Abstract
The bone marrow microenvironment contributes to the regulation of hematopoietic stem cell (HSC) function, though its role in age-associated lineage skewing is poorly understood. Here we show that dysfunction of aged marrow macrophages (Mφs) directs HSC platelet bias. Mφs from the marrow of aged mice and humans exhibited an activated phenotype, with increased expression of inflammatory signals. Aged marrow Mφs also displayed decreased phagocytic function. Senescent neutrophils, typically cleared by marrow Mφs, were markedly increased in aged mice, consistent with functional defects in Mφ phagocytosis and efferocytosis. In aged mice, interleukin-1B (IL-1B) was elevated in the bone marrow, and caspase-1 activity, which can process pro–IL-1B, was increased in marrow Mφs and neutrophils. Mechanistically, IL-1B signaling was necessary and sufficient to induce a platelet bias in HSCs. In young mice, depletion of phagocytic cell populations or loss of the efferocytic receptor Axl expanded platelet-biased HSCs. Our data support a model wherein increased inflammatory signals and decreased phagocytic function of aged marrow Mφs induce the acquisition of platelet bias in aged HSCs. This work highlights the instructive role of Mφs and IL-1B in the age-associated lineage skewing of HSCs, and reveals the therapeutic potential of their manipulation as antigeronic targets.
Keywords: Aging, Hematology
Keywords: Bone marrow, Hematopoietic stem cells
Defective efferocytic capacity of macrophages within the aged marrow microenvironment promotes the expansion of platelet-biased hematopoietic stem cells through IL-1β signaling.
Introduction
Dysfunction of the human hematopoietic system with age includes diminished immune response, marrow failure, and clonal selection (1). Aging is also associated with a general increase in tissue inflammation that remains largely unexplained (2). The mechanisms driving these characteristics of aged hematopoiesis have, to date, primarily been attributed to intrinsic hematopoietic stem cell (HSC) changes (3). With age, in both humans and mice, the phenotypic long-term HSC (LT-HSC) pool is expanded, and globally LT-HSCs differentiate preferentially toward the myeloid lineage (4–6). Multipotent HSCs with platelet bias were recently identified by a number of investigators describing their increased expression of von Willebrand factor (vWF) (7) and of αIIb integrin (CD41) (8, 9). Recent data demonstrate that aged murine HSCs also have increased cell surface expression of CD41 and vWF (8, 10). Notably, human aged HSCs display platelet (or megakaryocytic) bias (6, 11), suggesting that insights in mechanisms determining murine HSC platelet bias will not only improve our understanding of diseases attributed to the aging hematopoietic system, but also provide novel therapeutic approaches to hematopoietic dysfunction associated with advanced age.
Since the bone marrow microenvironment (BMME) critically regulates HSCs, whether it be considered instructive or enabling distinct HSC fates (12), unique characteristics of the aged BMME could contribute to HSC changes associated with age. In fact, in the Drosophila gonad, extrinsic signals from the niche contribute to stem cell aging (13), and mathematical models have suggested that non–cell-autonomous changes could drive this process in mammalian HSCs (14). While data have suggested that aged endothelial and mesenchymal BMME populations are abnormal and may participate in HSC aging (15–17), microenvironmental signals governing the megakaryocytic bias of aged HSCs remain unclear. Thus, we hypothesized that defects in critical BMME populations caused by age could lead to the expansion of platelet-biased HSCs.
We found that macrophages (Mφs) within the aged BMME could impose the megakaryocytic bias characteristic of aging in HSCs. Aged human and murine marrow Mφs had distinct transcriptional profiles compared with young Mφs, including an increased inflammatory activation signature. We identified increased interleukin-1B (IL-1B) mRNA in aged marrow Mφs and elevated caspase-1 activity in Mφs and neutrophils from aged bone marrow. Moreover, IL-1B signaling was necessary and sufficient to induce HSC bias and drive young hematopoietic stem and progenitor cells (HSPCs) to adopt an aged phenotype. While investigating the cause of this increase, we made the novel observation that aged marrow Mφs had a defect in efferocytosis — their ability to clear apoptotic cells. Clearance of apoptotic cells is a critical function of Mφs that prevents necrosis of dead cells and associated local inflammation (18, 19) and also triggers antiinflammatory responses in phagocytes (20). In young mice, removal of phagocytic cells or genetic loss of the efferocytic receptor Axl increased HSCs with megakaryocytic bias, suggesting that the efferocytic defect in aged marrow Mφs leads to the increase in IL-1B activation and signaling. Together these data define a novel mechanism within the aged BMME that enables a specific HSC fate.
Results
Megakaryocytic HSC bias characteristic of aged HSCs is imparted by aged marrow macrophages.
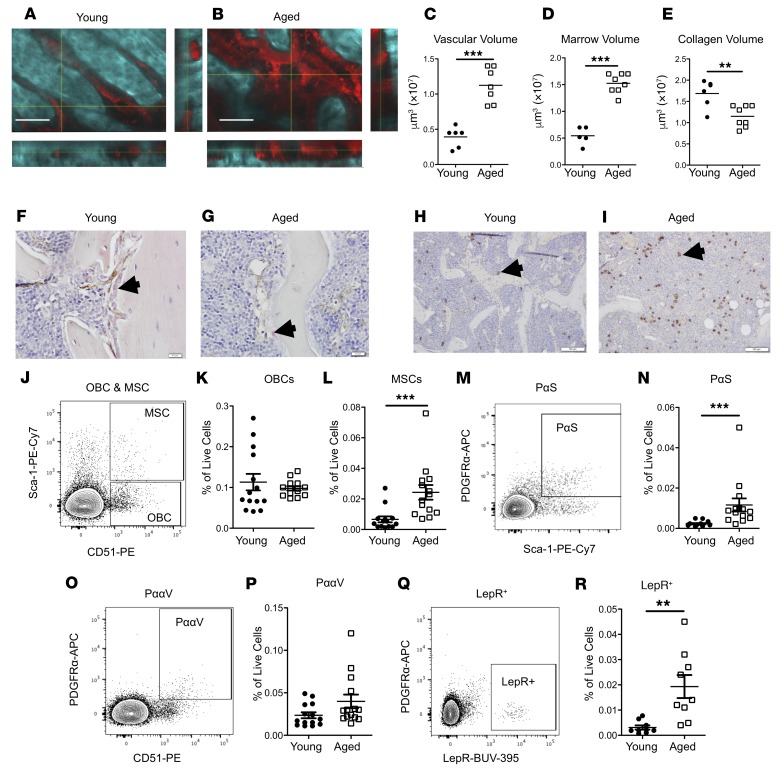
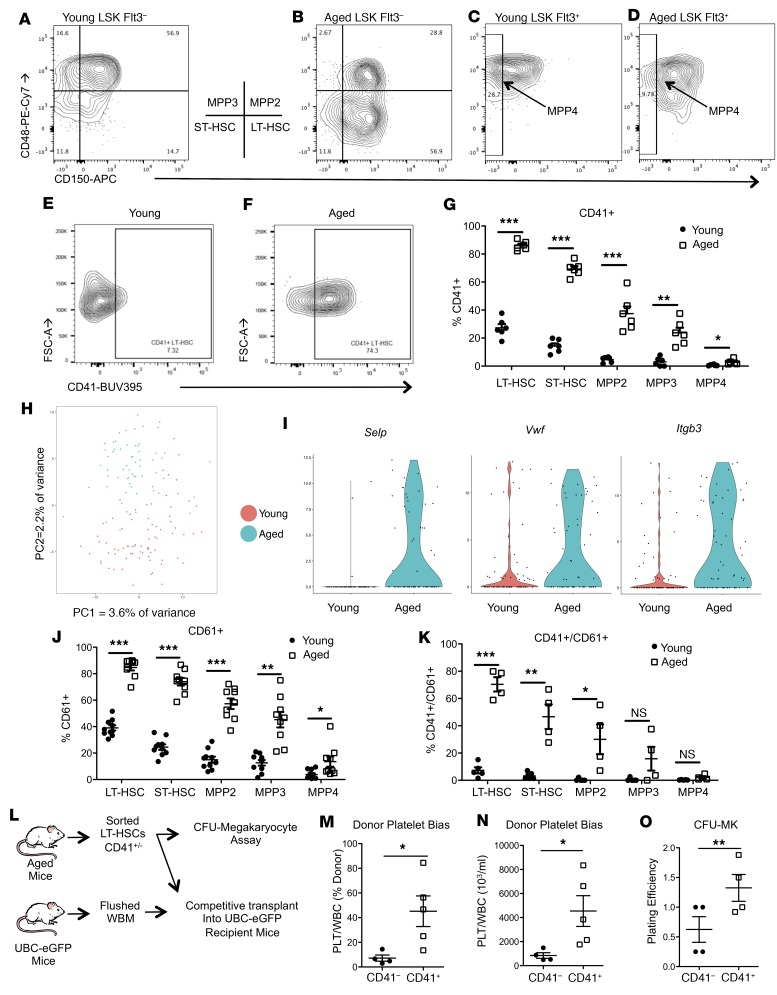
Intravital multiphoton microscopy allows for the visualization of the BMME while minimizing artifactual distortion of the marrow (21). In vivo aged (20–24 months old) compared with young (6–8 weeks old) C57BL/6J mice demonstrated global remodeling of the bone marrow (Figure 1, A and B), with decreased bone matrix, while bone marrow cavity and vascular volumes were increased (Figure 1, C–E). In aged mice, mature osteoblasts were decreased (Figure 1, F and G), while mesenchymal cell populations in the bone marrow were increased (Figure 1, H and I). Flow cytometric quantification for osteoblastic cells, multipotent stromal cells (MSCs), and MSC subsets previously demonstrated to be critical for HSC support, such as PDGFRa+SCA1+ (PαS) cells, PDGFRa/CD51 (PααV) cells, and leptin receptor–positive (LepR+) cells (22–25), confirmed the increase in MSC subpopulations (Figure 1, J–R), also recently reported by others (17). However, though vascular volume was increased (Figure 1C), an increase in bone marrow endothelial cells was not observed (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124213DS1). Therefore, the BMME is significantly altered by aging and could contribute to age-induced changes in hematopoiesis. In aged mice, we confirmed that the HSC pool was expanded (Supplemental Figure 2, A–E) and displayed myeloid and megakaryocytic skewing (Supplemental Figure 2, F–I), as was also reported in humans (4–6). We found that the cell surface expression of CD41 was increased not only in aged short-term HSCs and LT-HSCs, as previously reported (8, 10), but also on multipotent progenitor populations in aged mice (Figure 2, A–G). The increase in megakaryocytic bias of aged HSCs was also supported by an increase in megakaryocytic progenitors (Supplemental Figure 2, H and I). Consistent with these findings, platelet numbers were also significantly increased in the peripheral blood of aged mice, while red blood cells and hemoglobin concentrations were decreased, suggesting that the megakaryocytic skewing was not accompanied by an erythroid bias (Supplemental Figure 3). To identify additional markers of megakaryocytic bias in aged HSCs, single-cell RNA sequencing of phenotypic LT-HSCs (Lineage–SCA1+CKIT+CD150+CD48–Flt3–) from young and aged mice was performed. Principal component analysis demonstrated segregation of the transcriptional profiles of young compared with aged LT-HSCs (Figure 2H). Greater numbers of aged LT-HSCs displayed high expression of several genes found to be expressed in megakaryocyte-biased (Mk-biased) young LT-HSCs (7, 26), including Selp, Vwf, and Itgb3 (Figure 2I). The latter, encoding the protein CD61/integrin β3 , was also identified in recent global and single-cell transcriptional analysis of LT-HSCs from young and aged mice (4, 27, 28). Based on these transcriptional profiles, we next identified expansion of CD61+ cells within the Lineage–SCA1+CKIT+ (LSK) pool of aged mice (Figure 2J). A similar proportion of aged HSCs were also found to be positive for both CD41 and CD61 (Figure 2K). Therefore, cell surface expression of CD41 and CD61 is strongly associated with the aged megakaryocytic HSC bias. To determine whether CD41+ cell surface expression on LT-HSCs identifies LT-HSCs with megakaryocytic skewing, we sorted CD41+ or CD41– LT-HSCs from the marrow of aged mice, and transplanted them in competition with marrow from mice expressing enhanced GFP under the direction of the human ubiquitin C promoter (UBC-GFP mice; ref. 29), in order to quantify LT-HSC donor (GFP+) contribution to platelets. Recipient mice were also UBC-GFP (Figure 2L). Percentage donor and total platelet contribution was significantly increased in mice transplanted with CD41+ LT-HSCs (Figure 2, M and N). Consistent with increased megakaryocytic skewing, CD41+ LT-HSCs also demonstrated a statistically significant increase in generation of megakaryocytic CFUs in vitro (Figure 2O). Therefore, expression of CD41 by HSCs identifies a population of HSCs with increased megakaryocytic skewing.
Figure 1. Aged mice have remodeling of their bone marrow and expansion of MSC populations.
(A and B) Representative images of 2-photon intravital microscopy of calvaria; second harmonic generation (collagen; blue) and vasculature (70-kDa dextran; red). Scale bars: 100 μm. (C–E) Vascular (C), marrow (D), and collagen (E) volume (n = 5–7 mice per group). (F and G) Immunohistochemistry of osteocalcin (pink) and endomucin (brown). Scale bars: 10 μm. Black arrows indicate osteocalcin-positive osteoblasts. (H and I) Immunohistochemistry for LepR+ cells (brown). Scale bars: 100 μm. Black arrows indicate LepR+ perivascular cells. (J–R) Representative flow cytometry plots (J, M, O, and Q) and analysis quantification (K, L, N, P, and R) of marrow osteoblastic cells (OBCs: CD45–TER119–CD31–CD51+SCA1– cells, J and K), marrow multipotent stromal cell populations (MSCs: CD45–TER119–CD31–CD51+SCA1+ cells, J and L), PαS (CD45–TER119–CD31–CD140a+SCA1+ cells, M and N), PααV (CD45–TER119–CD31–CD140a+CD51+ cells, O and P) (n = 13–14 mice per group), and leptin receptor–positive (LepR+) (CD45–TER119–CD31–LepR+ cells, Q and R) (n = 9 mice per group). Each symbol represents an individual mouse; data represent mean ± SEM. P values, 2-tailed Student’s t test: **P < 0.01, ***P < 0.001.
Figure 2. Aged mice have expanded megakaryocyte-biased HSCs.
(A–G) Representative flow cytometry of CD41 expression and its quantification across LSK subsets in young and aged mice (n = 6 mice per group). (H) Principal component analysis (PCA) plot of single LT-HSCs from young and aged mice. Variance of each PCA is shown. (I) Violin plots of Selp (ANOVA P = 4.94 × 10–7), Vwf (P = 0.003051854), and Itgb3 (P = 6.38 × 10–5) expression from young and aged LT-HSCs (n = 76 young HSCs and 63 aged HSCs). (J and K) Quantification of CD61 (n = 9 mice per group) and of CD41/CD61 (n = 5 mice per group) expression across LSK subsets in young and aged mice. (L) Schematic representation of experimental design for analysis of megakaryocytic skewing potential of CD41+ LT-HSCs. (M and N) Donor platelet bias of sorted CD41+ (n = 5 recipients) versus CD41– (n = 4 recipients) LT-HSCs as percent donor and total number. (O) Quantification of in vitro megakaryocytic CFUs (CFU-MK) per 100 sorted CD41+ versus CD41– LT-HSCs (n = 4 per group). (G, J, K, and M–O). Each symbol represents an individual mouse; data represent mean ± SEM. P values, 2-tailed Student’s t test except as noted: *P < 0.05, **P < 0.01, ***P < 0.001.
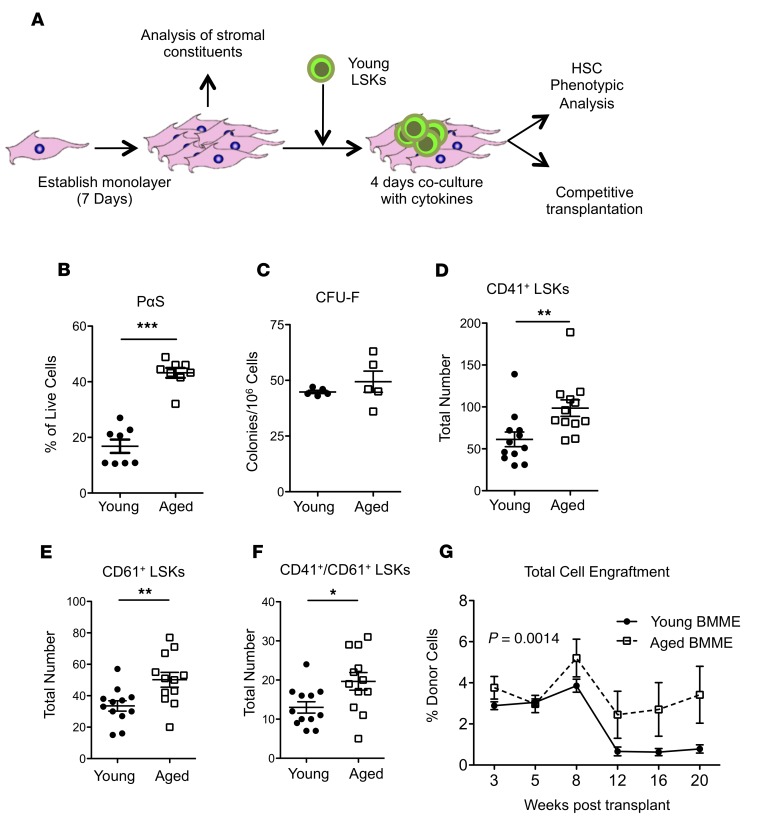
The age-dependent in vivo expansion of HSC-supportive MSC subsets was recapitulated ex vivo in cultures of aged BMME cells (Figure 3, A and B, and Supplemental Figure 4) that did not show an increase in MSC function (Figure 3C). Therefore, the ability of aged BMMEs to induce the megakaryocytic HSC bias was assessed by coculturing of BMME cells with HSC-enriched LSKs from young GFP+ mice (Figure 3A). Aged BMME cells significantly expanded the pool of phenotypic megakaryocytic-biased HSCs (Figure 3, D–F). The expanded LSK population included functional HSCs, evidenced by a significant increase in their engraftment from competitive reconstitution assays performed after coculture (Figure 3G), consistent with the known increase in self-renewal in megakaryocytic-biased HSCs (7, 8, 30). This coculture, however, did not induce myeloid bias (Supplemental Figure 5), also consistent with functional studies of megakaryocytic-biased HSCs (7, 26). This is in contrast with a previously reported decrease in engraftment capacity and myeloid bias of HSPCs cocultured with aged endothelial cells (31), suggesting that distinct cell components of the BMME differentially impact HSPCs in aged mice. These data support the hypothesis that signals from aged BMME cells can contribute to the megakaryocytic bias of aged HSCs.
Figure 3. Dysfunction in aged MSCs and regulation of HSCs by aged microenvironments.
(A) Schematic for in vitro coculture of HSCs and stromal cells. (B) Quantification of PαS cells from in vitro cultures derived from young and aged mice (n = 8 wells per group). (C) Quantification of fibroblastic CFUs (CFU-F) from young and aged mice (n = 5 young and aged mice). (D–F) Quantification of CD41 (D), CD61 (E), and CD41/CD61 (F) expression in LSKs grown on young or aged BMME cells (n = 12 young and aged per well). (G) Engraftment of HSCs grown on either young or aged BMME cells (n = 5 recipients per group). (B–F) Each symbol represents an individual mouse; data represent mean ± SEM. P values: (B–F) 2-tailed Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; (G) P = 0.0014 results from a 2-way ANOVA between young and aged longitudinally, **P < 0.01 between young and aged longitudinally.
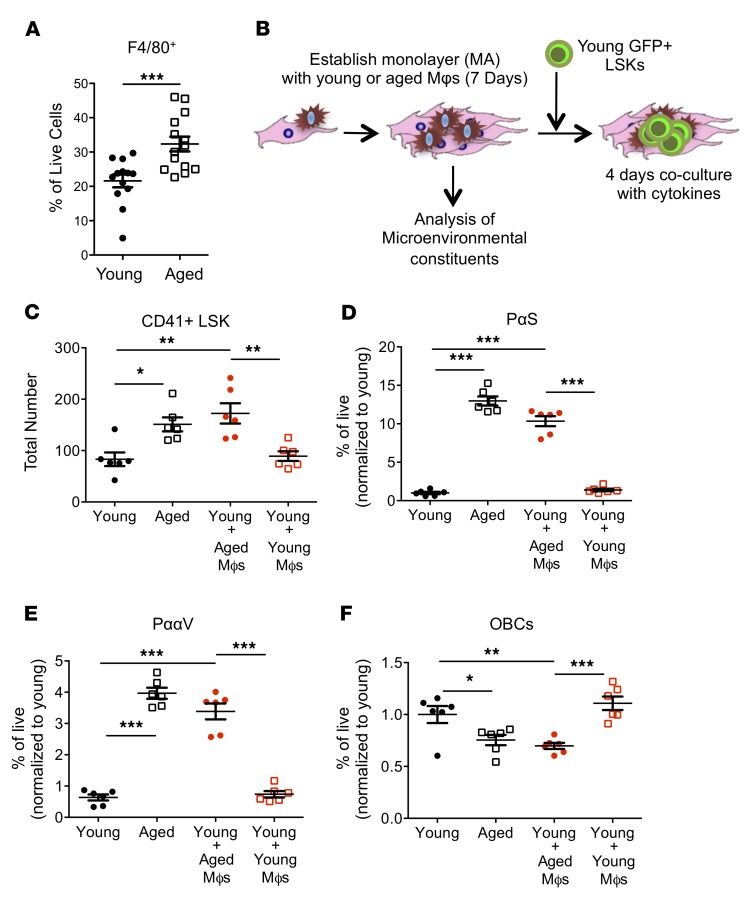
We next used a reductionist approach to identify the source within the aged BMME of signals responsible for HSC bias. Since in ex vivo BMME cells there was a large contribution of Mφs (Figure 4A), a population that was also implicated in HSC regulation (32–34), we investigated whether Mφs were responsible for the megakaryocytic bias imparted by the aged BMME. Sorted marrow Mφs from young or aged mice (Supplemental Figure 6) were added to young BMME cells (Figure 4B). The addition of aged marrow Mφs to young BMME cells induced a significant increase in CD41+ LSKs, similar to that produced by the total aged BMME compartment (Figure 4C). However, adding young Mφs did not increase CD41+ LSKs, suggesting that qualitative rather than quantitative changes in aged BMME Mφs drive the induction of megakaryocytic bias in HSCs, and that aged Mφs are sufficient to induce an aged phenotype even in the presence of young Mφs. The addition of aged BMME Mφs expanded HSC-supportive MSCs (Figure 4, D and E), consistent with the BMME changes observed in vivo (Figure 1, M–P) and ex vivo (Figure 3A), suggesting that aged marrow Mφs impact not only HSCs, but also mesenchymal populations known to regulate and support HSCs. Notably, more mature osteoblastic populations were decreased (Figure 4F) with the addition of Mφs from the aged BMME. These results demonstrate that aged BMME Mφs can induce a megakaryocytic bias in HSCs and an aged phenotype in young mesenchymal BMME cells.
Figure 4. Aged macrophages recapitulate age-related changes to HSCs and the BMME in vitro.
(A) F4/80+ cells in BMME cultures derived from young and aged marrow (n = 14 wells per group). (B) Schematic for addition of sorted Mφs to BMME coculture. (C) Expression of CD41 on GFP+ young LSKs after coculture with BMME cells with or without young or aged Mφs (n = 6 wells per group). (D–F) Quantification of PαS cells (D), PααV cells (E), and OBCs (F) as defined in Figure 1 (n = 6 wells per group). Each symbol represents an individual mouse; data represent mean ± SEM. P values: (A) 2-tailed Student’s t test, ***P < 0.001; (C–F) 1-way ANOVA with Tukey’s multiple-comparisons post-test, *P < 0.05, **P < 0.01, ***P < 0.001.
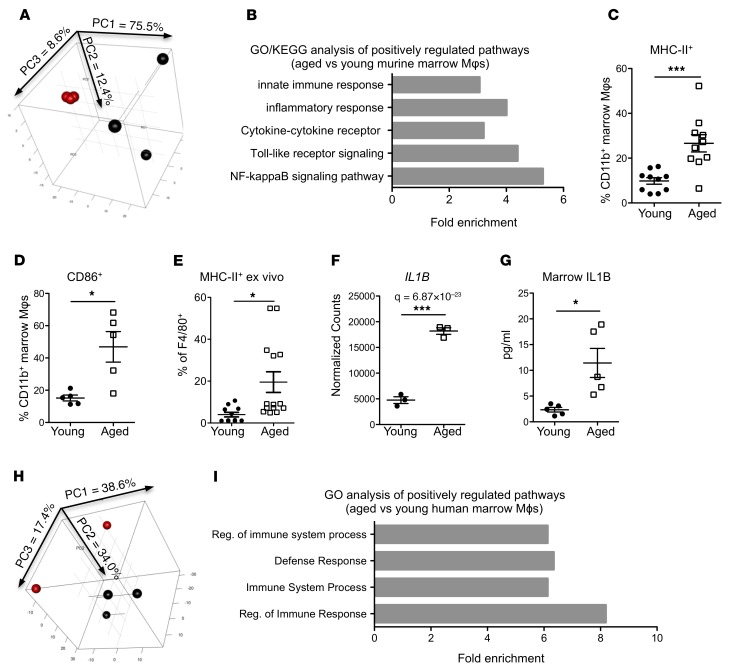
Aged marrow macrophages from both mice and humans have increased inflammatory activation.
To identify signals from aged BMME Mφs responsible for inducing the megakaryocytic bias in HSCs, we queried the transcriptional profiles of BMME Mφs from young and aged mice (Figure 5, A and B). Gene Ontology (GO) and KEGG Pathway analyses identified an increase in the inflammatory response in aged murine Mφs (Figure 5B), which we confirmed by an increase in phenotypic expression of MHC-II (Figure 5C) and of CD86 (Figure 5D) in aged BMME Mφs in vivo. This increase in inflammatory activation was also found in our ex vivo system (Figure 5E). Among the proinflammatory signals identified in aged murine Mφs, Il1B, previously reported to impact HSC myeloid bias (35), was upregulated in aged BMME Mφs from mice (Figure 5F). We also measured increased IL-1B in the marrow of aged mice compared with young mice (Figure 5G). In spite of significant divergence in the human samples (Figure 5H), the increases in immune activation and inflammatory signals were conserved in aged BMME Mφs (Supplemental Figure 7) from human marrow, as shown by gene enrichment analysis (Figure 5I). Reanalysis of published human microarray data identified transcriptional upregulation of the IL-1B receptor (Il1R1) in HSCs from elderly humans compared with young (GSE32719: log2 6.13 vs. 4.83, ANOVA P = 0.0029) (6). Therefore, IL-1B and activation of the IL1R1 are likely be relevant to both murine and human aged HSCs.
Figure 5. Aged marrow macrophages have a proinflammatory phenotype and elevated IL-1B expression.
(A) PCA plot of young (red) and aged (black) marrow Mφs isolated as shown in Supplemental Figure 3 (n = 3 mice per group). Variance of each PCA is shown on the axes. (B) Upregulated GO and KEGG categories in aged Mφs compared with young (n = 3 mice per group). See also Supplemental Tables 3 and 4 for details. (C and D) Expression of MHC-II (C) and CD86 (D) on Mφs from young and aged mice (n = 5–10 mice per group). (E) Expression of MHC-II in Mφs from BMME in vitro cultures (n = 11–14 wells per group). (F) Expression of Il1b in young and aged Mφs (n = 3 mice per group). (G) Quantification by ELISA of IL-1B protein in marrow of young and aged mice (n = 5 per group). (H) PCA plot of young (red, n = 2) and aged (black, n = 3) human marrow Mφs isolated as shown in Supplemental Figure 4. Variance of each PCA is shown on the axes. (I) Upregulated GO processes in aged human Mφs compared with young (n = 2–3 human marrow per group). Each symbol represents an individual mouse; data represent mean ± SEM. P values, 2-tailed Student’s t test: *P < 0.05, ***P < 0.001.
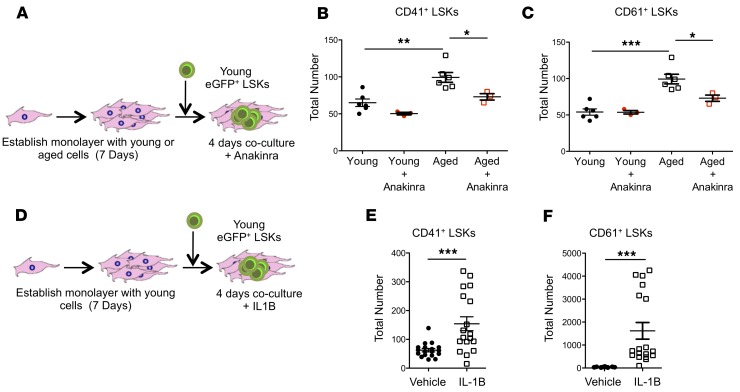
IL-1B signaling is necessary and sufficient to induce HSC bias imposed by the aged BMME.
We next assessed whether IL1R1-dependent signals are necessary and sufficient to mediate induction of platelet bias in HSCs by the BMME. The addition of the IL1R1 antagonist anakinra to aged BMME cells blocked the increase in CD41+ (Figure 6, A and B) and CD61+ (Figure 6C) young LSKs, thus mitigating the effect of the aged BMME on young HSCs. Addition of IL-1B to young BMME cells ex vivo could increase numbers of CD41+ and CD61+ LSKs (Figure 6, D–F). These data demonstrate that IL1R1-mediated signals are required to induce an aged phenotype consistent with megakaryocytic bias in HSCs, and suggest that the microenvironmental signals contributing to HSC aging could be attenuated. Moreover, IL-1B is sufficient to initiate megakaryocytic bias in young HSCs.
Figure 6. Exposure to IL-1B increases megakaryocytic HSCs ex vivo.
(A) Schematic of coculture LSKs and young or aged stromal cells with anakinra (rhIL1RA). (B and C) Expression of CD41 (B) and CD61 (C) on LSKs grown in vitro with young or aged BMME cells with or without anakinra (n = 3–6 wells per group). (D) Schematic of coculture LSKs and young or aged stromal cells with recombinant IL-1B. (E and F) Expression of CD41 (E) and CD61 (F) in LSKs grown in vitro in young BMME cultures with IL-1β (n = 18 wells per group). Each symbol represents an individual mouse; data represent mean ± SEM. P values: (B and C) 1-way ANOVA with Tukey’s multiple-comparisons post-test, *P < 0.05, **P < 0.01, ***P < 0.001; (E and F) 2-tailed Student’s t test, ***P < 0.001.
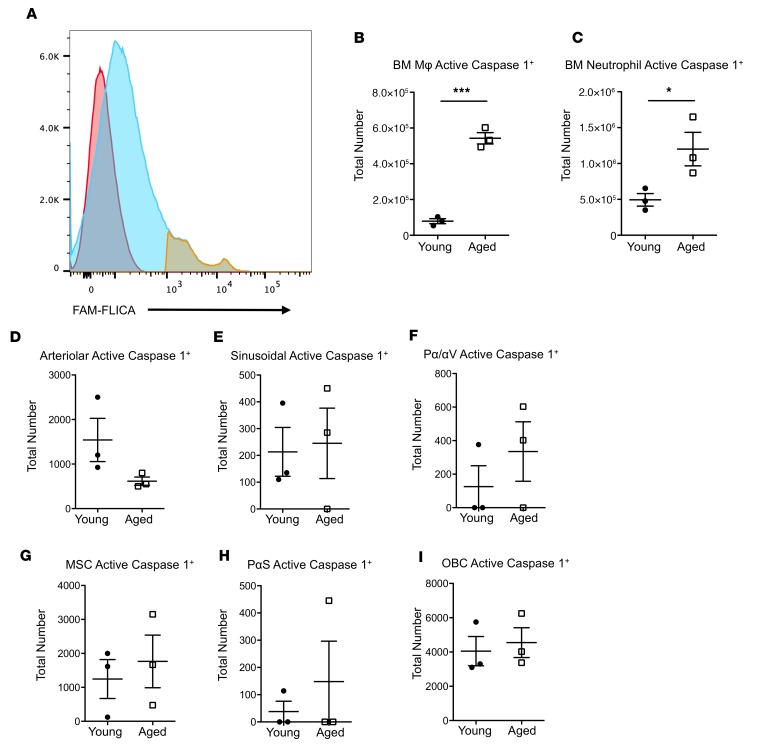
Potential sources of IL-1B in the aged BMME.
To elucidate potential sources of mature IL-1B in the marrow, we quantified caspase-1–active cells (Figure 7A), since mature IL-1B is generated by caspase-1–dependent cleavage of pro–IL-1B (36). We found a significant increase in the number of neutrophils and Mφs from aged mice with active caspase-1 (Figure 7, B and C), indicating that both populations represent potential sources of IL-1B in the aged BMME. There were no significant changes in key HSC-supportive BMME populations, such as mesenchymal, osteoblastic, and endothelial cell populations, with respect to caspase-1 activation in aged mice (Figure 7, D–I), suggesting that these BMME populations are not likely sources of the increased IL-1B protein in the marrow of aged mice.
Figure 7. Increased caspase-1 activity in marrow macrophages and neutrophils but not in other BMME populations in aged mice.
(A) Representative flow cytometry of bone marrow cells stained with FAM-FLICA (active caspase-1) (blue) and FAM-FLICA+ gate (yellow) overlaid onto a FAM-FLICA fluorescence minus one (FMO) control (red). (B and C) Number of marrow Mφs and neutrophils with caspase-1 activity from young and aged animals (n = 3 mice per group). (D–I) Number of microenvironmental cells with caspase-1 activity from young and aged animals. Populations quantified include arteriolar endothelial cells (CD45–TER119–CD31+SCA1+) (D), sinusoidal endothelial cells (CD45–TER119–CD31+SCA1+) (E), PααV cells (CD45–TER119–CD31–CD140a+CD51+) (F), MSCs (CD45–TER119–CD31–CD51+SCA1+) (G), PαS cells (CD45–TER119–CD31–CD140a+SCA1+) (H), and OBCs (CD45–TER119–CD31–CD51+SCA1–) (I) (n = 3 mice per group). Each symbol represents an individual mouse; data represent mean ± SEM. P values, 2-tailed Student’s t test: *P < 0.05, ***P < 0.001.
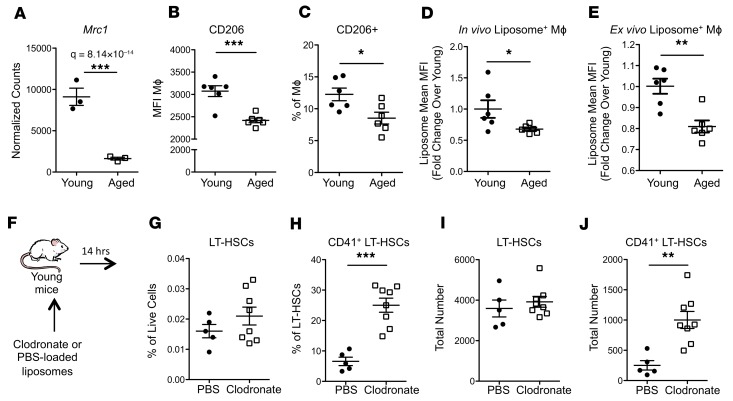
Aged BMME macrophages display defective phagocytosis.
The transcriptome of aged marrow Mφs from mice and humans showed an increase in inflammatory signals (Figure 5). Since it was recently reported that phagocytosis induces an antiinflammatory program in Mφs (37), we hypothesized that aged marrow Mφs may be defective in their ability to phagocytose. Consistent with this hypothesis, aged marrow Mφs exhibited decreased expression of the key scavenger receptor Mrc1/CD206 (37) (Figure 8A) consistent with reduced cell surface expression of this receptor (Figure 8B), and an overall decrease in the pool of CD206+ Mφs in the marrow of aged mice (Figure 8C). This phenotypic change correlated with a functional disruption of phagocytosis by aged BMME Mφs both in vivo (Figure 8D) and ex vivo (Figure 8E) as measured by the capacity of Mφs to phagocytose fluorescently labeled liposomes.
Figure 8. Aged marrow macrophages have defective phagocytosis and loss of macrophages is sufficient to drive premature megakaryocytic skewing of HSCs in vivo.
(A) Expression of scavenger receptor Mrc1 in young and aged marrow Mφs (n = 3 mice per group). (B and C) Quantification of cell surface expression of MRC/CD206 by mean fluorescence intensity (MFI) (B) and percentage of marrow Mφs (C) in marrow from young and aged mice (n = 6 mice per group). (D) Quantification of liposome uptake by marrow Mφs (n = 6 wells per group). (E) Quantification of liposome uptake by Mφs in BMME cultures (n = 6 wells per group). (F) Schematic of clodronate liposome administration and bone marrow analysis. (G–J) LT-HSC frequency (G), CD41 expression (H), total number (I), and total CD41+ (J) 14 hours after clodronate treatment (n = 5–8 mice per group). Each symbol represents an individual mouse; data represent mean ± SEM. P values, 2-tailed Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we tested whether loss of phagocytic cells could induce a megakaryocytic bias in the HSCs of young mice. Young mice were treated with clodronate-loaded liposomes (38) in order to deplete phagocytic Mφs (Figure 8F). In vivo treatment with clodronate-loaded liposomes (34, 38) expanded CD41+ LT-HSCs (Figure 8, G–J), suggesting that loss of phagocytic Mφs is sufficient to initiate BMME changes that induce HSC megakaryocytic bias in vivo. This is consistent with observations in a mouse model of aplastic anemia in which clodronate-mediated depletion of Mφs induced increased CD41+ LT-HSCs that exhibited robust platelet output in transplantation (39).
Aged BMME macrophages are defective in efferocytosis of senescent neutrophils.
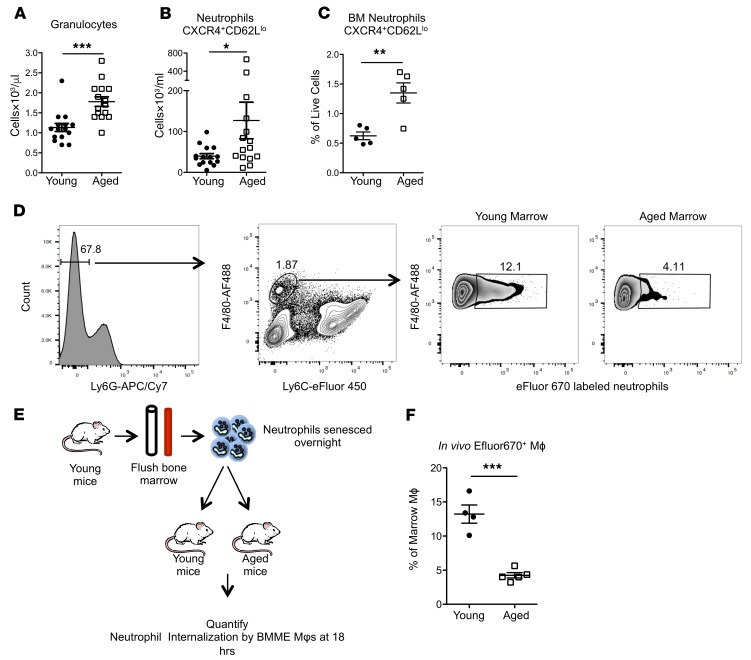
Neutrophils are short-lived and, when senescent, are removed from circulation at daily rates estimated to be 107 in mice and 1011 in humans (40). Since Mφs remove senescent neutrophils by phagocytosis (40), a phagocytic defect in aged marrow Mφs may increase senescent neutrophils in the circulation and in the marrow. Consistent with the important role of marrow Mφs in the clearance of senescent neutrophils (40, 41) and with our data showing that aged marrow Mφs have phagocytic defects, aged mice had increased total (Figure 9A) and senescent (CXCR4+CD62Llo) neutrophils in the circulation (Figure 9B) and in the bone marrow (Figure 9C), suggesting that marrow neutrophils may also contribute to the increase in marrow IL-1B in aged mice.
Figure 9. Impaired efferocytosis by macrophages in the bone marrow of aged mice.
(A) Quantification of peripheral blood granulocytes (n = 15 mice per group). (B and C) Quantification of senescent peripheral blood (B, n = 15 mice per group) and marrow (C, n = 5 mice per group) neutrophils. (D) Representative flow cytometry plots of young and aged marrow for the identification of Mφs that have phagocytosed senescent neutrophils. (E) Schematic of senescent neutrophil uptake experiment. (F) Quantification of senescent neutrophil engulfment in vivo by marrow Mφs 18 hours after injection (n = 4 mice per group). Each symbol represents an individual mouse; data represent mean ± SEM. P values, 2-tailed Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001.
Marrow Mφs remove and clear senescent neutrophils through efferocytosis, a form of specialized phagocytosis that minimizes the inflammatory consequences of apoptosis (20, 42). Given the phagocytic defects of marrow Mφs and the increase in senescent neutrophils in the marrow of aged mice, we sought to quantify engulfment of senescent neutrophils in young and aged mice (Figure 9, D and E, and ref. 43). We demonstrate that uptake of senescent neutrophils is significantly reduced in aged mice, supporting our hypothesis that aged Mφs have defective efferocytotic capacity (Figure 9F).
Loss of efferocytic receptor Axl is sufficient to drive premature myeloid-megakaryocytic bias in young HSCs.
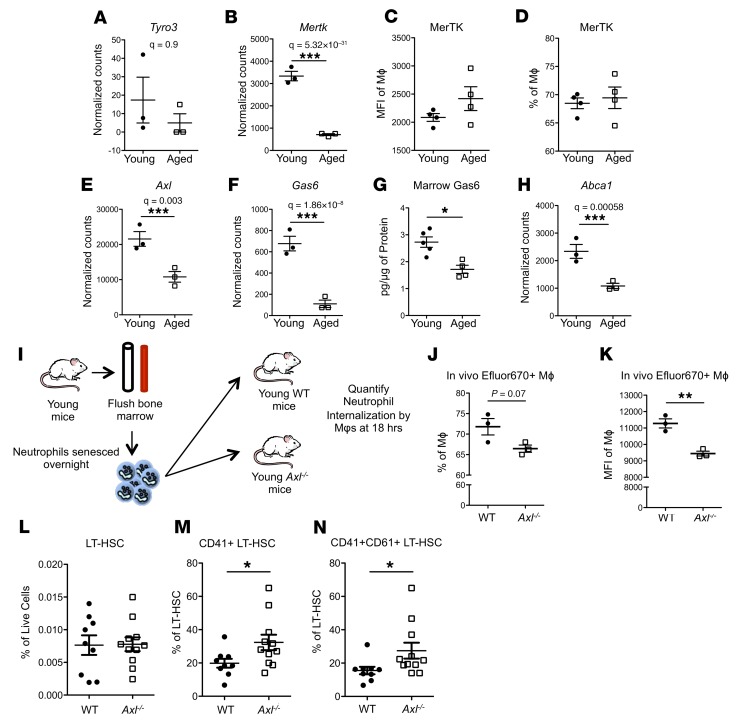
We next wanted to identify the mechanism responsible for the efferocytic defect in aged marrow Mφs. Efficient efferocytosis requires the activation of specialized tyrosine kinase receptors TYRO3, AXL, and MERTK (TAM receptors); however, these receptors may be differentially expressed in distinct phagocytic populations (44). Marrow Mφs from young mice had very low expression of Tyro3, consistent with previous data (44), and this level was not significantly changed in aged marrow Mφs (Figure 10A). The expression of Mertk (Figure 10B) was significantly decreased in aged marrow Mφs; however, the cell surface expression of MERTK was not changed in aged marrow Mφs (Figure 10, C and D), suggesting that MERTK may not be a critical mediator of the efferocytic defect displayed by aged marrow Mφs. However, Axl expression was significantly downregulated in aged marrow Mφs (Figure 10E). GAS6, the accessory protein that bridges binding of phosphatidyl serine to TAM receptors (45, 46) and was previously shown to be required to activate AXL (44), was significantly reduced in the marrow; and, consistent with this, transcriptional expression of Gas6 was significantly decreased in aged marrow Mφs, as was its protein level in the marrow of aged mice (Figure 10, F and G). Moreover, the expression of the efferocytic response gene Abca1 (Figure 10H and refs. 40, 42) was downregulated in aged BMME Mφs, consistent with the observed defective efferocytosis in aged marrow Mφs.
Figure 10. Genetic loss of efferocytic capacity is sufficient to drive premature aging phenotypes.
(A and B) Expression of the efferocytosis genes Tyro3 and Mertk in young and aged marrow Mφs (n = 3 mice per group). (C and D) Quantification of MERTK flow cytometric data, MFI (C) and percentage (D) of marrow Mφs. (E and F) Expression of the efferocytosis genes Axl and Gas6 in young and aged marrow Mφs (n = 3 mice per group). (G) Quantification by ELISA of the serum protein level for GAS6 (n = 4 young, 5 aged mice) in young compared with aged murine marrow. (H) Expression of Abca1 in young and aged marrow Mφs (n = 3 mice per group). (I–K) Schematic representation (I) and quantification (J and K) of efferocytic capacity of marrow Mφs from young WT and Axl–/– littermates 18 hours after injection, shown as percentage positive Mφs (J) and MFI (K) (n = 3 mice per experimental group). (L–N) Quantification of total LT-HSCs (L) and CD41+ (M) and CD41+/CD61+ (N) LT-HSCs from WT and Axl–/– mice (n = 9–11 mice per group). (A–N) Each symbol represents an individual mouse; data represent mean ± SEM. P values, 2-tailed Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001.
Therefore, we initiated studies with mice lacking the efferocytic receptor Axl (47) (Axl–/– mice) (48) to determine whether specifically blocking efferocytosis could mechanistically explain the age-associated increase in megakaryocytic bias in HSCs that correlates with reduced efferocytosis. Axl–/– mice were reported to have peritoneal Mφs with decreased phagocytic activity (44), and here we demonstrate that marrow Mφs in young Axl–/– mice exhibit defective engulfment of senescent neutrophils (Figure 10, I–K). If a defect in efferocytosis is sufficient to induce HSCs to acquire megakaryocytic skewing, young mice with genetic loss of Axl would be expected to have increased CD41+ LT-HSCs. Consistent with this, while the size of the LT-HSC pool remained unchanged (Figure 10L), young Axl–/– mice had increased CD41+ (Figure 10M) and CD41+CD61+ (Figure 10N) LT-HSCs, demonstrating that an efferocytic defect induced by loss of Axl is sufficient to induce megakaryocytic bias in young HSCs. LT-HSCs from young Axl–/– mice were superior to those from wild-type littermates in their ability to engraft in competitive transplantation assays (Supplemental Figure 8), consistent with the functional characteristics of HSCs with megakaryocytic skewing, and with the impact of the aged microenvironment on young HSCs (Figure 3, B–G).
Discussion
Our data define a previously unrecognized role for efferocytic defects in tissue-specific Mφs from aged marrow in regulating HSC megakaryocytic bias and provide evidence for the critical function of the BMME as a determinant of age-induced HSC lineage bias. We demonstrate a novel mechanism by which the marrow BMME induces age-dependent changes in HSCs through Mφs that display a proinflammatory phenotype and are defective in efferocytosis. Together, our data provide strong evidence that the BMME can indeed induce the Mk bias characteristic of aged HSCs, demonstrating an instructive role of the aged niche that could be broadly applied to the regulation of other tissue-specific stem cell populations. In addition, we demonstrate that the inflammatory signals in murine marrow Mφs are also recapitulated in the human bone marrow, suggesting that the mechanisms identified herein may contribute to the understanding of clonal selection and increased inflammation observed in humans with aging (1, 5, 49).
Recent reports have begun to elucidate the importance of BMME-derived signals in HSC aging, including osteopontin (16), sympathetic nervous system innervation (17), and endothelial cell aging (31). Our work identifies a previously unappreciated role for aged marrow Mφs in orchestrating the specific increase in HSC Mk skewing observed in aging. In defining marrow Mφs as a regulatory component of the HSC niche, our data align with reports suggesting that bone marrow Mφs support both physiological and pathological hematopoiesis (32–34).
While tissue Mφ heterogeneity is well described (50) even within a specific organ (51), specialization of marrow Mφs with respect to efferocytosis and immune activation during aging is poorly understood. Our data suggest that with aging there is a shift toward an inflammatory M1-like Mφ within the bone marrow, as production of the M1 markers MHC-II, CD86, and IL-1B is increased and the M2 Mφ marker CD206 is decreased in aged Mφs (52, 53). Conflicting data were previously reported regarding functional phagocytic changes in tissue Mφs with aging, with studies identifying decreased, unchanged, or increased function (54–61), with controversial results even within the bone marrow (62, 63). With respect to phagocytosis, we uncovered significant loss in phagocytic and efferocytic function in BMME Mφs using a physiologically relevant model. Phagocytic defects were recently described in a model of Alzheimer’s disease, a neurodegenerative disorder strongly associated with age. In that model, insufficient generation of phagocytic microglia resulted in their inability to clear amyloid plaques (64). In the context of this report, our data further support the notion that defects in phagocytosis in certain subsets of tissue-based Mφs may be a hallmark of advanced age. Additional data at the single-cell level are needed to determine whether the functional defect we uncovered in aged BMME Mφs is due to a global phagocytic impairment or to loss of a specific subpopulation of phagocytic Mφs within bone marrow.
The tyrosine kinase receptors MERTK, TYRO3, and AXL are essential for the ability of Mφs to engulf apoptotic cells. Transcriptional analysis of murine marrow Mφs showed that Tyro3 is expressed at low levels in marrow Mφs and it is not regulated by aging, while both Axl and Mertk expression is decreased with aging. However, flow cytometric analysis of MERTK cell surface expression did not show changes with aging, suggesting that MERTK may not be the critical TAM responsible for the loss of efferocytic capacity of aged marrow Mφs. Loss of Axl in the aging marrow is somewhat surprising given data showing Axl upregulation in the setting of a proinflammatory environment (44). In our studies, the decrease in Axl expression observed in aged marrow Mφs may represent an initiating defect that then induces inflammation secondary to the decrease in efferocytic function.
Increased Mk-biased HSCs in mice lacking only 1 of 3 TAM receptors that mediate efferocytosis (Axl–/– mice) were also somewhat unexpected since TAM receptors represent a redundant system; thus our findings point to a potentially important role of Axl in bone marrow Mφs. Given the previously reported dependence of Gas6 expression on AXL (44), a finding that is supported by our data, age-dependent decrease in Axl may also decrease transcriptional expression of Gas6, compounding efferocytic defects. In further support of an age-dependent loss of efferocytic function in marrow Mφs, we found age-dependent regulation of Mertk and Abca1, key genes rhythmically modulated in Mφs by circadian changes in clearance of senescent neutrophils (40). While our in vivo studies using Axl–/– mice cannot definitively rule out a potential role for Axl in other cell types, when taken together with our in vitro studies, they strongly support a role for Mφs in age-induced Mk bias. Future studies to investigate Axl regulation and function in marrow Mφs are warranted.
The elimination of apoptotic cells is a widely recognized and critical mechanism supporting organ growth and fitness, since apoptosis of cells occurs essentially in all tissues as part of normal development and homeostasis, and lack of apoptotic cell removal contributes to tissue pathology (65). Since it was recently reported that phagocytosis induces an antiinflammatory program in Mφs (37), we would expect that clodronate removal of phagocytic Mφs would selectively remove noninflammatory Mφs. Consequently, senescent neutrophils as well as nonphagocytic Mφs, higher in IL-1B production, would remain to cause skewing even of young HSCs in mice treated with liposomal clodronate. This is consistent with our observations using clodronate-loaded liposomes to deplete Mφs in young mice and further supported by the recent demonstration that Mφ depletion in a model of severe aplastic anemia elicits a population of CD41hi HSCs that exhibit robust platelet output (39).
The age-dependent decline in efferocytosis may induce HSC aging through remodeling of additional niche constituents, based on previous data demonstrating the impact of clearance of senescent neutrophils on rhythmic modulation of the HSC niche (40). In support of this, we show that the addition of aged Mφs to young BMME alters specific MSC populations in a manner characteristic of aged BMME. Because restoration of efferocytosis may be targeted (63), our data suggest the potential for remediation of age-induced microenvironmental defects. On the other hand, global inflammation causes depletion of platelets that may result in significant morbidity (26); thus the efferocytic defect and megakaryocytic HSC bias may be beneficial in the context of aging.
We focused on IL-1B as a key inflammatory mediator in the hematopoietic system based on our transcriptional analysis. Recent data also show that chronic pharmacologic dosing of IL-1B increases the number of myeloid-primed CD41+ HSCs (35). Moreover, IL-1B induces Mk maturation and platelet activation (66); however, its role in HSC megakaryocytic skewing was not previously reported. We have demonstrated that aged bone marrow Mφs have increased production of active IL-1B. Additionally, defective efferocytosis in aged Mφs leads to increased numbers of senescent neutrophils in the BMME, further contributing to increased IL-1B concentrations, as senescent neutrophils are a potent source of active IL-1B. Thus, defective clearance by marrow Mφs appears to increase IL-1B via 2 distinct mechanisms. We recapitulated the effect of aged BMME cells with the addition of IL-1B in young BMME cells, and demonstrated the critical role of IL-1B by blocking the effect of aged marrow microenvironmental cells on HSCs with treatment with an IL1R1 antagonist, pointing to a potential strategy for HSC rejuvenation. Together, the studies presented here identify IL-1B as a critical microenvironmental signal necessary and sufficient to induce HSC aging.
Collectively, our findings suggest that the efferocytic defect in aged niche Mφs drives an aged HSC phenotype, providing, to our knowledge, the first mechanistic explanation for megakaryocytic bias seen during aging. Our findings suggest that loss of efferocytosis in aged niche Mφs may increase local IL-1B both through increased production of IL-1B by aged marrow Mφs and by decreased engulfment of IL-1B–rich senesced neutrophils. Since efferocytosis has immunomodulatory roles by engulfing apoptotic cells and by modulating inflammatory signals in Mφs, bringing about resolution of inflammation (42), decreased engagement of efferocytotic signals in Mφs would be expected to enhance their production of IL-1B, increasing BMME IL-1B. IL-1B and activation of the IL1R1 were necessary and sufficient to recapitulate the phenotypic HSC bias, implicating this cytokine as the executor of aged marrow Mφ dysfunction (Supplemental Figure 9). These data illustrate how, given the emerging heterogeneity of the HSC niche, only complex genetic and biochemical studies of niche HSC interactions could provide novel and more specific therapeutic targets to prevent or reverse not only hematopoietic defects but also the increase in global inflammation associated with advanced age. Our studies provide an important framework to probe the precise mechanisms regulating Mφ function during aging, and open novel therapeutic avenues for rejuvenating aged HSCs.
Methods
Mice.
Male mice were maintained within the Vivarium facility at the University of Rochester School of Medicine and Dentistry. Aged C57BL/6J.NIA mice (20–30 months) were obtained from the National Institute on Aging (NIA) aged rodent colonies. Young (8–16 weeks) C57BL/6J were purchased from The Jackson Laboratory or bred in house. B6.SJL-Ptprca Pepcb/BoyJ mice, which express the CD45.1 congenic marker, were bred in house. UBC-GFP mice [C57BL/6-Tg(UBC-GFP)30Scha/J] were purchased from Charles River. Axl–/– were obtained from V.A. Korshunov’s laboratory, were previously described, and had been backcrossed onto the C57BL/6J background for more than 10 generations (48). Treatment with clodronate or PBS liposomes was done as previously described (38).
Complete blood counts.
Blood was collected from the submandibular plexus and collected in EDTA-coated tubes. The scil Vet abc Plus+ was used to determine complete blood counts.
Intravital imaging.
Mice were injected i.v. with 100 μl of 5- to 10-mg/ml 70-kDa dextran conjugated to Texas red (Invitrogen) before anesthesia. Animals were anesthetized with 5% isoflurane mixed with air, which was switched to 1%–2% during surgery and subsequent imaging. Calvaria were prepared for in vivo imaging as previously described (67). Intravital imaging was done using an Olympus FV1000-AOM multiphoton system. Fluorescence was collected with an Olympus XLPlanN 25× objective (NA 1.05) and detected with 2 proprietary external photomultipliers (Hamamatsu).
Image quantification.
Seventy-kilodalton dextran and second harmonic generation (SHG) volumes were calculated using Amira 5.4.5. Positive signal was determined by use of the threshold feature along each slice. This was further refined by use of the smooth and remove island feature. Marrow cavity volume was determined by subtraction of the volume of the SHG signal from the entire volume of the Z-stack.
Histology.
Tissue was prepared as previously described (49). The GBI Polink DS-RRt-Hu/Ms A Kit was used according to the manufacturer’s instructions (catalog DS211A-60). Endomucin (catalog 14-5851, eBioscience) was used at 1:500, and osteocalcin (catalog ALX-210-333-C100, Enzo Life Science) was used at 1:8000. The GBI Polink-2 HRP-Plus Goat DAB Kit (catalog D43-110) was used according to the manufacturer’s instruction to detect leptin receptor at 1:1000 (catalog AF497, R&D Systems).
Light microscopy.
Images were taken at room temperature using an Olympus BX50 microscope, Olympus DP25 camera, and ×100 pan oil immersion objective (NA 1.25). Images were also taken at room temperature on an Olympus BX41, Olympus DP70 camera, and ×20 UPlanFl objective (NA 0.5). Cellsens software (Olympus) was used to acquire images on both microscopes.
Flow cytometry.
Analysis of marrow cell populations was done as previously described (49) with the following changes. For marrow cell analysis, marrow was released by crushing with a mortar and pestle in 1× PBS with 2% heat-inactivated FBS (HyClone, lot AWB98615) and 10 U/ml of DNAse (catalog FERNEN0521, Thermo).
Isolation of peripheral blood mononuclear cells for analysis of senescent neutrophils was done as previously described (40). Analysis for hematopoietic, stromal, and Mφ cell populations was performed as previously described (refs. 38, 49; and Supplemental Tables 1 and 2 and Supplemental Figures 3 and 4). The FAM-FLICA caspase-1 Assay Kit (ImmunoChemistry Technologies) was used to detect caspase-1 activity. DAPI was used to discriminate live/dead cells (catalog D21490, Molecular Probes; final concentration, 2 μg/ml). Samples were run on an LSRII flow cytometer: 3 lasers, 355 nm, 488 nm, and 633 nm (BD Biosciences). Analysis was performed using FlowJo version 9.6.5 (Tree Star). The gating strategy used to identify populations enriched for cells of interest has been previously described (38, 49, 68, 69). Sorting was done on a FACSAria II with 405-, 488-, 532-, and 633-nm lasers (BD Biosciences).
CFU assays.
Colony-forming units (CFUs) from murine marrow cells were quantified as previously described (49). Marrow cell populations were plated at 1 × 106 cells per well in 6-well plates and grown at 37°C with 5% CO2 and 2% O2. For megakaryocyte progenitor assays, FACS-isolated HSCs were plated in MegaCult collagen media (StemCell Technologies) at a range of 100–500 HSCs per well and enumerated at 7 days of culture after acetylcholinesterase staining as previously described (70).
Human samples.
Human bone marrow samples were obtained from healthy volunteers by standard bone marrow aspiration procedures through iliac crest puncture. Human samples from young (<50 years old) and aged (>50 years old) human volunteers were filtered through a cell strainer and washed with PBS. A list of anti-human antibodies used is in Supplemental Table 1 along with the gating strategy for specific populations.
In vitro cell culture.
Marrow cells were plated at 3 × 106 cells per well in 6-well plates. Both populations were cultured in α-MEM media with no ascorbic acid, 10% heat-inactivated FBS, 1% penicillin and streptomycin, and 50 μM 2-mercaptoethanol. Cells were incubated in 5% CO2 and 2% O2. Medium was changed on day 4 to remove nonadherent and dead cells.
For addition of Mφs in cocultures (Figure 3), 2.1 × 105 F4/80+ Mφs from young and aged marrow cells were sorted by flow cytometry (Supplemental Table 1) and cultured simultaneously with young marrow cells in 5% CO2 and 2% O2.
On day 7, 500 Lin–SCA1+CKIT+ cells from young (6–12 weeks) UBC-GFP+ mice were sorted by flow cytometry (Supplemental Table 1) and added to the existing stromal cultures with the following recombinant murine cytokines at 25 ng/ml: SCF (catalog AF-250-03), Flt3-L (catalog 250-31L), and IL-11 (catalog 220-11; all from PeproTech). For addition of inflammatory cytokines (25 ng/ml), on day 7, IL-1B (catalog 211-11B) was added to the basal media before plating of LSKs. IL1R1 antagonism was done with IL1RA/Anakinra (Amgen) at 1500 ng/ml added on day 7. Cells were incubated undisturbed for an additional 4 days in 5% CO2 and 2% O2. On day 11, floating and adherent cells were collected for flow cytometry analysis. Adherent cells were removed by incubation of cells with 1× TrypLE (catalog 12605-010, Invitrogen). Floating and adherent cells were also collected on day 7 for phenotyping of stromal monolayers.
Competitive repopulation assay.
Recipient mice were given 10 Gy of radiation, split in two 5-Gy doses separated by 24 hours, as previously described (49). For transplant of LSK cocultures, cells from 1 well (floating and adherent) were mixed with 300,000 whole bone marrow cells from a CD45.1 competitor. In this scenario, residual hematopoietic cells from the coculture were CD45.2 and the competitor CD45.1, and the donor LSKs were GFP+, allowing us to distinguish all 3 populations.
For competitive bone marrow transplant of Axl–/–, 1 × 106 of both donor and competitor were transplanted. Donor cells expressed CD45.2, the competitor was CD45.1/CD45.2, and the recipients were CD45.1, allowing all 3 groups to be distinguished.
For analysis of platelet reconstitution, CD41+ and CD41– donor LT-HSCs (100 Lineage–Flt3–CD48–SCA1+CKIT+CD150+ cells per recipient mouse) were sorted from aged mice and combined with 1 × 106 competitor bone marrow harvested from UBC-GFP mice that express GFP in all tissues including platelets. Donor/competitor cells were transplanted into UBC-GFP recipient mice that were given 10 Gy of radiation, split in two 5-Gy doses. Proportions of GFP+ and GFP– platelets were determined by flow cytometry on whole peripheral blood sampled 3 weeks after transplantation and stained with anti-CD41 and -Ter119 antibodies (eBioscience) and analyzed on an LSRII (BD Biosciences) using FlowJo analysis software (Tree Star).
RNA isolation and generation of cDNA library.
Mφs were FACS-isolated directly in RLT Plus buffer (Qiagen). RNA extraction was performed with Qiagen RNeasy PLUS Micro kit following standard operating procedures. RNA quality was assessed using Agilent Bioanalyzer 2100. One nanogram of high-quality (RNA integrity number >8.0) total RNA from each sample was reverse-transcribed into cDNA using the Clontech SMART-Seq v4 Ultra Low Input RNA Kit. Final Illumina libraries were constructed using 150 pg of cDNA with the Illumina Nextera XT DNA Library Preparation Kit. Expression data sets are available as GEO GSE100907.
Transcriptional analysis of macrophage populations.
Raw reads generated from the Illumina HiSeq2500 sequencer were demultiplexed using configurebcl2fastq.pl version 1.8.4. Quality filtering and adapter removal were performed using Trimmomatic version 0.32 with the following parameters: “SLIDINGWINDOW:4:20 TRAILING:13 LEADING:13 ILLUMINACLIP:adapters.fasta:2:30:10 MINLEN:15” (71). Processed/cleaned reads were then mapped to the reference genome (mouse, GRCm38.p4; human, GRCh38.p3) using STAR_2.4.2a with the following parameters: “--twopassMode Basic --runMode alignReads --outSAMtype BAM SortedByCoordinate --outSAMstrandField intronMotif --outFilterIntronMotifs RemoveNoncanonical --outReadsUnmapped Fastx” (72). Gene-level read quantification was derived using htseq-count 0.6.1 with a GTF annotation file (mouse, Gencode M6; human, Gencode 23) and the following parameters: “-q -f bam -s no -r pos -i gene_name” (73). Differential expression analysis was performed using DESeq2-1.12.4 with an adjusted P value threshold of 0.05 within R version 3.3.0 (74). Heatmaps were created within R using the pheatmap package (R package version 0.7.7) given rlog-transformed expression values.
Single-cell RNA sequencing.
Single long-term hematopoietic stem cells (LT-HSCs) were captured on an integrated microfluidic chip (IFC) with 5–10 μm cell diameter using the Fluidigm C1 Single-cell AutoPrep system (Fluidigm Corp.). Cells were prestained with Calcein AM/EthD-1 LIVE/DEAD cell viability assay (Life Technologies) and loaded onto the IFC at a concentration of 500–700 cells/μl. Viable single-cell confirmation was performed with phase-contrast fluorescence microscopy to assess the number and viability of cells per capture site. Only single, live cells were included in the analysis. For RNA sequencing analysis, cDNAs were prepared “on-IFC” using the SMARTer Ultra Low RNA kit for Illumina (Clontech) following Fluidigm recommendations. Single-cell cDNA size distribution and concentration were assessed with PicoGreen (Life Technologies) and Agilent Bioanalyzer 2100 analysis (Agilent Technologies). Illumina libraries were constructed in 96-well plates using Illumina’s Nextera XT DNA Sample Preparation kit following the protocol supplied by Fluidigm. For each C1 experiment, a bulk RNA control and a negative control were processed in parallel, using the same reagent mixes as used on chip. Libraries were quantified by Agilent Bioanalyzer, using High Sensitivity DNA analysis kit, and also fluorometrically, using Qubit dsDNA HS Assay kits and a Qubit 2.0 Fluorometer (Life Technologies).
Analysis of single-cell RNA sequencing.
Read data were cleaned, aligned, and quantified in the same manner as the mouse RNA sequencing data. The R package scater_1.2.0 was used to load the data into R version 3.3.2 and perform initial filtering of genes (>0 reads in more than 5 cells) and cells (>0.5 million total reads per cell) (75). The scran_1.2.2 R package was used to compute sum factors with sizes 20, 40, and 60 before normalization with scater “normalize” function (75). The top 1000 highly variable genes were determined using the scran “trendVar” function with use.spikes=FALSE and “decomposeVar.” The R package Seurat_1.4.0.9 was then used to visualize the expression using the “VlnPlot” function and perform principal component analysis using the highly variable genes determined using scran (76).
GO and KEGG analysis.
The open-source Enrichr gene list enrichment analysis tool (77) was used for Gene Ontology (GO) and KEGG Pathway analysis. Significantly (padj < 0.05) differentially regulated genes were selected for analysis when the log2 fold change was ≤1 (downregulated, 508 genes) and >1 (upregulated, 570 genes). Gene annotation was performed within the GO terms “Biological Process/Direct” and KEGG Pathway analysis using a threshold count 2 and EASE 0.1. Enrichment values were displayed as fold enrichment for terms with FDR less than 0.05.
Gene set enrichment analysis.
Overlaps for selected genes were computed using gene set enrichment analysis (78). Significantly (padj < 0.05) differentially upregulated genes were selected for analysis (63 genes). Gene annotation was performed within the GO terms “Biological Process/Direct.” Top hits based on FDR (smallest to largest) were displayed as fold enrichment for terms with FDR less than 0.05. Fold enrichment was calculated as the ratio of the 2 proportions.
ELISA.
Sera and total bone marrow protein were collected from aged mice. Bone marrow cell lysates were homogenized with a pestle in a buffer containing IGEPAL CA-630 (Sigma-Aldrich) and proteinase inhibitors for protein analysis. ELISA for marrow IL-1B and GAS6 was performed in marrow serum using the commercially available Mouse IL-1β ELISA kit (ab197742, Abcam) and Mouse GAS6 ELISA Kit (ab155447, Abcam), respectively.
Phagocytosis assays.
For liposome uptake experiments, young and aged marrow cells were incubated with DOPC/CHOL Liposomes labeled with fluorescein-DHPE dye (FormuMax Scientific) for 1 hour. Cells were washed and stained for flow cytometric analysis. For in vivo neutrophil uptake experiments, phagocytosis was quantified via flow cytometry as described previously (79). Neutrophils were purified from young marrow cells using the EasySep Mouse Neutrophil Enrichment Kit (StemCell Technologies), following the The Big Easy protocol. Neutrophils were grown overnight at 37°C/5% CO2, labeled with efluor670 (catalog 65-0840-85, eBioscience), then retro-orbitally injected into young or aged mice, or into young (9–12 weeks old) Axl–/– and WT littermates. Eighteen hours after injection, marrow cells were harvested for flow cytometric analysis.
Accession codes.
Expression data sets are available in the NCBI’s Gene Expression Omnibus database (GEO GSE100907).
Statistics.
All data are presented as mean ± SEM. All analyses were made with GraphPad Prism software (version 5) using 2-tailed Student’s t test, Mann-Whitney nonparametric testing, or 1-way or 2-way ANOVA with Tukey’s multiple-comparisons post-test when appropriate. A P value less than 0.05 was considered significant.
Study approval.
All murine studies were performed in accordance with protocols approved by the institutional animal care and use committee, the University Committee on Animal Resources (University of Rochester, Rochester, New York, USA). Written informed consent was obtained from all human subjects. The study was approved by the institutional review board, the Research Subjects Review Board (University of Rochester), and conducted in accordance with the Declaration of Helsinki.
Author contributions
BJF, CMH, and SEL designed and performed experiments, analyzed data, and wrote the manuscript. MWL, KEM, and AJL performed experiments. JM and JA performed transcriptional analyses. A. McCabe, JNPS, JS, and FRE performed myeloid analysis. JLL provided human specimens. JP, ASP, and VAK participated in study design. A. McDavid provided statistical consultation. MRE contributed to study design and data analysis and performed experiments. MWB contributed to study design and performed experiments. KCM contributed to study design and data analysis. LMC designed and performed experiments, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Calvi laboratory for discussion; Lizz LaMere, Brandon J. Zaffuto, Daniel Byun, Rhonda J. Staversky, Mary A. Georger, Marlene Balys, Michelle Zanche, Xingping Zhang, Paivi Jordan, Maria Jepson, and Breandan Quinn for experimental support; and the University of Rochester Flow Cytometry and Genomics Cores for their assistance. Supported by the National Heart, Lung, and Blood Institute (F31 HL131184 to CMH), the National Institute of General Medical Sciences (K12 GM106997 to SEL and R01 GM105949 to KCM), the National Institute of Allergy and Infectious Diseases (U01 AI107276 to LMC and R01 AI114554 to MRE), the Department of Defense (Award W81XWH1810485 to LMC), the National Cancer Institute (R01 CA166280 to LMC and MWB and F32 CA180615 to BJF), the National Institute on Aging (R01 AG046293 to LMC and MWB), the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK098251 to JP), and funds from the Wilmot Cancer Institute.
Version 1. 04/18/2019
In-Press Preview
Version 2. 05/16/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(10):e124213. https://doi.org/10.1172/jci.insight.124213.
Contributor Information
Benjamin J. Frisch, Email: Benjamin_Frisch2@URMC.Rochester.edu.
Corey M. Hoffman, Email: choffman116@gmail.com.
Sarah E. Latchney, Email: sarah_latchney@urmc.rochester.edu.
Mark W. LaMere, Email: Mark_lamere@urmc.rochester.edu.
Jason Myers, Email: jason_myers@urmc.rochester.edu.
John Ashton, Email: John_Ashton@URMC.Rochester.edu.
Allison J. Li, Email: allison_li@urmc.rochester.edu.
James Palis, Email: James_Palis@urmc.rochester.edu.
Archibald S. Perkins, Email: archibald_perkins@urmc.rochester.edu.
Amanda McCabe, Email: Amanda.McCabe@childrens.harvard.edu.
Kathleen E. McGrath, Email: KathleenE_McGrath@urmc.rochester.edu.
Fatima Rivera-Escalera, Email: fatima_riveraescalera@urmc.rochester.edu.
Andrew McDavid, Email: andrew_mcdavid@urmc.rochester.edu.
Jane L. Liesveld, Email: jane_liesveld@urmc.rochester.edu.
Vyacheslav A. Korshunov, Email: Slava_Korshunov@urmc.rochester.edu.
Michael R. Elliott, Email: michael_elliott@urmc.rochester.edu.
Katherine C. MacNamara, Email: macnamk@mail.amc.edu.
Michael W. Becker, Email: michael_becker@urmc.rochester.edu.
Laura M. Calvi, Email: laura_calvi@urmc.rochester.edu.
References
- 1.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nat Rev Immunol. 2013;13(12):875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akunuru S, Geiger H. Aging, clonality, and rejuvenation of hematopoietic stem cells. Trends Mol Med. 2016;22(8):701–712. doi: 10.1016/j.molmed.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flach J, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florian MC, et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature. 2013;503(7476):392–396. doi: 10.1038/nature12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang WW, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci U S A. 2011;108(50):20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanjuan-Pla A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502(7470):232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 8.Gekas C, Graf T. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood. 2013;121(22):4463–4472. doi: 10.1182/blood-2012-09-457929. [DOI] [PubMed] [Google Scholar]
- 9.Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. 2019;20(5):303–320. doi: 10.1038/s41580-019-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernitz JM, Kim HS, MacArthur B, Sieburg H, Moore K. Hematopoietic stem cells count and remember self-renewal divisions. Cell. 2016;167(5):1296–1309.e10. doi: 10.1016/j.cell.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rundberg Nilsson A, Soneji S, Adolfsson S, Bryder D, Pronk CJ. Human and murine hematopoietic stem cell aging is associated with functional impairments and intrinsic megakaryocytic/erythroid bias. PLoS One. 2016;11(7):e0158369. doi: 10.1371/journal.pone.0158369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baryawno N, Severe N, Scadden DT. Hematopoiesis: reconciling historic controversies about the niche. Cell Stem Cell. 2017;20(5):590–592. doi: 10.1016/j.stem.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Pan L, et al. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell. 2007;1(4):458–469. doi: 10.1016/j.stem.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Rozhok AI, Salstrom JL, DeGregori J. Stochastic modeling indicates that aging and somatic evolution in the hematopoetic system are driven by non-cell-autonomous processes. Aging (Albany NY) 2014;6(12):1033–1048. doi: 10.18632/aging.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusumbe AP, et al. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature. 2016;532(7599):380–384. doi: 10.1038/nature17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidi N, et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J. 2017;36(10):1463. doi: 10.15252/embj.201796968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maryanovich M, et al. Adrenergic nerve degeneration in bone marrow drives aging of the hematopoietic stem cell niche. Nat Med. 2018;24(6):782–791. doi: 10.1038/s41591-018-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell. 2017;171(2):331–345.e22. doi: 10.1016/j.cell.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandivier RW, Henson PM, Douglas IS. Burying the dead: the impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease. Chest. 2006;129(6):1673–1682. doi: 10.1378/chest.129.6.1673. [DOI] [PubMed] [Google Scholar]
- 20.Arandjelovic S, Ravichandran KS. Phagocytosis of apoptotic cells in homeostasis. Nat Immunol. 2015;16(9):907–917. doi: 10.1038/ni.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Celso C, Lin CP, Scadden DT. In vivo imaging of transplanted hematopoietic stem and progenitor cells in mouse calvarium bone marrow. Nat Protoc. 2011;6(1):1–14. doi: 10.1038/nprot.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morikawa S, et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J Exp Med. 2009;206(11):2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenbaum A, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495(7440):227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinho S, et al. PDGFRα and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J Exp Med. 2013;210(7):1351–1367. doi: 10.1084/jem.20122252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas S, et al. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell. 2015;17(4):422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Grover A, et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat Commun. 2016;7:11075. doi: 10.1038/ncomms11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalczyk MS, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25(12):1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell Immunol. 2001;214(2):110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- 30.Umemoto T, et al. CD61 enriches long-term repopulating hematopoietic stem cells. Biochem Biophys Res Commun. 2008;365(1):176–182. doi: 10.1016/j.bbrc.2007.10.168. [DOI] [PubMed] [Google Scholar]
- 31.Poulos MG, et al. Endothelial transplantation rejuvenates aged hematopoietic stem cell function. J Clin Invest. 2017;127(11):4163–4178. doi: 10.1172/JCI93940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow A, et al. CD169+ macrophages provide a niche promoting erythropoiesis under homeostasis and stress. Nat Med. 2013;19(4):429–436. doi: 10.1038/nm.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow A, et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med. 2011;208(2):261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler IG, et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood. 2010;116(23):4815–4828. doi: 10.1182/blood-2009-11-253534. [DOI] [PubMed] [Google Scholar]
- 35.Pietras EM, et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol. 2016;18(6):607–618. doi: 10.1038/ncb3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 37.A-Gonzalez N, et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J Exp Med. 2017;214(5):1281–1296. doi: 10.1084/jem.20161375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCabe A, Zhang Y, Thai V, Jones M, Jordan MB, MacNamara KC. Macrophage-lineage cells negatively regulate the hematopoietic stem cell pool in response to interferon γ at steady state and during infection. Stem Cells. 2015;33(7):2294–2305. doi: 10.1002/stem.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCabe A, Smith JNP, Costello A, Maloney J, Katikaneni D, MacNamara KC. Hematopoietic stem cell loss and hematopoietic failure in severe aplastic anemia is driven by macrophages and aberrant podoplanin expression. Haematologica. 2018;103(9):1451–1461. doi: 10.3324/haematol.2018.189449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casanova-Acebes M, et al. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell. 2013;153(5):1025–1035. doi: 10.1016/j.cell.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang D, et al. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525(7570):528–532. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott MR, Koster KM, Murphy PS. Efferocytosis signaling in the regulation of macrophage inflammatory responses. J Immunol. 2017;198(4):1387–1394. doi: 10.4049/jimmunol.1601520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furze RC, Rankin SM. The role of the bone marrow in neutrophil clearance under homeostatic conditions in the mouse. FASEB J. 2008;22(9):3111–3119. doi: 10.1096/fj.08-109876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zagórska A, Través PG, Lew ED, Dransfield I, Lemke G. Diversification of TAM receptor tyrosine kinase function. Nat Immunol. 2014;15(10):920–928. doi: 10.1038/ni.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothlin CV, Carrera-Silva EA, Bosurgi L, Ghosh S. TAM receptor signaling in immune homeostasis. Annu Rev Immunol. 2015;33:355–391. doi: 10.1146/annurev-immunol-032414-112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemke G. Biology of the TAM receptors. Cold Spring Harb Perspect Biol. 2013;5(11):a009076. doi: 10.1101/cshperspect.a009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian M, et al. An AXL/LRP-1/RANBP9 complex mediates DC efferocytosis and antigen cross-presentation in vivo. J Clin Invest. 2014;124(3):1296–1308. doi: 10.1172/JCI72051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Q, et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 1999;398(6729):723–728. doi: 10.1038/19554. [DOI] [PubMed] [Google Scholar]
- 49.Balderman SR, et al. Targeting of the bone marrow microenvironment improves outcome in a murine model of myelodysplastic syndrome. Blood. 2016;127(5):616–625. doi: 10.1182/blood-2015-06-653113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157(4):832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabanyi I, Muller PA, Feighery L, Oliveira TY, Costa-Pinto FA, Mucida D. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell. 2016;164(3):378–391. doi: 10.1016/j.cell.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duluc D, et al. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood. 2007;110(13):4319–4330. doi: 10.1182/blood-2007-02-072587. [DOI] [PubMed] [Google Scholar]
- 53.Rőszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015;2015:816460. doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De La Fuente M. Changes in the macrophage function with aging. Comp Biochem Physiol A Comp Physiol. 1985;81(4):935–938. doi: 10.1016/0300-9629(85)90933-8. [DOI] [PubMed] [Google Scholar]
- 55.Fietta A, Merlini C, Dos Santos C, Rovida S, Grassi C. Influence of aging on some specific and nonspecific mechanisms of the host defense system in 146 healthy subjects. Gerontology. 1994;40(5):237–245. doi: 10.1159/000213591. [DOI] [PubMed] [Google Scholar]
- 56.Mancuso P, McNish RW, Peters-Golden M, Brock TG. Evaluation of phagocytosis and arachidonate metabolism by alveolar macrophages and recruited neutrophils from F344xBN rats of different ages. Mech Ageing Dev. 2001;122(15):1899–1913. doi: 10.1016/S0047-6374(01)00322-0. [DOI] [PubMed] [Google Scholar]
- 57.Swift ME, Burns AL, Gray KL, DiPietro LA. Age-related alterations in the inflammatory response to dermal injury. J Invest Dermatol. 2001;117(5):1027–1035. doi: 10.1046/j.0022-202x.2001.01539.x. [DOI] [PubMed] [Google Scholar]
- 58.Aprahamian T, Takemura Y, Goukassian D, Walsh K. Ageing is associated with diminished apoptotic cell clearance in vivo. Clin Exp Immunol. 2008;152(3):448–455. doi: 10.1111/j.1365-2249.2008.03658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch AM, et al. The impact of glial activation in the aging brain. Aging Dis. 2010;1(3):262–278. [PMC free article] [PubMed] [Google Scholar]
- 60.Hearps AC, et al. Aging is associated with chronic innate immune activation and dysregulation of monocyte phenotype and function. Aging Cell. 2012;11(5):867–875. doi: 10.1111/j.1474-9726.2012.00851.x. [DOI] [PubMed] [Google Scholar]
- 61.Njie EG, Boelen E, Stassen FR, Steinbusch HW, Borchelt DR, Streit WJ. Ex vivo cultures of microglia from young and aged rodent brain reveal age-related changes in microglial function. Neurobiol Aging. 2012;33(1):195.e1–195.12. doi: 10.1016/j.neurobiolaging.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linehan E, Dombrowski Y, Snoddy R, Fallon PG, Kissenpfennig A, Fitzgerald DC. Aging impairs peritoneal but not bone marrow-derived macrophage phagocytosis. Aging Cell. 2014;13(4):699–708. doi: 10.1111/acel.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnardottir HH, Dalli J, Colas RA, Shinohara M, Serhan CN. Aging delays resolution of acute inflammation in mice: reprogramming the host response with novel nano-proresolving medicines. J Immunol. 2014;193(8):4235–4244. doi: 10.4049/jimmunol.1401313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169(7):1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 65.Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14(3):166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beaulieu LM, et al. Interleukin 1 receptor 1 and interleukin 1β regulate megakaryocyte maturation, platelet activation, and transcript profile during inflammation in mice and humans. Arterioscler Thromb Vasc Biol. 2014;34(3):552–564. doi: 10.1161/ATVBAHA.113.302700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sipkins DA, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435(7044):969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schepers K, Hsiao EC, Garg T, Scott MJ, Passegué E. Activated Gs signaling in osteoblastic cells alters the hematopoietic stem cell niche in mice. Blood. 2012;120(17):3425–3435. doi: 10.1182/blood-2011-11-395418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 70.Palis J, Koniski A. Analysis of hematopoietic progenitors in the mouse embryo. Methods Mol Med. 2005;105:289–302. doi: 10.1385/1-59259-826-9:289. [DOI] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S, Pyl PT, Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park D, et al. Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature. 2011;477(7363):220–224. doi: 10.1038/nature10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.