2D cell culture (Figure S10), and antibodies against αvβ3 (vitronectin receptor) exhibit weaker fluorescence in 3D spheroid than in 2D cell culture. These results suggest the transformation of focal adhesion to fibrillar adhesion during cell spheroid formation, which is likely to evolve 3D-matrix adhesion.28,30 Collectively, these results support that intercellular instructed-assembly of small molecules effectively enhancing cell-cell interactions to form 3D cell spheroids.
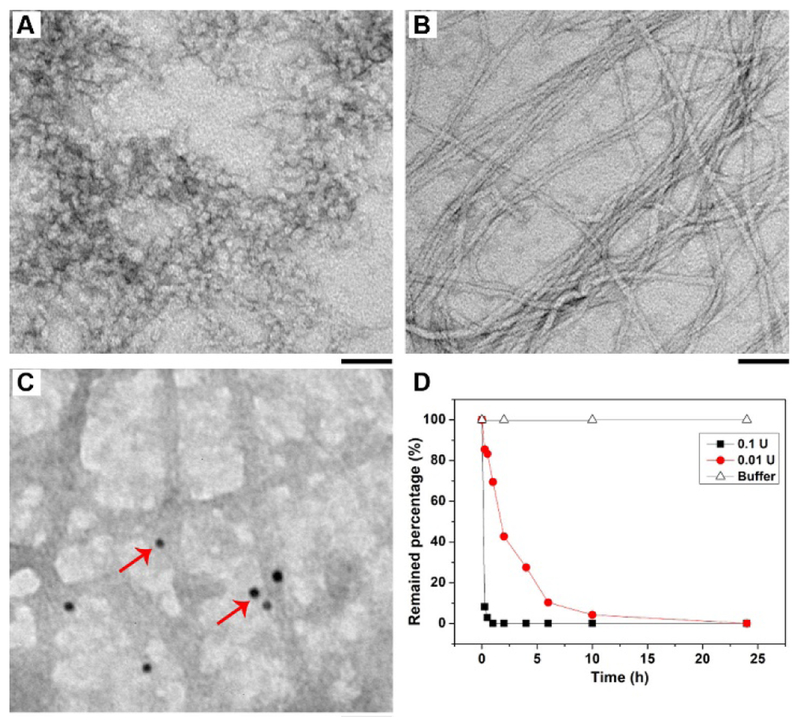
Transmission electron microscopy (TEM) helps infer the morphological transformation that likely occurs during intercellular instructed-assembly. pD1-B self-assembles to form relatively uniform nanoparticles (Figure 3A) with diameter of 7±2 nm in aqueous solution, which transform to nanofibers (Figure 3B) with several hundred nanometers in length and 7±2 in width upon the treatment of ectophosphatase (e.g., alkaline phosphatase (ALP)). Similar to pD1-B, adding pL1-B in aqueous solution results in nanoparticles with diameter of 5±2 nm, and then turn into filaments with 3±2 nm in width after addition of ALP (Figure S11). As a control, pD1 forms predominately worm like structures with about 36±2 nm in length and 6±2 nm in width (Figure S12), plus some nanospheres with diameter of 5±2 nm. Upon the addition of ALP, these worm like structures transform to short filaments with 58±2 in length and 5±2 in width (Figure S12). Directly dissolving D1-B in aqueous solution results in uniform nanofibers (Figure S13). These results not only indicate the influence of biotin on the morphology of pD1-B, but also confirm the morphology transition due to dephosphorylation. Such a dynamic, morphological change, reminiscing the cell-mediated dynamics of fibronectin (i.e., from globular to fibril structures during unfolding processes), is the key factor for inducing the cell spheroids.
Figure 3.
TEM images of pD1-B at concentration from 0.1 wt% in Tris-Cl buffer (pH = 7.4) (A) without or (B) with the treatment of ALP (1 U/mL). (C) TEM of gold-avidin (10 nm) stained fibers, red arrows point to the gold nanoparticles. Scale bar is 50 nm. (D) Dephosphorylation rate of pD1-B (200 μM) with or without the treatment of ALP (0.1 and 0.01 U/mL) in the Tris-buffer (pH=7.4).
Gold-avidin staining (Figure 3C and S14) indicates that biotin presents on the surface of nanofibers, being similar to a previous report,31 suggesting that biotin not only acts as a ligand for anchoring pD1-B on cell surface, but also provides additional hydrogen bonding for intercellular fibrillar interaction to reinforce ECM. Moreover, liquid chromatography mass spectrometry (LC-MS) reveals that the assemblies of pD1-B was dephosphorylated in different rates upon addition of different amounts of ALPs (Figure 3D). Specifically, 0.1 U/mL of ALP hydrolysis pD1-B is much faster than 0.01 U/mL of ALP does, with t½ to be 0.14 and 1.72 h for 0.1 U/mL and 0.01 U/mL of ALP, respectively. This result is consistent with the fact that pD1-B induces the spheroid formation of HS-5 cells, while hardly change the morphology of Saos-2 cells that express higher level of ALP (Figure S15).
In summary, this work describes that intercellular enzymatic formation of higher-order nanostructures of peptides for controlling the collective behavior of cells. Use only one type of precursor to induce cell spheroid formation, the strategy illustrated in this work is simpler and more general than previous works,32–33 which require two types of molecules (i.e., vancomycin and phosphopeptide). Similarly to the cases of heterotypic precursors, the rate of dephosphorylation likely dictates the formation of the peptide assemblies for forming cell spheroids. Introducing ligands (e.g., biotin) of cell surface receptor for instructed-assembly ensures locally controlling phase transition of assemblies on cell surface, which further influences the mechanical signaling locally for remodeling ECM11 to induce cell morphogenesis. Providing a facile strategy for in-situ constructing molecular assemblies to generate organoids, this work illustrates that spatiotemporal control of noncovalent (or supramolecular) interactions, and it ultimately may contribute to tissue engineering and stem cell therapies.34 Using other functional small molecules instead of biotin in instructed-assembly may lead to novel functions of supramolecular assemblies.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by NIH (CA142746), and NSF (DMR-1420382). ZF thanks the Dean’s fellowship and NIH (F99CA234746).
Footnotes
Supporting Information
Materials and detailed experimental procedures, and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests. The authors declare no competing financial interest.
REFERENCES
- (1).Clark EA; Brugge JS Integrins and signal transduction pathways: the road taken Science 1995, 268, 233. [DOI] [PubMed] [Google Scholar]
- (2).Engler AJ; Sen S; Sweeney HL; Discher DE Matrix elasticity directs stem cell lineage specification Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- (3).Lauffenburger DA; Horwitz AF Cell migration: A physically integrated molecular process Cell 1996, 84, 359. [DOI] [PubMed] [Google Scholar]
- (4).Sternlicht MD; Werb Z How matrix metalloproteinases regulate cell behavior Annu. Rev. Cell Dev. Biol. 2001, 17, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rowley JA; Madlambayan G; Mooney DJ Alginate hydrogels as synthetic extracellular matrix materials Biomaterials 1999, 20, 45. [DOI] [PubMed] [Google Scholar]
- (6).Wu W; Allen R; Gao J; Wang YD Artificial Niche Combining Elastomeric Substrate and Platelets Guides Vascular Differentiation of Bone Marrow Mononuclear Cells Tissue Eng. Part A 2011, 17, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Silva GA; Czeisler C; Niece KL; Beniash E; Harrington DA; Kessler JA; Stupp SI Selective differentiation of neural progenitor cells by high-epitope density nanofibers Science 2004, 303, 1352. [DOI] [PubMed] [Google Scholar]
- (8).Mortell KH; Gingras M; Kiessling LL Synthesis of Cell Agglutination Inhibitors by Aqueous Ring-Opening Metathesis Polymerization J. Am. Chem. Soc. 1994, 116, 12053. [Google Scholar]
- (9).Zhou J; Du X; Chen X; Xu B Adaptive Multifunctional Supramolecular Assemblies of Glycopeptides Rapidly Enable Morphogenesis Biochemistry 2018, 57, 4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kloxin AM; Kasko AM; Salinas CN; Anseth KS Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties Science 2009, 324, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hynes RO The extracellular matrix: not just pretty fibrils Science 2009, 326, 1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mao Y; Schwarzbauer JE Fibronectin fibrillogenesis, a cell-mediated matrix assembly process Matrix Biol. 2005, 24, 389. [DOI] [PubMed] [Google Scholar]
- (13).Wolanska KI; Morgan MR; Portland Press Limited: 2015.
- (14).Powers MJ; Griffith LG Adhesion-guided in vitro morphogenesis in pure and mixed cell cultures Microsc Res Techniq 1998, 43, 379. [DOI] [PubMed] [Google Scholar]
- (15).He H; Xu B Instructed-Assembly (iA): A Molecular Process for Controlling Cell Fate Bull. Chem. Soc. Jpn. 2018, 91, 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Feng Z; Wang H; Xu B Instructed assembly of peptides for intracellular enzyme sequestration J. Am. Chem. Soc. 2018, 140, 16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Razgulin A; Ma N; Rao JH Strategies for in vivo imaging of enzyme activity: an overview and recent advances Chem. Soc. Rev. 2011, 40, 4186. [DOI] [PubMed] [Google Scholar]
- (18).Ye DJ; Shuhendler AJ; Cui LN; Tong L; Tee SS; Tikhomirov G; Felsher DW; Rao JH Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo Nat. Chem. 2014, 6, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhang S; Holmes TC; DiPersio CM; Hynes RO; Su X; Rich A Self-complementary oligopeptide matrices support mammalian cell attachment Biomaterials 1995, 16, 1385. [DOI] [PubMed] [Google Scholar]
- (20).Sargeant TD; Rao MS; Koh C-Y; Stupp SI Covalent functionalization of NiTi surfaces with bioactive peptide amphiphile nanofibers Biomaterials 2008, 29, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Akasov R; Haq S; Haxho F; Samuel V; Burov SV; Markvicheva E; Neufeld RJ; Szewczuk MR Sialylation transmogrifies human breast and pancreatic cancer cells into 3D multicellular tumor spheroids using cyclic RGD-peptide induced self-assembly Oncotarget 2016, 7, 66119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Haq S; Samuel V; Haxho F; Akasov R; Leko M; Burov SV; Markvicheva E; Szewczuk MR Sialylation facilitates self-assembly of 3D multicellular prostaspheres by using cyclo-RGDfK(TPP) peptide Onco. Targets Ther. 2017, 10, 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Russell-Jones G; McTavish K; McEwan J; Rice J; Nowotnik D Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours J. Inorg. Biochem. 2004, 98, 1625. [DOI] [PubMed] [Google Scholar]
- (24).Zhang Y; Kuang Y; Gao Y; Xu B Versatile small-molecule motifs for self-assembly in water and the formation of biofunctional supramolecular hydrogels Langmuir 2010, 27, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Yang Z; Gu H; Fu D; Gao P; Lam JK; Xu B Enzymatic formation of supramolecular hydrogels Adv. Mater. 2004, 16, 1440. [Google Scholar]
- (26).Wang H; Feng Z; Wu D; Fritzsching KJ; Rigney M; Zhou J; Jiang Y; Schmidt-Rohr K; Xu B Enzyme-regulated supramolecular assemblies of cholesterol conjugates against drug-resistant ovarian cancer cells J. Am. Chem. Soc. 2016, 138, 10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chen J; Chen S; Zhao X; Kuznetsova LV; Wong SS; Ojima I Functionalized single-walled carbon nanotubes as rationally designed vehicles for tumor-targeted drug delivery J. Am. Chem. Soc. 2008, 130, 16778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cukierman E; Pankov R; Stevens DR; Yamada KM Taking cell-matrix adhesions to the third dimension Science 2001, 294, 1708. [DOI] [PubMed] [Google Scholar]
- (29).Gao Y; Shi J; Yuan D; Xu B Imaging enzyme-triggered self-assembly of small molecules inside live cells Nat. Commun. 2012, 3, 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pankov R; Cukierman E; Katz B-Z; Matsumoto K; Lin DC; Lin S; Hahn C; Yamada KM Integrin dynamics and matrix assembly: tensin-dependent translocation of α5β1 integrins promotes early fibronectin fibrillogenesis J. Biochem. Cell Biol. 2000, 148, 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Guler MO; Soukasene S; Hulvat JF; Stupp SI Presentation and recognition of biotin on nanofibers formed by branched peptide amphiphiles Nano Lett. 2005, 5, 249. [DOI] [PubMed] [Google Scholar]
- (32).Wang H; Shi J; Feng Z; Zhou R; Wang S; Rodal AA; Xu B An in situ dynamic continuum of supramolecular phosphoglycopeptides enables formation of 3D cell spheroids Angew. Chem., Int. Ed. 2017, 56, 16297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang H; Feng Z; Xu B Instructed-Assembly as Context‐Dependent Signals for Death and Morphogenesis of Cells Angew. Chem., Int. Ed. 2019, 58, 5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Madl CM; Heilshorn SC; Blau HM Bioengineering strategies to accelerate stem cell therapeutics Nature 2018, 557, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.