Abstract
The E3 ubiquitin ligase Parkin plays an important role in regulating clearance of dysfunctional or unwanted mitochondria in tissues, including the heart. However, whether Parkin also functions to prevent cardiac aging by maintaining a healthy population of mitochondria is still unclear. Here, we have examined the role of Parkin in the context of mitochondrial DNA (mtDNA) damage and myocardial aging using a mouse model carrying a proofreading-defective mtDNA polymerase γ (POLG). We observed both decreased Parkin protein levels and development of cardiac hypertrophy in POLG hearts with age; however, cardiac hypertrophy in POLG mice was neither rescued, nor worsened by cardiac-specific overexpression or global deletion of Parkin, respectively. Unexpectedly, mitochondrial fitness did not substantially decline with age in POLG mice when compared with that in WT mice. We found that baseline mitophagy receptor–mediated mitochondrial turnover and biogenesis were enhanced in aged POLG hearts. We also observed the presence of megamitochondria in aged POLG hearts. Thus, these processes may limit the accumulation of dysfunctional mitochondria as well as the degree of cardiac functional impairment in the aging POLG heart. Overall, our results demonstrate that Parkin is dispensable for constitutive mitochondrial quality control in a mtDNA mutation model of cardiac aging.
Keywords: Cardiology, Cell Biology
Keywords: Autophagy, Mitochondria
Parkin is dispensable for constitutive mitochondrial quality control in a mtDNA mutation model of cardiac aging.
Introduction
The occurrence of cardiovascular disease increases with advancing age, and age-related alterations in cardiomyocytes are a major contributor to the underlying pathogenesis. In particular, a decline in mitochondrial function is considered a major contributor to the cardiac aging process (1). Mitochondrial autophagy (mitophagy) is an important cellular quality control mechanism, and it ensures the existence of a healthy population of mitochondria in postmitotic cells. Studies have reported that aging is associated with reduced autophagy and mitochondrial quality control in the heart (2, 3), which can lead to accumulation of dysfunctional mitochondria. However, the molecular mechanisms responsible for the altered mitochondrial function and reduced quality control remain unclear.
The E3 ubiquitin ligase Parkin plays a critical role in marking dysfunctional mitochondria for degradation by autophagosomes (4). However, the exact functional role of Parkin-mediated mitophagy in the heart and other tissues is still unclear and under intense investigation. It was initially proposed that Parkin participates in the normal turnover of mitochondria in tissues. This notion originated from the accumulating evidence that mitochondrial defects play a central role in the pathogenicity of Parkinson’s disease (PD) and the fact that many patients diagnosed with familial PD have loss-of-function mutations in Park2 (5). In addition, muscle degeneration occurs in Parkin-null Drosophila due to accumulation of dysfunctional mitochondria and cell death (6). These studies indicated that the presence of Parkin was important for normal mitochondrial maintenance in cells. However, subsequent studies reported that Parkin deficiency in mice failed to recapitulate the behavioral signs and pathology of PD (7, 8). In addition, neither global nor cardiac-specific deletion of Parkin had any effect on cardiac structure or function in adult mice under normal conditions (9, 10). Rather, Parkin-mediated mitophagy was found to be rapidly activated in response to acute stress or altered metabolic demands in mouse hearts (10–12), suggesting that Parkin’s function in vivo is more complex than initially anticipated.
In this study, we have examined the role of Parkin in the context of mitochondrial DNA (mtDNA) damage and myocardial aging. mtDNA mutations accumulate with age in tissues in humans and animals, which can lead to decreased mitochondrial function (13–15). Mutant mice with accelerated mtDNA damage undergo premature aging, confirming a direct contribution of mitochondria in the aging process (16–18). The POLG mice are prone to age-dependent accumulation of mtDNA mutations and develop a broad spectrum of aging-like phenotypes, including cardiomyopathy. However, mitochondrial quality control and turnover in the heart have not been examined in this mouse model. Here, our findings demonstrate that Parkin plays a minor role in clearing dysfunctional mitochondria that have accumulated mtDNA damage due to the presence of a mutant POLG in vivo. Neither Parkin overexpression nor deficiency had any effect on the accelerated cardiac aging phenotype observed in POLG mice. Instead, our data show enhanced mitochondrial turnover via the mitophagy receptor pathway at baseline, which seems to limit the accumulation of dysfunctional mitochondria in the aging POLG hearts.
Results
Development of cardiac hypertrophy in POLG mice at 6 months of age.
Increased mtDNA mutations and mitochondrial dysfunction contribute to development of age-dependent cardiomyopathy (16, 17). To examine the relationship among Parkin, mtDNA damage, and cardiac aging, we utilized mice carrying a proofreading defective mtDNA polymerase γ (POLG) and evaluated their cardiac phenotype at 6 months of age. We found that, although POLG mice had a modest but significant increase in heart weight/body weight ratio compared with WT littermates (Figure 1A), they had similar cardiac structure and function to WT mice, as measured by echocardiography. WT and POLG mice had similar ejection fraction, fractional shortening, and left ventricular internal end-diastolic and -systolic dimensions (Figure 1, B and C).
Figure 1. Characterization of cardiac and mitochondrial functions in WT and POLG mice at 6 months of age.
(A) Heart weight/body weight (HW/BW) ratios of WT and POLG mice (n = 6–9, ***P < 0.001). Echocardiography showed similar (B) ejection fraction (EF) and fractional shortening (FS) and (C) left ventricular internal dimension at diastole (LVID;d) and systole (LVID;s) in WT and POLG hearts (n = 5–6). Assessment of mitochondrial respiration using isolated mitochondria from WT and POLG hearts show no differences in (D) state 3 respiration (ADP stimulated), (E) state 4 respiration (ADP depleted), (F) respiratory control ratio (RCR), or (G) maximal respiration rates (FCCP uncoupled) with substrates for complex I (pyruvate/malate and palmitoyl carnitine/malate) or II (succinate/rotenone) (n = 10). (H) Representative Western blot of proteins involved in mitochondrial oxidative phosphorylation. (I) Quantitation of proteins (n = 3). COX I, complex I subunit NDUFB8; COX II, complex II subunit 30 kDa; COX III, complex III subunit core 2; COX IV, complex IV subunit II; ATP Synthase, ATP synthase subunit α. Data are mean ± SEM (*P < 0.05). Statistical analysis was performed using Student’s t test.
Next, we investigated whether altered mitochondrial function contributed to the cardiac hypertrophy observed in POLG mice. Using mitochondria isolated from 6-month-old WT and POLG hearts, we assessed mitochondrial respiration in the presence of substrates for complex I (pyruvate/malate and palmitoyl carnitine/malate) or II (succinate/rotenone). WT and POLG mitochondria had similar state 3 (ADP-stimulated) and state 4 (ADP-depleted) respiratory rates for all 3 substrates (Figure 1, D and E). In addition, there were no significant differences in respiratory control ratio, an indicator of mitochondrial coupling, and uncoupled FCCP rates, a measurement of maximal electron transport chain capacity, for the different substrates (Figure 1, F and G). However, when examining levels of proteins involved in oxidative phosphorylation (OXPHOS) in WT and POLG hearts, we found a small but significant decrease in complex I and IV subunits (Figure 1, H and I). The complex I subunit is nuclear encoded (Ndufb8), while the complex IV subunit is mitochondrial encoded (Mtco1), suggesting that the POLG mutation affects subunits encoded by both genomes.
Parkin protein levels are reduced in POLG hearts at 6 months.
The E3 ubiquitin ligase Parkin plays an important role in clearing dysfunctional mitochondria, and studies have reported that Parkin is upregulated in response to mitochondrial stress in the heart (10, 19, 20). Interestingly, while Parkin mRNA levels were significantly increased at 6 months (Figure 2A), Parkin protein levels were significantly decreased both in whole heart lysates and at the mitochondria (Figure 2B). Thus, this suggests that Parkin-mediated mitophagy might be altered in POLG hearts. Studies have also found that Parkin is prone to misfolding and reduced solubility under certain cellular conditions, such as increased oxidative stress (21, 22). There is increased oxidative stress in the POLG hearts and overexpression of a mitochondrial targeted human catalase (mCAT) attenuates the development of the age-dependent cardiomyopathy observed in POLG mice (17). To examine the potential role of mitochondrial ROS in regulating Parkin levels, we crossed POLG mice with mCAT-transgenic mice to generate POLG mice with mCAT overexpression. However, the presence of mCAT did not restore Parkin levels in the hearts of POLG mice at 6 months (Figure 2C), suggesting that increased mitochondrial ROS is not responsible for the reduced levels of Parkin.
Figure 2. Characterization of Parkin expression and mitophagy in WT and POLG mice at 6 months of age.
(A) Real-time qPCR analysis of Park2 (Parkin) transcript levels (n = 3). (B) Representative Western blots and quantitation of Parkin in whole heart lysates and isolated mitochondrial fractions prepared from WT and POLG hearts (n = 3). (C) Representative Western blot and quantitation of Parkin in heart lysates prepared from WT, POLG, mCAT-transgenic, and POLG+mCAT-transgenic mice. mCAT, mitochondrial-targeted catalase (n = 3–5). Assessment of mitochondrial recruitment of Parkin in isolated myocytes from 6-month-old WT and POLG mice following treatment with rotenone (40 μM, 60 minutes). (D) Representative images of mCherry-Parkin and mitochondria stained with anti-Tom20 demonstrate translocation of Parkin to mitochondria in both WT and POLG myocytes after rotenone treatment. Scale bar: 20 μm. Original magnification ×63. (F) Line scan analysis using ImageJ software confirmed colocalization between mCherry-Parkin (red) and mitochondria (Tom20, green) after exposure to rotenone. Data are mean ± SEM (*P < 0.05; **P < 0.01). Statistical analysis was performed using Student’s t test or ANOVA.
To examine whether Parkin-mediated mitophagy was still functional in POLG myocytes, we assessed translocation of Parkin to dysfunctional mitochondria in myocytes isolated from 6-month-old WT and POLG mice. We found that mCherry-Parkin translocated to mitochondria after exposure to the complex I inhibitor rotenone (Figure 2, D and E), a known inducer of mitophagy in myocytes (23). We also observed an increase in the colocalization between mitochondria and autophagosomes in rotenone-treated WT and POLG myocytes, further confirming induction of mitophagy in the POLG myocytes (Supplemental Figure 1, A and 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127713DS1). In addition, we verified that autophagic flux was intact in POLG myocytes. Western blotting showed that LC3I and p62 levels were significantly reduced in POLG hearts (Supplemental Figure 1C). The p62 transcript levels were similar in WT and POLG myocytes (Supplemental Figure 1D), indicating that autophagic activity might be slightly enhanced in POLG hearts. However, evaluation of autophagic flux in isolated myocytes revealed similar flux in both WT and POLG cells (Supplemental Figure 1E). The proteasome is responsible for degrading the majority of intracellular proteins and plays a key function in cellular quality control. The 20S subunit in the proteasome is a multicatalytic protease with 3 types of enzymatic activity, described as trypsin-like, chymotrypsin-like, and caspase-like. To assess proteasomal activity, chymotrypsin-like, caspase-like, and trypsin-like activities were monitored in WT and POLG hearts. However, the proteasomal activities were similar in WT and POLG hearts (Supplemental Figure 1F). Overall, these findings suggest that the 2 main degradation pathways important in cellular quality control are not altered in the POLG hearts at 6 months of age.
Parkin deficiency or overexpression has no effect on the cardiac phenotype observed in aged POLG mice.
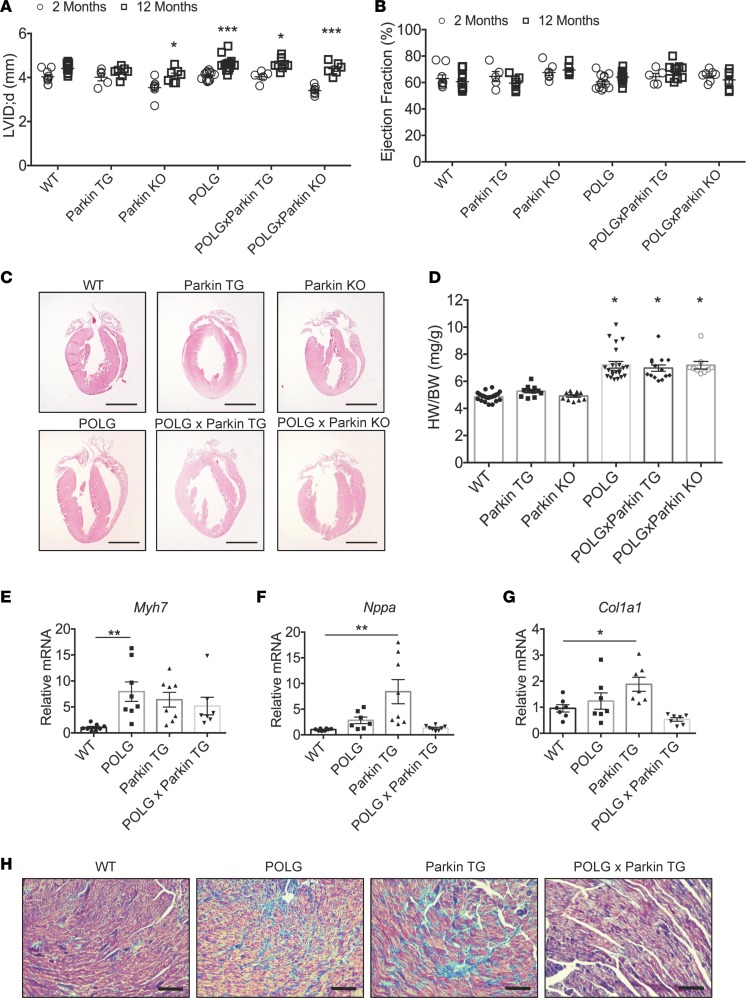
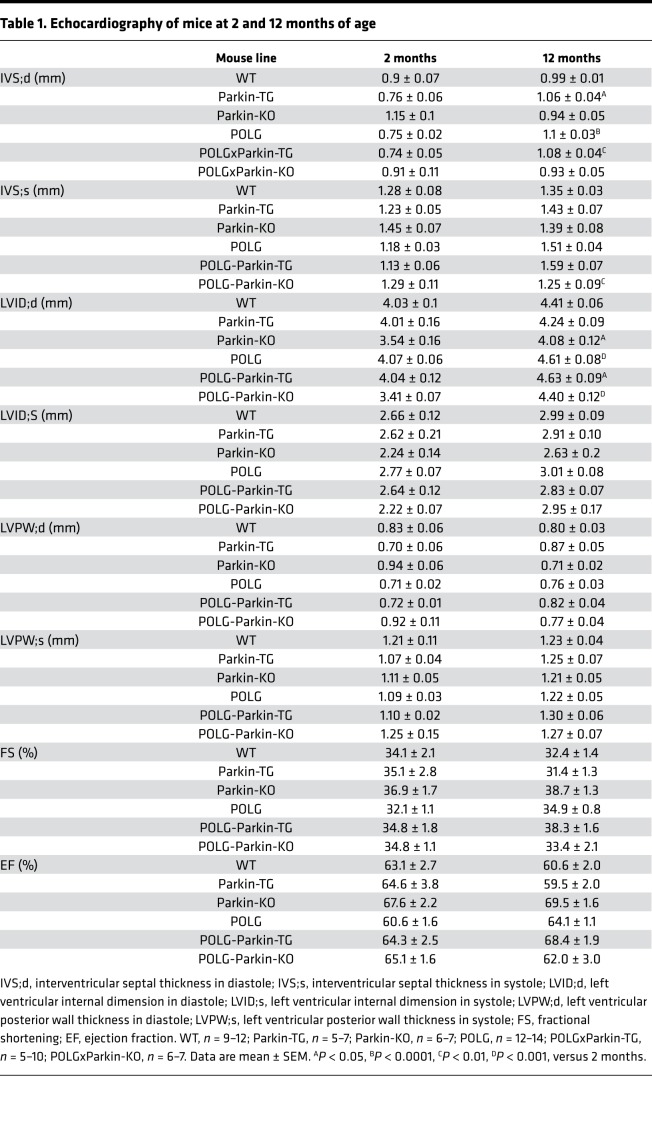
Because Parkin is important in clearing dysfunctional mitochondria, we investigated whether reduced Parkin-mediated mitophagy contributed to age-dependent mitochondrial defects and cardiac hypertrophy in POLG mice. We generated Parkin-deficient POLG (POLGxParkin-KO) mice to determine if the lack of Parkin would accelerate the cardiac aging phenotype observed in the mutant mice. Conversely, we generated mice with cardiac-specific overexpression of Parkin on the POLG background (POLGxParkin-Tg) to determine if restoring Parkin could rescue the accelerated cardiac aging phenotype caused by the mutated POLG. First, we performed echocardiography to evaluate cardiac structure and function in young (2 months) and aged (12 months) WT, Parkin-Tg, Parkin-KO, POLG, POLGxParkin-Tg, and POLGxParkin-KO mice. It has been well established that the heart undergoes structural changes with age with little effect on ventricular function (1). In agreement with this, echocardiography of young (2 months) and aged (12 months) mice revealed cardiac structural changes in the POLG mice with significant increased LV internal dimensions (Figure 3A) and interventricular septum thickness at 12 months (Table 1), confirming that these mice display accelerated cardiac aging. These structural changes were also evident in the POLG mice on the Parkin-deficient and Parkin-overexpressing backgrounds. In support of clinical studies suggesting that structural changes in the aging heart have little effect on resting systolic function (1), we observed no changes in ejection fraction (Figure 3B) and fractional shortening (Table 1), suggesting that baseline systolic function is unaltered in these mice at 12 months. All the data from the echocardiography analysis for the various mouse models are summarized in Table 1.
Figure 3. Characterization of heart function in WT, Parkin-TG, Parkin-KO, POLG, POLGxParkin-TG, and POLGxParkin-KO mice at 12 months of age.
(A) Echocardiogram analysis of left ventricular internal dimension in diastole (LVID;d) at 2 and 12 months of age and (B) ejection fraction at 2 and 12 months of age in WT (n = 5–10), Parkin-TG (n = 5–7), Parkin-KO (n = 6–7), POLG (n = 12–14), POLGxParkin-TG (n = 5–10), and POLGxParkin-KO mice (n = 6–7). (C) H&E staining of hearts. Scale bar: 2 mm. (D) Heart weight/body weight (HW/BW) ratios (n = 6–11). Real-time qPCR analysis of (E) β-myosin heavy chain (β-MHC/Myh7), (F) atrial natriuretic peptide (Nppa), and (G) collagen type 1 α 1 (Col1a1) transcript levels in hearts at 12 months (n = 7–8). (H) Representative images of Masson’s trichrome–stained hearts to visualize the extent of fibrosis. Scale bar: 100 μm. Data are mean ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). Statistical significance was calculated using ANOVA followed by Dunnett’s test for multiple comparison.
Table 1. Echocardiography of mice at 2 and 12 months of age.
In addition, we found that development of cardiac hypertrophy was similar in POLG, POLGxParkin-Tg, and POLGxParkin-KO mice following 12 months of aging (Figure 3, C and D). Examination of cardiac hypertrophy markers revealed significant increases in β myosin heavy chain (Myh7) in POLG mouse hearts (Figure 3E). Interestingly, although we did not observe increased heart size in Parkin-Tg mice at this age, we found a significant upregulation of atrial natriuretic factor (Nppa) in the hearts overexpressing the Parkin transgene alone (Figure 3F). We also found that collagen type 1 α 1 (Col1a1) mRNA levels were significantly increased in the Parkin-Tg mouse hearts (Figure 3G). Furthermore, Masson’s trichrome staining of the aged hearts revealed elevated levels of fibrosis in both POLG and Parkin-Tg mice, indicating that chronic elevation of Parkin expression is not healthy for the heart (Figure 3H). Overall, these findings suggest that Parkin plays a minimal role in preventing the accelerated cardiac aging process observed in the POLG mice.
Parkin-mediated mitophagy is reduced in aged POLG hearts.
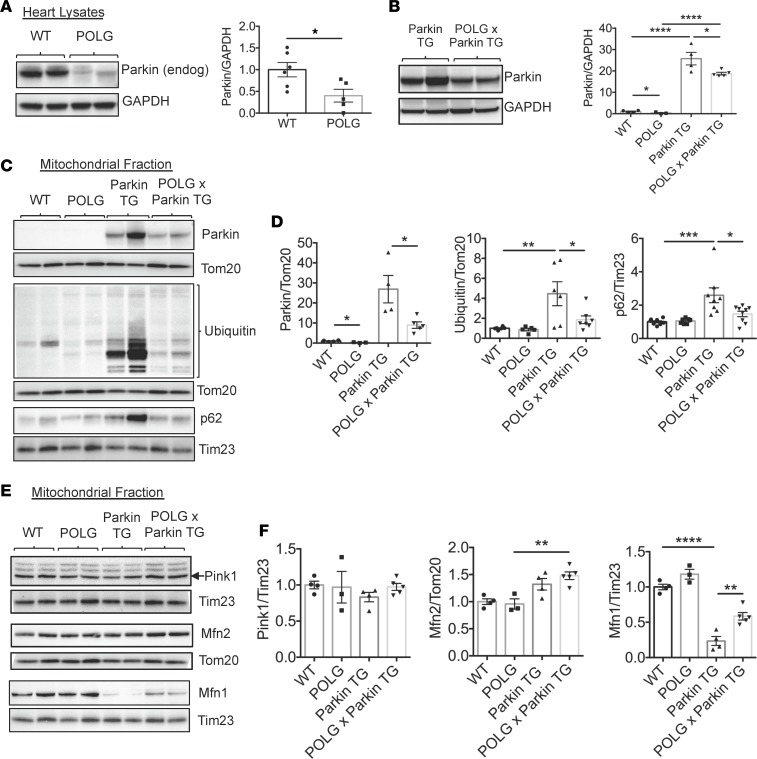
Next, we examined whether Parkin and mitophagy were altered at 12 months in the hearts of POLG mice. Parkin protein levels in the heart were still significantly reduced in POLG mice compared with WT mice (Figure 4A). Interestingly, Parkin protein levels were also significantly reduced in the POLG xParkin-Tg heart (Figure 4B). This prompted us to examine whether downstream targets of Parkin were altered in the POLG hearts. Parkin mediates mitophagy by translocating to mitochondria where it ubiquitinates outer membrane proteins (4). As previously reported (20), we found abundant levels of Parkin, protein ubiquitination, and the adaptor protein p62 in the mitochondrial fraction in Parkin-Tg hearts (Figure 4, C and D), indicating activation of Parkin-mediated mitophagy. We also found increased levels of Parkin in the cardiac mitochondrial fraction of POLGxParkin-Tg mice compared with POLG mice, which correlated with increased levels of protein ubiquitination and p62 (Figure 4, C and D), indicating that Parkin-mediated mitophagy is restored, although to a reduced extent compared with Parkin-Tg hearts. Thus, these data suggest that restoring Parkin-mediated mitophagy in the POLG hearts does not rescue the cardiac hypertrophy that develops with age in these mice.
Figure 4. Assessment of Parkin expression and mitophagy in aged hearts.
(A) Representative Western blot and quantitation of endogenous Parkin in lysates prepared from WT and POLG hearts at 12 months of age (n = 3–4). (B) Representative Western blot and quantitation of Parkin transgene expression in lysates prepared from Parkin-TG and POLGxParkin-TG hearts at 12 months of age (n = 4–5). (C) Representative Western blot analysis of proteins present in the mitochondrial fraction prepared from WT, POLG, Parkin-TG, and POLGxParkin-TG hearts at 12 months. (D) Quantitation of Parkin, ubiquitin, and p62 in the mitochondrial fraction (n = 6–9). (E) Representative Western blot analysis of proteins present in the mitochondrial fraction prepared from WT, POLG, Parkin-TG, and POLGxParkin-TG hearts at 12 months. (F) Quantitation of PTEN-induced kinase 1 (Pink1), mitofusin 2 (Mfn2), and mitofusin 1 (Mfn1) (n = 3–5). Data are mean ± SEM (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001). Statistical significance was calculated using Student’s t test or ANOVA followed by Dunnett’s test for multiple comparison.
While total levels of Parkin were significantly lower in POLGxParkin-Tg hearts compared with Parkin-Tg mice (~27% decrease), the difference in Parkin protein levels in mitochondrial fractions was substantially greater, around 67% lower in POLGxParkin-Tg hearts compared with Parkin-Tg mice. This led us to hypothesize that Parkin recruitment to mitochondria might be affected in POLG hearts. Pink1 and Mfn2 are both responsible for recruiting Parkin to mitochondria (24, 25). Therefore, we investigated if altered levels of Pink1 and Mfn2 were responsible for the reduced amount of mitochondrial Parkin in the POLGxParkin-Tg mice. We found that Pink1 and Mfn2 levels were not decreased in the mitochondrial fractions of POLG and POLGxParkin-Tg hearts (Figure 4, E and F). Mfn1 is a known Parkin substrate, and ubiquitination of Mfn1 leads to its degradation (26). We observed reduced Mfn1 levels in the Parkin-Tg and POLGxParkin-Tg mouse hearts (Figure 4, E and F); however, consistent with decreased levels of Parkin in the mitochondrial fraction (Figure 4D), degradation of Mfn1 was reduced in POLGxParkin-Tg hearts compared with Parkin-Tg hearts.
POLG hearts have increased mitochondrial turnover at baseline.
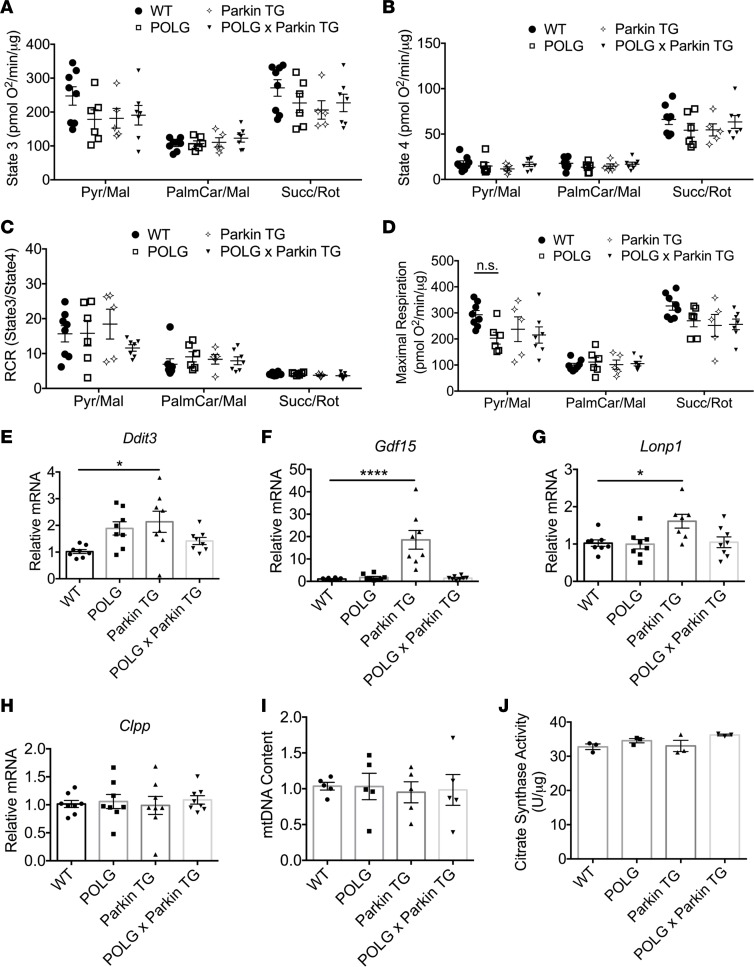
Next, we assessed if mitochondrial respiration in aged (12-month-old) POLG hearts was impaired with age and whether restoration of Parkin-mediated mitophagy preserved mitochondrial health in the aging hearts. Unexpectedly, mitochondrial respiration and health had not deteriorated as would be expected with aging in the POLG hearts. We found that POLG and POLGxParkin-Tg mitochondria showed no significant differences in state 3 (ADP-stimulated) and state 4 (ADP-depleted) respiratory rates when compared with WT mitochondria (Figure 5, A and B). In addition, there were no significant differences in respiratory control ratios and maximal respiration rates for the complex I and complex II substrates (Figure 5, C and D). To further evaluate the level of mitochondrial stress in the aged POLG mice, we examined activation of the mitochondrial unfolded protein response (UPRmt). The UPRmt is a transcriptional response activated by mitochondrial dysfunction to promote repair and cell survival (27). Interestingly, we found that expression of Ddit3 (also known as CHOP), a transcription factor that mediates the UPRmt, the myomitokine Gdf15, and mitochondrial proteases Lonp1 and Clpp were not increased in the aged POLG and POLGxParkin-Tg hearts compared with WT hearts (Figure 5, E–H). However, we found significantly elevated levels of Ddit3, Gdf3, and Lonp1 transcripts in Parkin-Tg hearts, indicative of increased mitochondrial stress. We also confirmed that the mitochondrial content did not differ in the various mouse hearts by assessing mtDNA content (Figure 5I) and mitochondrial citrate synthase activity (Figure 5J).
Figure 5. Evaluation of mitochondrial health in aged hearts.
Assessment of mitochondrial respiration using isolated mitochondria from WT, POLG, Parkin-TG, and POLGxParkin-TG hearts show no differences in (A) state 3 respiration (ADP stimulated), (B) state 4 respiration (ADP depleted), (C) respiratory control ratio (RCR), or (D) maximal respiration rates (FCCP uncoupled) with substrates for complex I (pyruvate/malate and palmitoyl carnitine/malate) or II (succinate/rotenone) (n = 10). (n = 5–8). Real-time qPCR analysis of transcript levels for mitochondrial unfolded protein response (UPRMito) markers to evaluate levels of mitochondrial stress in the aged heart: (E) DNA damage–induced transcript 3 Ddit3, (F) growth/differentiation factor 15 (Gdf15), (G) lon protease homolog (Lonp1), and (H) caseinolytic mitochondrial matrix peptidase proteolytic subunit (Clpp) in aged hearts (n = 7–8). (I) Mitochondrial DNA (mtDNA) content was assessed by qPCR using primers to the mitochondrial D-Loop and nuclear 18s (n = 5). (J) Quantification of mitochondrial citrate synthase activity in heart lysates (n = 3). Data are mean ± SEM (*P < 0.05; ****P < 0.0001). Statistical significance was calculated using ANOVA followed by Dunnett’s test for multiple comparison.
Finally, we used transmission electron microscopy (TEM) to examine POLG hearts at the ultrastructural level (Figure 6A and Supplemental Figure 2). Mitochondrial cristae structure was similar in all 4 mouse lines. Interestingly, TEM revealed the presence of enlarged mitochondria in POLG and POLGxParkin-Tg myocytes. Confocal imaging of mitochondria in heart sections confirmed the presence of enlarged mitochondria in POLG and POLGxParkin-Tg hearts (Figure 6B). This enlargement of mitochondria, termed megamitochondria, is often seen in tissues with age and in certain types of cardiomyopathy (28–30).
Figure 6. Identification of megamitochondria in aged POLG hearts.
(A) Representative transmission electron micrographs of WT, POLG, Parkin-TG, and POLGxParkin-TG heart tissue at 12 months. Scale bar: 1 μm. (B) Representative confocal micrographs of myocytes stained with an antibody against mitochondrial complex IV subunit 1 in longitudinal and transverse sections. Scale bar: 10 μm.
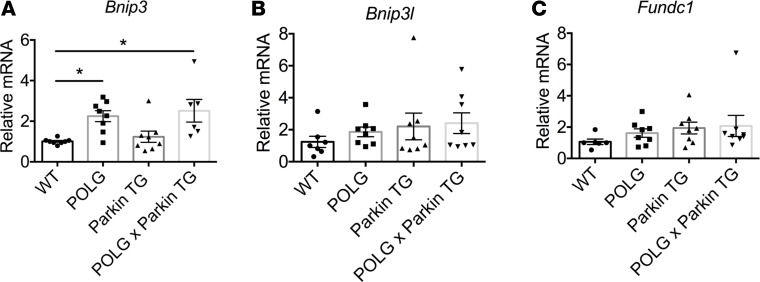
The lack of exacerbated mitochondrial impairment in the POLG hearts led us to investigate whether there was an increase in mitochondrial turnover in these hearts to replace dysfunctional mitochondria that had accumulated excessive mtDNA damage. Mitophagy can be mediated by the Pink1/Parkin pathway or via mitophagy receptors in the outer mitochondrial membrane. Because our findings suggested that Parkin-mediated mitophagy played a minimal role in POLG hearts at baseline, we examined whether mitophagy receptor levels were altered in POLG hearts. We found a significant increase in BNIP3 transcript levels in POLG and POLGxParkin-Tg hearts compared with WT and Parkin-Tg hearts (Figure 7A). There were no significant changes in BNIP3L/Nix and Fundc1 transcript levels (Figure 7, B and C).
Figure 7. Assessment of mitophagy receptors in aged hearts.
Real-time qPCR analysis was used to assess mRNA transcript levels of mitophagy receptors in hearts at 12 months of age: (A) BCL2-interacting protein 3 (Bnip3), (B) BCL2-interacting protein 3–like (Bnip3l), and (C) FUN14 domain–containing 1 (Fundc1). Bnip3 transcript levels were significantly increased in POLG and POLGxParkin-TG hearts when compared with WT (*P < 0.05, n = 8). Data are mean ± SEM. Statistical significance was calculated using ANOVA followed by Dunnett’s test for multiple comparison.
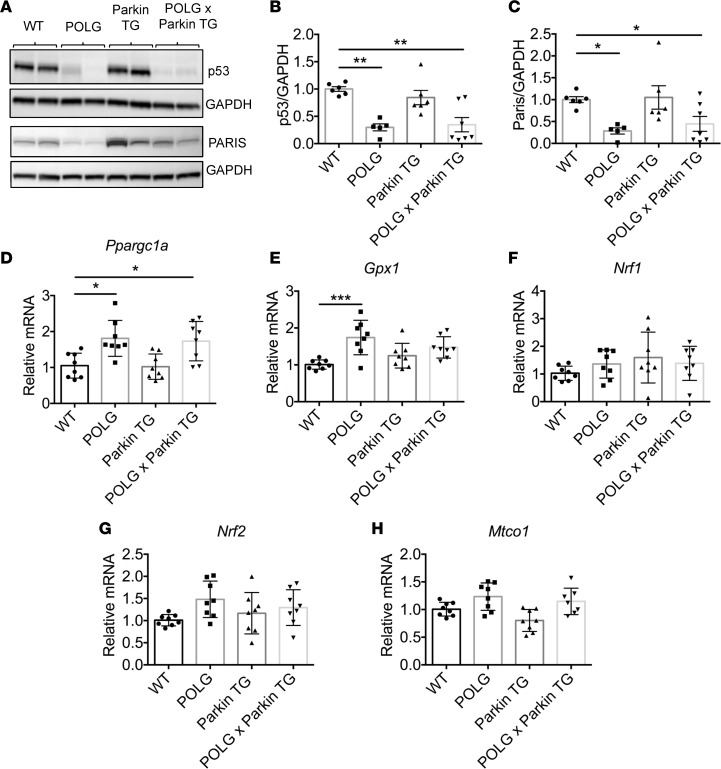
Increased p53 activity is associated with cellular senescence and aging, but recent studies have also identified p53 as a repressor of peroxisome proliferator–activated receptor γ coactivator 1α (PGC-1α), a master coregulator of mitochondrial biogenesis, in the heart (31). Examination of p53 levels in the aged hearts revealed that p53 protein levels were significantly reduced in the POLG and POLGxParkin-Tg hearts but not in Parkin-Tg hearts (Figure 8, A and B). Parkin-interacting substrate (PARIS) (also known as zinc finger protein 746) is ubiquitinated by Parkin targeting it for proteasomal degradation (32). PARIS is a transcriptional repressor that regulates the expression of PGC-1α (33). Similar to p53, PARIS protein levels were reduced in the POLG and POLGxParkin-Tg hearts but unaltered in Parkin-Tg hearts (Figure 8, A and C). Consistent with the reduced p53 and PARIS levels, we found significantly increased PGC-1α (Ppargc1a) transcript levels in POLG and POLGxParkin-Tg hearts compared with WT hearts (Figure 8D). PGC-1α regulates cellular antioxidant homeostasis and mitochondrial biogenesis by regulating gene expression of glutathione peroxidase 1 (Gpx1) and nuclear respiratory factors (Nrf1/2). There was a significant increase in Gpx1 expression in POLG hearts compared with WT hearts, while Nrf1 and 2 also trended higher in POLG hearts but did not reach statistical significance (Figure 8, E–G). In addition, levels of mitochondrially encoded cytochrome c oxidase I (Mtco1) trended higher in POLG and in POLGxParkin-Tg hearts, consistent with elevated levels mitochondrial biogenesis in these groups (Figure 8H). These findings suggest that the POLG hearts have increased levels of mitochondrial turnover at baseline.
Figure 8. Mitochondrial biogenesis is increased in aged POLG and POLGxParkin-TG hearts.
Assessment of tumor protein p53 and Parkin-interacting substrate (PARIS) protein levels in aged hearts. (A) Representative Western blots of p53 and PARIS. Quantitation of (B) p53 and (C) PARIS protein levels show a significant decrease in POLG and POLGxParkin-TG hearts when compared with WT at 12 months (n = 5–7). Real-time qPCR analysis to measure transcript levels of (D) peroxisome proliferator–activated receptor γ coactivator 1-α (Ppargc1a), (E) glutathione peroxidase 1 (Gpx1), (F) nuclear respiratory factor 1 (Nrf1), (G) nuclear factor, erythroid derived 2, like 2 (Nrf2), and (H) cytochrome c oxidase subunit 1 (Mtco1) in aged hearts (n = 8). Data are mean ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). Statistical significance was calculated using ANOVA followed by Dunnett’s test for multiple comparison.
Discussion
The results in this study provide what we believe to be important new insights into Parkin’s function in the heart. Although Parkin is well known to promote mitophagy of damaged mitochondria, our study shows that Parkin plays a minor role in clearing mitochondria with mtDNA damage to prevent the cardiac aging process. In addition, our data suggest that the POLG mutation leads to enhanced baseline mitochondrial turnover in the heart via the mitophagy receptor pathway, which may function to limit accumulation of dysfunctional mitochondria in the aging heart. Finally, our study shows that mtDNA damage directly contributes to development of cardiac hypertrophy with aging prior to mitochondrial respiratory defects, demonstrating the diverse functional role of mitochondria in cellular homeostasis.
The functional role of Parkin-mediated mitophagy in the normal maintenance of mitochondria in the heart has been questioned due to the lack of a cardiac phenotype in Parkin-KO mice under baseline conditions and the fact that patients with familial PD do not exhibit cardiac impairment (5, 10, 20, 34). Combined with the observation that cardiac Parkin is rapidly upregulated in mouse models suffering mitochondrial stress (10, 19, 20), these findings emphasize the importance of the Parkin pathway in the response to stress, rather than baseline mitochondrial maintenance. Our study supports this notion. However, it is possible that Parkin’s function in ensuring mitochondrial health might vary with the tissue. Another group recently generated POLGxParkin-KO mice to evaluate them in the context of PD, and they observed increased loss of DA neurons and motor defects in the aged POLGxParkin-KO mice when compared with POLG mice (35). This suggests different roles for Parkin-mediated mitophagy in brain and heart. This would also explain why patients with loss-of-function mutations in Parkin develop PD and not heart failure.
It is well known that the aged heart has lowered tolerance to stress, and ischemic insults result in larger infarcts and increased mortality after I/R (36). In addition, cellular quality control pathways, including autophagy, are reduced with age in various tissues (2, 3). Our study provides a link between mtDNA damage and reduced Parkin-mediated mitophagy as one of the potential causes underlying this increased susceptibility in the aging population. We and others have previously reported that Parkin is important in clearing mitochondria in response to myocardial ischemia (10, 12). In this study, we found that the POLG mutation contributed to decreased Parkin levels in the heart, suggesting that these hearts will have a reduced capability in clearing damaged mitochondrial after acute stress. Our findings also show that recruitment of Parkin to POLG mutant mitochondria is reduced and it is possible that elevated ROS levels affects Parkin activity. Parkin contains multiple cysteine residues that are susceptible to modification by oxidative stress, which can promote Parkin misfolding (21, 22). A growing body of evidence indicates that misfolding is a major mechanism of Parkin inactivation in neurons (21, 37–39). Although our study focused on Parkin in the heart, it also has important implications for other tissues. Loss-of-function mutations in Parkin have primarily been implicated in familial PD (40). However, hereditary PD is relatively rare and the majority of PD is sporadic and occurs in the aging population (5). mtDNA mutations also accumulate in neurons with age, and our results suggest a potential link among mtDNA damage, reduced Parkin levels, and the development of sporadic PD in older patients.
An unexpected finding in our study was that the mitochondria still appeared relatively healthy at 12 months of age despite the POLG mutation. Although the lack of a respiratory defect could potentially be due to loss of damaged mitochondria during the preparation, we do not believe that this is a major factor since the TEM did not indicate the presence of swollen mitochondria in the POLG hearts. In addition, a previous study reported that transgenic mice overexpressing a proofreading-deficient POLG (D181A) mutant in the heart accumulated mtDNA mutations at an accelerated rate but without an effect on mitochondrial respiration (41). Instead, the normal mitochondrial respiration in POLG hearts is likely due to 3 factors: (a) enhanced (Parkin-independent) mitochondrial turnover at baseline in POLG hearts, (b) a high tolerance (threshold) for mtDNA mutations in cardiac mitochondria, and (c) formation of megamitochondria, which might function to dilute the mtDNA damage.
First, we found that transcript levels of the mitophagy receptor BNIP3 were significantly increased in both POLG and POLGxParkin-Tg hearts, suggesting enhanced mitophagy receptor–mediated mitochondrial clearance in these hearts. This pathway has previously been reported to function in mitochondrial maintenance at baseline, and myocytes deficient in BNIP3/Nix have accelerated accumulation of dysfunctional mitochondrial in the heart with age (42). Removal of mitochondria must be combined with increased synthesis. Accordingly, markers of mitochondrial biogenesis were concurrently elevated in the POLG hearts, suggesting enhanced mitochondrial turnover at baseline. Dillon and colleagues previously reported that POLG mice develop sarcopenia and that increased expression of PGC-1α in skeletal muscle led to enhanced mitochondrial biogenesis and improved muscle function (43). Thus, the enhanced mitochondrial turnover at baseline in the POLG hearts might function to replace severely dysfunctional mitochondria with high levels of mtDNA damage. Second, the heart appears to be remarkably resistant to mtDNA mutations, and a very high proportion of mutations are needed before mitochondrial respiratory defects and cardiac dysfunction become evident (44). Each mitochondrion contains multiple genome copies and will harbor a mix of normal and mutated mtDNA. Once a threshold level of mtDNA mutational load has been reached, it will lead to a defect in respiration (44). The initial characterization of POLG mice showed that the frequency of mtDNA mutations in hearts at 5–6 months was about 3-fold higher compared with WT mice (16). Subsequently, it was reported that mtDNA deletions were about 10-fold higher in POLG hearts at 13–14 months (17). These studies did not directly evaluate mitochondrial function in these hearts, but our results suggest that the threshold effect is high in the heart. Finally, the presence of enlarged or megamitochondria has been observed in aged tissues, including the heart (45), and it is likely that the formation of megamitochondria in the POLG hearts contributes to the protection against mtDNA damage. Fusion of mitochondria leads to mixing of normal and mutated mtDNA (46). Thus, the dilution of the mutated DNA protects mitochondrial function and allows the myocytes to tolerate high levels of mtDNA mutations (47).
Another unexpected finding is that cardiac hypertrophy develops before any major respiratory defect is observed. This implies that other deleterious effects of the mtDNA mutations exist in myocytes. A similar observation was made by Zhang and colleagues, who reported that, although accumulation of mtDNA mutations in αMHC-POLG(D181A)-transgenic mice was accompanied by a severe cardiomyopathy, cardiac mitochondrial respiration was unaltered (41). In our study, ultrastructural analysis confirmed that the cardiac mitochondria in POLG mice had similar cristae density and structure as WT mice at this age, suggesting that the POLG mutation might effect another key function of mitochondria. Although mitochondria are mostly considered to be responsible for the generation of ATP, they also play a key role in the biosynthesis of macromolecules, such as lipids, heme, and iron-sulfur clusters (48, 49). More recent studies have reported that mitochondria also function as signaling organelles to regulate diverse cellular functions in cells (50), and it is possible that mtDNA mutations alter these additional mitochondrial functions in cells, possibly even at a lower threshold. However, additional studies are needed to focus on how mtDNA mutations alter these other mitochondrial functions in the cell and how this contributes to aging.
A number of intrinsic changes occur in the aging heart that increase susceptibility to insult, including reduced repair processes that are designed to deal with cellular damage. mtDNA damage has been proposed to directly contribute to cardiac hypertrophy and dysfunction in the aging population (1, 17), suggesting that therapies aimed at enhancing mitochondrial quality control could prevent or at least delay aging. Future studies should focus on unanswered questions, such as, do reduced Parkin levels due to mtDNA damage lead to increased susceptibility to injury in the aged heart? How does mtDNA damage in the heart contribute to development of hypertrophy in the absence mitochondrial respiratory defects? What specific mitochondrial functions are altered by mtDNA damage? Increased understanding of the aged heart will enable the development of therapies that prevent the development of heart failure or treat the failing heart.
Methods
Animals and generation of mouse lines.
Mice are on a C57bl/6J background and have been backcrossed for a minimum of 10 generations. POLGD257A mice were obtained from Tomas Prolla, University of Wisconsin-Madison, Madison, Wisconsin, USA (16). Parkin–/– mice were obtained from Jackson Laboratories and their cardiac phenotype has been previously characterized (10). The Parkin-Tg mice overexpress human Parkin under the transcriptional control of the cardiomyocyte-specific α-myosin heavy chain (α-MHC) promoter and were generated at the UCSD Mouse Transgenic Core.
Mice heterozygous for the POLGD257A mutation were crossed to generate mice with WT and mutant POLGD257A/D257A genotypes. Two additional mouse lines were also generated. Heterozygous POLGD257A mice were crossed with Parkin-Tg mice to obtain mice with WT, POLGD257A/D257A, Parkin-Tg, and POLGD257A/D257A/Parkin-Tg genotypes. Heterozygous POLGD257A mice were also crossed with Parkin–/– mice to obtain mice with WT, POLGD257A/D257A, Parkin–/–, and POLGD257A/D257A/Parkin–/– genotypes. These mice are referred to as WT, POLG, Parkin-TG, POLGxParkin-Tg, Parkin-KO, and POLGxParkin-KO throughout. Both male and female mice were used in all experiments. WT littermates were used as control in the experiments.
Adult mouse myocyte isolation.
Adult mouse cardiomyocytes were isolated from 6-month-old WT and POLG mice as previously described (19). For microscopy experiments, cardiomyocytes were plated on 35 mm glass-bottom dishes (MatTek) coated with laminin (10 μg/ml, Invitrogen) at a density of 100,000 cells per dish. Cells were allowed to adhere for 2 hours before infection with GFP-LC3, mCherry-Parkin, or β-Gal adenoviruses at a MOI of 100. Cells were incubated with adenoviruses for 3 hours, rinsed with plating medium, and then allowed to recover in plating medium for 18 hours before treatment.
For Western blot experiments, cardiomyocytes were plated on 60 mm laminin-coated tissue culture dishes at a density of 200,000 cells per dish. Cells were allowed to adhere overnight prior to treatment with Bafilomycin A1 (50 nM, Millipore) or vehicle control (DMSO, Sigma) for 18 hours. Cells were lysed in Triton X-100 lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail (Roche). Protein concentrations were determined by Bradford assay.
Quantitative PCR.
For gene expression assays, RNA was extracted from heart tissue using the RNeasy Fibrous Tissue Mini Kit (Qiagen). Genomic DNA was isolated from hearts using the GenElute Mammalian Genomic DNA Miniprep Kit (MilliporeSigma). cDNA was synthesized from 500 ng RNA using the QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer’s protocols. TaqMan primers for Ppargc1a, Park2, Myh7, Nppa, Col1a1, Sqstm1, Atg5, Atg7, Becn1, Rab7, Ddit3, Gdf15, Lonp1, Clpp, Bnip3, Bnip3l, Fundc1, Gpx1, Nrf1, Nrf2, Mtco1, D-Loop, and Rn18s were purchased from Life Technologies-Thermo Fisher Scientific. The TaqMan Universal Master Mix II was purchased from Applied Biosystems/Life Technologies. qPCR was carried out on a CFX96 Real-Time PCR Detection System (Bio-Rad). Relative amounts of mRNA were normalized to Rn18s, and fold change in gene expression was calculated using the 2(–ΔΔCt) method.
Subcellular fractionation and Western blot.
Hearts were minced in homogenization buffer containing 250 mM sucrose, 5 mM KH2PO4, 2 mM MgCl2, 10 mM MOPS, pH 7.4, 1 mM EGTA, 0.1% fatty acid-free BSA, and protease inhibitor cocktail (Roche). Hearts were homogenized by Polytron at 11,000 rpm, followed by 3–4 strokes using the Potter-Elvehjem Teflon tissue grinder to further homogenize the tissue. 1% Triton X-100 buffer was added to the homogenates and incubated on ice for 45 minutes to induce further cell lysis, followed by centrifugation at 20,000 g for 20 minutes to obtain cleared whole heart lysates.
To obtain the mitochondrial fraction, whole heart homogenates were centrifuged at 600 g for 5 minutes at 4°C to remove nuclear contamination and debris. The supernatant was transferred to a new tube, and the centrifugation step was repeated. The supernatant was collected and transferred to another tube and centrifuged at 3000 g for 10 minutes at 4°C. The resulting supernatant contained the cytosolic fraction, and the pellet contained the mitochondrial fraction. The mitochondrial pellet was washed in homogenization buffer and centrifuged at 3000 g for 10 minutes at 4°C; this wash step was performed 3 times. The resulting mitochondria pellet was resuspended and lysed in Triton X-100 lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail (Roche). Protein concentrations for whole heart lysates and mitochondrial fractions were determined by Bradford assay.
Samples were loaded and run on Invitrogen NuPAGE Bis-Tris gels. The membranes were probed with the following antibodies: Actin (1:1000; GeneTex, GTX109638), Catalase (1:1000; Athens Research and Technology, 01-05-030000), GAPDH (1:1000; GeneTex, GTX627408), LC3 (1:1000; Cell Signaling, 4108), MFN1 (1:1000; Santa Cruz Biotechnology, sc-50330), MFN2 (1:1000; MilliporeSigma, M6319), MitoProfile Total OXPHOS Rodent WB Antibody cocktail (1:1000; Abcam/MitoSciences, MS604), p62 (1:1000; Abcam, ab56416), PARIS (1:1000; EMD Millipore, MABN476), Parkin (1:1000; Cell Signaling, 4211), PINK1 (1:1000; Cayman, 10006283), Tim23 (1:1000; BD Biosciences, 611222), Tom20 (1:1000; Santa Cruz, sc-11415), Ubiquitin (1:200; Santa Cruz Biotechnology, sc-8017), and Tubulin (WB 1:2000; MilliporeSigma, T6074). Membranes were imaged using a ChemiDoc XRS+ System (Bio-Rad). See complete unedited blots in the supplemental material.
Mitochondrial respiration measurements of isolated mitochondria.
Intact mitochondria were isolated from 6-month-old and 12-month-old mouse hearts by investigators blinded to the genotype, and mitochondrial respiration was measured using the Seahorse XF96 Analyzer (Seahorse Bioscience – Agilent Technologies) following a modified protocol originally optimized in the Seahorse XF24 Analyzer (51). Hearts were homogenized using a Polytron followed by a Potter-Elvehjem Teflon tissue grinder in buffer containing 70 mM sucrose, 210 mM mannitol, 5 mM HEPES, 1 mM EGTA, and 0.5% fatty acid-free BSA. The homogenate was centrifuged at low speed (500 g for 10 minutes at 4°C). The supernatant was collected and centrifuged at 12,000 g for 10 minutes at 4°C to obtain a mitochondrial pellet. The pellet was washed in buffer and centrifuged at 12,000 g for 10 minutes at 4°C, and this wash step was performed twice. Finally, the pellet was resuspended in a small volume (~20 μl) of buffer, and the protein concentration was determined by Bradford assay. Mitochondria were plated on a 96-well microplate at densities of either 0.5 μg or 1 μg per well. Assay media (70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA, 0.2% BSA, and 4 mM ADP) containing different substrates were used to assess glucose metabolism (10 mM pyruvate and 1 mM malate), fatty acid oxidation (40 μM palmitoylcarnitine and 1 mM malate), and complex II–dependent oxygen consumption (10 mM succinate and 2 μM rotenone). Oligomycin (2 μM) was added to inhibit ATP synthase followed by 3 successive additions of 1.5 μM FCCP to obtain maximal respiration rate. Data were analyzed using Wave for Desktop (Seahorse Bioscience – Agilent Technologies).
Citrate synthase assay.
Citrate synthase activity was determined in heart tissue using the Citrate Synthase Assay Kit (Abcam), following the manufacturer’s instructions.
Proteasomal activity assay.
Hearts were homogenized in assay buffer (50 mM HEPES, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, 2 mM ATP, 5 mM DTT, pH 7.8) and centrifuged at 16,000 g for 10 minutes. The resulting supernatant was transferred to a new tube, and protein concentration was determined by Bradford assay. 10 μg protein was added per well of a clear-bottomed/black-walled 96-well plate (MilliporeSigma) along with 195 μl assay buffer and 5 μl of fluorescent substrate (chymotrypsin like, trypsin like, or caspase like, Enzo). Each sample was measured in triplicate. Plates were incubated for 60 minutes at 37°C, 5% CO2. Fluorescence was measured at A360ex/A460em on a fluorescent plate reader.
Echocardiography.
Echocardiography was performed as previously described using a Vevo770 In Vivo Micro-Imaging System with an RMV707B 15–45 MHz imaging transducer (VisualSonics Inc.) (10). Mice were maintained under light anesthesia through a nose cone (0.5%–1% isoflurane, 98%–99.5% O2). Mice were placed in a supine position on a recirculating water warming pad while acquiring measurements. Cardiac parameters were quantified using the VisualSonics software.
Histology.
Prior to tissue collection, mice were anesthetized with 100 mg/kg sodium pentobarbital. Hearts were perfused with 200 mM KCl to induce arrest at diastole before harvesting. Hearts were fixed in 10% neutral buffered formalin for 24 hours and transferred to 70% dehydrant alcohol for another 24 hours. Hearts were processed, embedded in paraffin, and cut into sections using a microtome (Leica Biosystems). To visualize general size and wall thickness, hearts were cut at the coronal plane to obtain a 4-chamber view. Sections were cut at 10-μm thickness and stained with H&E. To visualize fibrosis, sections were stained with Masson’s trichrome (MilliporeSigma). Sections were imaged using a Carl Zeiss Axio Cam ICc3 camera attached to a dissection microscope (Zeiss Stemi 2000-C). Images were taken at ×1.0 magnification.
Fluorescence microscopy.
Myocytes were treated with DMSO or 40 μM rotenone in plating medium for 1 hour. Cells were fixed in 4% paraformaldehyde and permeabilized in 0.2% Triton X-100 in PBS for 30 minutes at 37°C. Blocking and antibody applications were performed in 5% normal goat serum plus 0.2% Triton X-100 in PBS. Mitochondria were labeled using anti-Tom20 (1:100; Santa Cruz Biotechnology, sc-11415). Following antibody labeling, Hoechst 33342 (Life Technologies) was applied to label nuclei. Cells were imaged by fluorescence microscopy on a Carl Zeiss AxioObserver Z1 equipped with a motorized Z-stage and an ApoTome for optical sectioning. Z-stacks were acquired in ApoTome mode using a high-resolution AxioCam MRm digital camera, a ×63 Plan-Apochromat oil-immersion objective, and Zeiss AxioVision 4.8 software (Carl Zeiss). Pseudo-line scans were performed using ImageJ software (NIH).
Hearts were collected and processed as described for histology. Cross sections of paraffin-embedded hearts were cut at 5 μm and deparaffinized. Antigen retrieval was performed by boiling sections in 10 mM Citrate Buffer (Sodium citrate + 0.05% Tween20, made in-house) (pH 6.6) for 15 minutes. Blocking and antibody applications were performed in a mixture of 5% normal goat and 5% normal rabbit serum in PBS. Mitochondria were labeled with complex IV subunit 1 (IF 1:50; Novex/ThermoFisher Scientific, 459600). Stained sections were sealed with a cover slip and Vectashield hard set mounting media with DAPI (Vector Laboratories). Sections were imaged on a Zeiss LSM 880 Confocal with FAST Airyscan System. Three-color FAST Airyscan mode was used with ×63 Plan-Apochromat oil-immersion objective and Zen Black Zeiss acquisition and processing software.
TEM and morphometric analysis.
Adult mouse hearts were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer; post-fixed in 1% osmium tetroxide; then treated with 0.5% tannic acid, 1% sodium sulfate; cleared in 2-hydroxypropyl methacrylate; and embedded in LX112 (Ladd Research). Sections were mounted on copper slot grids coated with parlodion and stained with uranyl acetate and lead citrate for examination on a Philips CM100 electron microscope (FEI).
Statistics.
All values are expressed as mean ± SEM. Student’s 2-tailed t test was used to compare 2 sets of data. For data comparing more than 2 groups, statistical analyses were performed using 1-way or 2-way ANOVA, followed by Dunnett’s multiple comparison test (GraphPad Prism 7). P values of less than 0.05 were considered statistically significant.
Study approval.
All experimental procedures were performed in accordance with institutional guidelines and approved by the Institutional Animal Care and Use Committee of the University of California, San Diego.
Author contributions
ÅBG, BPW, and AMO designed the study, analyzed the experiments, and wrote the paper. BPW, AMO, RHN, and MQC designed, performed, and analyzed the majority of the experiments. ERM performed the confocal microscopy of heart sections. HW assisted with Western blotting and qPCR experiments. ASD and ANM performed the mitochondrial respiration experiments and subsequent analysis. All authors reviewed the results and approved the manuscript.
Supplementary Material
Acknowledgments
ÅBG is supported by NIH grants R21AG052280, R01HL138560, R01HL132300, and P01HL085577. BPW is supported by NIH grant T32HL007444. AMO is supported in part by the UCSD Graduate Training Program in Cellular and Molecular Pharmacology through an institutional training grant from the National Institute of General Medical Sciences grant T32GM007752 and National Institutes of Health NRSA Predoctoral Fellowship F31HL123309. ANM is supported by NIH grant R01NS087611 and a gift from Seahorse Biosciences/Agilent Technologies.
Version 1. 04/16/2019
In-Press Preview
Version 2. 05/16/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(10):e127713. https://doi.org/10.1172/jci.insight.127713.
Contributor Information
Benjamin P. Woodall, Email: bwoodall@ucsd.edu.
Amabel M. Orogo, Email: amabel.marie@gmail.com.
Rita H. Najor, Email: rnajor@ucsd.edu.
Melissa Q. Cortez, Email: mquinsay@gmail.com.
Eileen R. Moreno, Email: erg026@ucsd.edu.
Hongxia Wang, Email: 156244534@qq.com.
Ajit S. Divakaruni, Email: ADivakaruni@mednet.ucla.edu.
Anne N. Murphy, Email: anmurphy@ucsd.edu.
References
- 1.Strait JB, Lakatta EG. Aging-associated cardiovascular changes and their relationship to heart failure. Heart Fail Clin. 2012;8(1):143–164. doi: 10.1016/j.hfc.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marzetti E, Csiszar A, Dutta D, Balagopal G, Calvani R, Leeuwenburgh C. Role of mitochondrial dysfunction and altered autophagy in cardiovascular aging and disease: from mechanisms to therapeutics. Am J Physiol Heart Circ Physiol. 2013;305(4):H459–H476. doi: 10.1152/ajpheart.00936.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirakabe A, Ikeda Y, Sciarretta S, Zablocki DK, Sadoshima J. Aging and Autophagy in the Heart. Circ Res. 2016;118(10):1563–1576. doi: 10.1161/CIRCRESAHA.116.307474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Exner N, Lutz AK, Haass C, Winklhofer KF. Mitochondrial dysfunction in Parkinson’s disease: molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31(14):3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA. 2003;100(7):4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA. 2005;102(6):2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg MS, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278(44):43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 9.Song M, et al. Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. Circ Res. 2015;117(4):346–351. doi: 10.1161/CIRCRESAHA.117.306859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubli DA, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288(2):915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ, Dorn GW. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350(6265):aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6(6):e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai DF, et al. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119(21):2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2(4):324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 15.Wanagat J, Wolff MR, Aiken JM. Age-associated changes in function, structure and mitochondrial genetic and enzymatic abnormalities in the Fischer 344 x Brown Norway F(1) hybrid rat heart. J Mol Cell Cardiol. 2002;34(1):17–28. doi: 10.1006/jmcc.2001.1483. [DOI] [PubMed] [Google Scholar]
- 16.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 17.Dai DF, et al. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010;9(4):536–544. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 19.Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson ÅB. PINK1 Is Dispensable for Mitochondrial Recruitment of Parkin and Activation of Mitophagy in Cardiac Myocytes. PLoS ONE. 2015;10(6):e0130707. doi: 10.1371/journal.pone.0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song M, et al. Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. Circ Res. 2015;117(4):346–351. doi: 10.1161/CIRCRESAHA.117.306859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winklhofer KF, Henn IH, Kay-Jackson PC, Heller U, Tatzelt J. Inactivation of parkin by oxidative stress and C-terminal truncations: a protective role of molecular chaperones. J Biol Chem. 2003;278(47):47199–47208. doi: 10.1074/jbc.M306769200. [DOI] [PubMed] [Google Scholar]
- 22.Chung KK, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304(5675):1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 23.Frank M, et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta. 2012;1823(12):2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Dorn GW. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340(6131):471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8(1):e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191(7):1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callegari S, Dennerlein S. Sensing the Stress: A Role for the UPR. Front Cell Dev Biol. 2018;6:31. doi: 10.3389/fcell.2018.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vays VB, Eldarov CM, Vangely IM, Kolosova NG, Bakeeva LE, Skulachev VP. Antioxidant SkQ1 delays sarcopenia-associated damage of mitochondrial ultrastructure. Aging (Albany NY) 2014;6(2):140–148. doi: 10.18632/aging.100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García ML, Fernández A, Solas MT. Mitochondria, motor neurons and aging. J Neurol Sci. 2013;330(1-2):18–26. doi: 10.1016/j.jns.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 30.Hoppel CL, Tandler B, Fujioka H, Riva A. Dynamic organization of mitochondria in human heart and in myocardial disease. Int J Biochem Cell Biol. 2009;41(10):1949–1956. doi: 10.1016/j.biocel.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahin E, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin JH, et al. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144(5):689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116(3):615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustafsson ÅB, Dorn GW. Evolving and Expanding the Roles of Mitophagy as a Homeostatic and Pathogenic Process. Physiol Rev. 2019;99(1):853–892. doi: 10.1152/physrev.00005.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickrell AM, et al. Endogenous Parkin Preserves Dopaminergic Substantia Nigral Neurons following Mitochondrial DNA Mutagenic Stress. Neuron. 2015;87(2):371–381. doi: 10.1016/j.neuron.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boengler K, Schulz R, Heusch G. Loss of cardioprotection with ageing. Cardiovasc Res. 2009;83(2):247–261. doi: 10.1093/cvr/cvp033. [DOI] [PubMed] [Google Scholar]
- 37.LaVoie MJ, Cortese GP, Ostaszewski BL, Schlossmacher MG. The effects of oxidative stress on parkin and other E3 ligases. J Neurochem. 2007;103(6):2354–2368. doi: 10.1111/j.1471-4159.2007.04911.x. [DOI] [PubMed] [Google Scholar]
- 38.Meng F, et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Laar VS, et al. Glutamate excitotoxicity in neurons triggers mitochondrial and endoplasmic reticulum accumulation of Parkin, and, in the presence of N-acetyl cysteine, mitophagy. Neurobiol Dis. 2015;74:180–193. doi: 10.1016/j.nbd.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 41.Zhang D, Mott JL, Chang SW, Denniger G, Feng Z, Zassenhaus HP. Construction of transgenic mice with tissue-specific acceleration of mitochondrial DNA mutagenesis. Genomics. 2000;69(2):151–161. doi: 10.1006/geno.2000.6333. [DOI] [PubMed] [Google Scholar]
- 42.Dorn GW. Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3(4):374–383. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dillon LM, et al. Increased mitochondrial biogenesis in muscle improves aging phenotypes in the mtDNA mutator mouse. Hum Mol Genet. 2012;21(10):2288–2297. doi: 10.1093/hmg/dds049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370(Pt 3):751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boengler K, Kosiol M, Mayr M, Schulz R, Rohrbach S. Mitochondria and ageing: role in heart, skeletal muscle and adipose tissue. J Cachexia Sarcopenia Muscle. 2017;8(3):349–369. doi: 10.1002/jcsm.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakada K, Sato A, Hayashi J. Mitochondrial functional complementation in mitochondrial DNA-based diseases. Int J Biochem Cell Biol. 2009;41(10):1907–1913. doi: 10.1016/j.biocel.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24(12):761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Eisenberg-Bord M, Schuldiner M. Mitochatting - If only we could be a fly on the cell wall. Biochim Biophys Acta Mol Cell Res. 2017;1864(9):1469–1480. doi: 10.1016/j.bbamcr.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 50.Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell Metab. 2015;22(2):204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 51.Rogers GW, et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6(7):e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.