Abstract
The cytochrome P450 (CYP)4F2 gene is known to influence mean coumarin dose. The aim of the present study was to undertake a meta-analysis at the individual patients level to capture the possible effect of ethnicity, gene—gene interaction, or other drugs on the association and to verify if inclusion of CYP4F2*3 variant into dosing algorithms improves the prediction of mean coumarin dose. We asked the authors of our previous meta-analysis (30 articles) and of 38 new articles retrieved by a systematic review to send us individual patients’ data. The final collection consists of 15,754 patients split into a derivation and validation cohort. The CYP4F2*3 polymorphism was consistently associated with an increase in mean coumarin dose (+9% (95% confidence interval (CI) 7–10%), with a higher effect in women, in patients taking acenocoumarol, and in white patients. The inclusion of the CYP4F2*3 in dosing algorithms slightly improved the prediction of stable coumarin dose. New pharmacogenetic equations potentially useful for clinical practice were derived.
Coumarins have proved to be effective in the treatment of thromboembolic disease and despite the introduction of direct oral anticoagulants, they remain one of the most widely prescribed family of drugs worldwide.1
The narrow therapeutic index and high interindividual variability in therapeutic dose make coumarin therapy difficult to manage. Many studies have showed two genes, cytochrome P450 (CYP)2C9 and VKORC1, which are associated with variation in warfarin, phenprocoumon, and acenocoumarol maintenance doses requirement and have suggested some clinical benefits from genotype-guided dosing.2 On the basis of such data, the US Food and Drug Administration has updated the label for warfarin twice, advising that two variants in the CYP2C9 gene (C144R and I359L) and one in the Vitamin K epOxide Reductase Complex subunit 1 (VKORC1) gene (G-1639A) might be taken into consideration when initiating warfarin therapy (warfarin product labeling, US Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/009218s107lbl.pdf).
Although there have been contradictory results in randomized clinical trials (RCTs) about the utility of genotype-guided dosing of coumarin drugs when compared with either standard clinical care or clinical algorithms,3–5 a recent RCT in patients under-going elective hip or knee arthroplasty6 showed superiority of genetic dosing compared with clinical dosing. Some, but not all, meta-analyses have also shown an improvement in clinical end points, such as bleeding events.3,7–11 Moreover, none of the trials included in the meta-analyses included CYP4F2*3 polymorphism (1347C>T; c.1297G>A; p.Val433Met; rs2108622), whose effect on coumarin dose was discovered later when compared with CYP2C9 and VKORC1.
Our previous meta-analysis performed on aggregate data from 30 studies showed that CYP4F2 variation was associated with nearly 8% higher coumarin doses in T allele carriers. Indeed, a possible gene–gene interaction and an effect of race on the genetic effect were detected.12 Despite the low effect size, CYP4F2 is currently regarded as the third most influential genetic locus with respect to coumarin drug maintenance dose. Older studies, which compared pharmacogenetic algorithms with either clinical-based algorithms or fixed-dose approach, did show a possible improvement in prediction only in selected subgroups.13,14 The incorporation of CYP4F2 into existing models might improve the accuracy of dose prediction with coumarins.15,16 Recently, the Clinical Pharmacogenetics Implementation Consortium updated the guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing, including evidence from the published literature for the nonsynonymous variant CYP4F2*3 (1347C>T; c.1297G>A; p.Val433Met; rs2108622), which was found to be significantly associated with altered dose requirements for coumarin anticoagulants.2 In order to clarify the actual clinical utility of including the CYP4F2 polymorphism into pharmacogenetic dosing algorithms, some essential information is needed. Thus, we performed a meta-analysis at the individual patients level to understand the real effect size of this polymorphism and to test how much either a possible gene–gene interaction or the effect of ethnicity or other covariates could modify the pharmacogenetic association and prove to be useful in creating new pharmacogenetic equations. We hereby provide the largest meta-analysis of CYP2C9, VKORC1, and CYP4F2 polymorphisms affecting the dose of warfarin and acenocoumarol in samples collected from 25 different countries, including more than 15,000 participants treated with coumarin drugs. New pharmacogenetic equations potentially useful for clinical practice have been derived for different ethnic groups.
RESULTS
Characteristics of included studies
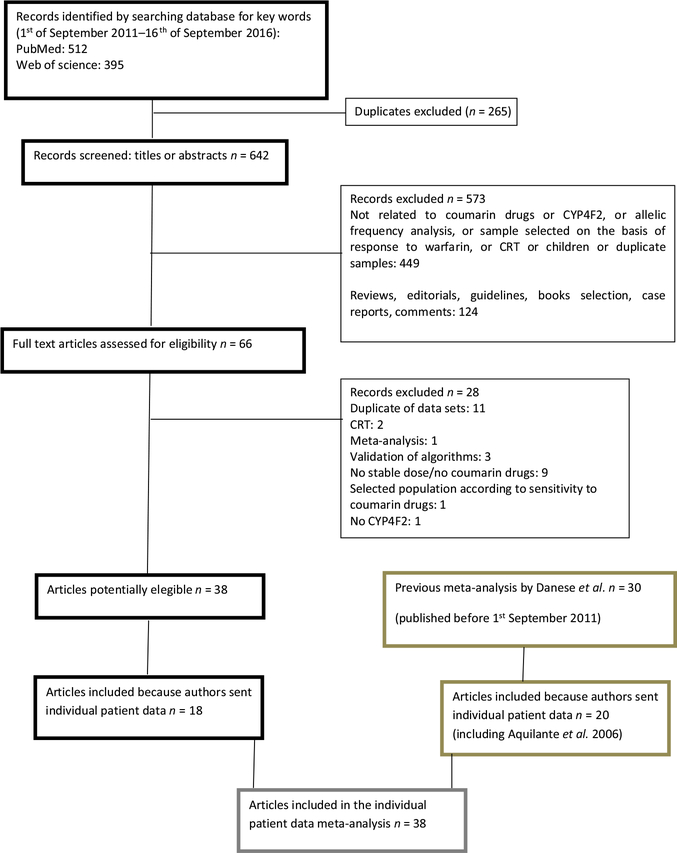
Starting from the 30 articles included in our previous meta-analysis (search from inception until August 2011), individual patient data were obtained from 19 studies.17–35 From one coauthor we obtained an additional dataset related to an article not previously included because no data about the CYP4F2 polymorphism were present in the original publication.36 From the group of 38 articles retrieved from the new search (from September 1, 2011, to September 14, 2016), individual patient data were obtained from 18 studies (Figure 1).15,16,37–52
Figure 1.
Flow diagram. CRT, controlled randomized trial; CYP4F2, cytochrome P450 F2.
Thus, 38 articles were included in the present work from authors who agreed to share individual patient data: 20 from the first systematic search and 18 from the second systematic search. Data from one study were divided into two distinct cohorts according to the main author’s subdivision of sample into discovery and validation cohorts.46 Moreover, data from two studies had been collected in one cohort.15,43 Finally, data from one study was divided into two cohorts: one cohort treated with acenocoumarol and the other with phenprocoumon treatment.44 This resulted in 39 cohorts that were considered for the meta-analysis, including a total of 15,754 patients. Characteristics of the individual studies are summarized in Table 1. Thirty-one cohorts examined the association between CYP4F2 polymorphism and the maintenance dose of warfarin; seven cohorts evaluated this association for acenocoumarol and one for phenprocoumon. Information on CYP4F2, VKORC1, and CYP2C9*3 genotyping were available for all 39 cohorts, whereas CYP2C9*2 genotype was recorded for 35 of the 39 cohorts (89.7%). All studies but one19 included both male and female participants with a minimum of 24% men. One study selected very elderly patients (mean age 86.7 years).35 Data on body mass index (BMI) and drugs known to potentially interfere with warfarin were available for 31 and 27 cohorts, respectively. All studies were published between 2006 and 2016.
Table 1.
Descriptive characteristics of studies included in the analysis
| First author (ref) | PY | Country | Ethnicity | Subjects, n | Men (%) | Age, years (mean ± SD) | Drug | INR target | Gene polymorphisms | Available confoundersa |
|---|---|---|---|---|---|---|---|---|---|---|
| Aquilante36 | 2006 | Florida (USA) | Whites (93%) Blacks (7%) | 344 | 300 (87%) | 69 ± 11 | Warfarin | 2.5–3.5 | CYP2C9*2, CYP2C9*3, CYP2C9*5, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment |
| Borgiani18 | 2009 | Italy | Whites | 141 | 75 (53%) | 69 ± 12 | Warfarin | 2.0–4.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, gender, indication for treatment |
| Borobia15 Tong43 | 2012–2016 | Spain | Whites (Spanish) | 679 | 345 (51%) | 68 ± 13 | Acenocoumarol | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Botton28 | 2011 | Brazil | Whites | 279 | 155 (57%) | 63 ±14 | Warfarin | 1.8–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2b | Age, BMI, gender, smoking, other drugs |
| Bourgeois45 | 2016 | UK | Whites | 217 | 119 (55%) | 71 ± 11 | Warfarin | 2.0–4.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Caldwell17 | 2008 | Wisconsin (USA) | Whites | 429 | 252 (59%) | 69 ± 11 | Warfarin | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, indication for treatment, other drugs |
| Cavallari29 | 2010 | Illinois (USA) | Blacks | 208 | 57 (27%) | 56 ± 16 | Warfarin | 2.0–4.0 | CYP2C9*2, CYP2C9*3, CYP2C9*5,VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Cen30 | 2010 | China | Asians | 221 | 103 (47%) | 45 ±12 | Warfarin | 1.5–3.0 | CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, other drugs |
| Cerezo-Manchado (a)46 | 2013 | Spain | Whites | 943 | 459 (49%) | 75 ± 9 | Acenocoumarol | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VKORC1,CYP4F2 | Age, gender, indication for treatment, other drugs |
| Cerezo-Manchado (b)46 | 2013 | Spain | Whites | 3,882 | 1,916 (49%) | 74 ± 10 | Acenocoumarol | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VK0RC1, CYP4F2b | Age, gender, indication for treatment, other drugs |
| Cha31 | 2010 | Japan | Asians | 440 | 293 (77%) | 68 ± 11 | Warfarin | 1.5–3.0 | CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, indication for treatment, other drugs |
| Gong32 | 2011 | UK | Whites (95%) Blacks (3%) Asians (2%) | 167 | 96 (57%) | 60 ± 18 | Warfarin | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, gender, indication for treatment |
| Hirai47 | 2015 | Japan | Asians | 217 | 143 (66%) | 68 ±10 | Warfarin | 1.5–3.0 | CYP2C9*2, CYP2C9*3, VK0RC1, CYP4F2 | Age, gender, indication for treatment |
| Isaza48 | 2010 | Colombia | Hispanic | 145 | 72 (50%) | 55 ± 15 | Warfarin | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2b | Age, BMI, gender, indication for treatment |
| Jimenez-Varo49 | 2014–2015 | Spain | Whites | 162 | 89 (55%) | 73 ± 9 | Acenocoumarol | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2b | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Kringen33 | 2011 | Norway | Whites | 105 | 87 (83%) | 60 ± 9 | Warfarin | 1.9–3.6 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, gender, other drugs |
| Lee50 | 2012 | Korea | Asians | 188 | 62 (33%) | 59 ± 10 | Warfarin | 2.0–3.0 | CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, other drugs |
| Lee27 | 2009 | China | Asians | 233 | 130 (56%) | 63 ± 13 | Warfarin | 1.7–3.0 | CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, indication for treatment, other drugs |
| Lubitz34 | 2010 | New York (USA) | Whites (68%) Blacks (20%) Asians (12%) | 155 | 97 (63%) | 69 ± 14 | Warfarin | 2.0–3.0 | CYP2C9*2, CYP2C9*3, CYP2C9*5, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Mazzaccara51 | 2013 | Italy | Whites | 256 | 142 (55%) | 67 ± 11 | Warfarin | 1.6–3.9 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, indication for treatment, other drugs |
| Ozer52 | 2013 | Turchia | Whites | 107 | 53 (50%) | 54 ± 14 | Warfarin | 1.5–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment |
| Pathare37 | 2012 | Oman | Asians | 188 | 88 (47%) | 51 ± 17 | Warfarin | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Pautas35 | 2010 | France | Whites | 272 | 65 (24%) | 87 ±6 | Warfarin | 2.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, gender, other drugs |
| Pavani38 | 2012 | India | Indians | 207 | 108 (52%) | 40 ± 13 | Warfarin | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender |
| Perez-Andreu19 | 2009 | Spain | Whites | 100 | 100 (100%) | 65 ±6 | Acenocoumarol | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age |
| Perini20 | 2010 | Brazil | Whites (50%) Brown (30%) Blacks (20%) | 390 | 186 (48%) | 54 ± 15 | Warfarin | 2.0–3.5 | CYP2C9*2, CYP2C9*3, CYP2C9*5,VKORC1, CYP4F2b | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Ramirez39 | 2012 | Tennessee (USA) | Whites | 1,029 | 586 (57%) | 65 ± 15 | Warfarin | 1.6–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Rathore16 | 2012 | India | Indians | 217 | 145 (67%) | 39 ± 12 | Acenocoumarol | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2b | Age, BMI, gender, smoking |
| Sagreiya21 | 2010 | California (USA) | Whites (75%) Asians (17%) Blacks (8%) | 101 | 58 (57%) | 64 ±15 | Warfarin | 1.8–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Shain22 | 2011 | Egypt | Egyptians | 188 | 84 (44%) | 48 ± 15 | Warfarin | 1.5–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment |
| Shendre40 | 2016 | Alabama (USA) | Whites (58%) Blacks (41%) Asians (1%) | 1,169 | 610 (52%) | 61 ± 16 | Warfarin | 1.8–3.2 | CYP2C9*2, CYP2C9*3, CYP2C9*5,VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Suriapranata23 | 2011 | Indonesia | Asians | 85 | 48 (56%) | 57 ± 11 | Warfarin | 1.5–2.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Tan41 | 2013 | China | Asians | 317 | 95 (30%) | 45 ± 10 | Warfarin | 1.8–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender |
| Tatarunas42 | 2014 | Lithuania | Whites | 189 | 118 (62%) | 65 ± 11 | Warfarin | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, other drugs |
| van Schie (a)44 | 2013 | Netherlands | Whites | 568 | 328 (58%) | 70 ± 11 | Other (phenprocoumon) | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, indication for treatment, other drugs |
| van Schie (b)44 | 2013 | Netherlands | Whites | 397 | 217 (55%) | 73 ±9 | Acenocoumarol | 2.0–3.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, indication for treatment, other drugs |
| Wells24 | 2010 | Canada | Whites | 246 | 136 (55%) | 61 ± 14 | Warfarin | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
| Zambon25 | 2011 | Italy | Whites | 371 | 231 (62%) | 73 ± 9 | Warfarin | 2.5 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment |
| Zhang26 | 2009 | UK | Whites | 202 | 120 (59%) | 66 ± 14 | Warfarin | 2.0–3.0 | CYP2C9*2, CYP2C9*3, VKORC1, CYP4F2 | Age, BMI, gender, smoking, indication for treatment, other drugs |
BMI, body mass index; CYP, cytochrome P450; INR, International Normalized Ratio; PY, publication year.
Covariates with < 20% of missing data are here indicated and used in the multivariate analysis.
CYP2F4 not in Hardy-Weinberg equilibrium.
Association between CYP4F2*3 polymorphism and stable coumarin dose
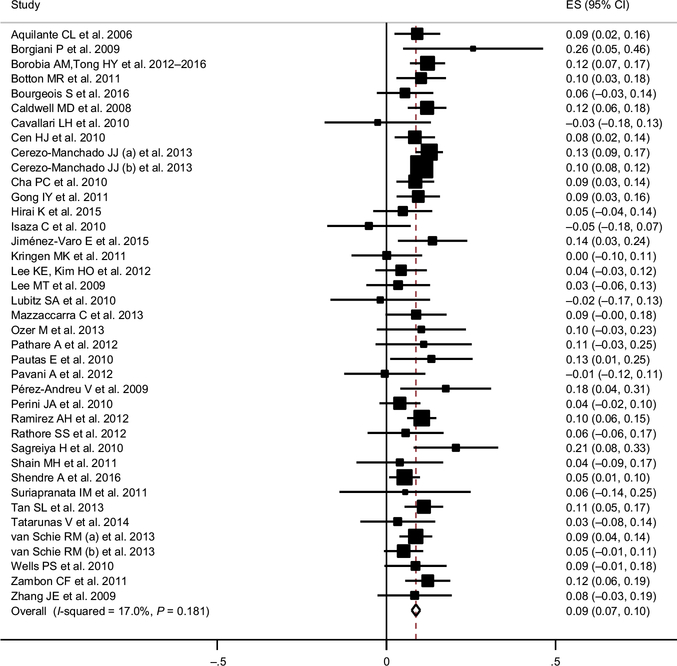
Figure 2 shows the forest plot for the difference in log dose of warfarin for subjects with at least one T allele (CT + TT) CYP4F2 as compared to wild-type (CC) subjects, according to a dominant model. The estimated effect size was 0.09 (95% confidence interval (CI) 0.07–0.10), corresponding to a 9% increase in mg/week (95% CI 7–10%). The funnel plot (see Figure S1) is compatible with no effect of bias on publication.
Figure 2.
Forest plot for the difference in logarithm of stable coumarin dose* for subjects with cytochrome P450 (CYP)4F2 polymorphism (CT + TT) compared with subjects with (CYP)4F2 wild-type (CC), according to dominant model.
CI=Confidence Intervals; ES=Estimate
* exp(ES) gives the relative percentage difference as weekly dose in mg
Separate estimates for CT and TT CYP4F2 genotypes are reported in Figure S2: the estimated effect size for CT vs. CC subjects is 0.07 (95% CI 0.06–0.08), corresponding to a 7% increase in mg/week; whereas for TT vs. CC subjects it is 0.17 (95% CI 0.15–0.19), corresponding to a 19% increase in mg/week. In Table 2, the analysis of the available subgroups highlights that the effect of the CYP4F2*3 polymorphism is significant in whites and Asians but not in blacks and other ethnic groups. Moreover, there was a significant difference by gender for the effect of the CYP4F2 polymorphism on coumarin dose (the effect is significantly higher in women) and by type of coumarin drugs (the effect was lower for warfarin as compared to acenocoumarol). No significant difference in the effects of smoking, target International Normalized Ratio (INR), adjustment for other drugs, consistency of genotype frequencies with the Hardy-Weinberg equilibrium, quality score, and other polymorphisms was found (Table 2). The figures for the different meta-analyses in subgroups are presented in Figures S3 and S4.
Table 2.
Subgroup analyses of association studies of the CYP4F2 polymorphism on coumarin dose requirements
| Variable | Subgroup (N studies) | Differencea (95% Cl) | I2 (Q test F value) | Meta regression P value |
|---|---|---|---|---|
| Ethnicity | Whites (26) | 0.10 (0.08–0.11) | 15% (0.25) | 0.002 |
| Asians (10) | 0.08 (0.05–0.11) | 0% (0.85) | 0.02 | |
| Blacks (5) | 0.05 (−0.04; 0.14) | 21% (0.28) | 0.36 | |
| Others (5)b | 0.01 (−0.05; 0.06) | 0% (0.72) | Reference | |
| Drug | Acenocoumarol (7) | 0.11 (0.09–0.13) | 13% (0.33) | 0.03 |
| Warfarin (31) | 0.08 (0.06–0.09) | 9% (0.33) | ||
| Sex | Males (39) | 0.07 (0.06–0.09) | 16% (0.20) | 0.03 |
| Females (38) | 0.10 (0.08–0.12) | 19% (0.16) | ||
| INR target | < 2.5 (11) | 0.08 (0.05–0.11) | 0% (0.79) | Reference |
| 2.5 (23) | 0.09 (0.07–0.11) | 22% (0.17) | 0.42 | |
| > 2.5 (18) | 0.08 (0.06–0.10) | 0% (0.49) | 0.93 | |
| Smoking | No (21) | 0.09 (0.07–0.11) | 0% (0.68) | 0.74 |
| Yes (12) | 0.07 (−0.02; 0.15) | 33% (0.12) | ||
| Other drugs considered | No (7) | 0.08 (0.03–0.12) | 47% (0.08) | 0.72 |
| Yes (32) | 0.09 (0.08–0.10) | 10% (0.31) | ||
| Hardy-Weinberg equilibrium | No (6) | 0.08 (0.04–0.12) | 53% (0.06) | 0.92 |
| Yes (33) | 0.09 (0.07–0.10) | 9% (0.33) | ||
| Quality score | < 5 | 0.08 (0.06–0.10) | 29% (0.11) | 0.46 |
| ≥ 5 | 0.09 (0.08–0.11) | 6% (0.39) | ||
| CYP2C9 | CYP2C9 *1*1 | 0.08 (0.07–0.10) | 18% (0.18) | 0.73 |
| CYP2C9 *1*2/*1*3/*2*2/*2*3/*3*3 | 0.09 (0.06–0.12) | 25% (0.10) | ||
| VK0RC11 | VK0RC1 GG | 0.08 (0.06–0.10) | 4% (0.40) | 0.13 |
| VKORC 1 AA/AG | 0.10 (0.08–0.11) | 10% (0.30) |
Data in bold are significant at P < 0.05.
CI, confidence interval; CYP, cytochrome P450; INR, International Normalized Ratio.
Difference in logarithm of stable coumarin dose of subjects with CYP4F2 polymorphism (CT + TT) compared with subjects with CYP4F2 wild-type (CC), according to dominant model.
Includes Indian, Egyptian, brown, and Hispanic.
Stable coumarin dose predictive model
Table 3 presents the predictive model for logarithm of stable coumarin dose according to patients’ clinical and genetics characteristics. As statistical test for model fit (R2) is reported for both the test and validation cohorts. Looking at our calculated model on the whole dataset, adjusted R2 was slightly higher for models including CYP4F2*3 polymorphisms than for models without CYP4F2*3 for all the ethnic groups except blacks (for warfarin dose, adjusted R2 for models with and without CYP4F2*3 polymorphism were, respectively, 0.51 and 0.50 for whites, 0.43 and 0.42 for Asians, and 0.27 and 0.27 for blacks). For cohorts that included black patients, addition of the CYP2C9*5 single-nucleotide polymorphism (SNP) to the models did not result in substantial improvement of the adjusted R2 (Table 3). Further prediction models also including concomitant drugs (amiodarone, etc.) and smoking habits are presented in Table S1.
Table 3.
Predictive model for logarithm of stable coumarin dose according to patients’ clinical and genetic characteristics
| WHITES | ||||||||
|---|---|---|---|---|---|---|---|---|
| Acenocoumarol | Warfarin | |||||||
| Parameter estimate (95% Cl) | P value | R2 test (N = 2,744) | R2 validation (N = 1,410) | Parameter estimate (95% Cl) | P value | R2latest (N = 3,016) | R2 validation (N = 1,532) | |
| Intercept | 4.069 (3.883; 4.256) | < 0.0001 | 0.33 | 0.28 | 3.981 (3.887; 4.075) | < 0.0001 | 0.51 | 0.52 |
| Agea | −0.014 (−0.015; −0.012) | < 0.0001 | −0.009 (−0.010; −0.008) | < 0.0001 | ||||
| BMIa | −0.002 (−0.006; 0.002) | 0.28 | 0.010 (0.008; 0.012) | < 0.0001 | ||||
| Male gender | 0.014 (−0.024; 0.052) | 0.47 | 0.123 (0.098; 0.148) | < 0.0001 | ||||
| Indication for treatmentb | 0.000 (−0.042; 0.042) | 0.98 | −0.043 (−0.069; −0.017) | 0.001 | ||||
| CYP2C9 *2 1-allele | −0.190 (−0.232; −0.147) | < 0.0001 | −0.231 (−0.261; −0.202) | < 0.0001 | ||||
| CYP2C9 *2 2-alleles | −0.359 (−0.484; −0.234) | < 0.0001 | −0.513 (−0.600; −0.426) | < 0.0001 | ||||
| CYP2C9 *3 1-allele | −0.394 (−0.446; −0.342) | < 0.0001 | −0.387 (−0.425; −0.350) | < 0.0001 | ||||
| CYP2C9 *3 2-alleles | −1.214 (−1.522; −0.907) | < 0.0001 | −1.316 (−1.502; −1.131) | < 0.0001 | ||||
| VK0RC1 AG | −0.291 (−0.332; −0.249) | < 0.0001 | −0.266 (−0.292; −0.240) | < 0.0001 | ||||
| VK0RC1 AA | −0.762 (−0.816; −0.708) | < 0.0001 | −0.666 (−0.704; −0.629) | < 0.0001 | ||||
| CYP4F2 CT | 0.018 (−0.022; 0.058) | 0.39 | 0.073 (0.047; 0.098) | < 0.0001 | ||||
| CYP4F2 TT | 0.100 (0.041; 0.159) | 0.0009 | 0.191 (0.147; 0.235) | < 0.0001 | ||||
| ASIANS | ||||||||
| Acenocoumarol | Warfarin | |||||||
| Parameter estimate | P value | R2 test (N=0) | R2 validation (N=0) | Parameter estimate | P value | R2 test (N = 292) | R2 validation (N = 146) | |
| Intercept | - | - | - | - | 3.484 (3.112; 3.855) | < 0.0001 | 0.45 | 0.42 |
| Agea | - | - | -0.005 (−0.008; −0.001) | 0.02 | ||||
| BMIa | - | - | 0.014 (0.004; 0.023) | 0.004 | ||||
| Male gender | - | - | 0.058 (−0.050; 0.167) | 0.29 | ||||
| Indication for treatmentb | - | - | −0.027 (−0.139; 0.084) | 0.63 | ||||
| CYP2C9*2 1-allele | - | - | −0.114 (−0.351; 0.124) | 0.35 | ||||
| CYP2C9*2 2-alleles | - | - | - | - | ||||
| CYP2C9*3 1-allele | - | - | −0.224 (−0.428; −0.020) | 0.03 | ||||
| CYP2C9*3 2-alleles | - | - | −1.065 (−1.717; −0.412) | 0.002 | ||||
| VK0RC1 AG | - | - | −0.422 (−0.574; −0.271) | < .0001 | ||||
| VK0RC1 AA | - | - | −0.827 (−0.975; −0.679) | < .0001 | ||||
| CYP4F2 CT | - | - | 0.117 (0.003; 0.231) | 0.04 | ||||
| CYP4F2 TT | - | - | 0.124 (−0.075; 0.324) | 0.22 | ||||
| BLACKS | ||||||||
| Acenocoumarol | Warfarin | |||||||
| Variable | Parameter estimate | P value | R2test (/V=0) | R2 validation (N = 0) | Parameter estimate | P value | R2 test (N = 534) | R2 validation (N = 288) |
| Intercept | - | - | - | - | 3.875 (3.692; 4.061) | < 0.0001 | 0.30 | 0.22 |
| Agea | - | - | −0.009 (−0.011; −0.006) | < 0.0001 | ||||
| BMIa | - | - | 0.010 (0.007; 0.015) | < 0.0001 | ||||
| Male gender | - | - | 0.152 (0.086; 0.219) | < 0.0001 | ||||
| Indication for treatmentb | - | - | −0.090 (−0.160; −.0183) | 0.01 | ||||
| CYP2C9*2 1-allele | - | - | −0.007 (−0.149;0.133) | 0.93 | ||||
| CYP2C9*2 2-alleles | - | - | - | - | ||||
| CYP2C9 *3 1-allele | - | - | −0.469 (−0.666; −0.270) | < 0.0001 | ||||
| CYP2C9 *3 2-alleles | - | - | - | - | ||||
| CYP2C9 *5 1-allele | - | - | −0.436 (−0.736; −0.137) | 0.005 | ||||
| CYP2C9 *5 2-alleles | - | - | - | - | ||||
| VK0RC1 AG | - | - | −0.284 (−0.585; −0.020) | 0.07 | ||||
| VK0RC1 AA | - | - | −0.281 (−0.588; −0.020) | < 0.0001 | ||||
| CYP4F2 CT | - | - | −0.0382 (−0.124; 0.050) | 0.40 | ||||
| CYP4F2 TT | - | - | 0.300 (−0.068; 0.664) | 0.11 | ||||
Statistical test for model fit (R2) is reported both for the test and validation cohorts. Due to significant heterogeneity, separate models are reported for different ethnic groups and drugs. BMI, body mass index; CI, confidence interval; CYP, cytochrome P450.
Estimate for 1 unit increase.
Estimate for the following indication for treatment: fibrillation/flutter, cardiomyopathy/left ventricular dilation, postorthopedic.
Beta coefficients for single-gene and gene–gene interaction are presented in Table 4 for each ethnicity and drug subgroups.
Table 4.
Beta coefficients (P values) for single genes and gene–gene interaction
| Ethnicity | Drug | N subjects (N studies) | CYP4F2 | CYP2C9 | VKORC1 | CYP4F2*CYP2C9 | CYP4F2*VKORC1 | CYP2C9*VKORC1 |
|---|---|---|---|---|---|---|---|---|
| Whites | Acenocoumarol | 4,154 (5) | 0.08 (0.0002) | −0.22 (< 0.0001) | −0.40 (< 0.0001) | −0.02 (0.51) | −0.03 (0.21) | −0.01 (0.79) |
| Warfarin | 4,548 (15) | 0.08 (0.0001) | −0.30 (< 0.0001) | −0.38 (< 0.0001) | −0.001 (0.96) | 0.02 (0.37) | −0.01 (0.55) | |
| Asians | Acenocoumarol | 0(0) | NE | NE | NE | NE | NE | NE |
| Warfarin | 438 (8) | 0.10 (0.34) | −0.26 (0.05) | −0.46 (< 0.0001) | 0.12 (0.36) | −0.08 (0.48) | −0.004 (0.98) | |
| Blacks | Acenocoumarol | 0(0) | NE | NE | NE | NE | NE | NE |
| Warfarin | 815 (5) | 0.04 (0.30) | −0.20 (0.0004) | −0.27 (< 0.0001) | 0.004 (0.97) | −0.02 (0.82) | 0.02 (0.83) | |
| Others | Acenocoumarol | 0(0) | NE | NE | NE | NE | NE | NE |
| Warfarin | 701 (7) | −0.08 (0.13) | −0.19 (0.003) | −0.27 (< 0.0001) | 0.07 (0.31) | 0.09 (0.13) | −0.05 (0.48) | |
| All | All | 11,435 (29) | 0.07 (< 0.0001) | −0.24 (< 0.0001) | −0.37 (< 0.0001) | 0.02 (0.21) | 0.02 (0.23) | −0.02 (0.12) |
Data in bold are significant at P < 0.05.
Ethnicity-specific and drug-specific models are adjusted by study, age, gender, body mass index, and indication for treatment. The final model is also adjusted by ethnicity and drug. For each gene, the reference category is the gene polymorphism according to the dominant model (heterozygous + variant homozygous vs. wt). For the analysis on blacks, CYP2C9 included, beyond *2 and *3, also *5 polymorphism. CYP, cytochrome P450; NE, not estimated.
The effect of potentially interacting drugs could be evaluated only on a subsample of the cohorts and is presented in Table S2. Patients taking amiodarone or drugs classified as CYP inhibitors required a lower dose, whereas patients taking CYP inducers required a higher dose of coumarin drugs. If the effect of the drugs was considered, the beta estimate for CYP4F2 and the other SNPs varied slightly but remained significant for most analyses. No significant interaction between SNPs and drugs were detectable apart from CYP2C9*2 and rifampin and all CYP inhibitors and all CYP inducers in white patients consuming acenocoumarol. Another weak but nominally significant interaction was present between CYP2C9*2 and statin or aspirin in black patients on chronic warfarin therapy (Table S2).
The comparison of R2 of our model with those calculated for two previously published models are reported in Table S3 and are basically comparable, ranging from 0.41−0.47 for whites, 0.44 for Asians, and from 0.23−0.33 for blacks.
DISCUSSION
In our previous meta-analysis on the effect of the CYP4F2 rs2108622 (1347C>T; c.1297G>A; p.Val433Met; CYP4F2*3), we found that the estimated effect size was nearly 10%. In this individual patient data meta-analysis, we have not only confirmed this finding in a larger cohort of primary studies that include all the available study-specific covariates but can add other important findings. Contrary to what was found in the first meta-analysis, a slight but significant effect of gender was identified such that men had a lower effect of the T allele when compared to women.
Indeed, a higher dose of coumarin drugs was needed in carriers of the T allele if they were whites or Asians but not in blacks or in other ethnic groups (Indians, browns from Brazil, and Egyptians), but the latter is probably a reflection of the lower sample size. We also identified differences between different coumarin drugs: patients taking acenocoumarol and carrying the T allele needed a higher dose of the drug when compared with patients taking warfarin and carrying the same polymorphism.
There was no effect of other possible important covariates, such as smoking, age, and indication for coumarin, and no interactions with the other relevant polymorphisms were found.
Evaluation of the beta estimate of the tested SNPs confirmed that the larger effect is due to the VKORC1 followed by CYP2C9, whereas CYP4F2 had a limited effect size.
Looking at primary studies, the large majority of them are in line with the results of the meta-analysis, and only 4 of the 39 have a central point of the estimate below the 0 line. Even the point estimate for the effect of CYP4F2 is not so different between primary studies. The extremes are the study performed by Borgiani et al.18 with a + 0.26 estimate and the one by Isaza et al.48 with a −0.05, which have a 95% CI that is around + 0.07, not far from our total effect size (slightly < 10%).
However, the funnel plot shows a certain asymmetry, almost significant when analyzed using Egger’s test. It is therefore possible that unpublished negative studies could affect the real estimate of the effect of the CYP4F2*3 polymorphism.
Differently from our previous meta-analysis, we could also add drugs as moderating parameters at least in some subgroups, and as expected, this evaluation decreased heterogeneity.
The functionality of the CYP4F2 polymorphism has been shown in relation to the production of 20-hydroxyeicosatetraenoic acid derived by arachidonic acid and in differences in mRNA production by liver cells in carriers of different alleles.53
The interaction of the CYP4F2 polymorphism with gender is not unexpected: also in other studies exploring other cardiovascular actions, some CYP polymorphisms have shown a differential effect in men and women probably due to an interaction with either androgens or estrogens.54 Even in animal models these differences are evident, at least for blood pressure determination.54
Due to our large sample size, we could calculate and subsequently validate different prediction models that included the effect of the CYP4F2*3, the other well-known polymorphisms of CYP2C9 and VKORC1, and the other covariates differentiating the effect of gender and ethnicity and obtaining discrete coefficient of determinations that indicate a good fit of the models. Other predictive pharmacogenetic equations estimating coumarin dose have been developed using large samples sizes,13,14 but both the International Warfarin Pharmacogenetics Consortium and the “Warfarin dosing” equations used only CYP2C9 and VKORC1 genetic variation to estimate warfarin dose and the R2 estimate for the final model (which also included amiodarone), obtaining values of 0.47 and 0.53, respectively. These results are in line with our data for white subjects, but our results are more generalizable because multiple cohorts from Europe were also included. In fact, Gage’s equation is derived from a more homogeneous group of patients collected in three centers in the United States (St. Louis, San Antonio, and Gainesville) with a fourth trial included in the validation cohort.13 By contrast, the International Warfarin Pharmacogenetics Consortium collected 21 research groups from 9 different countries and finally included only patients with a target INR between 2 and 3 (n = 5,052). Their final model was not divided according to ethnicity but instead the ethnicity variable was added in the model. Indeed, outlier patients were excluded from the final analysis. It is worth mentioning that the final sample size of our study is more than two times the previous studies for warfarin, and we have also calculated predictive models for acenocoumarol.
Even if newer anticoagulants have substantially changed clinical practice, especially in developed countries, the use of coumarin drugs is still widespread in the world, so that equations like the one derived from our study will be clinically useful for many years. The importance of genotype has been further shown in the Effective aNticoaGulation with factor xA next GEneration in Atrial Fibrillation-Thrombolysis In Myocardial Infarction (ENGAGE AF-TIMI) 48 trial, which compared the clinical efficacy of edox-aban, a direct oral anticoagulant, with warfarin in a prespecified genetic subanalysis. Stratification of patients according to CYP2C9 and VKORC1 polymorphisms revealed that of the three groups identified, normal responders, sensitive responders, and highly sensitive responders, the last group was found to spend a greater proportion of time over-anticoagulated compared with normal responders but only for the first 90 days of treatment.55
RCTs using not only the CYP2C9 and VKORC1 poly-morphism but also the CYP4F2 polymorphism have recently been performed. In nonvalvular atrial fibrillation, no apparent advantage was found for the group randomized to genotype base dose,56 but in a recent trial in patients aged 65 years or older initiating warfarin for elective hip or knee arthroplasty conducted at six US medical centers, genotyping reduced the combined risk of major bleeding, INR of 4 or greater, venous thromboembolism, or death.6
In another trial that compared a genotype-guided algorithm vs. physician management for the initiation of acenocoumarol, a higher proportion of patients in the genetic group reached and maintained a steady dose than patients randomized to routine practice when starting oral anticoagulation.57
Limitations and strengths of the study
Our individual data meta-analysis has limitations. First, although we applied a sensitive search strategy for the retrieval of potentially eligible studies, we cannot rule out the possibility that some relevant studies might not have been included. Indeed, not all the potentially eligible studies were added to the meta-analysis because the authors did not share individual patients’ data. Second, adjustment for certain covariates, such as amiodarone, was possible in only a limited sample of patients. The quality score of the included studies was heterogeneous, ranging from 3−7 (median: 5), but this did not affect CYP4F2*3–coumarin dose association, because we found no statistically significant difference in the estimates for studies with lower and higher quality score. Finally, our genotyping-based algorithms in blacks have low predictivity even including the CYP2C9*5 poly-morphism, probably because we could not include more variants in CYP2C9 that were demonstrated to be especially important in this ethnic group.2 Because the exclusion of specific CYP2C9 variants from the dosing algorithm in blacks can lead to overdosing, we would recommend against the use of the specific dosing algorithms in patients of African ancestry2 until more specific algorithms have been developed.
Strengths of our collaborative study are the large sample size with several ethnic groups allowing for generalizability of the results and the possibility to have equations not only for warfarin but also for acenocoumarol based on a quite large sample size. The heterogeneity was low possibly because most of the variables associated with mean coumarin dose have been considered in our models.
CONCLUSION
In conclusion, we have undertaken the largest individual patient data meta-analysis, including the CYP4F2 polymorphism, in patients taking warfarin or other coumarin drugs. Our data show that the CYP4F2 rs2108622 polymorphism affects the dose requirements of these drugs in whites and Asians but not in blacks or other ethnic groups. We also provide reliable prediction models, which can guide physicians to estimate the stable dose of warfarin according to genotypes, anthropometric and demographic factors, ethnicity, and the use of other drugs.
Regardless, because RCTs that tested genetic prediction models with the CYP4F2*3 SNP showed contradictory results,6,56 the utility of these models in clinical practice needs to be established in further RCTs before their widespread utilization in clinical settings.
METHODS
Search strategy and eligibility criteria
The 30 articles included in our previous meta-analysis were considered all potentially eligible for the present study.12 To expand our search to articles published after the date fixed for final inclusion in the previous meta-analysis, we searched Medline and Web of Science from September 1, 2011, to September 14, 2016, by applying the same search algorithm used previously (see Supplementary Material S1) and found 38 additional studies that could potentially be included (see Figure 1) according to the inclusion criteria (see Supplementary Material S1). All 68 studies evaluated for inclusion were clinical cohort or cross-sectional studies that have performed genotyping of CYP4F2 in combination with CYP2C9 (at least one of the two variants of interest) and/or VKORC1 in coumarin-treated patients. As per our previous study, we considered the following polymorphisms: rs2108622 (1347C>T; 1297G>A; p.Val433Met; CYP4F2*3) for CYP4F2, rs1799853 (430C>T) and rs1057910 (1075A>C) for CYP2C9 (also known as CYP2C9*2 and CYP2C9*3), and rs9923231 (−1639 G>A) for VKORC1. In relation to the latter variant, we also included data from studies that used the two alternative polymorphisms: rs9934438 (1173C>T) in the VKORC1 gene, which is in complete linkage disequilibrium with the reference polymorphism and rs10871454 (−1168C>T) located in the Syntaxin 4 A-(STX4A) gene, flanking the VKORC1 gene, which showed a linkage disequilibrium of 0.99 with the reference polymorphism.
In our previous analysis, consistent with published studies, the performance of our regression was low, especially in black subjects, in which an effect of other SNPs especially in CYP2C9 is considered important. Thus, in the five cohorts in which at least the CYP2C9*5 variant was available we repeated the analysis by adding this polymorphism.
Data collection
We asked the first/last or corresponding authors of the retrieved primary studies to participate in a collaborative meta-analysis on individual patient data. Authors who were willing to collaborate were finally included if their original database contained the following mandatory data for single patients: gender, age, race, genotypes, indication for coumarin therapy, INR target, type of coumarin used, and maintenance dose. Additional information on body weight, height, and use of interacting drugs were also recorded when available. Each cohort has been assigned to one single study unless otherwise specified. For studies containing overlapping samples we considered the first published study or the one that enrolled the largest number of patients. Data were harmonized into a pooled database. Two researchers (E.D. and M.M.) cross-checked trial details provided by the authors against published articles. Any inconsistencies were discussed with the original trialists, and corrections were made when appropriate. As for our previous meta-analysis, we graded the quality of epidemiologic studies in general, applying items taken from the Newcastle–Ottawa Quality Assessment Scale for Cohort Studies indicators specific to the quality of genetic association studies, and indicators specific for coumarin (e.g., stable anticoagulation). Quality assessment also included departure from Hardy-Weinberg equilibrium that was calculated by the χ2 test in controls. We applied a scale with a maximum score of seven points (see Supplementary Material S1 for details).
Statistical analysis
Two-stage analysis for the association between CYP4F2*3 polymorphism and stable coumarin dose.
We calculated study-specific estimates, with 95% CIs, for the difference in log dose of coumarin for subjects with at least one CYP4F2 T allele (CT + TT) compared to wild-type (CC) subjects, according to a dominant model. Separate estimates for CT and TT genotypes were also calculated as a sensitivity analysis. These study-specific estimates were obtained by fitting general linear models with log dose of coumarin as the dependent variable and CYP4F2*3 polymorphism as the independent variable. All the models were adjusted for available study-specific covariates, including age, gender, race, BMI, smoking status, indication for coumarin treatment, INR target, concomitant drugs, CYP2C9*2 and *3 polymorphisms, and VKORC1 polymorphism.
Following the two-stage analysis approach, we pooled study-specific estimates with random-effects models, using the DerSimonian and Laird method (see Supplementary Methods S1). We evaluated homogeneity among study-specific estimates by the Q statistic and I2, which represents the percentage of total variation across studies that is attributable to heterogeneity rather than to chance (see Supplementary Methods S1). We performed metaregression analysis to assess the influence on Summary Estimates of different study features: type of drugs (acenocoumarol/warfarin), gender, ethnicity (whites/Asians/blacks/others), INR target (< 2.5/2.5/> 2.5), current smoking status, study adjustment for concomitant drugs (yes/no), deviation from Hardy-Weinberg equilibrium, quality score (< 5/≥ 5), CYP2C9*2/*3 (wild-type/any polymorphism), and VKORC1 (wild-type/any poly-morphism). When significant differences according to specific study factors were suggested by metaregression, stratified analyses were performed for CYP4F2*3–coumarin dose association on subgroups of significant factors.
We assessed possible participation bias by drawing funnel plots and by Egger’s test (see Supplementary Methods S1).
The P values < 0.05 were considered statistically significant for all the tests apart from the Q statistic, where P values < 0.10 were considered statistically significant. The analysis was carried out using the SAS (SAS Institute Inc, Cary, NC) version 9.4 and STATA (StataCorp, College Station, TX) version 13 software.
Stable coumarin dose predictive model.
Due to significant differences in coumarin dose and CYP4F2*3 association for different drugs and ethnic groups, the individual data analysis on the pooled dataset was always reported for each type of drug (acenocoumarol/warfarin) and for each ethnic group.
For each ethnic and drug subgroup, we randomly chose 2/3 of patients as the “derivation cohort” for developing dose-prediction models, whereas the remaining 1/3 of the patients constituted the “validation cohort,” which was used for testing the final selected model. In order to keep a large sample size for prediction model construction, we included covariates that were available in the majority of studies (Table 1): age, BMI, gender, indication for treatment, CYP4F2*3, CYP2C9*2, *3, and *5 (for blacks), and VKORC1 polymorphisms, by using general linear models with log dose of coumarin as dependent variable. To use an additive genetic model, we coded the number of variant alleles at each locus as 0, 1, or 2. Sensitivity analyses were also conducted on the whole cohort of subjects by including further available covariates collected in a smaller number of studies (concomitant drugs, especially amiodarone, and smoking status) to assess their role in stable coumarin dose prediction. The coefficient of determination (R2) was calculated both for the main prediction model on the “derivation cohort” and for models included in sensitivity analyses. We applied the scores obtained from the main prediction model to the validation dataset and also calculated the R2.
For the sake of comparison, we also applied scores obtained from two previously published models for warfarin dose prediction13,14 to our validation cohort and converted the scores to units of mg/week. In order to correctly compare our proposed model with each of the two previously published models, R2 was calculated on the subset of subjects for whom both scores could be calculated on the basis of available data. In order to assess the importance of CYP4F2*3 on warfarin dose prediction in our data, we also compared dose predictions from our pharmacogenetic model, including CYP4F2*3 in the whole dataset with that from our model excluding CYP4F2*3 by using the adjusted R2 as defined by Darlington (see Supplementary Methods S1).
Gene–gene and gene–drug interactions were investigated by adding an interaction term to the main prediction model fitted on the whole cohort of subjects (for each drug/ethnicity subgroup) in order to have the largest sample size to test for interaction. Moreover, we performed subgroup analyses according to the use or not of specific concomitant drugs, to evaluate whether the change in coumarin dose associated with specific gene poly-morphisms were modified by concomitant drugs.
The assumption of exchangeability for this analysis was briefly discussed in the Supplementary Methods S1. The P values < 0.05 were considered statistically significant. The analyses were carried out using SAS version 9.4 software. The SAS code is available as Supplementary Material S1.
Supplementary Material
Figure S1. Funnel plot for association studies of the CYP4F2 polymorphism on coumarin dose.
Figure S2. Forest plot for the difference in logarithm of stable coumarin dose* for subjects with (a) CYP4F2 polymorphism (CT) compared to subjects with CYP4F2 wild-type (CC), (b) CYP4F2 polymorphism (TT) compared with subjects with CYP4F2 wild-type (CC).
Figure S3. Forest plot for the difference in logarithm of stable coumarin dose* for subjects with CYP4F2 polymorphism (CT + TT) compared to subjects with CYP4F2 wild-type (CC), according to dominant model and stratified by (A) ethnicity; (B) drug; (C) gender.
Figure S4. Forest plot for the difference in logarithm of stable coumarin dose* for subjects with CYP4F2 polymorphism (CT + TT) compared with subjects with CYP4F2 wild-type (CC), according to dominant model and stratified by (A) CYP2C9; (B) VKORC1.
Table S1. Predictive models for logarithm of INR dose according to patients’ clinical and genetics characteristics: sensitivity analysis including different models.
Table S2. Effect of concomitant drugs on warfarin dose and genetic polymorphisms of CYP4F2 and CYP2C9 genes: gene− drug interaction and subgroup analyses.
Table S3. Statistical test for model fit (R2) of two previously published models for warfarin dose prediction (Gage 2008, Klein 2009) in comparison with the model presented here in Table 3 (“new model”): application to a subset of subjects from the validation cohort for whom both scores could be calculable on the basis of the available information.
Supplementary Methods S1. Methods.
Supplementary Material S1. Discussion and SAS code.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Coumarin drugs have a narrow therapeutic index, but single-nucleotide polymorphisms (SNPs) in the cytochrome P450 (CYP2)C9 and VKORC1 genes may help in predicting the dose.
WHAT QUESTION DID THIS STUDY ADDRESS?
Do genetic algorithms, including the CYP4F2*3 SNP, perform better than old ones in predicting mean coumarin dose?
WHAT DOES THIS STUDY ADD TO OUR KNOW LEDGE?
In this single-patient meta-analysis, we confirm that CYP4F2*3 influences mean coumarin dose especially in women, in patients taking acenocoumarol, and in white patients but with a low effect size.
HOW MIGHT THIS CHANGE CLINICAL PHA RMACOLOGY OR TRANSLATIONAL SCIENCE?
New pharmacogenetic equations potentially useful for clinical practice have been derived for different ethnic groups.
FUNDING
P.D. is supported by British Heart Foundation (BHF) grant RG/14/5/30893; this study forms part of the research themes contributing to the translational research portfolio of Barts Cardiovascular Biomedical Research Centre, which is funded by the National Institute for Health Research (NIHR). C.F. was supported by a grant of the CARIVERONA foundation for the project “towards a tailored interpretation of the individual variability of the response to the drug”.
Footnotes
SUPPORTING INFORMATION
Supplementary information accompanies this paper on the Clinical Pharmacology & Therapeutics website (www.cpt-journal.com).
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
References
- 1.Lim GB. Milestone 2: warfarin: from rat poison to clinical use. Nat Rev Cardiol. 10.1038/nrcardio.2017.172. [e-pub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JA et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided war-farin dosing: 2017 update. Clin. Pharmacol. Ther 102, 397–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stergiopoulos K & Brown DL Genotype-guided vs clinical dosing of warfarin and its analogues: meta-analysis of randomized clinical trials. JAMA Intern. Med 174, 1330–1338 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Pirmohamed M et al. A randomized trial of genotype-guided dosing of warfarin. N. Engl. J. Med 369, 2294–2303 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Kimmel SE et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med 369, 2283–2293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gage BF et al. Effect of genotype-guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty. JAMA 318, 1115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X et al. Clinical benefits of pharmacogenetic algorithm-based warfarin dosing: meta-analysis of randomized controlled trials. Thromb. Res 135, 621–629 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Franchini M, Mengoli C, Cruciani M, Bonfanti C & Mannucci PM Effects on bleeding complications of pharmacogenetic testing for initial dosing of vitamin K antagonists: a systematic review and meta-analysis. J. Thromb. Haemost 12, 1480–1487 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Shi C et al. Pharmacogenetics-based versus conventional dosing of warfarin: a meta-analysis of randomized controlled trials. PLoS One 10, e0144511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belley-Cote EP et al. Genotype-guided versus standard vitamin K antagonist dosing algorithms in patients initiating anticoagulation. A systematic review and meta-analysis. Thromb. Haemost 114, 768–777 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Wang Z-Q et al. Pharmacogenetics-based warfarin dosing algorithm decreases time to stable anticoagulation and the risk of major hemorrhage. J. Cardiovasc. Pharmacol 65, 364–370 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Danese E et al. Impact of the CYP4F2 p.V433M polymorphism on coumarin dose requirement: systematic review and meta-analysis. Clin. Pharmacol. Ther 92, 746–756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gage B et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin. Pharmacol. Ther 84, 326–331 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium IWP et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med 360, 753–764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borobia AM et al. An acenocoumarol dosing algorithm using clinical and pharmacogenetic data in Spanish patients with thromboembolic disease. PLoS One 7, e41360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathore SS et al. Therapeutic dosing of acenocoumarol: proposal of a population specific pharmacogenetic dosing algorithm and its validation in north Indians. PLoS One 7, e37844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldwell MD et al. CYP4F2 genetic variant alters required warfarin dose. Blood 111, 4106–4112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borgiani P et al. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics 10, 261–266 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Andreu V et al. Pharmacogenetic relevance of CYP4F2 V433M polymorphism on acenocoumarol therapy. Blood 113, 4977–4979 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Perini JA, Struchiner CJ, Silva-Assunção E & Suarez-Kurtz G Impact of CYP4F2 rs2108622 on the stable warfarin dose in an admixed patient cohort. Clin. Pharmacol. Ther 87, 417–420 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Sagreiya H et al. Extending and evaluating a warfarin dosing algorithm that includes CYP4F2 and pooled rare variants of CYP2C9. Pharmacogenet. Genomics 20, 407–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahin MHA et al. Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet. Genomics 21, 130–135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suriapranata IM et al. Genetic factors associated with patient-specific warfarin dose in ethnic Indonesians. BMC Med. Genet 12, 80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells PS et al. A regression model to predict warfarin dose from clinical variables and polymorphisms in CYP2C9, CYP4F2, and VKORC1: derivation in a sample with predominantly a history of venous thromboembolism. Thromb. Res 125, e259–e264 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Zambon C-F et al. VKORC1, CYP2C9 and CYP4F2 genetic-based algorithm for warfarin dosing: an Italian retrospective study. Pharmacogenomics 12, 15–25 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Zhang JE et al. Effects of CYP4F2 genetic polymorphisms and haplotypes on clinical outcomes in patients initiated on warfarin therapy. Pharmacogenet. Genomics 19, 781–789 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Lee MTM et al. Genetic determinants of warfarin dosing in the Han-Chinese population. Pharmacogenomics 10, 1905–1913 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Botton MR, Bandinelli E, Rohde LEP, Amon LC & Hutz MH Influence of genetic, biological and pharmacological factors on warfarin dose in a Southern Brazilian population of European ancestry. Br. J. Clin. Pharmacol 72, 442–450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavallari LH et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin. Pharmacol. Ther 87, 459–464 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Cen H-J et al. CYP4F2 rs2108622: a minor significant genetic factor of warfarin dose in Han Chinese patients with mechanical heart valve replacement. Br. J. Clin. Pharmacol 70, 234–240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cha P-C et al. Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum. Mol. Genet 19, 4735–4744 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Gong IY et al. Prospective evaluation of a pharmacogenetics-guided warfarin loading and maintenance dose regimen for initiation of therapy. Blood 118, 3163–3171 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Kringen MK et al. Genetic variation of VKORC1 and CYP4F2 genes related to warfarin maintenance dose in patients with myocardial infarction. J. Biomed. Biotechnol 2011, 739751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lubitz SA et al. Comparative performance of gene-based warfarin dosing algorithms in a multiethnic population. J. Thromb. Haemost 8, 1018–1026 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pautas E et al. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin. Pharmacol. Ther 87, 57–64 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Aquilante C et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin. Pharmacol. Ther 79, 291–302 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Pathare AV et al. Warfarin pharmacogenetics: polymorphisms of the CYP2C9, CYP4F2, and VKORC1 loci in a genetically admixed Omani population. Hum. Biol 84, 67–77 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Pavani A et al. Optimization of warfarin dose by population-specific pharmacogenomic algorithm. Pharmacogenomics J. 12, 306–311 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Ramirez AH et al. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics 13, 407–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shendre A et al. Race-specific influence of CYP4F2 on dose and risk of hemorrhage among warfarin users. Pharmacotherapy 36, 263–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan S-LL et al. Cytochrome P450 oxidoreductase genetic polymorphisms A503V and rs2868177 do not significantly affect warfarin stable dosage in Han-Chinese patients with mechanical heart valve replacement. Eur. J. Clin. Pharmacol 69, 1769–1775 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Tatarunas V et al. The effect of CYP2C9, VKORC1 and CYP4F2 polymorphism and of clinical factors on warfarin dosage during initiation and long-term treatment after heart valve surgery. J. Thromb. Thrombolysis 37, 177–185 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Tong HY et al. A new pharmacogenetic algorithm to predict the most appropriate dosage of acenocoumarol for stable anticoag-ulation in a mixed Spanish population. PLoS One 11, e0150456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Schie RMF, Aoussar A, van der Meer FJM, Boer A. de & Maitland-van der Zee AH Evaluation of the effects of single-nucleotide polymorphisms in CYP3A4 and CYP4F2 on stable phenprocoumon and acenocoumarol maintenance doses. J. Thromb. Haemost 11, 1200–1203 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Bourgeois S et al. A multi-factorial analysis of response to warfa-rin in a UK prospective cohort. Genome Med. 8, 2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerezo-Manchado JJ et al. Creating a genotype-based dosing algorithm for acenocoumarol steady dose. Thromb. Haemost 109, 146–153 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Hirai K et al. Plasma vitamin K concentrations depend on CYP4F2 polymorphism and influence on anticoagulation in Japanese patients with warfarin therapy. Thromb. Res 135, 861–866 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Isaza CA et al. Factores genéticos y ambientales asociados con la respuesta a warfarina en pacientes colombianos. Biomedica 30, 410–420 (2010). [PubMed] [Google Scholar]
- 49.Jiménez-Varo E, Cañadas-Garre M, Garcés-Robles V, Gutiérrez-Pimentel MJ & Calleja-Hernández MÁ Extrapolation of acenocoumarol pharmacogenetic algorithms. Vascul. Pharmacol 74, 151–157 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Lee K-E et al. Effects of CYP4F2 gene polymorphisms on warfarin clearance and sensitivity in Korean patients with mechanical cardiac valves. Ther. Drug Monit. 34, 275–282 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Mazzaccara C et al. Warfarin anticoagulant therapy: a Southern Italy pharmacogenetics-based dosing model. PLoS One 8, e71505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Özer M et al. Impact of genetic factors (CYP2C9, VKORC1 and CYP4F2) on warfarin dose requirement in the Turkish population. Basic Clin. Pharmacol. Toxicol 112, 209–214 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Zhang JE et al. Effect of genetic variability in the CYP4F2, CYP4F11, and CYP4F12 genes on liver mRNA levels and warfarin response. Front. Pharmacol 8, 323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fava C, Ricci M, Melander O & Minuz P Hypertension, cardiovascular risk and polymorphisms in genes controlling the cytochrome P450 pathway of arachidonic acid: a sex-specific relation? Prostaglandins Other Lipid Mediat. 98, 75–85 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Mega JL et al. Genetics and the clinical response to warfarin and edoxaban: findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet (London, England) 385, 2280–2287 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Pengo V et al. A randomized trial of pharmacogenetic warfarin dosing in naïve patients with non-valvular atrial fibrillation. PLoS One 10, e0145318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cerezo-Manchado JJ et al. Genotype-guided therapy improves initial acenocoumarol dosing: results from a prospective randomised study. Thromb. Haemost 115, 117–125 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Funnel plot for association studies of the CYP4F2 polymorphism on coumarin dose.
Figure S2. Forest plot for the difference in logarithm of stable coumarin dose* for subjects with (a) CYP4F2 polymorphism (CT) compared to subjects with CYP4F2 wild-type (CC), (b) CYP4F2 polymorphism (TT) compared with subjects with CYP4F2 wild-type (CC).
Figure S3. Forest plot for the difference in logarithm of stable coumarin dose* for subjects with CYP4F2 polymorphism (CT + TT) compared to subjects with CYP4F2 wild-type (CC), according to dominant model and stratified by (A) ethnicity; (B) drug; (C) gender.
Figure S4. Forest plot for the difference in logarithm of stable coumarin dose* for subjects with CYP4F2 polymorphism (CT + TT) compared with subjects with CYP4F2 wild-type (CC), according to dominant model and stratified by (A) CYP2C9; (B) VKORC1.
Table S1. Predictive models for logarithm of INR dose according to patients’ clinical and genetics characteristics: sensitivity analysis including different models.
Table S2. Effect of concomitant drugs on warfarin dose and genetic polymorphisms of CYP4F2 and CYP2C9 genes: gene− drug interaction and subgroup analyses.
Table S3. Statistical test for model fit (R2) of two previously published models for warfarin dose prediction (Gage 2008, Klein 2009) in comparison with the model presented here in Table 3 (“new model”): application to a subset of subjects from the validation cohort for whom both scores could be calculable on the basis of the available information.
Supplementary Methods S1. Methods.
Supplementary Material S1. Discussion and SAS code.