Abstract
Immune checkpoint blockade (ICB) immunotherapy induces potent antitumor immunity across multiple solid tumors, although few patients respond well to this therapy. An emerging biomarker for predicting responsiveness to ICB immunotherapy is tumor mutational burden (TMB). Although several surrogate biomarkers, including deficient mismatch repair, TP53/KRAS mutations, and comutations in DNA damage response pathways, have been shown to be effective for predicting the response to checkpoint blockade immunotherapy, each is positive for only a small cohort of candidates, and many potential responders to ICB are inevitably missed. Here, we found that titin (TTN), which is frequently detected in solid tumors, is associated with increased TMB and correlated with objective response to ICB. In 7 public clinical cohorts, all patients with mutated TTN showed longer progression-free survival or overall survival than those with wild-type status. Furthermore, an improved objective response rate and higher TMB were identified in cohorts with accessible information. Identification of TTN mutation as a predictor of improved outcomes in response to ICBs provides a clinically feasible assessment for estimating TMB and ICB therapy outcomes.
Keywords: Therapeutics
Keywords: Bioinformatics, Cancer immunotherapy
Genome-scale screening analysis demonstrates that Titan mutation indicates higher tumor mutational burden and predicts favorable response to immune checkpoint blockade across multiple solid tumors.
Introduction
Following the approval of ipilimumab, multiple immune checkpoint blockade (ICB) therapies have greatly advanced the clinical management of solid tumors. Tumor cells disrupt the immune equilibrium and progress or metastasize through several suppressive mechanisms, including by activating immune regulatory checkpoints that negatively regulate antitumor immunity (1). ICBs reinvigorate the cytolytic potential of cytotoxic T cells by interrupting malfunctioning “self-tolerance” signaling and reinforcing effective elimination of tumor cells. Although the response rate to ICBs varies substantially in different tumor types, the proportion of patients who are sensitive to ICB administration is generally 20%–30% in an unselected population for most solid tumor types (2). Identifying patients with a greater likelihood of benefiting from these antibodies is an increasingly important area of investigation.
Immunohistochemical identification of programmed cell death ligand 1 (PD-L1) expression was the first FDA-approved biomarker for patients’ favorable responsiveness to ICBs (3). However, the undetermined positivity threshold and spatiotemporal heterogeneity of PD-L1 expression severely complicate its clinical application (4, 5). Categorization of the tumor immune microenvironment of patients also results in different clinical outcomes of ICB therapies (6–8). The tumor immune microenvironment contains numerous components that contribute to cytolytic activity, including T cell–derived effector molecules, immune cell infiltration, and immune-related signaling pathways (9, 10). However, poorly reproducible measurement approaches and the lack of clinical validation in prospective trials prevent the identification of candidates for ICB treatment. The relationship between the tumor mutational burden (TMB) and clinical efficacy indicates the potential of TMB as a predictive biomarker in nearly all types of solid tumors (2, 11). Initially, whole-exome sequencing (WES) was performed to obtain a direct measurement method of nonsynonymous somatic mutations (12, 13). However, there are 3 main limitations to its wide implementation: (a) large amounts of nucleic material are required for sequencing, which are difficult to obtain by routine biopsies; (b) no consensus threshold has been determined for identifying responders; and (c) the test platform has not been standardized (14). Thus, the development of alternative biomarkers that can reflect the mutational burden status is necessary to accurately estimate TMB.
Several surrogate biomarkers of TMB have been developed and found to be related to the clinical response to ICB treatment. Deficient mismatch repair (dMMR) results in an exceptionally high number of somatic mutations (15). The significance of dMMR in tumor immunotherapy was demonstrated through retrospective assessments, which revealed a 24.5-fold increase in TMB and 40% higher objective response rate (ORR) than those in patients with proficient mismatch repair disease (16). Furthermore, Le and colleagues demonstrated that large proportions of mutant neoantigens in patients with dMMR cause sensitivity to ICBs, regardless of the origin of cancer (17). TP53/KRAS–comutated tumors exhibited substantially increased mutational burdens and a notable clinical benefit from anti–programmed cell death 1 (anti–PD-1) blockade (18). Wang and colleagues recently reported that comutations in specific DNA damage response (DDR) pathways may be a clinically convenient approach for estimating the TMB and may predict superior survival outcomes of patients treated with ICB therapy (19). However, broad implementation of these surrogate biomarkers in the routine setting remains challenging. For candidates intended for immunotherapy, the positivity rate for any of these biomarkers was found to be remarkably lower than the actual response rate in unselected patients treated with ICB therapies, suggesting that many patients are missed.
In this study, genome-scale screening analysis of a pan-solid tumor cohort was conducted to evaluate the relationship between the single gene mutation status and corresponding median mutation count in individuals with and without a specific mutation. By examining a discovery cohort containing 34 solid tumor types and 7 independent validation cohorts from clinical trials treated with anti–PD-1/PD-L1 or anti–cytotoxic T lymphocyte–associated antigen 4 (anti–CTLA-4), we found that the mutation of titin (TTN), with an average frequency of 29.68% in solid tumors, could stratify TMB into higher/lower groups and represent different clinical responses to ICB monotherapy. The development of TTN mutation as a surrogate TMB biomarker provides an alternative indicator for identifying responders to ICB treatment.
Results
TTN mutation predicts higher TMB and correlates with the response rate to ICB.
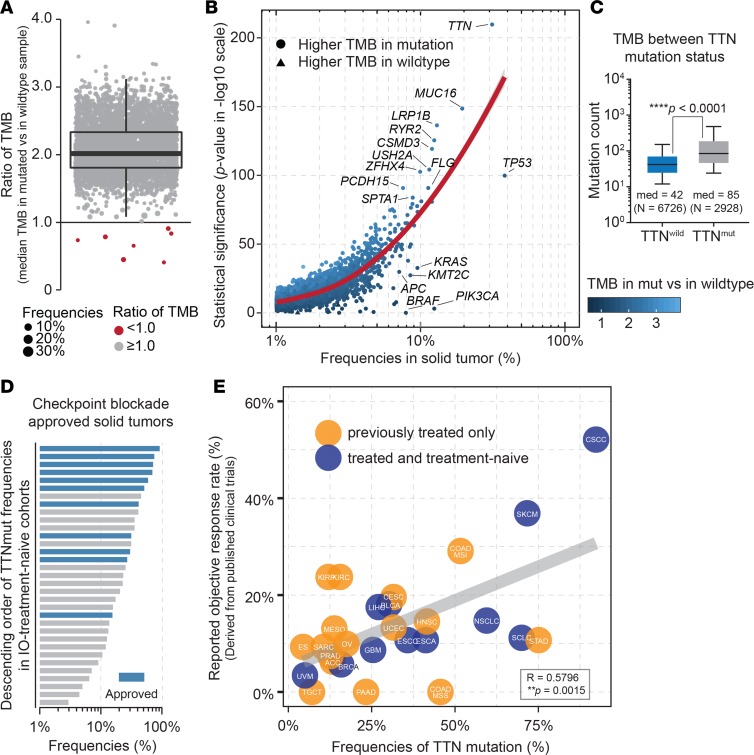
Frequently mutated genes were found to be associated with a higher mutational load in gastric cancer. Thus, we speculated whether a single gene mutation was sufficient to alter TMB in pan-solid tumors. To investigate, we performed comprehensive screening analysis to compare the TMB between patients with or without a specific mutated gene on a genomic scale. A total of 4246 genes (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127901DS1), which showed at least 1% frequency across 34 solid tumor types (Supplemental Table 2), were derived from The Cancer Genome Atlas (TCGA) (20) database using cBioportal tools (21, 22). Unexpectedly, for nearly all genes, we found a significantly higher TMB in patients with a mutated gene than in their wild-type counterparts (Figure 1A and Supplemental Table 1). To assess the robustness of the correlation between TMB and gene mutation status, the statistical significance (P values) of the median TMB between patients with or without mutation was calculated for each gene (Figure 1B). Analysis of the occurrence frequencies and corresponding significance identified that (a) the frequency of each gene was positively associated with the robustness of varying TMB disparity; (b) consistent with a previous report, TP53 and KRAS were highly robust genes; and (c) TTN, which was the second most frequently mutated gene in solid tumors, showed the most robust statistical difference among all screened genes and directly showed an approximately 2-fold higher TMB than that in wild-type individuals (median TMB was 85 for patients with mutation and 42 for wild-type patients; ****P < 0.0001; Mann-Whitney U test; Figure 1C).
Figure 1. TTN mutation is related to higher TMB and an increased response rate to ICB.
(A) The ratio of the median TMB between patients with or without mutation for a specific gene is shown. Red dots indicate a lower TMB for patients with the mutation; gray dots indicate a higher TMB for patients with the mutation. Box plot shows median with first and third quantiles. (B) Two-tailed Mann-Whitney U test was performed to compare the TMB between patients with or without the mutation. The frequencies of genes are plotted against their significance in the U test. Color gradient indicates ratio of TMB in A; red curve indicates fitting line in Loess regression. (C) TMB is shown based on TTN mutation status. Statistics based on 2-tailed Mann-Whitney U test. Box plot shows median with min to max range. (D) Frequencies of TTN mutation. Solid blue indicates tumor types approved for treatment with ICB. (E) Frequencies of mutated TTN for each tumor type with their reported ORR to ICB. Treatment settings, such as previously treated or treatment naive, are shown according to the enrolled criteria in clinical trials. R, Pearson’s coefficient of correlation.
After identifying the TTN/TMB correlation, we next examined whether the TTN mutation was related to the favorable efficacy of ICB treatment. First, we assessed the frequencies of TTN mutation across solid cancers and found that the percentage of mutated TTN varied across different tumor types, with “enrichment” among those that generally showed a better response to ICB treatment, including cases of cutaneous squamous cell carcinoma (23), stomach adenocarcinoma (24), skin cutaneous melanoma (25, 26), and lung cancer (27, 28), and less detection among tumors that are typically insensitive to immunotherapy, such as uveal melanoma (29) and testicular germ cell cancer (ref. 30, Figure 1D, and Supplemental Figure 1). A significant relationship was established between TMB and ORR of patients treated with ICBs across multiple solid cancers (2). To confirm this positive correlation between TTN and favorable responsiveness, we plotted the percentage of TTN mutations and the reported ORR for anti–PD-1/PD-L1/CTLA-4 monotherapy in unselected patients (Figure 1E). We observed a significant positive correlation between the reported ORR and frequencies of TTN mutation for each tumor type (r = 0.5796; **P = 0.0015; Pearson’s correlation), supporting the clinical relevance of TTN mutation for predicting a beneficial response to ICB treatment of solid tumors.
Higher TMB correlates with favorable clinical outcomes of patients with mutated TTN.
Given the finding that the baseline frequency of TTN mutations is correlated with a higher TMB and better responsiveness to immunotherapies, we evaluated whether the mutation of TTN could be used to discriminate between different TMBs and response rates of the patients. We examined 7 cohorts with accessible clinical characteristics and genomic information in the validation test (31–37). Consistent with our previous findings, validation analysis of the TMB and ORR across different TTN mutations revealed a 1.83- to 5.54-fold higher TMB (only predicted neoantigen available for Hellman cohort, ref. 32) and 1.38- to 2.90-fold increased ORR in patients with mutated TTN, supporting the clinical relevance of TTN mutation in subjects administered ICB therapies (Supplemental Figure 2, A–F).
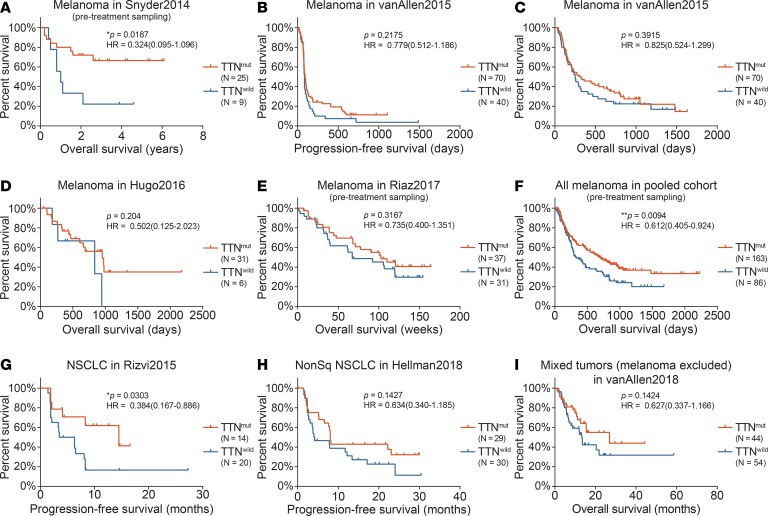
We next investigated whether a greater response rate translates into a prolonged survival benefit for individuals with mutant TTN. First, we performed survival analysis for patients not administered immunotherapies to determine whether the TTN mutation conveyed intrinsic survival superiority with solid tumors. Pan-cancer analysis showed that rather than a survival benefit, patients with TTN mutation exhibited a relatively poor prognosis for disease-free survival (***P = 1.6 × 10–6; HR = 1.181; log-rank test) and overall survival (OS, ***P = 2.2 × 10–11; HR = 1.137; log-rank test) (Supplemental Figure 3). Further, melanoma, lung adenocarcinoma, and lung squamous cell cancer, which were most widely tested in clinical trials of ICB administration, showed similar results for both disease-free survival and OS (Supplemental Figure 4). These data demonstrate that TTN mutation does not confer an intrinsic survival benefit to treatment-naive patients treated with ICB.
Four melanoma cohorts, treated with anti–PD-1 (nivolumab and pembrolizumab) or anti–CTLA-4 (ipilimumab and tremelimumab) antibodies, showed a survival benefit in patients with TTN mutation (HR = 0.324, and *P = 0.0187 for ref.
33; HR = 0.779, and P = 0.2175 for progression-free survival [PFS], and HR = 0.825, and P = 0.3915 for OS for ref. 34; HR = 0.502, and P = 0.204 for ref. 35; HR = 0.735, and P = 0.3167 for ref. 36) (Figure 2, A–E). Considering the limited size of some of the trials, we analyzed their merged cohorts including 163 patients with TTN mutation and 86 wild-type controls. We identified substantially prolonged OS (691 days for TTN-mutated and 325 days for wild-type individuals; *P = 0.0113; log-rank test) and decreased event hazard (HR = 0.612; 95% CI, 0.405–0.924) in patients with mutated TTN, further supporting the predictive value of TTN mutation in guiding ICB therapies (Figure 2F).
Figure 2. Survival analysis.
Survival analysis for TTN mutation status was performed for patients treated with ICBs. Kaplan-Meier survival plots, P value of log-rank test, and hazard ratios with 95% confidence interval (CI) are shown. To reflect the baseline of TTN mutation, only genomic data from samples obtained prior to ICB administration were used. (A–F) Patients with melanoma. (G and H) Patients with NSCLC. (I) Solid tumors with mixed tissue origins.
Next, we extended the survival analysis to cohorts with lung cancer or other solid tumor types. The Rizvi cohort (31) enrolled 34 patients with non–small cell lung cancer (NSCLC) who were treated with pembrolizumab. Patients with wild-type TTN showed damaged survival (median OS = 4.9 months for wild-type patients and 14.5 months for patients with TTN mutation; *P = 0.0303; log-rank test) and increased event hazard (HR = 0.384; 95% CI, 0.167–0.886, mutated vs. wild-type), unlike their TTN-mutated counterparts (Figure 2G). The same tendency was identified in the lung adenocarcinoma cohort that included patients treated with anti–PD-1/anti–CTLA-4 combinatorial immunotherapy (median PFS = 8.0 months for patients with mutation and 4.1 months for wild-type patients; P = 0.1427; log-rank test. HR = 0.634; 95% CI, 0.340–1.185) (Figure 2H). Finally, when the analysis was extended to a cohort including multiple solid tumor types (patients with melanoma were excluded), we observed a 2-fold OS change (median OS = 26.91 months for patients with mutation and 13.22 months for wild-type patients; P = 0.1424; log-rank test. HR = 0.627; 95% CI, 0.337–1.166) in the TTN-mutated group, suggesting the potential of this mutation as a generally applicable marker for predicting ICB response in solid tumors (Figure 2I).
TTN mutation distinguishes responders from candidates with unfavorable features.
Currently, the responsiveness to ICB treatment cannot be reliably determined by a unique biomarker alone. Therefore, we investigated whether TTN mutation can identify responders among patients whose clinical features seem to represent inferior sensitivity to ICB administration in clinical trials. The survival outcomes of patients with differences in PD-L1 expression (measured by immunohistochemistry), TMB, and smoking status were determined in relation to the efficacy of ICB therapies. For patients showing strong PD-L1 expression, showing TMB of 243 or greater, or having current smoker status, the observed HR suggested a prolonged PFS from ICB in the mutated TTN group (Supplemental Figure 5, A–C). In patients who had weak or no PD-L1 expression, who had TMB of less than 243, or who were former smokers or never smokers, a consistent survival benefit was observed in the mutated TTN group (Supplemental Figure 5, D–E) (HR = 0.395, and P = 0.0645 for weak or no PD-L1 expression; HR = 0.336, and P = 0.056 for TMB < 243; and HR = 0.386, and P = 0.073 for never/former smoker). These findings suggest that TTN mutation can serve as a complementary biomarker for rescuing responders from among patients who were missed using other validated biomarkers.
Potential performance of TTN mutation as a biomarker for ICB treatment.
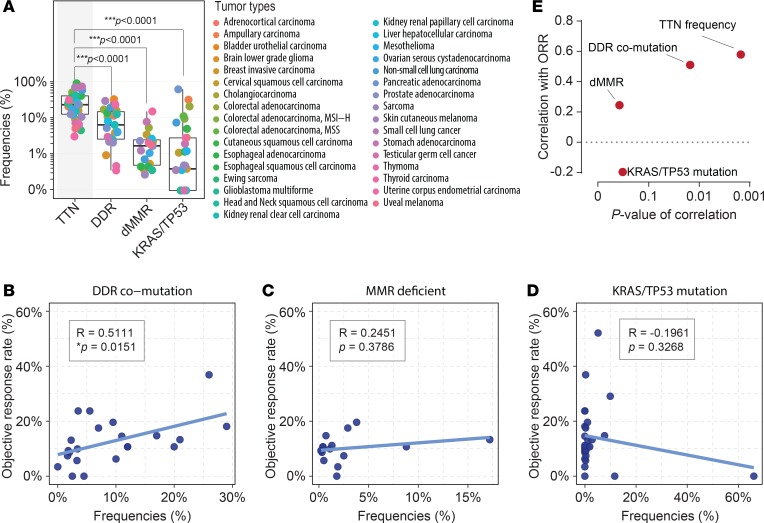
A reasonable predictive biomarker that is clinically useful should balance the specificity and sensitivity of performance. An extremely low positivity of a biomarker may prevent identification of responders and result in unsatisfactory sensitivity of prediction. The positivity of established predictive biomarkers for immunotherapy, including comutations in the DDR signaling pathway (19), dMMR (17), and TP53/KRAS comutation (18), have been widely evaluated. Exploring the frequency of each genomic biomarker (Figure 3A), we observed a substantially higher occurrence of TTN mutation (23.33%, median frequencies) than DDR comutation (5.50%, ***P < 0.0001), dMMR (1.55%, ***P < 0.0001), and TP53/KRAS comutation (0.3%, ***P < 0.0001). Furthermore, the potential performance of these 3 genomic biomarkers was examined against ORR (Figure 3, B–D). Despite the significant ORR observed for DDR comutation, the coefficient of correlation was still lower than that of the TTN mutation (r = 0.5111; *P = 0.0151; Pearson’s correlation; Figure 3B). Partially because of the lower positivity, the percentages of both dMMR (r = 0.2451; P = 0.3786; Pearson’s correlation; Figure 3C) and TP53/KRAS comutation (r = –0.1961; P = 0.3268; Pearson’s correlation; Figure 3D) failed to correlate with the reported ORR significantly. By comparing their performance with the occurrence frequency of TTN mutation, we found the highest correlation with the ICB response rate and high significance (P value) for TTN mutation, suggesting that TTN mutation can be used as a predictive biomarker in further clinical trials (Figure 3E).
Figure 3. Correlation between ORR and genomic predictive biomarkers in ICB therapies.
(A) Frequencies of biomarkers are shown and labeled according to each tumor type. Statistics were based on a 2-tailed Mann-Whitney U test. Box plot shows median with min to max range. (B–D) ORR to ICBs versus the frequencies of (B) DDR comutation, (C) dMMR, and (D) TP53/KRAS comutation for each tumor type. Tumor types without accessible data were excluded. R, coefficient of correlation; P value, Pearson’s correlation. (E) Correlation coefficient and statistical significance (P value) for biomarkers.
Length of exon contributes to the predictive value of TTN.
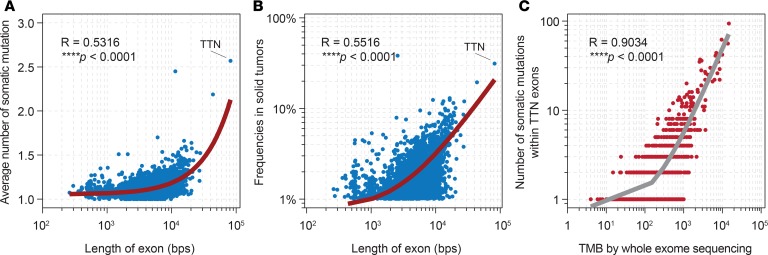
It is unreasonable to speculate that almost every frequently mutated gene is involved in the generation of somatic mutations in a cellular biological mechanism. Thus, we hypothesized that genes with larger size would show higher likelihood of randomly accumulating somatic mutations. To test this, we analyzed whether the exon length was correlated with the number of somatic mutations in each gene. We found that the length of exons for each gene correlated with the somatic mutations within them (Figure 4A; r = 0.5316; ****P < 0.0001). In fact, TTN showed the longest exon among the whole genome and harbored the highest number of mutations (Figure 4A). Furthermore, we identified a positive correlation between the frequency of occurrence and the length of exons for genes with 1% or greater prevalence (Figure 4B; r = 0.5516; ****P < 0.0001), implying that the longest length of exon is responsible for the frequently detected mutation of TTN. To further support the “randomly accumulating” hypothesis, we directly evaluated the relationship between the TMB and the total count of somatic mutations in the TTN exon in patients with TTN-mutant solid tumors. These analyses revealed a tightly positive correlation (Figure 4C; r = 0.9034; ****P < 0.0001) and strongly suggested the predictive value of TTN mutation status was attributed to its longest length of exon.
Figure 4. Mechanistic analysis.
(A) Length of exons versus the mean number of somatic mutations in the exon region for each gene. (B) Length of exons versus the frequency of occurrence in solid tumors for each gene. (C) Correlation between somatic mutations in the TTN exon and the TMB estimated by WES. R, Pearson’s correlation; curve, fitness lines.
Discussion
The immune system exerts antitumor activity that relies on effective recognition of novel neopeptides by intratumor cytotoxic T cells. Numerous studies have established mathematic models for describing the neoantigen/T cell interaction and have successfully improved the estimation of immune activity and clinical outcomes of ICB immunotherapies (38–40). However, a consensus approach for identifying recognizable neoantigens for antitumor activity has not been established. Nonsynonymous somatic mutation leads to the expression of a wide variety of non-native neoantigens that are targeted by tumor-specific cytotoxic T cells (41). Several studies have shown TMB is associated with both the response rate and survival benefit to checkpoint blockade immunotherapy in various tumor types (2, 11). In the current study, we demonstrated that the presence of nonsynonymous somatic mutation in TTN was associated with a higher TMB and may be a predictive biomarker for ICBs. The mutated status of TTN can identify a patient population that may benefit from ICB.
Accurate assessment of TMB may play a pivotal role in the clinical management of ICB immunotherapy (5). However, the implementation of TMB evaluation in clinical practice remains challenging. The sampling methods, amount of nucleic material, different sequencing platforms, varying bioinformatics analysis strategies, threshold definition, and costs are among the limitations that prevent broad implementation of TMB assessment. Thus, an alternative clinically applicable indicator of TMB is necessary for identifying candidates who may benefit from immunotherapy. Rather than quantifying the exact number of nonsynonymous somatic mutations directly by WES, the mutation status of a specific panel of genes may act as a surrogate for estimating TMB by categorizing patients into groups with significantly different TMBs and thus opposite likelihoods of response to therapies directed at blocking inhibitory immune checkpoint molecules (42). Some oncogene KRAS/TP53 comutations are related to higher nonsilence mutation counts, potent immunogenicity, and favorable responses to pembrolizumab administration in patients with lung adenocarcinoma (18). However, our results revealed that the TP53/KRAS comutation was nonexistent in several tumor types, and thus it cannot be applied for identifying responders to ICB treatment in pan-solid tumor cancers. Deficient expression of mismatch repair machinery increases the levels of tumor cell–encoded, mutation-associated neoantigens, which may be recognized by immune systems (43). PD-1 blockade has been demonstrated to be effective for treating patients with dMMR disease regardless of the origin of tumor cells (17). However, except for a few cancers, such as endometrial carcinoma (44), the positivity rate of dMMR is substantially low in most solid tumors. In NSCLC, the positivity rate is less than 1% and much lower than the ORR to anti–PD-1/anti–PD-L1 antibodies in unselected patients, in either first-line or second-line settings (28, 45). A functional deficiency in the DDR system results in a hypermutated genotype (46). Using a multiple linear regression approach, Wang identified comutations in homologous recombination repair-mismatch repair or homologous recombination repair-base excision repair to serve as indicators of a higher TMB in pan-cancer analysis (19). Data from several clinical trials have validated the correlation among DDR comutations, improved response rates, and increased survival. The positivity rate of DDR comutation was markedly higher than that of KRAS/TP53 and dMMR; however, sequencing of a panel of genes is complex and inconvenient in the clinic.
Titin is a structural protein in striated muscles (47). Mutations in this gene are related to familial hypertrophic cardiomyopathy (48), and autoantibodies against titin have been detected in patients with the autoimmune disease scleroderma (49). No studies have demonstrated that TTN is directly involved in regulating DDR or other biological processes that may result in the overgeneration of somatic mutations. Therefore, we speculated whether the predictive significance of TTN mutation was driven by a statistical mechanism. We found that the frequencies of mutated genes were obviously positively correlated with the length of their exons, and TTN showed the longest length of exon within the whole genome. Moreover, we also observed the highest number of mutated sites in the TTN exon among all genes, supporting that longer exon increased likelihood of accumulating more mutations. Coincidentally, Muc16, which is frequently mutated and encodes the cancer antigen CA-125, was demonstrated to be associated with a higher TMB and favorable survival outcome of patients with gastric cancer (50). Moreover, Yosef found that in addition to Muc16, most (210 of 212) frequently mutated genes (defined as >1%) in gastric cancer were associated with a significant tumor mutation load (51). The observations in Muc16, which was the second-longest gene in the genome, also support the correlation between higher mutational burden and the mutated status of a gene with a long exon.
Taken together, our analysis revealed the predictive significance of TTN mutation for estimating mutational load in pan-solid tumors. Because the average mutation burden is highly tumor type dependent, the mutated status of TTN may overcome the problem of an undetermined cutoff value for TMB to identify potential responders to ICB treatment. The clinical relevance of TTN mutation was validated in numerous cohorts treated with ICB therapies, including melanoma and NSCLC cohorts, and in a merged cohort containing multiple types of solid tumors. Compared with the other 3 available genomic biomarkers, the frequency of TTN mutation showed the highest correlation with the response rate to ICBs for individual tumor types. However, there were 3 main limitations to our study: (a) despite the long exon contributing to the predictive values of TTN, the biological role of TTN in the production of somatic mutations cannot be completely ruled out; (b) improved survival of patients with TTN mutation was observed in 7 cohorts using retrospective data; and (c) because of the lack of public data, our results could not be validated in a large cohort with mixed solid tumor types. Overall, our findings suggest TTN mutation is an alternative predictive biomarker for ICB therapies; however, further validation of these results is required in prospective clinical trials.
Methods
Data source.
The genomic alterations from WES and clinical characteristics for 34 solid tumor types were retrieved from TCGA database via cBioportal CGDS-R package (21). For the survival analysis, genomic data, survival follow-up, and baseline information, such as smoking status, nonsynonymous mutation courts, response evaluation or PD-L1 immunochemistry, were downloaded from publicly accessible publications reporting trials administrated with anti–PD-1/PD-L1 or anti–CTLA-4 antibodies.
Statistics.
Two-tailed Mann-Whitney U tests were performed for comparison of the nonparametric data set. We performed a χ2 test to test whether the sampling distribution was equal for 2 groups. Regression lines in scatter plots were plotted with Gaussian regression. The unsupervised clustering for tumor sample and gene expression/signaling pathway was generated with Pearson’s correlation-based hierarchical clustering. Survival outcomes were measured with OS, disease-free survival, or PFS according to the accessibility for each cohort. Events were defined as death of any cause, confirmation of disease recurrence, or confirmation of disease progression, respectively. Kaplan-Meier survival curves were generated to show the different survival outcomes between TTN-mutant and wild-type status. Objective response was defined as complete response and partial response according to the criteria in RECIST1.1 and reported in each publication. A P value less than 0.05 was considered significant.
Mechanism analysis.
Somatic mutations of all patients in TCGA were retrieved from Broad GDAC Firehose with the version as stddata__2016_01_28. Gene models and length of exon regions were retrieved from Ensembl Genes 87 with the human genome version as GRCh37. Considering that the covered genomic regions are almost the same for WES panels used in TCGA, the number of somatic mutations classified as missense, nonsense, and indels was used as the TMB for each patient. The number of somatic mutations on TTN was counted for each patient as the mutation burden of this gene. For each gene, the average number of somatic mutations and mutation frequency were calculated. Three scatter plots were drawn to compare length of exon region and average number of somatic mutations, length of exon region and mutation frequency, and TMB and mutation burden of TTN. Pearson’s correlation coefficient was calculated for each scatter plot.
Study approval.
All the genomic data and clinical information were retrieved from publicly accessible data.
Author contributions
BZ and JW conceived and designed the experiments. QJ, NH, JH, and BZ analyzed and interpreted data. QJ and BZ wrote the manuscript.
Supplementary Material
Acknowledgments
We are thankful for the financial support from National Nature Science Foundation of China (81222031 and 81620108023 to BZ).
Version 1. 05/16/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(10):e127901. https://doi.org/10.1172/jci.insight.127901.
Contributor Information
Qingzhu Jia, Email: jiaqingzhu0801@outlook.com.
Jun Wang, Email: ggjun2005@126.com.
Ning He, Email: hening@gloriousmed.com.
Ji He, Email: he.ji@genecast.com.cn.
Bo Zhu, Email: bo.zhu@tmmu.edu.cn.
References
- 1.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 2.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia Q, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9(1):5361. doi: 10.1038/s41467-018-07767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Teng F, Kong L, Yu J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 2016;9:5023–5039. doi: 10.2147/OTT.S105862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 7.Sharma P, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017;18(3):312–322. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- 8.Ayers M, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18(1):10. doi: 10.1186/s12943-018-0928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meléndez B, Van Campenhout C, Rorive S, Remmelink M, Salmon I, D’Haene N. Methods of measurement for tumor mutational burden in tumor tissue. Transl Lung Cancer Res. 2018;7(6):661–667. doi: 10.21037/tlcr.2018.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalmers ZR, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allgäuer M, et al. Implementing tumor mutational burden (TMB) analysis in routine diagnostics-a primer for molecular pathologists and clinicians. Transl Lung Cancer Res. 2018;7(6):703–715. doi: 10.21037/tlcr.2018.08.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther. 2018;189:45–62. doi: 10.1016/j.pharmthera.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong ZY, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, et al. Comutations in DNA damage response pathways serve as potential biomarkers for immune checkpoint blockade. Cancer Res. 2018;78(22):6486–6496. doi: 10.1158/0008-5472.CAN-18-1814. [DOI] [PubMed] [Google Scholar]
- 20.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migden MR, et al. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med. 2018;379(4):341–351. doi: 10.1056/NEJMoa1805131. [DOI] [PubMed] [Google Scholar]
- 24.Kang YK, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10111):2461–2471. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 25.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schachter J, et al. Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006) Lancet. 2017;390(10105):1853–1862. doi: 10.1016/S0140-6736(17)31601-X. [DOI] [PubMed] [Google Scholar]
- 27.Reck M, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 28.Vokes EE, et al. Nivolumab versus docetaxel in previously treated advanced non-small-cell lung cancer (CheckMate 017 and CheckMate 057): 3-year update and outcomes in patients with liver metastases. Ann Oncol. 2018;29(4):959–965. doi: 10.1093/annonc/mdy041. [DOI] [PubMed] [Google Scholar]
- 29.Algazi AP, et al. Clinical outcomes in metastatic uveal melanoma treated with PD-1 and PD-L1 antibodies. Cancer. 2016;122(21):3344–3353. doi: 10.1002/cncr.30258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adra N, et al. Phase II trial of pembrolizumab in patients with platinum refractory germ-cell tumors: a Hoosier Cancer Research Network Study GU14-206. Ann Oncol. 2018;29(1):209–214. doi: 10.1093/annonc/mdx680. [DOI] [PubMed] [Google Scholar]
- 31.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellmann MD, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843–852.e4. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riaz N, et al. Tumor and microenvironment evolution during immunotherapy with Nivolumab. Cell. 2017;171(4):934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao D, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50(9):1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghorani E, et al. Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann Oncol. 2018;29(1):271–279. doi: 10.1093/annonc/mdx687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rech AJ, et al. Tumor immunity and survival as a function of alternative neopeptides in human cancer. Cancer Immunol Res. 2018;6(3):276–287. doi: 10.1158/2326-6066.CIR-17-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Łuksza M, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551(7681):517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon S, et al. PD-1 expression conditions T cell avidity within an antigen-specific repertoire. Oncoimmunology. 2016;5(1):e1104448. doi: 10.1080/2162402X.2015.1104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizvi H, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36(7):633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riaz N, Morris L, Havel JJ, Makarov V, Desrichard A, Chan TA. The role of neoantigens in response to immune checkpoint blockade. Int Immunol. 2016;28(8):411–419. doi: 10.1093/intimm/dxw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson GS, et al. MMR deficiency is common in high-grade endometrioid carcinomas and is associated with an unfavorable outcome. Gynecol Oncol. 2013;131(2):309–314. doi: 10.1016/j.ygyno.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Carbone DP, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciferri A, Crumbliss AL. The assembling and contraction mechanisms of striated muscles. Front Chem. 2018;6:570. doi: 10.3389/fchem.2018.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerull B, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30(2):201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 49.Ohyama K, et al. Proteomic profiling of antigens in circulating immune complexes associated with each of seven autoimmune diseases. Clin Biochem. 2015;48(3):181–185. doi: 10.1016/j.clinbiochem.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Pasche B, Zhang W, Chen K. Association of MUC16 mutation with tumor mutation load and outcomes in patients with gastric cancer. JAMA Oncol. 2018;4(12):1691–1698. doi: 10.1001/jamaoncol.2018.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maruvka YE, Haradhvala NJ, Getz G. Analyzing frequently mutated genes and the association with tumor mutation load. JAMA Oncol. 2019;5(4):577. doi: 10.1001/jamaoncol.2019.0127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.