Abstract
Hematopoietic stem cells (HSCs) have numerous therapeutic applications including immune reconstitution, enzyme replacement, regenerative medicine, and immunomodulation. The trafficking and persistence of these cells after administration is a fundamental question for future therapeutic applications of HSCs. Here, we describe the safe and efficacious labeling of human CD34+ HSCs with a novel, self-delivering perfluorocarbon 19F magnetic resonance imaging (MRI) tracer, which has recently been authorized for use in a clinical trial to track therapeutic cells. While various imaging contrast agents have been used to track cellular therapeutics, the impact of this MRI tracer on HSC function has not previously been studied. Both human CD34+ and murine bone marrow (BM) HSCs were effectively labeled with the MRI tracer, with only a slight reduction in viability, relative to mock-labeled cells. In a pilot study, 19F MRI enabled the rapid evaluation of HSC delivery/retention following administration into a rat thigh muscle, revealing the dispersal of HSCs after injection, but not after surgical implantation. To investigate effects on cell functionality, labeled and unlabeled human HSCs were tested in in vitro colony forming unit (CFU) assays, which resulted in equal numbers of total CFU as well as individual CFU types, indicating that labeling did not alter multipotency. Cobblestone assay forming cell precursor frequency was also unaffected, providing additional evidence that stem cell function was preserved after labeling. In vivo tests of multipotency and reconstitution studies in mice with murine BM containing labeled HSCs resulted in normal development of CFU in the spleen, compared to unlabeled cells, and reconstitution of both lymphoid and myeloid compartments. The lack of interference in these complex biological processes provides strong evidence that the function and therapeutic potential of the HSCs are likely maintained after labeling. These data support the safety and efficacy of the MRI tracer for clinical tracking of human stem cells.
Keywords: Cell tracking, Magnetic resonance imaging (MRI) tracer, Contrast agent, Preclinical safety, Stem cell therapy
INTRODUCTION
Hematopoietic stem cells (HSCs) have a range of therapeutic applications, from hematopoietic reconstitution following radiation and chemotherapy for leukemia and lymphoma to therapeutic replacement for sickle cell anemia and enzyme replacement (41). Recently, HSCs have been applied as a putative treatment to reverse the effects of ischemic injury (21) and chronic liver disease (34) and for treatment of demyelination (33) with promising results. Evaluation of the homing and engraftment of HSCs in clinical studies has largely been limited to the use of clinical end points and ex vivo analysis of peripheral blood or bone marrow (BM) (15). As applications for HSCs to nonhematopoietic disorders increase, studies in a clinical setting of cell trafficking posttransplant becomes a critical issue (8).
Recently, fluorine-19 magnetic resonance imaging (19F MRI) tracking of perfluorocarbon (PFC)-labeled cells has emerged as a noninvasive method for enumerating therapeutic cells, such as T-cells, dendritic cells, and stem cells, in vivo and with clinical potential (1,4,5,31,38). A review of these methodologies is given in reference (36). Since there is little native fluorine in biological tissue, 19F MRI enables the nonambiguous detection of labeled cells against a null background. The 19F MRI results are overlaid onto conventional, anatomical 1H MRI acquired in the same imaging session, which is unaffected by the presence of the 19F label. Thus, coregistration is a facile process, providing a highly specific method for tracking therapeutic cells in a three-dimensional anatomical context. 19F-based cell tracking has been greatly enabled by the development of a self-delivering tracer, which does not require exogenous transfectants or electroporation (18). This feature is particularly advantageous for human CD34+ cells, which are generally resistant to transfection and susceptible to the toxic effects of such techniques when used to deliver DNA (24,44).
Clinical translatability of cell tracking agents requires two levels of safety testing: The ideal tracer must be safe for the patient and must not alter the function or therapeutic index of the cellular products. While the former can be established for the reagent in general, the modulation of cellular function by the tracking agent may be validated for each therapy. The 19F MRI tracer used herein was previously evaluated for its effect on the function of dendritic cells, demonstrating comparable cytokine production, surface marker expression, and biological activities in vitro (12). The effects of the label on multipotent cells, such as HSCs, have not previously been reported. Here, we describe the use of a 19F MRI tracer to label human CD34+ and murine BM HSCs using both in vitro and in vivo tests to evaluate cellular function. To our knowledge, this is the first study that has tested whether a PFC-based MRI label affects the functionality of HSCs in vivo. Results of the in vivo tests of multipotency and short-term reconstitution with labeled murine BM, along with in vitro assays, provide strong evidence that this label does not alter the function of HSCs.
MATERIALS AND METHODS
Cell Sources
Frozen, magnetic bead isolated, human BM-derived CD34+ hematopoietic cells were purchased from Stemcell Technologies, Inc. (Vancouver, BC) and the National Disease Research Interchange (Philadelphia, PA). Mobilized peripheral blood (MPB) CD34+ cells were purchased from All Cells (Emeryville, CA). Frozen murine BM was purchased from Astarte Biologicals (Redmond, WA) for use in in vitro labeling efficiency studies.
Labeling
Frozen human CD34+ cells or murine BM mono-nuclear cells were thawed in a 37°C water bath and washed twice prior to labeling. All washes and labeling steps were conducted using phosphate-buffered saline (PBS) with 0.5% fetal bovine serum (FBS) or OptiMEM (Invitrogen, Carlsbad, CA) as noted, with the exception of the first wash of thawed human CD34+ cells in media containing DNase (Stemcell Technologies). Thawed, washed cells were immediately incubated with 3–10 mg/ml Cell Sense or a dual-mode fluorescent version of the Cell Sense 19F MRI tracer (CS-1000 or CS-ATM-DM Green, respectively; Celsense, Inc., Pittsburgh, PA) for 4 h. Mock-labeled cells were set up in parallel cultures, without the addition of the MRI tracer. After labeling, cells were washed two to three times to remove excess labeling agent prior to further analysis. Label uptake was measured by 19F NMR and/or flow cytometry [fluorescein isothiocyanate (FITC)-MRI tracer reagent only].
19F Nuclear Magnetic Resonance (NMR) Spectroscopy
To determine labeling efficiency of MRI tracer-labeled HSCs, cell pellets containing 7 to 10 million cells were lysed with 125 μl of 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO). A standard 19F reference solution was added to the lysate consisting of 125 μl of 0.1% (vol/vol) trifluoroacetic acid (TFA) and sufficient manganese chlo-ride (Sigma Aldrich) to match the TFA and contrast agent magnetic relaxation time, and the mixture was placed in a glass NMR tube. 19F NMR analysis was carried out using a Bruker AVANCE spectrometer operating at 282.4 MHz equipped with a Bruker 5-mm high-resolution broad band observe (BBO) probe. Typical scan parameters were 32 scans, spectral range of −60 to −100 ppm, and a recycle delay of 1.5 s. To measure tracer-labeled HSCs in tissue, dissected muscles were placed into 10-mm NMR tubes with a TFA reference enclosed in a glass capillary, and 19F NMR was conducted on a Bruker AVANCE spectrometer operating at 470.6 MHz equipped with a high-resolution 10-mm PABBO BB probe (similar parameters as above, except 512 averages, were acquired). One-dimensional 19F spectra were obtained, where both the MRI tracer and TFA appear as single narrow resonances, having a chemical shift difference of −15.58 ppm. The ratio of the integrated areas under these two peaks was used to calculate the mean labeling efficiency (LE) 19F/cell in cell pellets, typically ranging from 1011 to 1012, as previously described (18,37). The total number of HSCs retained in each of the muscles was calculated as the total 19F divided by the LE (19F/cell).
In Vitro and In Vivo MRI
Cells were labeled as outlined above and then resuspended either in PBS for injection or in a 5:1 matrigel (BD Biosciences): culture media mixture for plug formation. The MRI tracer-labeled cells in PBS suspension in a tube or contained in the matrigel plug were first scanned alongside 19F phantoms containing dilutions of the tracer agent. All samples were positioned near isocenter in a 7-T Bruker Biospec MRI scanner (Bruker Biospin, Billerica, MA). A 50-mm diameter dual-tuned (1H/19F) transmit/receive volume coil (m2m Imaging Corp., Cleveland, OH) was used to acquire all images to ensure uniform sensitivity and thereby enable accurate quantification. High-resolution 1H RARE (rapid acquisition with relaxation enhancement) (13) images were first acquired for positioning and coregistration of the 19F scans. The 1H RARE acquisition parameters were as follows: transmission/echo time (TR/TE) = 5,000/8 ms, slice thickness = 1 mm, field of view (FOV) = 7 cm, matrix = 128 × 128, and RARE factor = 4. A lower resolution, coregistered, spin density-weighted 19F image was acquired to detect the labeled cells using TR/TE = 5,000/8 ms, RARE factor = 32, 100 averages, matrix = 64 × 64, slice thickness = 5 mm.
An adult female (180 g) Sprague–Dawley rat (Charles River, Wilmington, MA) was anesthetized with 1.2 g/kg urethane administered by intraperitoneal (IP) injection. Once anesthetized, a longitudinal incision was made in the lateral surface of right thigh, and the matrigel plug was implanted between the vastus lateralis and biceps femoris muscles and sutured. Silk sutures were used to retain the position of the matrigel during the scanning session while also contributing no additional image artifacts. The rat was placed positioned near isocenter in the 7-T scanner, and coronal images were acquired using a 1H RARE sequence with TR/TE = 6,000/8 ms, slice thickness = 1 mm, FOV = 7 cm, matrix = 128 × 128, RARE factor = 4, and 1 average. For 19F, RARE was also used with TR/TE = 6,000/8 ms, slice thickness = 5 mm, FOV = 7 cm, matrix = 64 × 64, RARE factor = 32, and 500 averages. The total acquisition time for the in vivo 19F image acquisition was ~100 min. The rat was then sacrificed and scanned for 19F overnight (12 h). Similar in vivo scanning parameters (100 min) were used to image a separate rat injected intramuscularly into the biceps femoris with 1.2 × 107 labeled CD34+ HSCs in suspension using a 26-g needle.
Image Analysis and Quantification
Voxel Tracker™ software was employed to quantify the 19F signal in the acquired images. The software accounts for the non-Gaussian noise distribution of the MRI images as described previously (37) and allows facile quantification of even low signal-to-noise ratio (SNR) images. Briefly, signal from a region of interest (ROI) was compared to the signal generated from the known concentration of fluorine in the references, allowing estimation of the number of fluorine atoms in the ROI. In the case of in vitro data, the known cell number allowed calculation of the labeling efficiency (fluorine per cell). In the case of in vivo data, the calculated labeling efficiency allowed an estimate of the number of cells present in the ROI.
Flow Cytometry Viability Studies
Labeled cells were compared with mock-labeled controls, and viability and yields were determined by trypan blue exclusion. Viability (%) = live cells/(live cells + dead cells) × 100%. Yield (%) = live cells (postlabeling)/total # cells added (prelabeling) × 100%. To measure apoptosis, nonyl acridine orange (NAO) was added to labeled and mock-labeled cells for 5 min, followed by flow cytometric analysis.
Label Uptake Studies
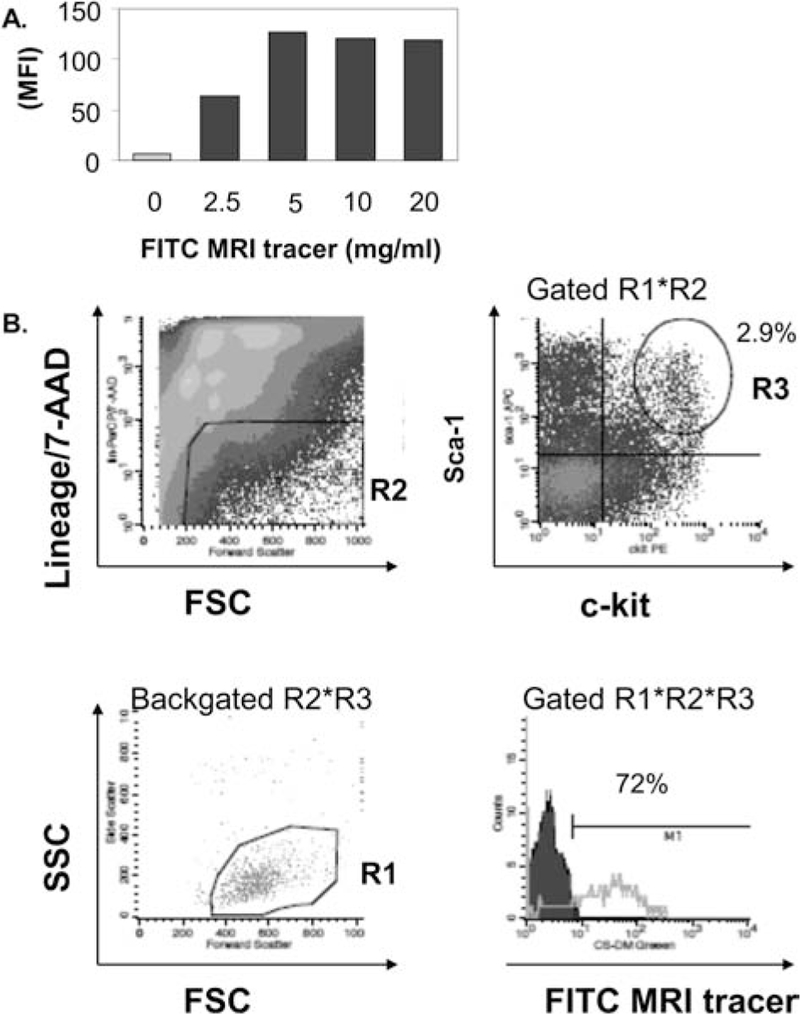
For human CD34+ cells, uptake of FITC-MRI tracer was analyzed in the FL-1 channel, and both percentage of cells containing label and the mean fluorescence intensity were measured. For studies of MRI tracer uptake in murine BM, multicolor flow cytometry was employed to determine uptake of CS-ATM-DM Green by hematopoietic progenitor cells. FITC MRI tracer-labeled murine BM and mock-labeled murine BM cells were incubated with primary allophycocyanin (APC)-conjugated anti-stem cell antigen-1 (Sca-1), phycoerythrin (PE)-conjugated anti-c-kit antibodies, and biotinylated antibodies to lineage (lin) markers (CD3, CD11b, CD45R, TER119, and Gr1; all antibodies are from e-Bioscience, Inc., San Diego, CA), followed by an incubation with secondary streptavidin–peridinin–chlorophyll protein complex (PerCP; BD Biosciences, San Jose, CA) and then viability dye 7-aminoactinomycin D (7-AAD, e-Bioscience). Samples were analyzed using a FACSCalibur flow cytometer and CellQuest software (BD Biosciences). The electronic gating strategy included selection of live cells by forward scatter and side scatter (R1), exclusion of dead 7-AAD+ and PerCP-labeled lin+ cells in the FL-3 channel (R2), and gating double-positive c-kit+ Sca-1+ BM cells [FL-2 and FL-4 channels, respectively, (R3)] to examine live, 7-AAD– lin– Sca-1+ c-kit+ cells for the presence of CS-ATM-DM Green label (detected in FL-1 channel).
In Vitro Colony Forming Cell (CFC) Assay
Labeled or mock-labeled human CD34+ BM cells were resuspended at 1 × 103 cells/ml in complete Methocult Classic media (Stemcell Technologies) and plated in triplicate in 35-mm untreated culture dishes. Dishes were incubated in humidified trays in a humid incubator at 37°C, 5% CO2 for 14 days prior to enumeration. Colonies were counted independently by two investigators via microscopy and gridded scoring dishes.
In Vitro Cobblestone Area Forming Cell (CAFC) Assay
Labeled or mock-labeled human CD34+ cells were plated at 1:3 serial dilutions (18000–7 cells per well) over a confluent stromal cell layer, which supports CAFC growth (AFT024 cells treated with mitomycin C and plated on 1% gelatin-treated 96-well tissue culture clusters) (26,43). Cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Mediatech) containing 15% serum supplemented with human interleukin (IL)-3 and granulocyte macrophage-colony stimulating factor (GM-CSF; 10 ng/ml each) and 0.1 mMβ-mercaptoethanol, refeeding weekly for 5 weeks. Wells were then scored for positive cobble-stone appearing colonies by microscopy, conducted independently by two investigators. The negative fraction of wells were graphed for each CD34+ cell dilution and compared across labeling conditions.
In Vivo Colony Forming Unit-Spleen (CFU-S) Assay and Reconstitution Assay
All murine studies were conducted in accordance with animal welfare regulations by TGen Drug Development (TD2), a nonprofit contract research organization and subsidiary of TGen, (Phoenix, AZ). Female, 10- to 12-week-old C57BL/6 mice were provided with acidified drinking water, housed in microisolator cages, and irradiated with 9.5 gray (Gy) using an RS 2000 X-ray Biological Irradiator (Rad Source Technologies, Inc., Suwanee, GA). Total BM prepared from nonirradiated littermates was labeled with MRI tracer, as described above, and myeloablated hosts were injected intravenously with 1.5 × 105 cells (labeled or mock-labeled) in 0.2 ml PBS within 3 h of irradiation. Control animals not receiving transplantations were euthanized at day 11 due to moribund condition, and spleens were collected. On day 12 posttransplant, BM-treated mice appeared healthy and were euthanized prior to splenectomy. Spleens were placed in Telly’s fixative and then scored for macroscopic hematopoietic colonies using a dissection microscope. For in vivo reconstitution studies, irradiated host animals (n = 6) were transplanted with MRI tracer labeled BM cells (1 × 107). After 30 days, blood was collected into tubes containing ethylenediaminetetraacetic acid (EDTA) as an anticoagulant. Complete blood count (CBC) was measured with an automatic hematology analyzer (Abaxis VetScan HM5).
Statistical Analysis
Student’s two-tailed paired t test was used in comparisons of labeled versus unlabeled cells in in vitro studies, using a value of p = 0.05 for significance, under the implicit assumption of normally distributed data. Spleen CFU counts from recipients of labeled and unlabeled cells were compared using an unpaired t test with and without Bonferroni correction. All errors reported in plots, tables, and text are standard deviations.
RESULTS
Labeling Efficiency and Detection
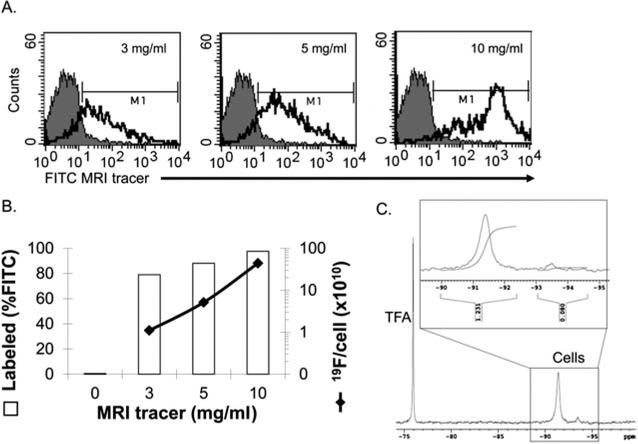
To establish that HSCs were effectively labeled with the MRI tracer, CD34+ cells isolated from human adult BM were cultured with the FITC-MRI tracer at varying doses. Uptake was measured by flow cytometry (Fig. 1A, B) and confirmed by 19F NMR (Fig. 1B, C). Cell uptake was found to be dose dependent, with an average uptake of 4.4 × 1011 19F atoms/cell at 10 mg/ml reagent.
Figure 1.
Labeling efficiency of CD34+ hematopoietic stem cells (HSCs) with 19F MRI tracer. (A) Human bone marrow (BM) CD34+ cells were incubated with increasing doses of fluorescein isothiocyanate-magnetic resonance imaging (FITC-MRI) tracer to optimize labeling efficiency by flow cytometry. Autofluorescence in control cells is represented by the gray-filled histograms and the green fluorescence in MRI tracer labeled cells is indicated by the unfilled histograms (one of three experiments with similar results). (B) Average labeling efficiency versus increasing dose of MRI tracer. Left axis, open bars: percentage of the CD34+ cell population containing label as measured by flow cytometry fluorescent signal above background (denoted by M1 in A); right axis, solid line, perfluorocarbon (PFC) label uptake measured by 19F nuclear magnetic resonance (NMR). (C) Representative NMR spectra of 10 mg/ml MRI tracer labeled CD34+ cells corresponding to a 4.4 × 1011 19F atoms/cell labeling efficiency (one of three experiments with similar results). An inset depicts a typical integration curve, with the peak at −91 ppm used for labeling efficiency calculations.
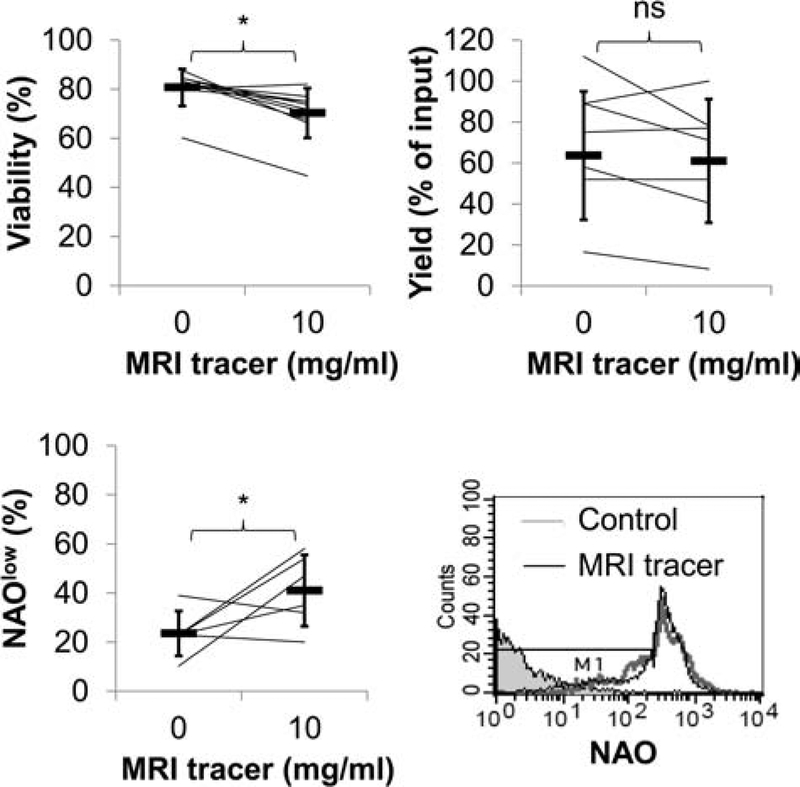
The effect of labeling on cell viability was determined using several methods. A slight decrease in viability (on average up to 10% less, p < 0.05) was measured by Trypan blue exclusion (Fig. 2). NAO staining (a probe for the presence of cardiolipin) was used to measure mitochondrial membrane integrity as an indicator of apoptosis. Loss of cardiolipin (cells with low levels of NAO staining) was observed indicating increased rates of apoptosis in labeled cells versus mock-labeled cells (p < 0.05). Yields of CD34+ cells were not significantly different between labeled and unlabeled control cells.
Figure 2.
CD34+ cellular viability is modestly reduced with MRI tracer. Results represent the paired analysis of repeated experiments assessing viability by Trypan blue, apoptosis by nonyl acridine orange (NAO) expression and yields after CD34+ cells were cultured with and without MRI tracer (10 mg/ml). Averages are indicated in heavy horizontal bars for viability (n = 10), NAO expression (n = 6), and yields (n = 7). Histograms of flow cytometric analysis of NAO staining in MRI tracer-labeled and unlabeled cells as well as unstained cells in shaded histogram. Cells undergoing apoptosis (NAOlow) are detected by a decrease in fluorescence (region defined by M1).
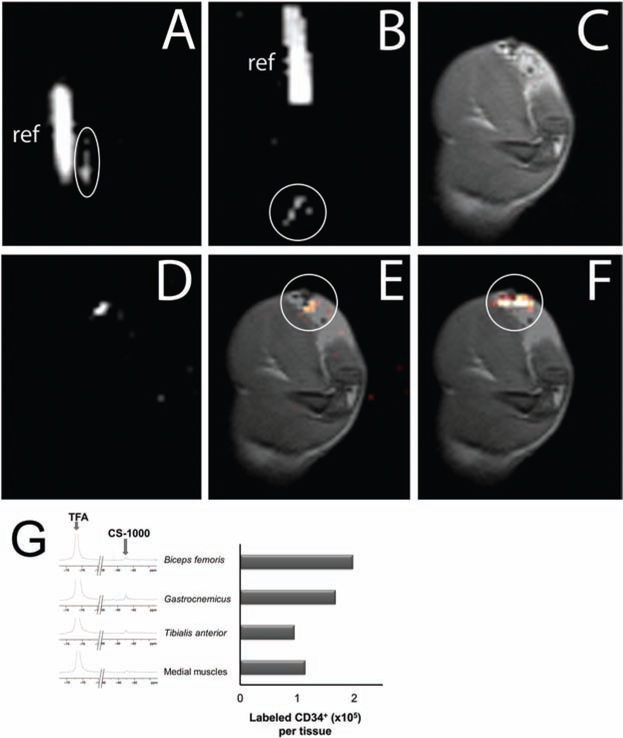
A preliminary imaging study was performed to validate that MRI tracer-labeled human CD34+ cells are detectable in vivo. Labeled human CD34+ cells were administered to a rat by direct intramuscular injection of suspended cells or surgical implantation of a matrigel plug into the thigh for in vivo imaging by 1H and 19F (Fig. 3). Both the suspension and matrigel plug of labeled cells were visible in ex vivo 19F MRI scans prior to implantation (Fig. 3A, B). Two hours postdelivery, cells implanted within the matrigel plug were readily detectable by 19F MRI (Fig. 3C–F). Calculations using Voxel Tracker software confirmed that the number of cells detected in the surgical implantation by MRI was approximately equal to the number of cells encapsulated prior to delivery, indicating their retention within the matrigel plug. In contrast, cells delivered by intramuscular injection were not detected under similar scanning conditions (data not shown), suggesting that the cells were dispersed from the site of injection (biceps femoris). To confirm this finding, the thigh was dissected, and individual muscles or groups of muscles near and distal to the site of injection were analyzed ex vivo by NMR spectroscopy (Fig. 3G). Results unexpectedly showed that all of muscles evaluated contained sparse numbers of labeled cells (on average approximately 50 cells/mm3), well below the sensitivity required for imaging, and cumulatively represented less than 5% of the total cells injected, consistent with the rapid dispersal of cells from the site of injection.
Figure 3.
Imaging delivery of PFC-labeled human CD34+ cells in vivo with 19F/1H MRI. (A, B) In vitro 19F MRI of labeled CD34+ cells prior to administration: 15 × 106 cells in 1× PBS suspension for injection (A) and 14 × 106 CD34+ cells in a Matrigel support for implantation (B) (n = 2). Both labeled cells (circled) and reference tubes (“ref”) are in the field of view. In vitro images employ nonlinear brightness contrast adjustments to distinguish the cell signal from background and reference signal from saturation simultaneously. (C–F) In vivo MRI approximately 2 h after surgical implantation of the cell-laden Matrigel plug. (C) Proton image of the rat thigh anatomy with the hydrogel implant clearly visible as a hypointense region at the top right of the image. (D) 19F image (100-min acquisition) of the same field of view. (E) Coregistered 1H and 19F image overlay (C, D), with the 19F rendered in a “hot-iron” color scale (circled). Image quantification confirmed approximately 14 × 106 ± 2 × 106 cells remain in the region of implant. (F) Overlay of postsacrifice, 19F scan of implanted cells (circled) on 1H anatomical image. (G) Ex vivo 19F NMR spectroscopy was conducted with dissected thigh muscles to evaluate the delivery of labeled CD34+ suspension cells injected using a syringe. Analyzed muscles included the site of injection (biceps femoris), muscles proximal to the injection site (gastrocnemius and tibialis anterior), and a set muscles distal to the injection site (“medial muscles” including portions of the semitendenous, gracilis, semimebranous, and adductor magnus). Spectra are depicted beside a chart of the quantification of transplanted cell numbers in each tissue. TFA, trifluoroacetic acid; CS-1000, Cell Sense tracer.
Functional Assessments
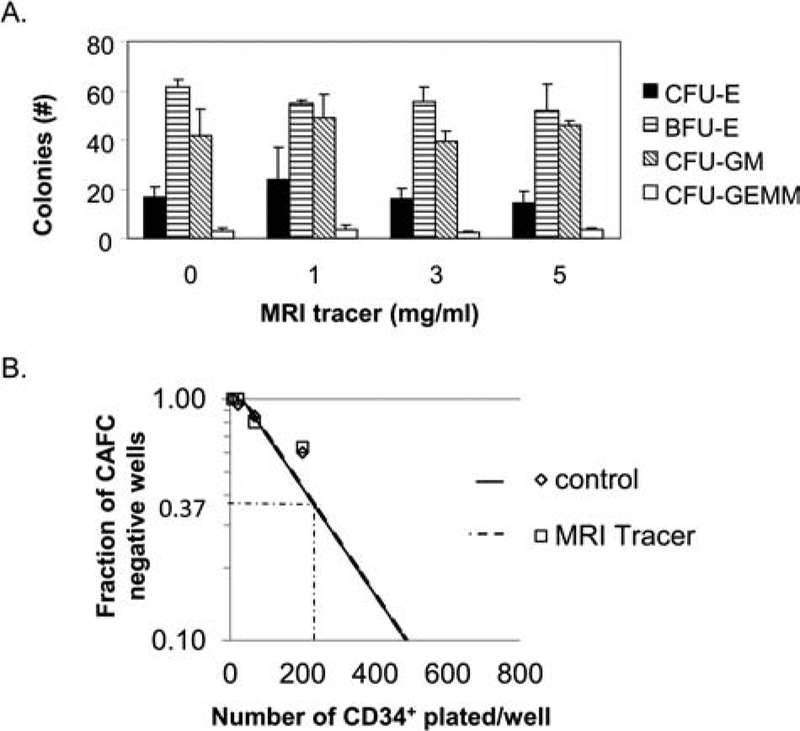
As the application of cell tracers to study the behavior of therapeutic cells requires them to retain the properties of the cells of interest, a number of studies were employed to evaluate the function of the MRI tracer-labeled HSCs. Clonogenic CFU assays have been adapted to study the toxicity of a variety of drugs and environmental toxicants in vitro and have high predictive value (9,32). To evaluate the impact of the MRI tracer on multipotency of CD34+ cells, labeled and unlabeled HSCs were assessed in CFU assays (Fig. 4A). Importantly, all CFU types observed in the unlabeled conditions were also found at all labeling concentrations studied. Furthermore, the numbers of total colony-forming units (CFU), as well as individual CFU types, were not significantly different between cultures initiated with either labeled or unlabeled cells, indicating that labeling did not alter their multipotent potential. Stem cell repopulation activity may be examined through cobblestone area forming cell (CAFC) assay (6) as a surrogate measure of reconstitution in vivo. Control and MRI tracer-labeled CD34+ cells assessed in CAFC assays indicated a precursor frequency of 1/169 versus 1/168 (labeled vs. unlabeled) in CD34+ cells, demonstrating no effect of labeling on this functional activity (Fig. 4B).
Figure 4.
MRI tracer preserves differentiation and self-renewal of human CD34+ cells in vitro. (A) Human bone marrow CD34+ HSCs were incubated with or without increasing amounts of the MRI tracer for 4 h prior to seeding in methocellulose in triplicate for colony-forming unit (CFU) assays. Results show the average CFU per 1,000 progenitor cells/dish, and error bars indicate standard deviations. One of two independent studies with identical findings is depicted. (B) Limiting dilution of human bone marrow CD34+ HSCs (unlabeled and MRI tracer-labeled, 10 mg/ml), 20 replicate wells per condition, were cultured with stromal cells in the presence of serum, interleukin (IL)-3, and granulocyte macrophage-colony-stimulating factor (GM-CSF) for 5 weeks and then observed for the formation of cobblestone areas (cobblestone area forming cell, CAFC, assay). The frequency of negative wells plotted against the number of cells added per culture provides a graphical depiction of the Poisson equation to calculate precursor frequency in the treated and untreated cells. CFU-E, CFU-erythroid; BFU-E, blast forming unit-erythroid; CFU-GM, CFU-granulocyte, macrophage; CFU-GEMM, CFU-granulocyte, erythroid, macrophage, megakaryocyte.
The ability of HSC progenitors to reconstitute and differentiate can be assessed by CFU-S and studying the reconstitution of blood cell populations in vivo. Previously, this approach has been used to study the myelotoxicity of radiolabeled cellular products on bone marrow (17). To minimize potential artifacts in a xenograft system with human HSCs, syngeneic murine bone marrow was used as the source of HSCs for in vivo experiments designed to address the impact of labeling on hematopoietic reconstitution. Optimization of labeling of murine BM (media, temperatures, dose, time) resulted in efficient labeling of murine BM cells (Fig. 5A) with an average of 2.50 × 1011 19F atoms/cell (n = 3). Multicolor flow cytometry was used to verify uptake in murine lin−Sca-1+c-kit+ murine HSCs (30) and indicated that the majority of HSCs contained the MRI tracer (72.7 ± 0.6%, n = 3) (Fig. 5B).
Figure 5.
MRI tracer labels murine bone marrow HSCs. (A) Increasing doses of FITC-MRI tracer was incubated with murine bone marrow mononuclear cells and then evaluated by flow cytometry. (B) Labeling of murine HSCs present in bone marrow was confirmed by flow cytometry. Gating strategy for enumeration of labeled HSCs is depicted and included selection of live cells by forward scatter (FSC) and side scatter (SSC) (R1), exclusion of dead 7-aminoactinomycin D (7-AAD)+ and peridinin-chlorophyll-protein complex (PerCP)-labeled lineage (lin)+ cells in the FL-3 channel (R2), and gating double-positive c-kit+ stem cell antigen (Sca)-1+ BM cells in FL-2 and FL-4 channels, respectively (R3), to examine live, 7-AAD− lin− Sca-1+ c-kit+ cells for the presence of FITC-MRI tracer (detected in FL-1 channel), lower right plot. One of three independent experiments with similar results is shown. Backgated R2*R3 scatter plot is provided to verify selection of the correct HSC population in lower left plot. MFI, mean fluorescence intensity.
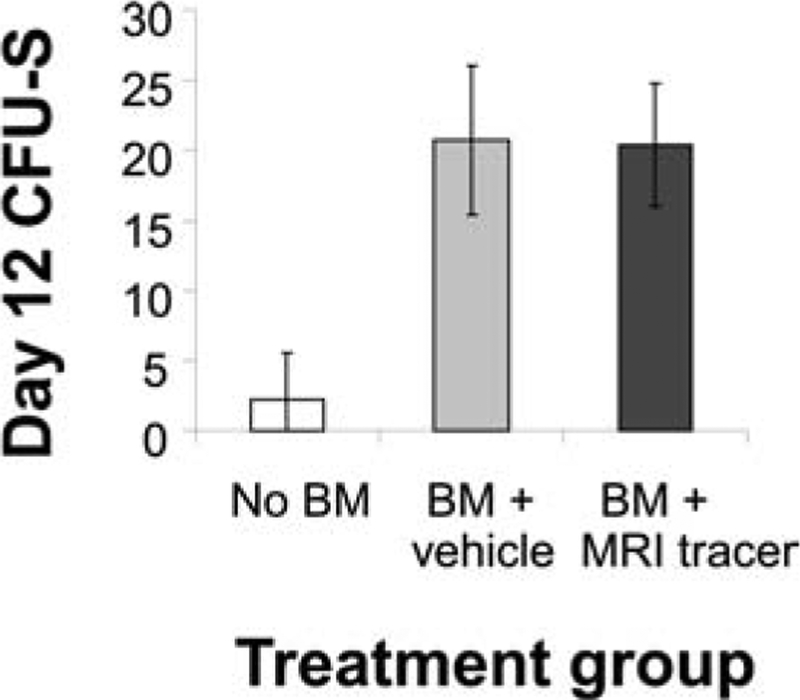
Day 12 CFU-S assays were used to enumerate murine HSCs with long-term repopulating activity in vivo. Approximately 20.4 ± 4.4 CFU-S/150,000 cells were found in tracer-labeled BM, equal to the numbers of CFU-S in mice transplanted with unlabeled BM (20.8 ± 5.3); in contrast, significantly fewer colonies (small and sparse) were formed in nontransplanted recipients (2.3 ± 3.3) (Fig. 6). To confirm the hematopoietic reconstitution potential of MRI tracer labeled murine BM, a cohort of mice were transplanted with 1 × 107 labeled BM cells, containing labeled HSCs. Blood samples were obtained and submitted for CBC panels on day 30 and compared to normal murine values (Table 1). Analysis of blood samples indicated that all cell lineages were present including lymphocytes, monocytes, and neutrophils, consistent with the ability of the MRI tracer-labeled BM cells to effectively differentiate and reconstitute the host animals. In two of six animals, platelet levels were below the normal range, although the cause of this deficiency could not be determined from this study. White blood cell counts, red blood cell counts, and hematocrit were all within normal ranges. Overall, these results indicate that the HSC activity in vivo is not impaired following labeling with the MRI tracer studied here.
Figure 6.
HSC activity is preserved in vivo with the addition of MRI tracer. Myeloablated murine hosts were transplanted with labeled or unlabeled syngeneic bone marrow, with untransplanted mice serving as a negative control (n = 8 per cohort). On day 12 (day 11 for untransplanted recipients), spleens were harvested and CFU-S enumerated. Results represent the average CFU-S per animal. Significantly fewer numbers of colonies were measured in untransplanted recipients compared to labeled or unlabeled transplanted recipients (p < 0.0001 for each comparison). No significant differences in colony number were found between labeled and unlabeled cells when analyzed by unpaired t test (with or without Bonferroni correction).
Table 1.
Analysis of Peripheral Blood
| Recipients of Labeled BM | |||||
|---|---|---|---|---|---|
| Blood Count Parameters | Abbreviation | Units | Normal Ranges | Ave | SD |
| White blood cells | WBC | ×103/μl | 6–15 | 8.2 | 1.45 |
| Lymphocytes | LYM | ×103/μl | 3.4–7.44 | 5.73 | 0.64 |
| Monocytes | MON | ×103/μl | 0–0.6 | 0.35 | 0.14 |
| Neutrophils | NEU | ×103/μl | 0.5–3.8 | 2.12 | 1.08 |
| Lymphocytes | LY | % | 57–93 | 70.7 | 7.64 |
| Monocytes | MO | % | 0–7 | 4.35 | 1.84 |
| Neutrophils | NE | % | 8–18 | 24.92 | 7.43 |
| Red blood cells | RBC | ×106/μl | 7–12 | 9.84 | 0.16 |
| Hemoglobin | HGB | g/dl | 12.2–16.2 | 14.57 | 0.58 |
| Hematocrit | HCT | % | 35–45 | 42.45 | 1.74 |
| Mean corpuscular volume | MCV | fl/cell | 45–55 | 43.17 | 0.98 |
| Mean corpuscular | |||||
| hemoglobin | MCH | pg/cell | 11.1–12.7 | 14.82 | 0.56 |
| Mean cell hemoglobin | |||||
| concentration | MCHC | % | 22.3–32 | 34.38 | 1.86 |
| Red cell distribution width | RDWc | % | N/A | 19.55 | 0.33 |
| Platelet count | PLT | ×103/μl | 200–450 | 238* | 175 |
| Plateletcrit | PCT | % | N/A | 0.17 | 0.11 |
| Mean platelet volume | MPV | fl | N/A | 7.13 | 0.70 |
| Platelet distribution width | PDWc | % | N/A | 33.35 | 4.71 |
An automated analyzer was used for complete blood count (CBC) of murine blood samples. Experimental values are compared to normal murine control values established for the hematology analyzer. Ave, average; SD, standard deviation; BM, bone marrow.
Two animals had levels of platelets below the normal range.
DISCUSSION
A critical parameter for evaluating the safety and efficacy of cellular therapeutics is the homing, distribution, and fate of transplanted cells (11,45). However, limited tools are available clinically to enable cell tracking (36,39). 19F MRI with a PFC tracer provides a potential means of assessing posttransplant retention in vivo in the clinic, as well as in preclinical studies. At a fundamental level, all imaging methods for cell tracking require that an imaging agent be loaded into the cells to render them distinct from background in the imaging modality. Not only do these probes need to be safe to the patient systemically, but the tracers also need to preserve the therapeutic efficacy of the product itself. Because the differentiation and fate of HSCs is well understood, it was used here as a model for studying the effects of an MRI tracer on stem cell function. Our results indicate that this MRI tracer may be used to label HSCs for detection by 19F MRI with only a modest decrease in viability and without affecting their function and differentiation in vitro and in vivo. In addition, the pilot MRI study of the intramuscular delivery of HSCs highlights a known problem that may impair the effective clinical implementation of cellular therapeutics—even when cells appear to be delivered to a site of action, their retention and ultimate distribution is uncertain (16,25,42). This not only may decrease the efficacy of a cellular product, but off-target effects become a greater concern. 19F MRI is a tool that may be used to address these questions.
While cell tracking in preclinical models has grown rapidly, efforts to track human cells clinically have lagged, due in part to the lack of safe and effective agents for this purpose (39). As optical approaches are limited by depth penetration, the development of agents for clinical cell tracking has focused on MRI, positron emission tomography (PET), or single photon emission computed tomography (SPECT). The latter two methodologies are sensitive techniques that rely on the detection of a gamma rays emitted from a radioactive probe, limiting the longevity of the imaging study. Thus, the long-term grafting and persistence of cells cannot be measured with these techniques. Furthermore, for stem cells, the prospect of using inherently genotoxic labels (i.e., radioactive probes) reduces overall enthusiasm for PET and SPECT because of potential concerns of increased tumorigenicity and other cellular effects (17,29).
MRI has the advantage of providing high-resolution spatial information and allowing labeled cells to be monitored for extended times following transplantation. Importantly, no radiation is associated with MRI contrast agents. To date, a small number of clinical trials have used paramagnetic particles to track therapeutic cells (7). While superparamagnetic iron oxide (SPIO) and other paramagnetic particles can be phagocytosed, nonphagocytic cells require transfection agents or electroporation, thus potentially hindering the use of SPIO for labeling of CD34+ HSCs, as they are sensitive to these techniques. However, previous studies have shown under certain conditions, SPIO or other magnetic particles can be used to label HSCs (2,3,14,23). The impact of labeling with SPIO on HSC function is controversial: While some report no alterations (2,3,14,23), in other cases the inhibition of differentiation has been observed in in vitro CFU assays (27), and effects have been observed on the migration, viability, and differentiation of other stem cells in some studies (10,22,28). Few published studies evaluate the effects of MRI tracers on HSC function in vivo. A direct comparison of AC133+ hematopoietic cells with and without ferumoxide labeling followed by transfer in vivo results in similar differentiation and incorporation into the vasculature of orthotopic implanted human glioma in rats (Ali Arbab, personal communication). However, to our knowledge, the hematopoietic potential of HSCs labeled with internalized contrast agents has not been previously studied in vivo.
19F MRI is unique in that the detection of the probe is specific to the labeled cells due to the paucity of 19F naturally occurring in the body. Thus, unlike metal ion contrast agents, the images obtained are highly specific and interpretation is straightforward; moreover, any pathological or anatomical features that are typically imaged by 1H MRI are unaltered by the presence of the 19F tracer. However, MRI is an inherently insensitive technique. In this study, approximately 100,000 cells per voxel could be visualized with an SNR of 2.5. Literature reports of the limit of sensitivity suggest even greater sensitivity (as few as 2,000 cells/voxel) (4,5). Sensitivity will vary based on the coil used, acquisition parameters, magnet strength, and cell-labeling efficiency. It should be noted that HSCs, due to their small physical size and highly quiescent character, contain 10- to 100-fold less MRI tracer than dendritic cells (5,12), yet data on cell retention/delivery is demonstrated.
19F MRI has previously been used to track cultured human cord blood mononucleated stem/progenitor cells in animal models, some of which expressed markers of both HSCs and endothelial-like precursor cells, including CD34 and AC133 (31). Functional tests of this heterogeneous cellular population labeled with PFC tracers (perfluorooctyl bromide or perfluoro[15]-crown-5 ether) indicated no effect on phenotype, viability, or receptor-mediated endocytosis (31). Our present study furthers the characterization of 19F MRI tracer-labeled cells by using a unique PFC formulation that is optimized for MRI, a highly purified HSC population for in vitro studies, and utilizing gold standard tests for the functional activity of HSCs in vitro and in vivo. The MRI tracers used in our study (CS-1000 and CS-ATM-DM Green) have a proprietary formulation that enables uptake by a wide variety of cell types without the use of transfection agents and have been found to label dendritic cells, T-cells, B-cells, NK cells, platelets, adipose-derived stem cells, bone marrow mesenchymal stem cells, epithelial cells and precursors, neural progenitor cells, oligodendrocyte precursor cells, macrophages [(4,5,12,19,20) and data not shown], and now hematopoietic stem cells. These reagents are optimized for detection by MRI, having a short T1/T2 ratio and essentially a single 19F NMR peak, giving rise to enhanced sensitivity over prior formulations (35). Relevant to the safe application to cellular therapeutics in patients, the CS-1000 tracer has also been found to be nonmutagenic in both Ames and mammalian chromosomal aberration assays and has no adverse effects detected in vivo even with acute administration to rodents at 100-fold the expected clinical dose, supporting its safety profile and applicability for clinical use (data not shown). Recently, investigators at the University of Pittsburgh Cancer Institute have received authorization from the US Food and Drug Administration (FDA) to incorporate this MRI tracer in a phase I clinical trial to track a dendritic cell-based vaccine in cancer patients (40).
The main focus of this study was the labeling and effect of an MRI tracer agent on the therapeutic cells themselves. Specifically, we investigated a PFC tracer agent on the viability and functional differentiation of stem cells, both in vitro and in vivo. HSCs were labeled without the use of exogenous transfection agents at levels rendering them detectable by MRI. In vitro experiments showed labeled cells had a slight decrease in viability but were similar to unlabeled cells in functional tests including differentiation. The lack of change in CAFC precursor frequency in labeled versus unlabeled cells also provided evidence that the renewal capacity of HSCs was not significantly affected. Furthermore, in vivo experiments demonstrated that labeled HSCs retained their ability to contribute to hematopoiesis, as assessed by CFU-S and reconstitution. While the quantitative CFU-S assay provided evidence that the labeled cells were equivalent to unlabeled BM cells, the survival, engraftment, and reconstitution of mice treated with labeled BM cells provided strong evidence that MRI tracer preserves the functional capacity of HSCs to proliferate and differentiate in vivo. While there was no a priori in vitro evidence that any functional impact would be observed, the cellular assays indicated no alteration in cell function, further supporting the validity of the in vitro assessments of cellular function. During the review process, an independent group evaluated this same MRI tracer for labeling human neural stem cells, which resulted in a similar conclusion (4).
19F MRI tracers may provide valuable clinical information related to the efficacy of therapies, enabling bench-to-bedside translational efforts to improve clinical cellular products. In the future PFC MRI tracers could also be incorporated into clinical studies of the delivery, homing, and/or retention of stem cell products. The in vitro and in vivo data reported here are directly relevant to the safety of this PFC tracer on the efficacy and function of a given cellular product, HSCs, and represents data that could be included in IND applications for cellular therapeutics involving 19F MRI cell tracking.
ACKNOWLEDGMENTS:
We thank Dr. Eric Ahrens for critical reading of the manuscript and Dr. Ali Arbab for helpful discussion. Support was provided in part by the National Institutes of Health (RO1 CA134633). We acknowledge the National Disease Research Inter change (NDRI), with support from NIH Grant 5 U42 RR006042, for supplying human CD34+ bone marrow cells. B. Helfer, A. Balducci, C. O’Hanlon, and A. Wesa are employees of Celsense, Inc., and hold stock and/or unexercised stock options in Celsense, Inc. The authors declare no conflict of interest.
REFERENCES
- 1.Ahrens ET; Flores R; Xu HY; Morel PA In vivo imaging platform for tracking immunotherapeutic cells. Nat. Biotechnol 23(8):983–987; 2005. [DOI] [PubMed] [Google Scholar]
- 2.Arbab AS; Yocum GT; Kalish H; Jordan EK; Anderson SA; Khakoo AY; Read EJ; Frank JA Efficient magnetic cell labeling with protamine sulfate complexed to ferumoxides for cellular MRI. Blood 104(4):1217–1223; 2004. [DOI] [PubMed] [Google Scholar]
- 3.Arbab AS; Yocum GT; Rad AM; Khakoo AY; Fellowes V; Read EJ; Frank JA Labeling of cells with ferumoxides-protamine sulfate complexes does not inhibit function or differentiation capacity of hematopoietic or mesenchymal stem cells. NMR Biomed. 18(8):553–559; 2005. [DOI] [PubMed] [Google Scholar]
- 4.Boehm-Sturm P; Mengler L; Wecker S; Hoehn M; Kallur T In vivo tracking of human neural stem cells with 19F magnetic resonance imaging. PLoS One 6(12): e29040; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonetto F; Srinivas M; Heerschap A; Mailliard R; Ahrens ET; Figdor CG; de Vries IJ A novel (19)F agent for detection and quantification of human dendritic cells using magnetic resonance imaging. Int. J. Cancer 129(2):365–373; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breems DA; Blokland EA; Neben S; Ploemacher RE Frequency analysis of human primitive haematopoietic stem cell subsets using a cobblestone area forming cell assay. Leukemia 8(7):1095–1104; 1994. [PubMed] [Google Scholar]
- 7.Bulte JW In vivo MRI cell tracking: Clinical studies. AJR. Am. J. Roentgenol 193(2):314–325; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavakis E; Urbich C; Dimmeler S Homing and engraftment of progenitor cells: A prerequisite for cell therapy. J. Mol. Cell. Cardiol 45(4):514–522; 2008. [DOI] [PubMed] [Google Scholar]
- 9.Clarke E; Pereira C; Chaney R; Woodside S; Eaves AC; Damen J Toxicity testing using hematopoietic stem cell assays. Regen. Med 2(6):947–956; 2007. [DOI] [PubMed] [Google Scholar]
- 10.Crabbe A; Vandeputte C; Dresselaers T; Sacido AA; Verdugo JM; Eyckmans J; Luyten FP; Van Laere K; Verfaillie CM; Himmelreich U Effects of MRI contrast agents on the stem cell phenotype. Cell Transplant. 19(8):919–936; 2010. [DOI] [PubMed] [Google Scholar]
- 11.Fink DW Jr.. FDA regulation of stem cell-based products. Science 324(5935):1662–1663; 2009. [DOI] [PubMed] [Google Scholar]
- 12.Helfer BM; Balducci A; Nelson AD; Janjic JM; Gil RR; Kalinski P; M. d. V. IJ; Ahrens ET; Mailliard RB. Functional assessment of human dendritic cells labeled for in vivo (19)F magnetic resonance imaging cell tracking. Cytotherapy 12(2):238–250; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennig J; Nauerth A; Friedburg H RARE imaging—a fast imaging method for clinical mr. Magn. Reson. Med 3(6):823–833; 1986. [DOI] [PubMed] [Google Scholar]
- 14.Hinds KA; Hill JM; Shapiro EM; Laukkanen MO; Silva AC; Combs CA; Varney TR; Balaban RS; Koretsky AP; Dunbar CE Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allows magnetic resonance imaging of single cells. Blood 102(3):867–872; 2003. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann M; Wollert KC; Meyer GP; Menke A; Arseniev L; Hertenstein B; Ganser A; Knapp WH; Drexler H Monitoring of bone marrow cell homing into the infarcted human myocardium. Circulation 111(17):2198–2202; 2005. [DOI] [PubMed] [Google Scholar]
- 16.Hou D; Youssef EA; Brinton TJ; Zhang P; Rogers P; Price ET; Yeung AC; Johnstone BH; Yock PG; March KL Radiolabeled cell distribution after intramyocardial, intracoronary, and interstitial retrograde coronary venous delivery: Implications for current clinical trials. Circulation 112(9 Suppl):I150–156; 2005. [DOI] [PubMed] [Google Scholar]
- 17.Hoyes KP; Wadeson PJ; Murby B; Sharma HL; Cowan RA; Lord BI Bone marrow toxicity in mice treated with indium-114m-labelled blood cells. J. Exp. Clin. Cancer Res 20(4):505–510; 2001. [PubMed] [Google Scholar]
- 18.Janjic JM; Srinivas M; Kadayakkara DKK; Ahrens ET Self-delivering nanoemulsions for dual fluorine-19 MRI and fluorescence detection. J. Am. Chem. Soc 130(9):2832–2841; 2008. [DOI] [PubMed] [Google Scholar]
- 19.Kadayakkara DK; Beatty PL; Turner MS; Janjic JM; Ahrens ET; Finn OJ Inflammation driven by overexpression of the hypoglycosylated abnormal mucin 1 (MUC1) links inflammatory bowel disease and pancreatitis. Pancreas 39(4):510–515; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadayakkara DK; Janjic JM; Pusateri LK; Young WB; Ahrens ET In vivo observation of intracellular oximetry in perfluorocarbon-labeled glioma cells and chemotherapeutic response in the CNS using fluorine-19 MRI. Magn. Reson. Med 64(5):1252–1259; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller LH Bone marrow-derived aldehyde dehydrogenase-bright stem and progenitor cells for ischemic repair. Congest. Heart Fail 15(4):202–206; 2009. [DOI] [PubMed] [Google Scholar]
- 22.Lee JK; Lee MK; Jin HJ; Kim DS; Yang YS; Oh W; Yang SE; Park TS; Lee SY; Kim BS; Jeun SS Efficient intracytoplasmic labeling of human umbilical cord blood mesenchymal stromal cells with ferumoxides. Cell Transplant. 16(8):849–857; 2007. [DOI] [PubMed] [Google Scholar]
- 23.Lewin M; Carlesso N; Tung CH; Tang XW; Cory D; Scadden DT; Weissleder R Tat peptide-derivatized magnetic nanoparticles allow in vivo tracking and recovery of progenitor cells. Nat. Biotechnol 18(4):410–414; 2000. [DOI] [PubMed] [Google Scholar]
- 24.Li LH; McCarthy P; Hui SW High-efficiency electrotransfection of human primary hematopoietic stem cells. FASEB J. 15(3):586–588; 2001. [DOI] [PubMed] [Google Scholar]
- 25.Martens TP; Godier AF; Parks JJ; Wan LQ; Koeckert MS; Eng GM; Hudson BI; Sherman W; Vunjak-Novakovic G Percutaneous cell delivery into the heart using hydrogels polymerizing in situ. Cell Transplant. 18(3):297–304; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore KA; Ema H; Lemischka IR In vitro maintenance of highly purified, transplantable hematopoietic stem cells. Blood 89(12):4337–4347; 1997. [PubMed] [Google Scholar]
- 27.Niemeyer M; Oostendorp RA; Kremer M; Hippauf S; Jacobs VR; Baurecht H; Ludwig G; Piontek G; Bekker-Ruz V; Timmer S; Rummeny EJ; Kiechle M; Beer AJ Non-invasive tracking of human haemopoietic CD34(+) stem cells in vivo in immunodeficient mice by using magnetic resonance imaging. Eur. Radiol 20(9):2184–2193; 2010. [DOI] [PubMed] [Google Scholar]
- 28.Nohroudi K; Arnhold S; Berhorn T; Addicks K; Hoehn M; Himmelreich U In vivo MRI stem cell tracking requires balancing of detection limit and cell viability. Cell Transplant. 19(4):431–441; 2010. [DOI] [PubMed] [Google Scholar]
- 29.Nowak B; Weber C; Schober A; Zeiffer U; Liehn EA; von Hundelshausen P; Reinartz P; Schaefer WM; Buell U Indium-111 oxine labelling affects the cellular integrity of haematopoietic progenitor cells. Eur. J. Nucl. Med. Mol. Imaging 34(5):715–721; 2007. [DOI] [PubMed] [Google Scholar]
- 30.Osawa M; Hanada K; Hamada H; Nakauchi H Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273(5272): 242–245; 1996. [DOI] [PubMed] [Google Scholar]
- 31.Partlow KC; Chen J; Brant JA; Neubauer AM; Meyerrose TE; Creer MH; Nolta JA; Caruthers SD; Lanza GM; Wickline SA 19F magnetic resonance imaging for stem/progenitor cell tracking with multiple unique perfluorocarbon nanobeacons. FASEB J. 21(8):1647–1654; 2007. [DOI] [PubMed] [Google Scholar]
- 32.Pessina A; Malerba I; Gribaldo L Hematotoxicity testing by cell clonogenic assay in drug development and pre-clinical trials. Curr. Pharm. Des 11(8):1055–1065; 2005. [DOI] [PubMed] [Google Scholar]
- 33.Prasad VK; Kurtzberg J Cord blood and bone marrow transplantation in inherited metabolic diseases: Scientific basis, current status and future directions. Br. J. Haematol 148(3):356–372; 2010. [DOI] [PubMed] [Google Scholar]
- 34.Salama H; Zekri AR; Zern M; Bahnassy A; Loutfy S; Shalaby S; Vigen C; Burke W; Mostafa M; Medhat E; Alfi O; Huttinger E Autologous hematopoietic stem cell transplantation in 48 patients with end-stage chronic liver diseases. Cell Transplant. 19(11):1475–1486; 2010. [DOI] [PubMed] [Google Scholar]
- 35.Srinivas M; Cruz LJ; Bonetto F; Heerschap A; Figdor CG; de Vries IJ Customizable, multi-functional fluoro-carbon nanoparticles for quantitative in vivo imaging using 19F MRI and optical imaging. Biomaterials 31(27):7070–7077; 2010. [DOI] [PubMed] [Google Scholar]
- 36.Srinivas M; Heerschap A; Ahrens ET; Figdor CG; de Vries IJ (19)F MRI for quantitative in vivo cell tracking. Trends Biotechnol. 28(7):363–370; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivas M; Morel PA; Ernst LA; Laidlaw DH; Ahrens ET Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn. Reson. Med 58:725–734; 2007. [DOI] [PubMed] [Google Scholar]
- 38.Srinivas M; Turner MS; Janjic JM; Morel PA; Laidlaw DH; Ahrens ET In vivo cytometry of antigen-specific t cells using F-19 MRI. Magn. Reson. Med 62(3):747–753; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terrovitis JV; Smith RR; Marban E Assessment and optimization of cell engraftment after transplantation into the heart. Circ. Res 106(3):479–494; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd DM Celsense, Pitt trial to ‘see’ cells fight cancer. Pittsburgh Post-Gazette; Pittsburgh, PA; 2011. [Google Scholar]
- 41.Treleaven J; Barrett AJ Hematopoietic stem cell transplantation in clinical practice. Edinburgh, New York: Elsevier; 2008. [Google Scholar]
- 42.Verdijk P; Aarntzen EH; Lesterhuis WJ; Boullart AC; Kok E; van Rossum MM; Strijk S; Eijckeler F; Bonenkamp JJ; Jacobs JF; Blokx W; Vankrieken JH; Joosten I; Boerman OC; Oyen WJ; Adema G; Punt CJ; Figdor CG; de Vries IJ Limited amounts of dendritic cells migrate into the T-cell area of lymph nodes but have high immune activating potential in melanoma patients. Clin. Cancer Res 15(7):2531–2540; 2009. [DOI] [PubMed] [Google Scholar]
- 43.Wineman J; Moore K; Lemischka I; Muller-Sieburg C Functional heterogeneity of the hematopoietic microenvironment: Rare stromal elements maintain long-term repopulating stem cells. Blood 87(10):4082–4090; 1996. [PubMed] [Google Scholar]
- 44.Wu MH; Smith SL; Danet GH; Lin AM; Williams SF; Liebowitz DN; Dolan ME Optimization of culture conditions to enhance transfection of human CD34+ cells by electroporation. Bone Marrow Transplant. 27(11):1201–1209; 2001. [DOI] [PubMed] [Google Scholar]
- 45.Zhang SJ; Wu JC Comparison of imaging techniques for tracking cardiac stem cell therapy. J. Nucl. Med 48(12):1916–1919; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]