Abstract
Biomechanical forces and endothelial-mesenchymal transition (EndoMT) are known to mediate valvulogenesis. However, the relative contributions of myocardial contractile and hemodynamic shear forces remain poorly understood. We integrated 4D light-sheet imaging of transgenic zebrafish models with moving-domain computational fluid dynamics to determine effects of changes in contractile forces and fluid wall shear stress (WSS) on ventriculobulbar (VB) valve development. Augmentation of myocardial contractility with isoproterenol increased both WSS and Notch1b activity in the developing outflow tract (OFT) and resulted in VB valve hyperplasia. Increasing WSS in the OFT, achieved by increasing blood viscosity through EPO mRNA injection, also resulted in VB valve hyperplasia. Conversely, decreasing myocardial contractility by Tnnt2a morpholino oligonucleotide (MO) administration, 2,3-butanedione monoxime treatment, or Plcγ1 inhibition completely blocked VB valve formation, which could not be rescued by increasing WSS or activating Notch. Decreasing WSS in the OFT, achieved by slowing heart rate with metoprolol or reducing viscosity with Gata1a MO, did not affect VB valve formation. Immunofluorescent staining with the mesenchymal marker, DM-GRASP, revealed that biomechanical force–mediated Notch1b activity is implicated in EndoMT to modulate valve morphology. Altogether, increases in WSS result in Notch1b- and EndoMT-mediated VB valve hyperplasia, whereas decreases in contractility result in reduced Notch1b activity, absence of EndoMT, and VB valve underdevelopment. Thus, we provide developmental mechanotransduction mechanisms underlying Notch1b-mediated EndoMT in the OFT.
Keywords: Cardiology, Development
Keywords: Cardiovascular disease, Embryonic development
Myocardial contractile force promotes Notch1b-mediated endocardial cell differentiation to modulate valvular leaflet formation.
Introduction
Cardiac valve development is a critical step in the initiation of an efficient unidirectional flow system (1). Abnormalities of cardiac valves, including bicuspid aortic valves, aortic stenosis, and pulmonic stenosis, are frequently seen in congenital heart disease (CHD), affecting up to 2% of live births (2). Valve formation is known to be initiated by endocardial cushion formation and an endothelial-mesenchymal transition (EndoMT), followed by valvular cell proliferation and subsequent remodeling of the valvular leaflets (3). In addition to genetic factors, these events are tightly regulated by epigenetic cues, particularly biomechanical forces imparted by hemodynamic shear stress and myocardial contraction (4–6). Fluid flow reversal has been identified as a key hemodynamic factor that activates the mechanosensitive gene klf2a, a transcription factor in the Krüppel-like factor family (7). Fluid flow, rather than myocardial contractility, is considered to regulate the development of atrioventricular (AV) valves (6), whereas myocardial contractility was reported as the key factor in an earlier study (5), underscoring the challenge of uncoupling the fluid and solid mechanical forces acting on valve leaflet development.
Although technical limitations hamper the evaluation of these biomechanical forces in mammalian development, zebrafish (Danio rerio) embryos, with their optical transparency and short developmental time, have proved to be a genetically tractable model for mechanotransduction studies of cardiac valvulogenesis (8). Of their 2 cardiac valves, the AV valve has been the most studied, and the ventriculobulbar (VB) valve, which forms between the ventricle and the bulbus arteriosus, has received less attention. These valves are structurally similar to mammalian valves, and the developmental cellular events of valvulogenesis are largely conserved among species (1). Previous studies have focused on the role of fluid shear stress in valve development; however, there remains a paucity of data elucidating the relative importance of contractile and hemodynamic shear forces in the outflow tract (OFT) of the developing zebrafish embryo (5, 9), and the downstream signaling pathways and effects on VB valve development remain largely unknown.
One target of mechanosignaling in the OFT is the Notch pathway, which is highly conserved and regulates developmental events, including cell fate specification, cell proliferation, and apoptosis (10). During cardiovascular development, Notch signaling is known to mediate EndoMT (11, 12) and ventricular chamber development (13), and mutations in the Notch pathway cause defects of the aortic valve and myocardial trabeculation (10, 14–17). Changes in hemodynamic forces, such as low shear or disturbed flow, induce differential expression of endothelial Notch signaling genes (18). Additionally, myocardial contractile forces have been shown to activate endocardial Notch signaling in the developing zebrafish heart (19). In this context, we have sought to assess how alterations in myocardial contractile versus hemodynamic shear forces regulate Notch-mediated VB valve morphology.
In the present study, we used a custom-designed 4D light-sheet imaging system (selective-plane illumination microscopy, SPIM) to image transgenic zebrafish lines in vivo (Figure 1 and refs. 17, 20, 21). These images were coupled with moving-domain computational fluid dynamic (CFD) modeling (20, 22, 23) to quantify the relative importance of contractile versus hemodynamic shear forces to activate Notch1b/EndoMT signaling to mediate VB valve morphogenesis. Our results suggest that myocardial contractile forces are essential to the initial formation of the VB valve, while hemodynamic shear stresses contribute to VB valve leaflet growth.
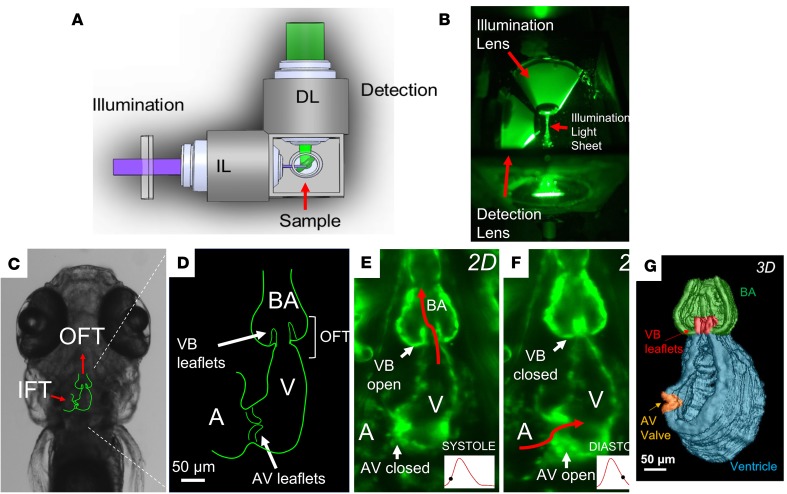
Figure 1. 4D light-sheet imaging of zebrafish embryos for assessment of myocardial contractility and valvular morphology.
(A) Schematic diagram and (B) photograph of the orthogonal optical pathway for single-sided illumination and dual-channel detection of the sample. (C) Outline of cardiac anatomy superimposed on a bright-field image from a zebrafish embryo at 5 days after fertilization (dpf) indicating orientation of the inflow tracts (IFTs) and OFTs (red arrows). (D) Schematic diagram of the embryonic heart (coronal section) highlighting the atrioventricular (AV) and ventriculobulbar (VB) valves, atrium, ventricle, OFT, and bulbus arteriosus. (E and F) Light-sheet fluorescence microscopy image of the embryonic heart at 5 dpf, showing (E) the open VB valve during systole and (F) the open AV valve during diastole. (G) 3D reconstruction of the embryonic heart during systole with VB and AV valves shown in red and orange, respectively.
Results
Pharmacological modulation of cardiac structure and function.
To modulate cardiac hemodynamic shear force, we treated embryos with a selective β1-receptor antagonist to reduce heart rate (metoprolol tartrate), a myofibrillar ATPase inhibitor to decrease myocardial contractility and subsequent heart rate (2,3-butanedione monoxime [BDM]), or a nonselective β-receptor agonist to increase myocardial contractility (isoproterenol), starting at 24 hours after fertilization (hpf). Embryos treated with metoprolol and isoproterenol showed no gross abnormalities in cardiac morphology at 48 hpf (Figure 2, A, B, and D), whereas those treated with BDM showed marked accumulation of precordial fluid (edema), associated with pooling of red blood cells in the sinus venosus (Figure 2C).
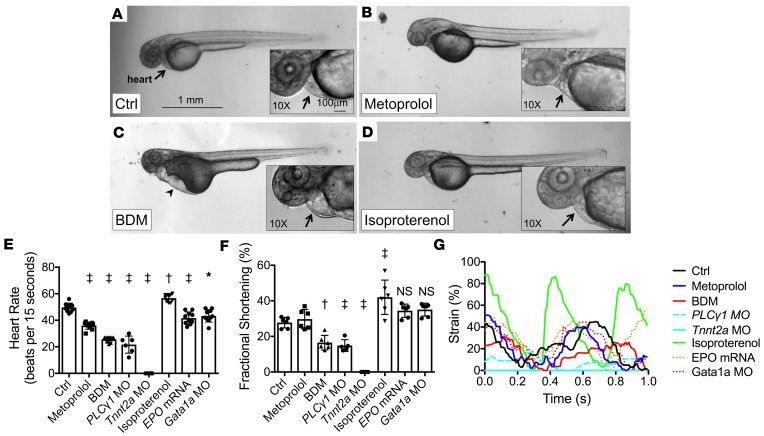
Figure 2. Effects of pharmacological interventions and genetic modification on cardiac hemodynamics in zebrafish embryos.
(A–D) Bright-field microscopic images (original magnification, ×2) of Tg(fli1a:GFP) zebrafish embryos treated with (A) control vehicle, (B) metoprolol, (C) 2,3-butanedione monoxime (BDM), and (D) isoproterenol at 48 hpf. Precordial edema (arrowhead) with pooling of red blood cells in the sinus venosus is seen in the BDM-treated embryo (arrow). Scale bar: 1 mm. (E) HRs at 48 hpf and (F) FS measurements at 56 hpf in response to pharmacological interventions and genetic modifications (n = 10 for HR measurements; n = 6 for FS measurements, except n = 5 for PLC1 MO group). Data are presented as mean ± SD; *P < 0.01; †P < 0.001; ‡P < 0.0001; 1-way ANOVA with Dunnett’s multiple-comparisons test. (G) Representative strain measurements for the listed treatment groups of embryonic hearts at 56 hpf.
Both metoprolol and BDM reduced the mean heart rates (HRs) by 27.4% and 48.4%, respectively (Figure 2E), whereas isoproterenol increased mean HR by 7.5%. PLCγ1 morpholino oligonucleotide (MO) injection reduced mean HR by 56.3%, though notably, there was only atrial contraction (no ventricular contraction) present in this group. Tnnt2a MO injection resulted in no heartbeat. EPO mRNA and Gata1a MO injections reduced the mean HRs by 15.9% and 12.7%, respectively.
Metoprolol did not affect ventricular fractional shortening (FS) as compared with controls, whereas BDM, PLCγ1 MO, and Tnnt2a MO significantly decreased FS (Figure 2F) and ventricular strain (Figure 2G). Isoproterenol increased FS and ventricular strain. EPO mRNA and Gata1a MO modestly increased FS, but compared to the control group, this slight increase did not achieve predefined statistical significance (P = 0.11 for EPO mRNA and P = 0.07 for Gata1a MO). Metoprolol, EPO mRNA, and Gata1a MO did not significantly affect ventricular strain compared to the control group (Figure 2G).
We determined ventricular ejection fraction (EF) using 4D SPIM-acquired imaging (Table 1; n = 4 per group). Results showed that (a) metoprolol had no significant effect on EF (71% ± 5% vs. control at 74% ± 4%, P = 0.98), (b) BDM significantly decreased EF (55% ± 8%, P = 0.001 vs. control), and (c) isoproterenol significantly increased EF (89% ± 4%, P = 0.01 vs. control). Although the ESV was smaller in the isoproterenol group, there were otherwise no significant differences in ESV, EDV, and stroke volume among the groups (Table 1). BDM produced a trend toward decreased stroke volume (P = 0.07 vs. control). Altogether, these results indicate that isoproterenol and BDM affected myocardial contractility, while metoprolol did not significantly affect contractility.
Table 1. Effects of pharmacological treatments and genetic modifications on ventricular hemodynamics.
Furthermore, we performed the following genetic modifications: (a) Gata1a MO injection to reduce hematopoiesis and subsequent blood viscosity and wall shear stress (WSS); (b) EPO mRNA injection to increase erythropoiesis, viscosity, and WSS; (c) NICD mRNA injection to induce Notch activity; (d) PLCγ1 MO injection to inhibit ventricular contractility alone; and (e) Tnnt2a MO injection to inhibit both atrial and ventricular contractility. The Gata1a MO, EPO mRNA, and NICD mRNA groups demonstrated similar contractility to the control group (Table 1). Meanwhile, as expected, the PLCγ1 MO and Tnnt2a MO groups demonstrated significantly reduced stroke volumes and contractility compared with the control group.
Pharmacological modulation of hemodynamic shear force in the OFT.
Intraventricular velocity streamlines (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124460DS1) and endocardial WSS parameters at 56 hpf (Figure 3 and Supplemental Figure 2) were reconstructed by time-dependent CFD simulation (22, 23) with the moving domain based on SPIM-derived 4D images. Effects of isoproterenol, metoprolol, and BDM treatments (n = 5 per group; Figure 3, C and D) on fluid velocity were compared in the ventricles and OFT of embryonic hearts (Figure 3, A and B). Compared with the control group, isoproterenol significantly increased the average velocity through the OFT (P = 0.0003), while metoprolol and BDM significantly decreased the average velocity (P = 0.018 and P = 0.009, respectively). EPO mRNA and Gata1a MO injections did not significantly affect the OFT velocity.
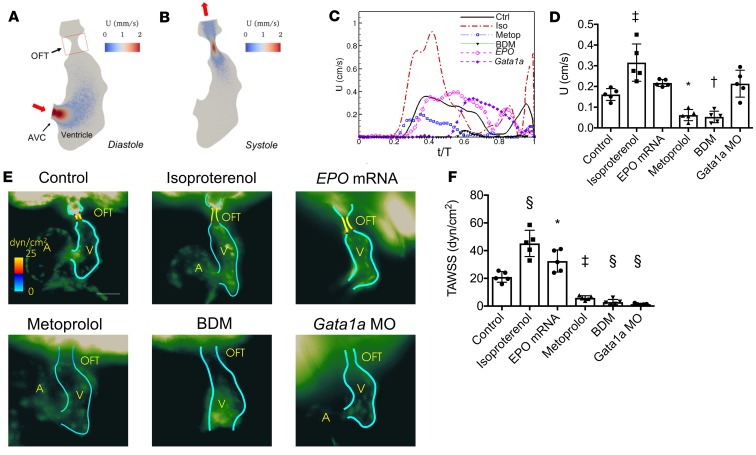
Figure 3. Moving-domain 2D CFD quantification of velocities and WSS in the developing OFT.
(A and B) Velocity (U) profiles during (A) diastole and (B) systole of a control embryo at 56 hpf. (C) Representative average velocities in the OFT of embryos at 56 hpf. (D) Average OFT velocities in the various treatment groups (n = 5 per group). (E) Representative endocardial border profiles of the time-averaged WSS (TAWSS) in the OFT of Tg(fli1a:GFP) embryos at 56 hpf, where (A) and (V) label the atria and ventricles. (F) Comparison of the TAWSS among the treatment groups (n = 5 per group). Data are represented as mean ± SD; *P < 0.05; †P < 0.01; ‡P < 0.001; §P < 0.0001; 1-way ANOVA with Dunnett’s multiple-comparisons test.
Next, WSS was averaged over 1 cardiac cycle to calculate the time-averaged WSS (TAWSS) in the OFT (Figure 3, E and F). Compared with the control group, isoproterenol increased, but metoprolol and BDM decreased, TAWSS (n = 5 per group; P < 0.0001 for isoproterenol; P = 0.0009 for metoprolol; P = 0.0002 for BDM; Figure 3F). Increasing blood viscosity with EPO mRNA injection increased TAWSS in the OFT, while decreasing viscosity with Gata1a MO injection decreased TAWSS.
We further assessed fluctuating WSS (FWSS), time-averaged WSS gradient (TAWSSG), and oscillatory shear index (OSI) in the OFT (Supplemental Figure 2). Although the trends in both FWSS and TAWSSG were similar to those observed in TAWSS, there were no significant differences in OSI in response to any of the treatments.
Contractile force modulation of valve leaflet morphology.
To determine whether cardiac contractile force modulates VB valve leaflet morphology, we treated the embryos with pharmacological modifications. SPIM imaging revealed that control and p53 MO–injected embryos developed normal VB valves by 5 dpf (Figure 4, A and B, and Supplemental Video 1). Metoprolol (which reduced TAWSS but preserved normal myocardial contractility) had no effect on VB valve size (Figure 4C), whereas BDM treatment (which reduced both TAWSS and myocardial contractility) resulted in the absence of VB valve leaflets (Figure 4D and Supplemental Video 2). Tnnt2a MO injection (causing loss of myocardial contractility) resulted in an underdeveloped OFT and the absence of AV or VB valve leaflets (Figure 4E), and Plcγ1 MO injection (causing loss of ventricular contractility but preserved atrial contractility) led to similar findings (Supplemental Figure 3). Conversely, isoproterenol (which increased TAWSS and increased myocardial contractility) resulted in elongated, hyperplastic VB valve leaflets (Figure 4F and Supplemental Video 3). Patency of the valve was demonstrated by imaging unidirectional flow of dsRed-labeled blood cells in Tg(gata1:dsRed) lines. Finally, depletion of the endocardial lining in the clochesk4-mutant (cloche-mutant) line resulted in the absence of VB valve leaflet formation (Supplemental Figure 4). Thus, enhanced myocardial contractility and the resulting increase in TAWSS caused VB valve hyperplasia, and reduced cardiac contractility inhibited VB valve formation.
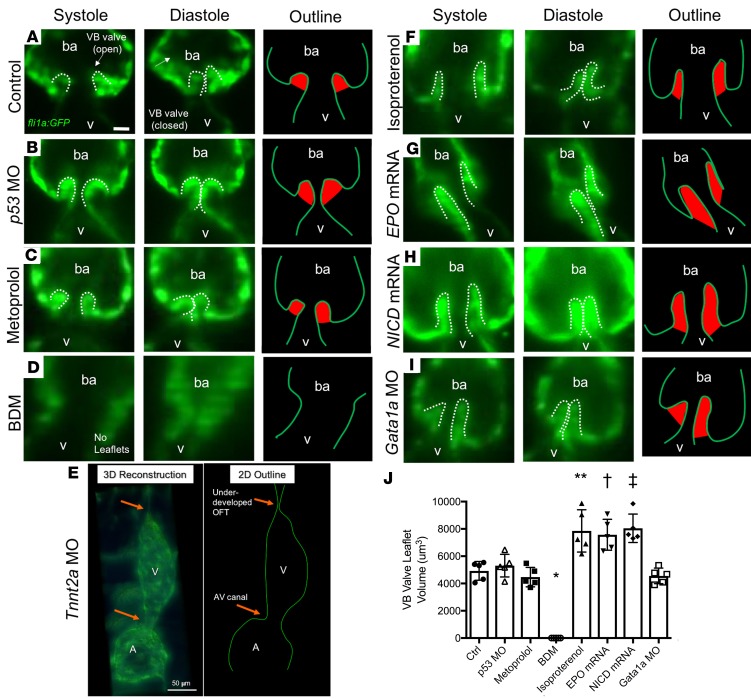
Figure 4. Effects of changes in hemodynamic shear force on VB valve leaflet formation.
Selective-plane illumination microscopy (SPIM) images of VB valves during ventricular systole and diastole at 5 dpf, with corresponding outline of the endocardium/endothelium (with valve leaflets highlighted in red), in Tg(fli1a:GFP) zebrafish embryos. (A) Control (untreated) embryo. (B) Vehicle control p53 MO–injected embryo showing similar size of VB valvular leaflets. (C) Metoprolol-treated embryo showing similar size of VB valvular leaflets despite reduction in HR. (D) BDM-treated embryo showing absence of VB valvular leaflets in response to significantly decreased HR and contractility. (E) Tnnt2a MO–injected embryo showing absence of VB leaflets in response to complete inhibition of myocardial contractile forces. 3D reconstruction at 5 dpf in a Tg(fli1a:GFP) zebrafish reveals an absence of cardiac looping accompanied by an underdeveloped OFT. The 2D schematic outline delineates the endocardial borders. (F) Isoproterenol-treated embryo showing prominence of VB valvular leaflets in response to increased HR and contractility. (G) EPO mRNA–injected embryo showing prominence of VB valve leaflets in response to increased blood viscosity with concomitant increase in shear stress. (H) NICD mRNA–injected embryo showing hyperplastic VB valve leaflets. (I) Gata1a MO–injected embryo showing normal leaflet morphology despite reductions in blood viscosity and endocardial shear stress. (J) Quantification of the VB valve leaflet volumes after 3D reconstruction (n = 5 per group). Scale bar: 10 μm in all except 50 μm in E. Data are presented as mean ± SD (*P < 0.0001, and **P = 0.0001 compared with Ctrl. †P = 0.003, and ‡P = 0.0003 compared with p53 MO. One-way ANOVA with Dunnett’s multiple-comparisons test with respective control groups). ba, bulbus arteriosus.
Hemodynamic force modulation of valve leaflet morphology.
To determine whether hemodynamic shear force modulates VB valve leaflets, we microinjected EPO mRNA to increase erythropoiesis, with resulting increases in viscosity and WSS. Results showed hyperplastic VB valve leaflets in the Tg(fli1a:GFP) line (Figure 4G). The same enlargement occurred in response to NICD mRNA microinjection (Figure 4H and Supplemental Video 4), which increased Notch activity in the Tg(tp1:GFP) Notch reporter line. However, VB valve leaflets were normal in size in response to Gata1a MO microinjection (which reduced hematopoiesis, resulting in reduced blood viscosity) (Figure 4I). As a corollary, VB valve leaflet size also increased significantly in response to isoproterenol, EPO mRNA, and NICD mRNA (Figure 4J). Thus, augmented WSS promoted VB valve leaflet enlargement, whereas reduced WSS had no effect on leaflet size.
Cardiac contractile force modulation of Notch1b activity.
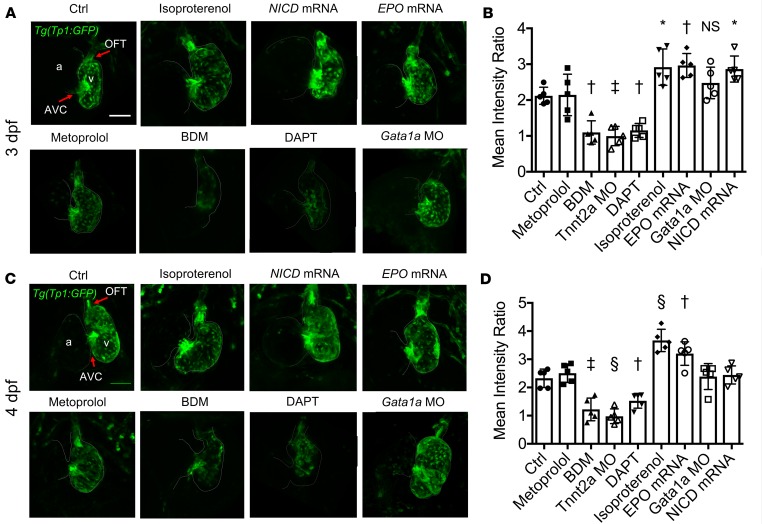
To demonstrate the role of contractile force–mediated Notch1b activity in VB valve morphogenesis, we assessed Notch1b signaling using tp1 expression as a reporter in Tg(tp1:GFP) embryos. Results showed Notch1b activity was prominent in both the OFT and AV canal (AVC) at 3 dpf (Figure 5, A and B) and 4 dpf (Figure 5, C and D). Notch1b activity in the OFT was significantly attenuated in response to BDM, DAPT (a γ-secretase inhibitor to inhibit cleavage of the Notch intracellular domain, NICD), and Tnnt2a MO microinjection (Figure 5), as well as Plcγ1 MO microinjection (Supplemental Figure 3).
Figure 5. Effects of changes in hemodynamic shear force on Notch1b activity in the developing OFT.
Confocal microscopy images (maximum-intensity projections) of the OFT and AVC in the Tg(tp1:GFP) Notch1b reporter zebrafish line at (A) 3 dpf and (C) 4 dpf with the indicated treatments. Scale bar: 50 μm. Notch1b activity was prominent in both the OFT and AVC in control embryos. Notch1b activity was more prominent in the OFT in response to isoproterenol, as well as with increasing blood viscosity with EPO mRNA microinjection and with upregulation of Notch activity with NICD mRNA microinjection. Notch1b activity in the OFT was nearly absent in response to BDM and DAPT treatments. (B and D) 3D quantification of the mean intensity of Notch1b activity in the OFT (normalized to activity in the respective ventricle) at (B) 3 dpf and (D) 4 dpf was significantly higher with isoproterenol treatment and EPO mRNA injection, consistent with these groups having more prominent VB valvular leaflets. Additionally, Notch1b activity in the OFT was significantly lower with BDM and DAPT treatments (n = 5 per group). Data are presented as mean ± SD; *P < 0.05; †P < 0.01; ‡P < 0.001; §P < 0.0001; 1-way ANOVA with Dunnett’s multiple-comparisons test.
Notch1b activity was accentuated in response to isoproterenol and EPO mRNA microinjection in the OFT, but it was not affected by Gata1a MO microinjection. The mean intensity of Notch1b activity in the OFT (normalized to activity in the respective ventricle) based on 3D reconstruction of SPIM-derived images (Figure 5, B and D) tended to increase with NICD mRNA microinjection at 3 dpf but less so at 4 dpf (Figure 5, B and D), consistent with the anticipated duration of the effects of mRNA microinjection. These findings suggest that both contractile force and hemodynamic shear stress modulate Notch1b activity in the OFT.
Attempts to rescue VB valve leaflet formation in Tnnt2a MO–injected (Supplemental Figure 5A), Plcγ1 MO–injected (Supplemental Figure 5B), and BDM-treated embryos (Supplemental Figure 5C) with NICD mRNA injection were unsuccessful, and no VB valve leaflets were present at 5 dpf in these groups.
Contractile force modulation of EndoMT in the OFT.
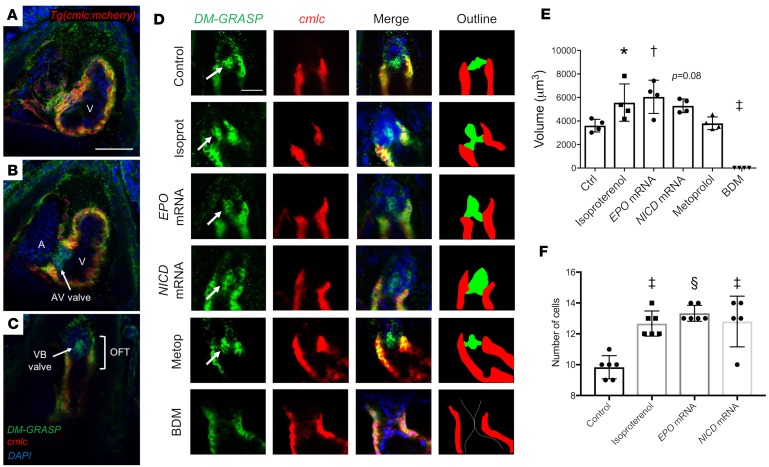
Expression of the cell surface adhesion molecule, DM-GRASP, is considered a marker for the EndoMT occurring when endocardial cells differentiate into valve-forming cells in Tg(cmlc:mCherry) embryos at 4 dpf (24). In the ventricular myocardium (Figure 6A), confocal images revealed immunoreactivity for DM-GRASP at the intercellular borders of the cmlc+ cardiomyocytes. In the AVC (Figure 6B), DM-GRASP was prominent in the AV valve leaflets where cmlc+ cardiomyocytes were absent. In the OFT (Figure 6C), DM-GRASP immunoreactivity was also prominent in the VB valve leaflets where myocardial cmlc was absent. Thus, DM-GRASP immunoreactivity identified differentiated endocardial cells in both AV and VB valve leaflets.
Figure 6. Effects of hemodynamic modulation on endocardial cell differentiation during VB valve formation in the OFT.
Immunostaining was performed against DM-GRASP, a cell surface adhesion protein expressed by differentiated valve-forming endocardial cells, in Tg(cmlc:mCherry) zebrafish embryos at 4 dpf. (A–C) Confocal imaging (original magnification, ×20) demonstrates sections through the (A) ventricle, (B) AVC, and (C) OFT of a control embryo. Green fluorescent protein (GFP) signal is observed between the cmlc+ myocardial cells, as well as in the AV and VB valve leaflets, which lack a cmlc signal, suggesting these are differentiated endocardial/endothelial cells. Scale bar: 50 μm. (D) Confocal images through the OFT of embryos subjected to the indicated treatments. Arrows denote DM-GRASP+, cmlc– valve leaflets. Scale bar: 25 μm. The outline denotes the DM-GRASP+, cmlc– valve leaflets in green and the cmlc+ myocardium in red. (E) Quantification of the volumes of DM-GRASP+, cmlc– valve leaflets in the indicated treatment groups (n = 4 per group). (F) Quantification of the cell counts in the DM-GRASP+, cmlc– valve leaflets in the indicated treatment groups (n = 6 per group, except n = 5 for the NICD mRNA group). Data are presented as mean ± SD; *P < 0.05; †P < 0.01; ‡P < 0.001; §P < 0.0001; 1-way ANOVA with Dunnett’s multiple-comparisons test.
To determine the effects of pharmacological interventions on endocardial cell differentiation in the OFT, the volumes of DM-GRASP+, cmlc– regions in the OFT were quantified (n = 4 per group, Figure 6D). Isoproterenol increased the volume of the DM-GRASP+, cmlc– region (P = 0.04 vs. control). Similarly, increasing WSS in the OFT with EPO mRNA injection increased the volume (P = 0.008 vs. control), and increasing Notch signaling with NICD mRNA injection resulted in a similar trend (P = 0.08). Metoprolol had no significant effect (P = 0.98 vs. control; Figure 6E). BDM-mediated attenuation of myocardial contractility resulted in no DM-GRASP+, cmlc– cells in the OFT (P = 0.0003 vs. control; Figure 6, D and E). Overall, EndoMT, identified by the volume of the DM-GRASP+, cmlc– regions in the OFT, was increased by isoproterenol and EPO mRNA, inhibited by BDM, and unaffected by metoprolol, suggesting that the effects of these interventions on valve morphogenesis are through EndoMT.
To determine whether the increase in VB valve leaflet sizes in the isoproterenol, EPO mRNA, and NICD mRNA groups was a result of cellular hyperplasia, the number of cells in the VB valve leaflets was evaluated (Figure 6F). Isoproterenol treatment resulted in increased cell number in the VB valve leaflet at 4 dpf compared with control (P = 0.0002), as did EPO mRNA (P < 0.0001) and NICD mRNA treatments (P = 0.0002). Thus, the increases in VB valve leaflet sizes that resulted from increased WSS and Notch1b signaling were the result of VB valve leaflet hyperplasia.
Discussion
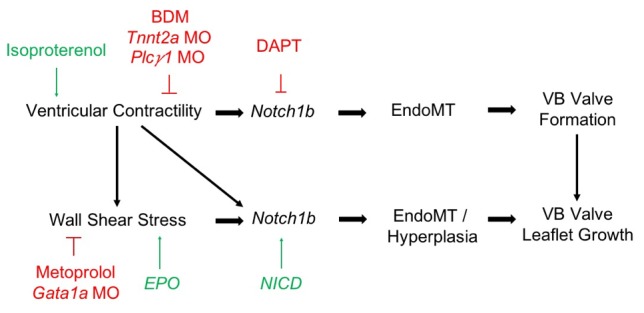
Since the seminal study by Hove et al. (4), several investigators have demonstrated the critical role of fluid shear stress and Notch signaling in the proper development of the heart (6, 7, 25–27). In the present study, we sought to distinguish the roles of solid versus fluid forces in Notch1b-mediated valve morphogenesis in the ventricular OFT, using 4D SPIM and moving-domain CFD in a zebrafish model of valvular development. To our knowledge, this study is the first to evaluate the mechanotransduction of VB valve development, because prior studies have focused on the AV valve. Results suggest that myocardial contractile force is the predominant regulator of the initial development of the VB valve, through a process that is, at least partially, Notch independent. Meanwhile, hemodynamic shear stress coordinates with contractility to regulate Notch1b-mediated EndoMT, leading to VB valve leaflet growth and, in conditions of augmented shear stresses, hyperplasia. However, the relative contributions of contractility and shear stress remain to be fully decoupled. These proposed mechanisms are summarized in Figure 7.
Figure 7. Schematic of proposed mechanism by which myocardial contractile and hemodynamic shear forces coordinate to promote Notch1b- and EndoMT-mediated valve formation in the developing OFT.
Our findings show Notch1b activity to be prominent in the valve-forming regions (AVC and OFT) of the embryonic heart (Figure 4), consistent with previous studies (28). Although our CFD analysis demonstrates that these are the regions experiencing the highest flow velocities and thus highest WSS, we found that reduction of WSS alone with preserved myocardial contractility (with metoprolol treatment or Gata1a MO injection) did not affect Notch1b activity in the OFT, similar to previously observed results (20, 28). However, Notch1b activity was attenuated in response to BDM-, Tnnt2a MO-, and PLCγ1 MO-mediated reduction in contractile force, leading to the failure of valve formation. As such, myocardial contractile forces (rather than hemodynamic shear forces) may be required to promote Notch1b-mediated valve formation.
We further elucidated how hemodynamic shear forces modulate endocardial Notch1b activity in the OFT. Increasing Notch signaling by transient ectopic expression of N1IC, which encodes the active intracellular domain of Notch1, was reported to induce hypercellularity in AV valves of zebrafish (11). Corroborating these findings, EPO mRNA–injected embryos, which have increased blood viscosity and thereby experience increased WSS, demonstrate similarly increased Notch1b activity and VB valve hyperplasia. Further, isoproterenol also significantly augmented TAWSS-mediated Notch1b activity, resulting in hyperplasia of the VB valves. However, decreasing viscosity and WSS with Gata1a MO injection had no significant effect on Notch1b activity or VB valve morphology, similar to that observed in the metoprolol-treated embryos (Figure 2F). This finding was consistent with a previous study showing no significant effect of Gata1a MO injection on AV valve formation (28). Only those treatments that inhibited myocardial contractility (BDM, Tnnt2a MO, Plcγ1 MO) impaired VB valve formation. Although hemodynamic shear forces play a lesser role in initial valve formation, they play a prominent role in promoting VB valve growth, and augmentation of WSS (through increased myocardial contractility or increased blood viscosity) results in increased Notch1b activity and subsequent VB valve hyperplasia.
The present work assesses the relative importance of contractile versus hemodynamic forces on the VB valve, which is analogous to the human aortic valve. Although the 2 valves may have developmental similarities, differences are found in mice, including spatiotemporal differences in VEGF signaling (29), differential responses to Notch inhibition (16), differing contributions from the neural crest (15), and differing hemodynamic forces in the 2 regions (30). The coordination between contractility and flow modulates valve development and remodeling following endocardial cushion formation (4, 27). The Notch1 pathway is critical to both early valve development and post-EndoMT aortic valve development via TNF-α in mice (16). Valve endothelial cell–specific (VEC-specific) inhibition of Notch1 signaling was reported to promote hyperplastic valves, whereas VEC-specific overexpression of Notch1 resulted in hypoplastic valves (16, 31). However, modulation of Notch1 activity in valve interstitial cells had no effects on valve development (31, 32). Although some studies suggest that Notch1 plays an inhibitory role in valve cell proliferation, this inconsistent finding may be species specific (11).
In our study, the relative importance of myocardial contractility and WSS in Notch1b-mediated VB valve development was not necessarily binary. Further increases in contractility and WSS led to valve leaflet hyperplasia, in part mediated by increased endocardial differentiation into valve-forming cells via EndoMT (Figure 6). Rugonyi et al. showed that cardiac contractility and hemodynamic load increase EndoMT in the OFT of developing chick embryos (33). Butcher and Merryman et al. further demonstrated shear stress– and myocardial contractility–mediated EndoMT in vitro (34, 35). The absence of endocardial cilia may prime these valve-forming regions to undergo TGF-β–mediated EndoMT (36, 37). The central role of endocardial cell signaling in valve development is recapitulated in the absence of valve formation in our endocardial endothelial lining–depleted, cloche-mutant embryos (Supplemental Figure 4). The cloche mutants failed to undergo cardiac looping in preparation for valvulogenesis. Similar to BDM-treated embryos, the cloche mutants developed significant precordial edema. This observation confirmed WSS-mediated Notch activity in the endocardial endothelium modulates the initiation of cardiac trabeculation and valve formation (15, 17, 20).
Other flow-sensitive signaling pathways play a role in valve morphogenesis. The miRNA miR-21 is reported to participate in flow-mediated valve development. Fraser, Vermot, and colleagues have demonstrated that the flow reversal-mediated transcription factor, klf2a, an upstream regulator of Notch1b signaling in zebrafish embryos, regulates valve development (7, 28) via modulation of fibronectin synthesis (26) and mechanosensitive ion channels (7). Cyclic mechanical loading also promotes valve remodeling and elongation (38). Despite changes in VB valve morphology in response to different pharmacological treatments, our moving-domain CFD analysis revealed no significant difference in OSI (Supplemental Figure 2). However, the high variability seen in the measured OSI values in the OFT may suggest the need to enhance our model’s temporal resolution to capture flow reversal characteristics within this region.
Notably, attempts to rescue VB valve leaflet formation in Tnnt2a MO–injected, PLCγ1 MO–injected, and BDM-treated embryos with NICD mRNA coinjection were unsuccessful (Supplemental Figure 5). In previous studies, despite evidence of the rescued expression of downstream targets Efrin2a and NRG1 (19), Notch1 activation was unable to rescue myocardial trabeculation in Tnnt2a MO–injected embryos (17). Taken together, these experiments underscore myocardial contractility as a driving force in VB valve leaflet formation, which may signal through both Notch-dependent and Notch-independent pathways.
The strengths of the current study include the high spatiotemporal resolution imaging of our 4D SPIM technique, which allows for visualization of the dynamic VB valve motion that is not well visualized on alternative imaging modalities and for quantification of valve leaflet volumes. To our knowledge, this is the first study to assess VB valve volume in response to hemodynamic modulation. Although the use of 2D moving-domain CFD modeling linked 3D hemodynamic forces (time and 2D plane) with mechanotransduction underlying AV valve morphogenesis, further increases in our computing capacity would enhance the use of 4D CFD (time and 3D space) to evaluate the hemodynamic microenvironment in the developing OFT. Nonetheless, the OFT WSS values obtained in our moving-domain CFD model system are consistent with previous studies (9).
In summary, integration of 4D light-sheet acquired imaging with the transgenic zebrafish system provides a computational basis to determine the mechanotransduction mechanisms underlying Notch-mediated valvular pathologies. Our data support the notion that myocardial contractility contributes to initial VB valve formation independently of hemodynamic shear stresses, while hemodynamic shear stresses promote endothelial Notch1b-mediated EndoMT in the OFT. This mechanotransduction pathway for VB valve development and hyperplasia has translational implications for dysmorphic valves in human CHDs.
Methods
Zebrafish maintenance. Adult zebrafish were raised in the aquarium system located in the vivarium of the UCLA. Fish were maintained with filtered fresh water under a 14-hour light/10-hour dark cycle. The following transgenic zebrafish lines were used: Tg(fli1a:GFP), Tg(gata1:dsRed), Tg(tp1:GFP), and Tg(cmlc:mCherry). David Traver at UCSD and Nathan Lawson at the University of Massachusetts Medical School (Worcester, Massachusetts, USA) provided the Tg(tp1:GFP) line (UCLA Zebrafish Core Facility). The fli1a promoter–driven enhanced GFP is expressed predominantly in vascular endothelial and endocardial cells. Gata1 is an erythroid-specific transcription factor allowing for visualization of red blood cells with red fluorescent protein. The Epstein-Barr virus terminal protein 1–transgenic (tp1-transgenic) zebrafish belong to a Notch reporter line containing 2 RbpJκ binding sites for NICD, thereby reporting regional Notch1b activation (39). Embryos from the transgenic zebrafish were transferred to a Petri dish and incubated at 28.5°C. To maintain zebrafish embryo transparency, the medium was supplemented with 0.003% phenylthiourea to suppress pigment formation as previously reported (20). We also used the cloche-mutant zebrafish line, in which the endocardium is absent (40).
Chemical treatment and genetic manipulation to modulate cardiac hemodynamics. At 24 hpf, isoproterenol hydrochloride (100 μM, MilliporeSigma) was applied to E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4 in ddH2O) to increase HR and contractility (41), while metoprolol tartrate (100 μM, MilliporeSigma) or BDM (10 mM, MilliporeSigma) was applied to reduce HR and contractility (5). MOs (GeneTools) were designed against the ATG of Gata1a (5′-CTG- CAAGTGTAGTATTGAAGATGTC-3′), Tnnt2a (5′-CGCGTGGA-CAGATTCAAGAGCCCTC-3′), and Plcγ1 (5′-AGAGCGTCCTCCTGA CCTTGATGAG-3′). The morpholinos were resuspended in nuclease-free water and injected at 8 ng/nl, 4 ng/nl, and 2 ng/nl, respectively, at the 1- to 2-cell stages. Gata1a is a transcription factor that is essential for erythroid differentiation, and microinjection of embryonic zebrafish with Gata1a MO has been shown to reduce hematopoiesis and viscosity by 90% (28, 42). Microinjection with Tnnt2a MO inhibits the expression of cardiac troponin T (Tnnt2a), resulting in a noncontractile atrium and ventricle (43). Microinjection with Plcγ1 MO recapitulates the dead beat mutation, resulting in selective inhibition of ventricular myocardial contractility (44). EPO mRNA (20 pg/nl) was injected at the 1-cell stage to increase blood viscosity by increasing hematopoiesis (45). NICD mRNA was injected at the 1-cell stage to promote the ectopic overexpression of Notch signaling as previously described (46). DAPT (100 μM, MilliporeSigma), a γ-secretase inhibitor that inhibits the cleavage of NICD from the Notch receptor, was diluted in 1% DMSO and added to E3 medium to inhibit Notch signaling at 24 to 72 hpf (17).
4D cardiac SPIM imaging. To visualize the dynamic cardiac architecture and valvular leaflet morphology and motion, we integrated our in-house 4D SPIM imaging system (Figure 1) with postprocessing synchronization as previously described (17). Briefly, using the SPIM technique, we scanned approximately 50 sections from the rostral to the caudal end of the zebrafish heart. Each section was captured with 50 lateral (or cross-sectional) planes at 10-ms exposure/frame via an sCMOS camera (Hamamatsu, Flash 4.0). The thickness of the light-sheet was tuned to approximately 1 μm to provide a high axial resolution for adequate reconstruction of the 3D cardiac morphology, and the axial scanning was set to 2 μm for lossless digital sampling according to the Nyquist sampling principle (47). To synchronize with the cardiac cycle, we established the cardiac periodicity frame to frame by comparing the pixel intensity from peak systole (smallest ventricular volume) to end diastole (largest ventricular volume) (20, 48, 49). We processed reconstructed 4D images using Amira software (FEI). To measure the VB valve leaflet volume, a 3D image stack was selected in midsystole with the VB valve open, and the volumes of the leaflets were measured from base to tip.
Cardiac hemodynamic measurements. Based on SPIM-derived images with a nongated, 4D synchronization computational algorithm, the ESV and EDV were measured in Tg(fli1a:GFP) zebrafish embryos at 56 hpf using Amira software. The EF was calculated as (EDV – ESV)/(EDV × 100). The stroke volume was calculated as EDV – ESV. HR was measured by counting the number of beats over a 15-second period.
To measure 2D ventricular diameter change, we used SPIM imaging of embryos at 56 hpf. The captured images were segmented to create the 2D moving boundary conditions with 600 nodes, and the nodes were guided to replicate cardiac wall motion captured by SPIM segmentation. Customized MATLAB code (MathWorks) was used to calculate the strain as defined by the time-dependent changes in displacement (D) between the individual time steps as described previously (17, 22):
(Equation 1)
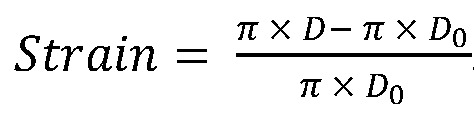
where D0 represents the initial displacement at time = 0, and D at time = Δt. FS was calculated by measuring the changes in displacement between end diastole and end systole:
(Equation 2)
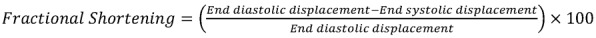
Moving-domain CFD modeling. Moving-domain CFD modeling of zebrafish embryos was used as described previously (22, 23). Briefly, the individual developmental stages were captured using SPIM technique, and video frames were processed using ImageJ (NIH). A custom program written in MATLAB (MathWorks) was then used to create 2D segmentations of the endocardial boundary and an unstructured mesh representing the blood volume. Points were selected on the image to trace the boundary of the endocardium, and then the MMG2D software (50) was used to create a point cloud. A Delaunay triangulation algorithm was then used to generate an unstructured triangulated mesh. The segmentation step was performed with the heart at end systole. A nonrigid, deformable image registration technique was used to extract the endocardial boundary’s motion (20, 23, 51, 52). Using this approach, a source image (e.g., at end systole) was registered to a target image (e.g., at end diastole) by minimizing a similarity function, and a cubic B-spline transformation was used to deform the control points on the source image during registration. A Laplacian-based smoothing operator weighted by a regularization coefficient was added to the similarity function to ensure that the deformations were smooth and nonintersecting. These computed deformations were then used to morph the initial segmented endocardium boundary, and the process was repeated sequentially for all images throughout the cardiac cycle. Based on the wall motion determined by image registration over the cardiac cycle, the triangulated mesh was deformed accordingly during the CFD simulation as described below (20).
We modeled blood flow in a moving domain as incompressible Newtonian fluid governed by the Navier-Stokes equation written in arbitrary Lagrangian-Eulerian formulation as follows (20):
(Equation 3)
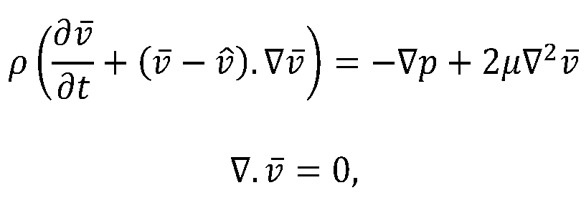
where 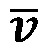 and p are the fluid velocity and pressure, respectively;
and p are the fluid velocity and pressure, respectively; 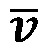 is the endocardial wall velocity; and ρ and μ are the fluid density and viscosity, respectively. We solved the above equations using the open-source svFSI solver from the SimVascular project, a parallelized finite element solver that uses stabilized linear finite elements for spatial discretization, uses the stable and second-order accurate generalized-α method for time integration, and uses a modified Newton-Raphson method for the linearization of the nonlinear Navier-Stokes equation (53). The solver has been thoroughly validated (22, 54) and was earlier used to simulate cardiac hemodynamics in zebrafish embryos (22, 23) and in studies of congenital heart disease in humans (55). The velocities were previously validated with velocities of red blood cell flow obtained via particle imaging velocimetry (22). The computed velocity field was then postprocessed to extract WSS defined as:
is the endocardial wall velocity; and ρ and μ are the fluid density and viscosity, respectively. We solved the above equations using the open-source svFSI solver from the SimVascular project, a parallelized finite element solver that uses stabilized linear finite elements for spatial discretization, uses the stable and second-order accurate generalized-α method for time integration, and uses a modified Newton-Raphson method for the linearization of the nonlinear Navier-Stokes equation (53). The solver has been thoroughly validated (22, 54) and was earlier used to simulate cardiac hemodynamics in zebrafish embryos (22, 23) and in studies of congenital heart disease in humans (55). The velocities were previously validated with velocities of red blood cell flow obtained via particle imaging velocimetry (22). The computed velocity field was then postprocessed to extract WSS defined as:
(Equation 4)
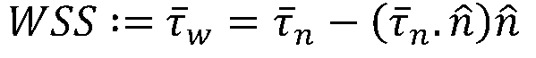
where, 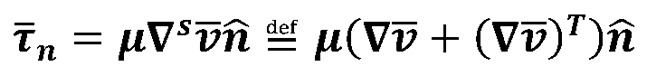 is the stress vector aligned along the normal to the endocardium boundary with a unit normal
is the stress vector aligned along the normal to the endocardium boundary with a unit normal 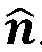 . We then computed WSS-derived metrics, including (a) TAWSS, which is WSS averaged over the entire cardiac cycle; (b) OSI, which indicates the change in the direction of the shear vector during the cardiac cycle; (c) TAWSSGs, which capture the spatial variation of WSS along the endocardial surface; and (d) FWSS, which determines the dynamic range of the WSS magnitude experienced by the endocardial layer during the cardiac cycle. The mathematical expressions for these quantities
. We then computed WSS-derived metrics, including (a) TAWSS, which is WSS averaged over the entire cardiac cycle; (b) OSI, which indicates the change in the direction of the shear vector during the cardiac cycle; (c) TAWSSGs, which capture the spatial variation of WSS along the endocardial surface; and (d) FWSS, which determines the dynamic range of the WSS magnitude experienced by the endocardial layer during the cardiac cycle. The mathematical expressions for these quantities
are given below:
(Equation 5)
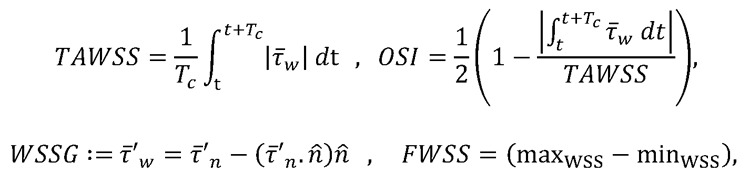
where, 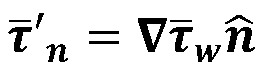 and Tc is the cardiac cycle duration. For this study, we computed the parameters only in the OFT region and ignored the remaining ventricular surface.
and Tc is the cardiac cycle duration. For this study, we computed the parameters only in the OFT region and ignored the remaining ventricular surface.
Localization and quantification of Notch1b activity. The Tg(tp1:GFP) Notch reporter line was used to visualize and quantify endocardial Notch1b signaling. At 3 dpf, embryos exposed to the indicated treatments were randomly selected to image cardiac Notch1b activity using either confocal microscopy (Zeiss) or SPIM. Confocal images of Notch1b activity and the endocardial/endothelial layers were acquired at 2-μm intervals in the z axis and superimposed to visualize the distribution and intensity of Notch1b signaling. SPIM-derived images were reconstructed into 3D images using Amira software, and the mean intensity of the GFP signal was measured in the OFT. To account for variations in GFP signal intensities between samples, each OFT mean intensity was normalized to the mean GFP intensity of the ventricle from the same embryo.
Whole-mount fluorescence immunostaining. At 4 dpf, whole-mount zebrafish embryos were immunostained with mouse monoclonal antibodies against Zn5 and Zn8 (1:10; Developmental Studies Hybridoma Bank), which recognizes the cell surface adhesion molecule DM-GRASP, with Alexa Fluor 488–conjugated anti–mouse IgG antibody (1:200, Abcam) as the secondary antibody. Samples were stained with DAPI (1:3000) for nuclear visualization, and immunostained samples were imaged by confocal microscopy (TCS SP8, Leica Microsystems), using a ×20 objective.
Statistics. All values are expressed as mean ± SD unless otherwise indicated. A comparison of multiple mean values was performed by 1-way ANOVA, and statistical significance among multiple groups was determined using Dunnett’s multiple-comparisons procedure. A P value of less than 0.05 was deemed statistically significant.
Study approval. All experiments with zebrafish were performed in compliance with and with the approval of a UCLA Institutional Animal Care and Use Committee protocol.
Author contributions
JJH, CC, JC, and MIC performed zebrafish breeding, embryo microinjections, and SPIM imaging. VV designed and performed CFD analysis. JJH, KIB, CC, and JC performed confocal imaging and immunostaining experiments. JJH, CC, MIC, J. Lam, SS, and JW performed image analysis and hemodynamic measurement. J. Lee, CCC, and YD designed the SPIM system and 4D synchronization algorithm. JJH wrote the manuscript. JL, YT, LLD, ALM, and TKH designed, supervised, revised, and supported the study.
Supplementary Material
Acknowledgments
The authors would like to thank David Traver and Nathan Lawson for providing the Tg(tp1:GFP) line. This study was supported by grants from the NIH (HL111437, HL129727, HL81753, and GM17-543 to TKH and T32HL007895 to JJH), an American Heart Association Post-Doctoral Fellowship Award (to JJH), and awards from the UCLA Specialty Training and Advanced Research program and Claude D. Pepper Older American Independence Center at UCLA to JJH.
Version 1. 04/11/2019
In-Press Preview
Version 2. 05/16/2019
Electronic publication
Funding Statement
Post-Doctoral Fellowship Award
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(10):e124460. https://doi.org/10.1172/jci.insight.124460.
Contributor Information
Jeffrey J. Hsu, Email: JJHsu@mednet.ucla.edu.
Vijay Vedula, Email: vvedula@stanford.edu.
Kyung In Baek, Email: Qorruddls122@gmail.com.
Cynthia Chen, Email: cynthiachen@ucla.edu.
Junjie Chen, Email: roy.chen0404@gmail.com.
Man In Chou, Email: choumanin@gmail.com.
Jeffrey Lam, Email: jlam343@gmail.com.
Shivani Subhedar, Email: s.subhedar02@gmail.com.
Jennifer Wang, Email: jennifer.wang24@gmail.com.
Yichen Ding, Email: ycding@g.ucla.edu.
Chih-Chiang Chang, Email: Changc4@ucla.edu.
Juhyun Lee, Email: juhyun.lee@uta.edu.
Linda L. Demer, Email: ldemer@mednet.ucla.edu.
Yin Tintut, Email: ytintut@mednet.ucla.edu.
Alison L. Marsden, Email: amarsden@stanford.edu.
Tzung K. Hsiai, Email: thsiai@mednet.ucla.edu.
References
- 1.Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105(5):408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39(12):1890–1900. doi: 10.1016/S0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circ Res. 2004;95(5):459–470. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hove JR, Köster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421(6919):172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 5.Bartman T, et al. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2004;2(5):E129. doi: 10.1371/journal.pbio.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalogirou S, Malissovas N, Moro E, Argenton F, Stainier DY, Beis D. Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc Res. 2014;104(1):49–60. doi: 10.1093/cvr/cvu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heckel E, et al. Oscillatory flow modulates mechanosensitive klf2a expression through trpv4 and trpp2 during heart valve development. Curr Biol. 2015;25(10):1354–1361. doi: 10.1016/j.cub.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 8.Yalcin HC, Amindari A, Butcher JT, Althani A, Yacoub M. Heart function and hemodynamic analysis for zebrafish embryos. Dev Dyn. 2017;246(11):868–880. doi: 10.1002/dvdy.24497. [DOI] [PubMed] [Google Scholar]
- 9.Jamison RA, Samarage CR, Bryson-Richardson RJ, Fouras A. In vivo wall shear measurements within the developing zebrafish heart. PLoS One. 2013;8(10):e75722. doi: 10.1371/journal.pone.0075722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9(1):49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 11.Timmerman LA, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noseda M, et al. Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circ Res. 2004;94(7):910–917. doi: 10.1161/01.RES.0000124300.76171.C9. [DOI] [PubMed] [Google Scholar]
- 13.D’Amato G, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18(1):7–20. doi: 10.1038/ncb3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 15.Luxán G, D’Amato G, MacGrogan D, de la Pompa JL. Endocardial Notch signaling in cardiac development and disease. Circ Res. 2016;118(1):e1–e18. doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, et al. Notch-Tnf signalling is required for development and homeostasis of arterial valves. Eur Heart J. 2017;38(9):675–686. doi: 10.1093/eurheartj/ehv520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, et al. 4-Dimensional light-sheet microscopy to elucidate shear stress modulation of cardiac trabeculation. J Clin Invest. 2016;126(5):1679–1690. doi: 10.1172/JCI83496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahnsen ED, Trindade A, Zaun HC, Lehoux S, Duarte A, Jones EA. Notch1 is pan-endothelial at the onset of flow and regulated by flow. PLoS One. 2015;10(4):e0122622. doi: 10.1371/journal.pone.0122622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samsa LA, Givens C, Tzima E, Stainier DY, Qian L, Liu J. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development. 2015;142(23):4080–4091. doi: 10.1242/dev.125724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, et al. Spatial and temporal variations in hemodynamic forces initiate cardiac trabeculation. JCI Insight. 2018;3(13):96672. doi: 10.1172/jci.insight.96672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Y, et al. Integrating light-sheet imaging with virtual reality to recapitulate developmental cardiac mechanics. JCI Insight. 2017;2(22):97180. doi: 10.1172/jci.insight.97180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, et al. Moving domain computational fluid dynamics to interface with an embryonic model of cardiac morphogenesis. PLoS One. 2013;8(8):e72924. doi: 10.1371/journal.pone.0072924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vedula V, Lee J, Xu H, Kuo CJ, Hsiai TK, Marsden AL. A method to quantify mechanobiologic forces during zebrafish cardiac development using 4-D light sheet imaging and computational modeling. PLoS Comput Biol. 2017;13(10):e1005828. doi: 10.1371/journal.pcbi.1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132(18):4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 25.Steed E, Boselli F, Vermot J. Hemodynamics driven cardiac valve morphogenesis. Biochim Biophys Acta. 2016;1863(7 pt B):1760–1766. doi: 10.1016/j.bbamcr.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Steed E, Faggianelli N, Roth S, Ramspacher C, Concordet JP, Vermot J. klf2a couples mechanotransduction and zebrafish valve morphogenesis through fibronectin synthesis. Nat Commun. 2016;7:11646. doi: 10.1038/ncomms11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestel J, Ramadass R, Gauvrit S, Helker C, Herzog W, Stainier DY. Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development. 2016;143(12):2217–2227. doi: 10.1242/dev.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermot J, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7(11):e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stankunas K, Ma GK, Kuhnert FJ, Kuo CJ, Chang CP. VEGF signaling has distinct spatiotemporal roles during heart valve development. Dev Biol. 2010;347(2):325–336. doi: 10.1016/j.ydbio.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, et al. Hemodynamics and ventricular function in a zebrafish model of injury and repair. Zebrafish. 2014;11(5):447–454. doi: 10.1089/zeb.2014.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koenig SN, Bosse K, Majumdar U, Bonachea EM, Radtke F, Garg V. Endothelial Notch1 is required for proper development of the semilunar valves and cardiac outflow tract. J Am Heart Assoc. 2016;5(4):e003075. doi: 10.1161/JAHA.115.003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacGrogan D, et al. Sequential ligand-dependent notch signaling activation regulates valve primordium formation and morphogenesis. Circ Res. 2016;118(10):1480–1497. doi: 10.1161/CIRCRESAHA.115.308077. [DOI] [PubMed] [Google Scholar]
- 33.Midgett M, López CS, David L, Maloyan A, Rugonyi S. Increased hemodynamic load in early embryonic stages alters endocardial to mesenchymal transition. Front Physiol. 2017;8:56. doi: 10.3389/fphys.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahler GJ, Frendl CM, Cao Q, Butcher JT. Effects of shear stress pattern and magnitude on mesenchymal transformation and invasion of aortic valve endothelial cells. Biotechnol Bioeng. 2014;111(11):2326–2337. doi: 10.1002/bit.25291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sewell-Loftin MK, et al. Myocardial contraction and hyaluronic acid mechanotransduction in epithelial-to-mesenchymal transformation of endocardial cells. Biomaterials. 2014;35(9):2809–2815. doi: 10.1016/j.biomaterials.2013.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ten Dijke P, Egorova AD, Goumans MJ, Poelmann RE, Hierck BP. TGF-β signaling in endothelial-to-mesenchymal transition: the role of shear stress and primary cilia. Sci Signal. 2012;5(212):pt2. doi: 10.1126/scisignal.2002722. [DOI] [PubMed] [Google Scholar]
- 37.Egorova AD, et al. Lack of primary cilia primes shear-induced endothelial-to-mesenchymal transition. Circ Res. 2011;108(9):1093–1101. doi: 10.1161/CIRCRESAHA.110.231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gould RA, et al. Cyclic mechanical loading is essential for ac1-mediated elongation and remodeling of the embryonic mitral valve. Curr Biol. 2016;26(1):27–37. doi: 10.1016/j.cub.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126(10):898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stainier DY, Weinstein BM, Detrich HW, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121(10):3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 41.et al. ZebraBeat: a flexible platform for the analysis of the cardiac rate in zebrafish embryos. Sci Rep. 2014;4:4898 [Google Scholar]
- 42.Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8(1):109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6(5):e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rottbauer W, et al. VEGF-PLCγ1 pathway controls cardiac contractility in the embryonic heart. Genes Dev. 2005;19(13):1624–1634. doi: 10.1101/gad.1319405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paffett-Lugassy N, et al. Functional conservation of erythropoietin signaling in zebrafish. Blood. 2007;110(7):2718–2726. doi: 10.1182/blood-2006-04-016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baek KI, et al. Ultrafine particle exposure reveals the importance of FOXO1/Notch activation complex for vascular regeneration. Antioxid Redox Signal. 2018;28(13):1209–1223. doi: 10.1089/ars.2017.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fei P, et al. Cardiac light-sheet fluorescent microscopy for multi-scale and rapid imaging of architecture and function. Sci Rep. 2016;6:22489. doi: 10.1038/srep22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liebling M, Forouhar AS, Gharib M, Fraser SE, Dickinson ME. Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences. J Biomed Opt. 2005;10(5):054001. doi: 10.1117/1.2061567. [DOI] [PubMed] [Google Scholar]
- 49.Mickoleit M, et al. High-resolution reconstruction of the beating zebrafish heart. Nat Methods. 2014;11(9):919–922. doi: 10.1038/nmeth.3037. [DOI] [PubMed] [Google Scholar]
- 50. Dobrzynski C, Frey P. Anisotropic delaunay mesh adaptation for unsteady simulations. In: Garimella RV, ed. Proceedings of the 17th International Meshing Roundtable. Berlin, Germany: Springer Berlin Heidelberg; 2008:177–194. [Google Scholar]
- 51. Myronenko A. MIRT – Medical Image Registration Toolbox for Matlab. MIRT website. https://sites.google.com/site/myronenko/research/mirt Accessed April 18, 2019.
- 52. Myronenko A. Non-rigid image registration regularization, algorithms and applications [dissertation]. Portland, Oregon, USA: Oregon Health and Science University; 2010. [Google Scholar]
- 53.Esmaily-Moghadam M, Bazilevs Y, Marsden AL. A bi-partitioned iterative algorithm for solving linear systems arising from incompressible flow problems. Comput Methods Appl Mech Eng. 2015;286:40–62. doi: 10.1016/j.cma.2014.11.033. [DOI] [Google Scholar]
- 54.Arbia G, et al. Numerical blood flow simulation in surgical corrections: what do we need for an accurate analysis? J Surg Res. 2014;186(1):44–55. doi: 10.1016/j.jss.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 55.Marsden AL, Esmaily-Moghadam M. Multiscale modeling of cardiovascular flows for clinical decision support. Appl Mech Rev. 2015;67(3):030804. doi: 10.1115/1.4029909. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.