Abstract
Copy number increase or decrease of certain dosage-sensitive genes may cause genetic diseases with distinct phenotypes, conceptually termed genomic disorders. The most common cause of Pelizaeus-Merzbacher disease (PMD), an X-linked hypomyelinating leukodystrophy, is genomic duplication encompassing the entire proteolipid protein 1 (PLP1) gene. Although the exact molecular and cellular mechanisms underlying PLP1 duplication, which causes severe hypomyelination in the central nervous system, remain largely elusive, PLP1 overexpression is likely the fundamental cause of this devastating disease. Here, we investigated if adeno-associated virus–mediated (AAV-mediated) gene-specific suppression may serve as a potential cure for PMD by correcting quantitative aberrations in gene products. We developed an oligodendrocyte-specific Plp1 gene suppression therapy using artificial microRNA under the control of human CNP promoter in a self-complementary AAV (scAAV) platform. A single direct brain injection achieved widespread oligodendrocyte-specific Plp1 suppression in the white matter of WT mice. AAV treatment in Plp1-transgenic mice, a PLP1 duplication model, ameliorated cytoplasmic accumulation of Plp1, preserved mature oligodendrocytes from degradation, restored myelin structure and gene expression, and improved survival and neurological phenotypes. Together, our results provide evidence that AAV-mediated gene suppression therapy can serve as a potential cure for PMD resulting from PLP1 duplication and possibly for other genomic disorders.
Keywords: Neuroscience, Therapeutics
Keywords: Gene therapy, Neurological disorders
An AAV-mediated gene suppression therapy using gene-specific artificial miRNA can serve as a potential treatment for Pelizaeus-Merzbacher disease resulting from PLP1 duplication.
Introduction
Dosage-sensitive genes require precise copy numbers in the genome for their proper physiological function (1). Copy number changes in these genes — even an increase of just a single copy resulting from a segmental duplication — can cause pathological malfunction that leads to disease phenotypes, collectively termed genomic disorders (1). PLP1 (encoding proteolipid protein 1) is one such dosage-sensitive gene; a single copy gain or loss results in hypomyelinating leukodystrophy of the central nervous system (CNS) called Pelizaeus-Merzbacher disease (PMD) (2–4). Common clinical features of PMD can be recognized within the first year of life as hypotonia, nystagmus, and delayed developmental milestones, especially in motor function. Thereafter, spasticity, ataxia, and choreoathetotic movements may become more prominent (2). Most patients are wheelchair users and require assistance throughout their lives. PMD is caused by mutations in the PLP1 gene (4–6), which encodes a major CNS myelin protein. Because PLP1 is located on Xq22.1, PMD is inherited in an X-linked recessive pattern. Distinct types of mutations, including point mutations, genomic duplications, and deletions, have been shown to cause a spectrum of disease phenotypes including PMD and a milder allelic disease, spastic paraplegia type 2 (SPG2) (7). Currently, no definitive cure for PMD/SPG2 is available.
Genomic duplication encompassing the entire PLP1 gene is the most frequent (60%–70%) cause of PMD (2). However, very little is known with respect to why PLP1 duplication, presumably resulting in overexpression of PLP1, leads to profound CNS hypomyelination in humans. Plp1-Tg mice, the animal models for PMD with PLP1 overexpression, exhibit dosage-dependent premature arrest of myelination and apoptotic cell death at the terminal stage of oligodendrocyte differentiation (2, 8–11). Previous studies in Plp1-Tg mice have led to the hypothesis that overexpressed Plp1 accumulates within late endosomes/lysosomes coupled with sequestration of cholesterol and these aggregates may potentially disturb the membrane trafficking of cholesterol or galactosylceramide that is necessary for normal myelination (12–14).
Although the exact molecular and cellular mechanisms underlying the PLP1 duplication that causes severe CNS hypomyelination are not completely understood, PLP1 overexpression is likely the fundamental cause of this devastating disease and can therefore be exploited as a primary therapeutic target. Because PLP1 overexpression is largely confined to oligodendrocytes, we examined the possibility of a gene therapy approach by suppressing Plp1 mRNA expression specifically in the oligodendrocytes of the mouse brain. We chose this approach because the range of Plp1 suppression levels to accomplish both safety and efficacy is presumably broadly based on the findings of clinical genotype-phenotype studies. Patients with PLP1-null mutations commonly show much milder phenotype than other types of mutations including PLP1 duplication (15, 16). This means that even if the gene suppression therapy leads to an exclusive efficiency that completely diminishes Plp1 expression, we would still expect some therapeutic effects rather than adverse events.
As a suitable platform for PMD gene suppression therapy, we considered an adeno-associated virus (AAV) vector because of its safety with regard to low genomic integration, high efficiency, and prolonged expression in the CNS (17). However, oligodendrocytes are known to be an inefficient target cell lineage for AAV transduction, even under high-power ubiquitous promoters such as CAG and U6, for most conventional AAV serotypes (18). To overcome these issues and to develop an AAV vector capable of efficient oligodendrocyte-specific gene suppression, we employed an artificial microRNA (miRNA) expression system (19), which enables expression of a gene-specific siRNA from an miRNA-flanking cassette placed in the 3′ UTR of the cDNA of the fluorescent protein Venus (20). We placed the miRNA expression system under the control of an RNA polymerase II–driven oligodendrocyte-specific promoter, specifically, the human 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP) gene promoter. Direct injection of the AAV vector harboring Plp1-targeting artificial miRNA into the brain of Plp1-Tg mice resulted in significant therapeutic effects in lifespan and behavior phenotype; histological rescue including alleviating cytoplasmic accumulation of Plp1, improving the decrease in mature oligodendrocyte number and dysmyelination, mitigating astrogliosis/inflammation; and molecular changes including rescuing myelin gene and protein expression. Together, we successfully developed an AAV platform and provide evidence for the proof of concept that AAV-mediated gene suppression therapy can serve as a treatment for PMD resulting from PLP1 duplication.
Results
Construction of an scAAV vector that enables widespread oligodendrocyte-specific transgene expression in cerebral white matter.
To develop an AAV vector capable of oligodendrocyte-specific gene suppression, we employed an artificial miRNA expression system (19) under the control of the human CNP promoter (Figure 1), which we optimized for transcriptional activity and size to fit in the self-complementary AAV (scAAV) vector (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125052DS1).
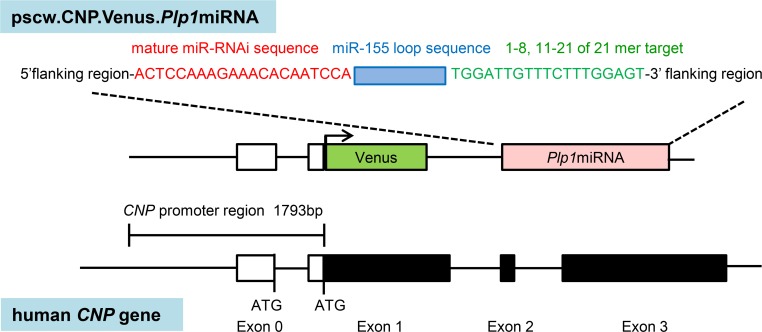
Figure 1. Schematic diagram of the self-complementary AAV (scAAV) backbone plasmid (pscw.CNP.Venus.Plp1miRNA) vector.
The scAAV backbone vector containing human CNP promoter, the coding sequence of fluorescent protein Venus, and the artificial miRNA sequence. The human CNP promoter was a 1.8-kilobase 5′ flanking sequence of the human CNP gene. The engineered miRNA cassette was placed in the 3′ UTR of the Venus sequence and consisted of a 5′ flanking region derived from miR155, a reverse complement of the 21-nucleotide target sequence, a 19-nucleotide loop sequence derived from miR155, nucleotides 1–8 and 11–21 of the sense target sequence, and a 3′ flanking region derived from miR155.
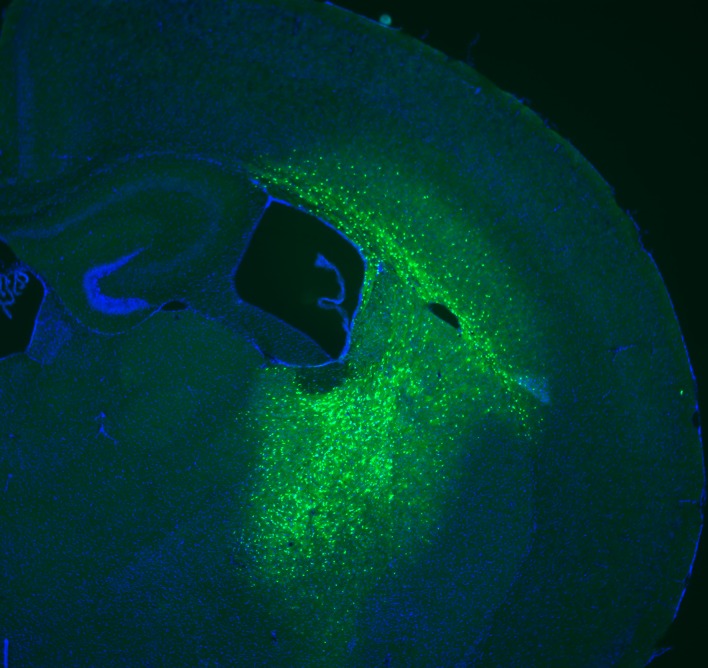
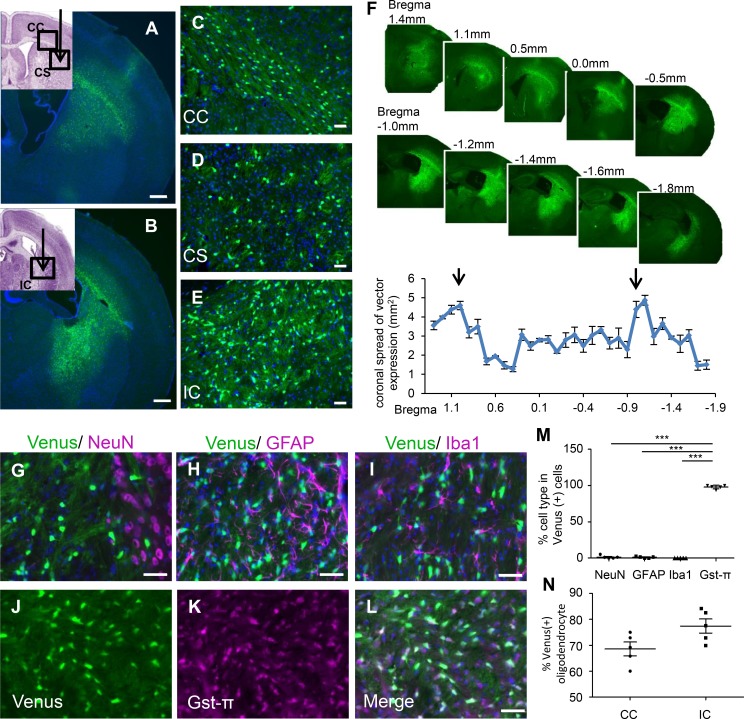
First, we examined whether this scAAV vector harbors high transduction ability for oligodendrocyte-specific gene expression. We generated pscw.AAV.CNP.Venus.miRneg, which contains an miRNA sequence targeting no known vertebrate gene in the scAAV backbone plasmid (21), and used it as a negative control in our subsequent studies. After chimeric AAV1/2 packaging (22) using AAV-293 cells, we obtained the scAAV vector (scAAV.CNP.Venus.miRneg) with a titer of 1.2 × 1012 vector genome copies/ml. We stereotactically injected 1 μl of the scAAV vector in the right corpus striatum (CS) and internal capsule (IC), where oligodendrocytes are enriched, of WT mouse brains at postnatal day 10 (P10). We found the highly selective and specific expression in oligodendrocytes after AAV injection at P10, but not at P0, P3, and P6, as previously reported (23). One week after injection (P17), we determined the distribution of the AAV vector by measuring the Venus-positive transduced area in a series of coronal sections of the mouse brains and observed widespread transduction in all directions (n = 5, Figure 2, A–F). The maximum Venus-positive area was 4.87 mm2 in the IC, approximately 20% area of the hemisphere (bregma –1.2 mm, Figure 2F). Furthermore, immunostaining with cell-type-specific markers revealed that very few Venus-positive cells colocalized with the protein neuronal nuclei (NeuN, a marker of neurons; Figure 2, G and M), glial fibrillary acidic protein (GFAP, a marker of astrocytes; Figure 2, H and M), ionized calcium binding adaptor molecule 1 (Iba1, a marker of microglia; Figure 2, I and M), or NG2 (a marker of oligodendrocyte progenitor cells [OPCs], Supplemental Figure 2, A–D), but almost all colocalized with a mature oligodendrocyte marker, glutathione S-transferase π (Gst-π) (Figure 2, J–L and M). In the center of the AAV-infected area in the corpus callosum (CC) and IC, approximately 70% and 80% of Gst-π–positive mature oligodendrocytes were transduced, respectively (Figure 2N). These findings suggest that widespread, oligodendrocyte-specific, and highly efficient transgene expression was achieved in our human CNP promoter–driven AAV vector.
Figure 2. Widespread oligodendrocyte-specific transgene expression in cerebral white matter following scAAV.CNP.Venus.miRneg vector injection.
One week prior to the analysis, scAAV.CNP.Venus.miRneg vector was injected into the right corpus striatum (CS) and internal capsule (IC) of P10 WT mice (n = 5). Representative low-magnification images in the corpus callosum (CC) (A) and IC (B), as well as high-magnification images in the CC (C), CS (D), and IC (E) demonstrate the extent of scAAV-derived reporter protein Venus expression. Cell nuclei were stained with DAPI (blue). (A and B figure inset) The schematic diagrams show the injection points (indicated by arrows) of the scAAV vector and the areas observed using a fluorescence microscope (indicated by boxes) in the CC (A inset), CS (A inset), and the IC (B inset). (F) Quantification of Venus expression areas in coronal brain sections taken from mice (n = 5) administered the scAAV vector. The x axis shows the relative distance from bregma and the arrows represent the injection points. (G–L) Immunostaining with cell-type-specific markers indicated that AAV-derived Venus expression rarely overlapped with NeuN-positive neurons (G), GFAP-positive astrocytes (H), or Iba1-positive microglia (I), but was mainly present in Gst-π–positive oligodendrocytes (J–L). (M) Quantification of the percentages of each cell type in Venus-positive cells (n = 5 mice, 4 sections per mouse). (N) Quantification of the percentage of Venus-positive oligodendrocytes relative to Gst-π–positive total mature oligodendrocytes in the center of AAV infection areas (n = 5 mice, 4 sections per mouse). Scale bars: 500 μm (A and B) and 50 μm (C–L). Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test and 2-tailed Student’s t test. ***P < 0.001.
Interestingly, the replacement of the human CNP promoter with a high-power ubiquitous CAG promoter resulted in Venus expression predominantly in neurons, but rarely in oligodendrocytes (Supplemental Figure 3), as also reported in a different system (24). Therefore, a cell-type-specific promoter such as the human CNP promoter was essential to accomplish the highly efficient and specific transduction of the oligodendrocytes.
Artificial Plp1-miRNA efficiently inhibited Plp1 mRNA and protein expression.
To develop a therapy targeting PLP1 duplication, we employed the RNA interference–mediated (RNAi-mediated) artificial miRNA approach (19, 25). A mouse Plp1 mRNA–targeting artificial Plp1-miRNA sequence was designed, selected, and inserted into the scAAV backbone plasmid vector (pscw.CAG.Venus.Plp1miRNA). We verified its knockdown efficiency in vitro by cotransfection with a mouse Plp1 cDNA expression plasmid vector in HeLa cells, followed by Plp1 protein immunostaining, quantitative reverse transcription PCR (RT-qPCR), and Western blotting, obtaining sufficient knockdown efficiencies at both the mRNA and protein levels (Supplemental Figure 4). We then replaced the CAG promoter with the CNP promoter to obtain pscw.CNP.Venus.Plp1miRNA for the following studies.
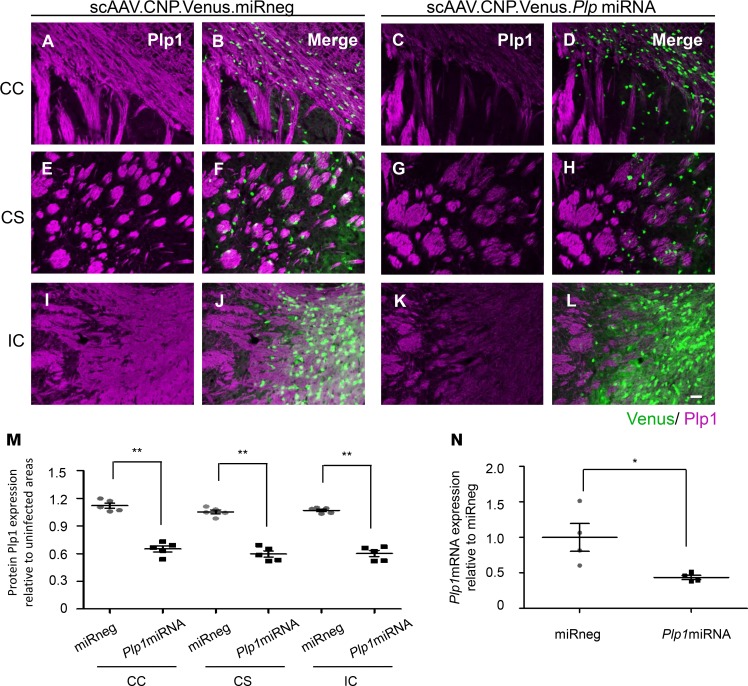
Next, we proceeded to the packaging of the chimeric AAV1/2 serotype to obtain scAAV.CNP.Venus.Plp1miRNA. We injected scAAV vectors in the right CS and IC of the WT mice at P10. One week later, mice were sacrificed and subjected to Plp1 immunostaining to examine the gene suppression efficiency in vivo. Quantification of the fluorescence intensity of endogenous Plp1 between infected areas (Venus-positive) and uninfected areas (Venus-negative) revealed significantly lower Plp1 fluorescence intensities in the infected areas, with approximately 50% reduction in the CC, CS, and IC. Meanwhile, no such changes were observed in negative control scAAV.CNP.Venus.miRneg-treated mice (Figure 3, A–M). To evaluate the Plp1 suppression efficiency at the mRNA level, we separated the Venus-positive oligodendrocytes from the mouse brains using fluorescence-activated cell sorting (FACS), and then analyzed them by RT-qPCR. Compared with the negative control, the Plp1 mRNA level was reduced to less than half in scAAV.CNP.Venus.Plp1miRNA-infected oligodendrocytes (Figure 3N). These findings indicated that the scAAV.CNP.Venus.Plp1miRNA effectively lowered the Plp1 expression in the mouse brain by approximately half at both protein and mRNA levels.
Figure 3. scAAV vector harboring artificial Plp1-miRNA effectively suppressed Plp1 mRNA and protein expression.
Coronal sections of P14 WT mice brains with Plp1 immunostaining in the CC (A–D), CS (E–H), and IC (I–L). The Plp1 expression in Venus-positive areas are detected in WT mice treated with scAAV.CNP.Venus.miRneg (A, B, E, F, I, and J) and scAAV.CNP.Venus.Plp1miRNA (C, D, G, H, K, and L). Images are representative of 5 mice per group. Scale bar: 50 μm. (M) Quantification of the Plp1 fluorescence intensities in scAAV-infected areas relative to corresponding AAV-uninfected areas in the contralateral hemisphere (n = 5 mice per group, 3 sections per site in each mouse). (N) Plp1 mRNA expression in Venus-positive oligodendrocytes from mice treated with scAAV.CNP.Venus.miRneg or scAAV.CNP.Venus.Plp1miRNA isolated by fluorescence-activated cell sorting. Mean relative expression of Plp1 mRNA in the scAAV.CNP.Venus.miRneg-treated group was set to 1 (n = 4 samples per group; in each sample, Venus-positive cells from 3 mice were pooled). Statistical significance was determined using 2-tailed Student’s t test. *P < 0.05; **P < 0.01.
To examine the effect of artificial Plp1-miRNA on off-target transcript silencing, we measured the expression of mRNA for Rhoq, Bcl11b, Robo1, Rbms3, Naca, Bbs4, Ica1l, and Ano2, which had 60% or more homology with the Plp1-miRNA seed complement sequences, using the same RNA from FACS-separated Venus-positive oligodendrocytes by RT-qPCR. Two genes with physical locations proximal or distal to Plp1 in the genome, Rab9b and Glra4, were also examined for potential position effect. Five genes (Bcl11b, Ica1l, Ano2, Rab9b, and Glra4) showed essentially no detectable mRNA expression in oligodendrocytes. No significant difference was observed in the mRNA expression of the remaining genes (Rhoq, Robo1, Rbms3, Naca, and Bbs4) between scAAV.CNP.Venus.miRneg- and scAAV.CNP.Venus.Plp1miRNA-infected oligodendrocytes (Supplemental Figure 5A). These findings suggest that the artificial Plp1-miRNA did not show apparent off-target effects on potential candidate genes. We also examined 6 oligodendrocyte-associated genes, Sox10, Olig2, Pdgfra, Cspg4, Cnp, and Mbp, and found no significant difference in expression levels (Supplemental Figure 5B), suggesting that at least in WT oligodendrocytes, Plp1 downregulation does not affect expression of other myelin genes.
scAAV.CNP.Venus.Plp1miRNA treatment extended the lifespan and improved growth in Plp1-Tg mice.
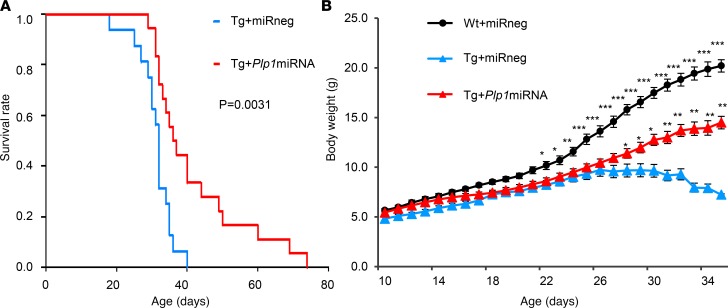
Having established the highly efficient Plp1 suppression of the artificial miRNA–harboring AAV vector in vivo, we proceeded to investigate if this AAV treatment can mitigate the disease phenotypes of Plp1-Tg mice, a model for PMD caused by PLP1 duplication (9). Plp1-Tg mice start to show fine truncal tremors around P14, followed by paroxysmal dystonia occurring around P21. Almost all Plp1-Tg mice die due to severe dystonic cramps, which eventually cause respiratory failure by P40. The scAAV.CNP.Venus.Plp1miRNA-treated Plp1-Tg mice (Tg+Plp1miRNA) lived for a mean of 42.0 days (ranging from 29 to 74 days, Figure 4A), which was significantly longer than negative control-treated Plp1-Tg mice (Tg+miRneg, with a mean of 31.1 days ranging from 18 to 40 days, P = 0.0031).
Figure 4. scAAV.CNP.Venus.Plp1miRNA treatment extended the lifespan and improved weight gain of Plp1-Tg mice.
(A) Survival curves of Tg+miRneg mice (black line, n = 16 mice) and Tg+Plp1miRNA mice (gray line, n =18 mice). Data from WT mice are not shown because no mouse died during the course of the study regardless of treatment. Statistical significance was determined using the Kaplan-Meier method combined with a generalized Wilcoxon test. (B) Body weight changes are shown for Wt+miRneg mice (open circle, n = 9 mice), for Tg+miRneg mice (closed triangle, n = 16 mice), and for Tg+Plp1miRNA mice (gray triangle, n = 18 mice). Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
In addition, Tg+Plp1miRNA showed improved bodyweight gain in comparison with Tg+miRneg from P28 (Figure 4B, P < 0.05). These findings suggest that the direct injection of scAAV.CNP.Venus.Plp1miRNA into the cerebral white matter can enhance the survival and growth of Plp1-Tg mice.
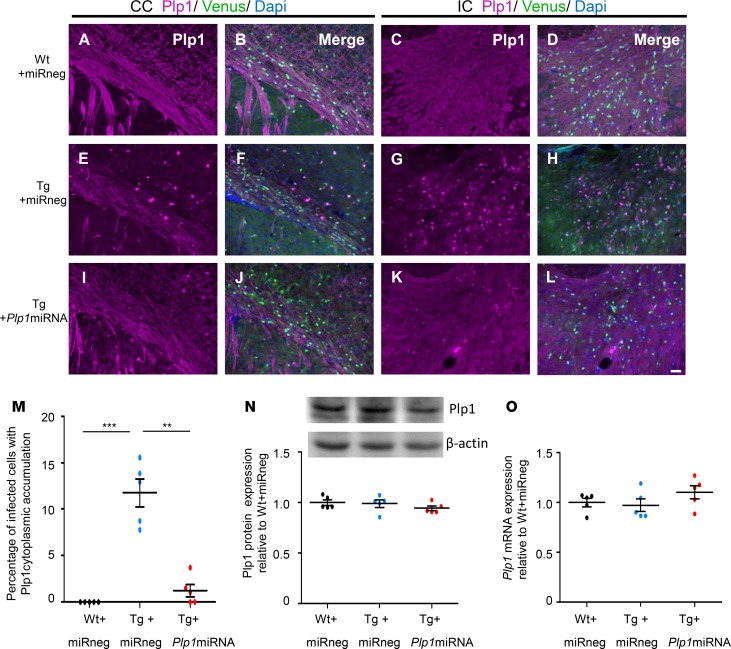
scAAV.CNP.Venus.Plp1miRNA treatment alleviated cytoplasmic accumulation of Plp1 protein in the oligodendrocytes.
Normal Plp1 expression predominates in the myelin fibers (Figure 5, A–D), while overexpressed Plp1 causes abnormal accumulation in the perinuclear cytoplasm to exhibit a punctiform appearance of the oligodendrocyte cell bodies in Plp1-Tg mice (Figure 5, E–H), as previously reported (9, 12, 26). We found this cytoplasmic Plp1 accumulation colocalized with an endoplasmic reticulum marker Bip and late endosome/lysosome marker Lamp1 (Supplemental Figure 6), as also reported in a different Plp1-Tg mouse line (12). Notably, this cytoplasmic accumulation was already present presymptomatically at P10 (Supplemental Figure 7), which means that this cellular pathology preceded the initiation of treatment. In Tg+Plp1miRNA mice, the number of Venus-positive oligodendrocytes with Plp1 punctiform accumulation decreased significantly at P25 (Figure 5, I–M); only uninfected oligodendrocytes showed cytoplasmic accumulation. These findings suggest that artificial miRNA–mediated suppression of Plp1 expression effectively mitigates the cellular pathology caused by Plp1 overexpression.
Figure 5. scAAV.CNP.Venus.Plp1miRNA treatment alleviated cytoplasmic accumulation of Plp1 protein in the oligodendrocytes of Plp1-Tg mice.
Coronal sections of P25 mouse brains with Plp1 immunostaining in the CC (A, B, E, F, I, and J) and IC (C, D, G, H, K, and L). The Plp1 expression in Venus-positive oligodendrocytes was detected in Wt+miRneg mice (A–D), Tg+miRneg mice (E–H), and Tg+Plp1miRNA mice (I–L). Scale bar: 50 μm. (M) Proportion of the cells showing Plp1 cytoplasmic accumulation in total Venus-positive cells in the CC and IC (n = 5 mice per group). (N) Representative Western blot analysis of Plp1 expression in scAAV-injected brain tissue of WT and Plp1-Tg mice. β-Actin was used as an internal control. Quantification of the intensity of the Plp1 bands is shown in the graph below (n = 5 mice per group). (O) Quantification of the relative expression of Plp1 mRNA in scAAV-injected brain tissue of WT and Plp1-Tg mice (n = 5 mice per group). Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test. **P < 0.01; ***P < 0.001.
To monitor the Plp1 expression level in these processes, we examined the Plp1 mRNA and protein expression level in the AAV-infected regions. No significant difference in Plp1 mRNA or protein expression was found among Wt+miRneg, Tg+miRneg, and Tg+Plp1miRNA mice (Figure 5, N and O). In fact, a previous study reported that the Tg-derived increase of Plp1 expression was only detectable in premyelinating embryos (E16), but not in the postnatal brains at P19 (9). We speculated that the difference in mature oligodendrocyte number and cellular state among these 3 groups may complicate the interpretation of gross Plp1 expression. Therefore, we next sought to examine the number of mature oligodendrocytes in mice treated with the AAV.
scAAV.CNP.Venus.Plp1miRNA treatment increased the mature oligodendrocyte number and myelin protein expression, and reduced degeneration of oligodendrocytes in Plp1-Tg mice.
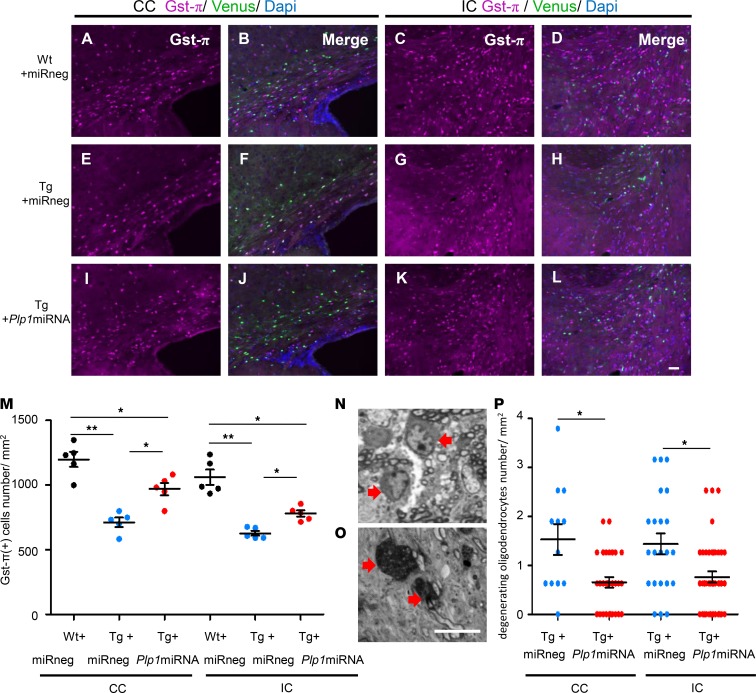
Arrested maturation, degeneration, and cell death of oligodendrocytes have been reported to occur in the Plp1-Tg mouse brain (9, 27). To determine if AAV-mediated Plp1-miRNA treatment can reverse the reduction of oligodendrocytes, we examined the number of Gst-π–positive mature oligodendrocytes by immunostaining. In Tg+miRneg mice, mature oligodendrocytes were reduced in number compared with Wt+miRneg mice (Figure 6, A–H and M; P < 0.01). However, in Tg+Plp1miRNA mice, the number of mature oligodendrocytes significantly increased (Figure 6, I–L and M; P < 0.05), suggesting that the Plp1-suppressing AAV treatment effectively rescued the number of mature oligodendrocytes of Plp1-Tg mice within 2 weeks. Interestingly, the NG2-positive OPCs showed an opposite trend to that of mature oligodendrocytes. In Tg+miRneg mice, the numbers of OPCs were increased compared with Wt+miRneg mice, which was mitigated in Tg+Plp1miRNA mice (Supplemental Figure 2). The increase of OPCs in Tg mice may be a compensatory reaction to loss of mature oligodendrocytes. At this point, it is unknown if these changes in the OPC number immediately contribute to the therapeutic effect of AAV treatment. Nevertheless, these changes are at least directed to normalizing the OPC numbers in the brain.
Figure 6. scAAV.CNP.Venus.Plp1miRNA treatment increased the mature oligodendrocyte population and decreased the degenerating oligodendrocyte population in Plp1-Tg mice.
Coronal sections of P25 mouse brains with mature oligodendrocyte marker Gst-π immunostaining in the CC (A, B, E, F, I, and J) and IC (C, D, G, H, K, and L) in Wt+miRneg mice (A–D), Tg+miRneg mice (E–H), and Tg+Plp1miRNA mice (I–L). Scale bar: 50 μm. (M) Quantification of the densities of Gst-π–positive oligodendrocytes (n = 5 mice per group, 3 sections per site in each mouse). (N and O) Representative images of normal oligodendrocytes (N, arrow) and degenerating oligodendrocytes (O, arrow) in the toluidine blue–stained brain sections from Plp1-Tg mice. Degenerating oligodendrocytes contain numerous vacuoles and highly condensed chromatin. Scale bar: 10 μm. (P) Quantification of the densities of degenerating oligodendrocytes. Toluidine blue–stained images of the CC and IC from P25 Wt+miRneg mice (n = 2), Tg+miRneg mice (n = 2), and Tg+Plp1miRNA mice (n = 3). Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test and 2-tailed Student’s t test. *P < 0.05; **P < 0.01.
Since Venus-positive AAV-transduced cells are all postmitotic mature oligodendrocytes, the increase in mature oligodendrocytes likely resulted from the prevention of apoptotic cell death. To investigate this possibility, we examined the toluidine blue–stained semi-thin–sliced brain samples of Tg+miRneg mice and observed a number of oligodendrocytes showing dark irregular nuclei with highly condensed chromatin, suggestive of degenerating oligodendrocytes (Figure 6O; as a comparison, Figure 6N shows normal oligodendrocytes). No such cells were found in WT mice. We measured the number of these degenerating cells in the CC and IC, and found that Tg+Plp1miRNA mice showed a significant reduction in degenerating cells in both regions in comparison with Tg+miRneg mice (Figure 6P). To confirm if this degeneration is mediated by apoptosis, we performed a TUNEL assay to quantify apoptotic cells. We observed that the TUNEL-positive cells were almost all positive (93.05% ± 3.39%) for the mature oligodendrocyte marker Gst-π (Supplemental Figure 8, A–D). In comparison with Wt+miRneg mice, Tg+miRneg mice showed an increase in number of the TUNEL-positive cells (Supplemental Figure 8, E–L; P < 0.001), which was mitigated in Tg+Plp1miRNA mice (Supplemental Figure 8, M–R; nonsignificant trend in the CC and significant difference in the IC, P < 0.01). These findings suggest that the reduction of oligodendrocytes in Tg mice likely resulted from apoptotic cell death, which was mitigated by the scAAV.CNP.Venus.Plp1miRNA treatment.
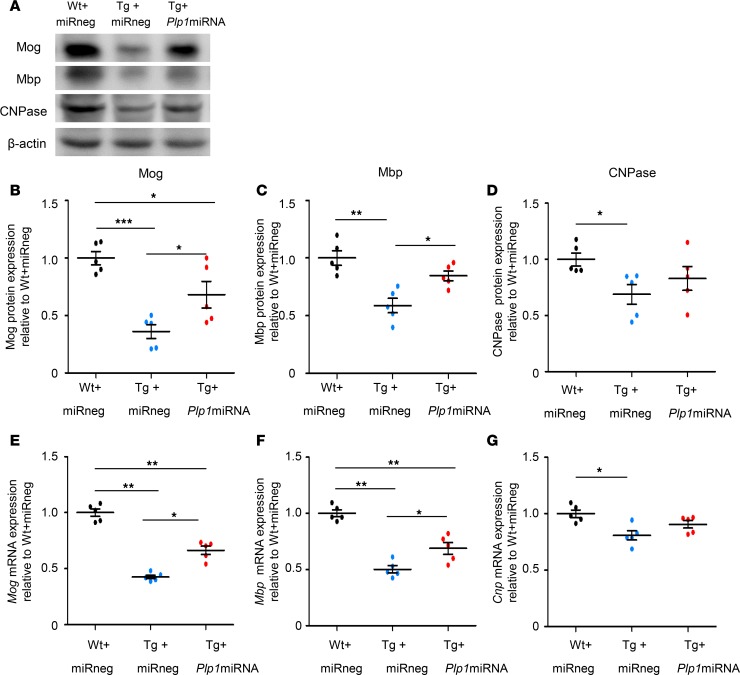
We next determined whether the expression of myelin proteins, which were produced by mature oligodendrocytes, changed simultaneously from treatment. The expression levels of myelin oligodendrocyte glycoprotein (MOG), myelin basic protein (MBP), and 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase) were assessed by immunoblot analysis using lysates of AAV-infected regions dissected from mouse brains (Figure 7A). All 3 proteins showed significantly lower expression in Tg+miRneg than in Wt+miRneg mice (Figure 7, B–D); MOG and MBP significantly increased in Tg+Plp1miRNA compared with Tg+miRneg mice (Figure 7, B–D). There was no significant difference for CNPase between these 2 groups, but a similar trend was shown in Tg+Plp1miRNA compared with Tg+miRneg mice. We also evaluated the mRNA levels of Mog, Mbp, and Cnp, and observed a similar pattern of changes (Figure 7, E–G). Together, these findings suggest that the artificial miRNA–mediated Plp1 suppression promoted the survival of oligodendrocytes by mitigating the degradation of oligodendrocytes, resulting in an enhancement of the myelin protein levels and gene expression in Plp1-Tg mice.
Figure 7. scAAV.CNP.Venus.Plp1miRNA treatment increased the expression of myelin proteins and their mRNA levels in Plp1-Tg mice.
(A) Representative Western blot analysis of Mbp Mog, and CNPase expression in AAV-infected brain regions of P25 WT and Plp1-Tg mice. β-Actin was used as an internal control. Quantification of the intensity of the Mog, Mbp, and CNPase signals is shown in B, C, and D, respectively (n = 5 mice per group). Quantification of the relative expression of Mog (E), Mbp (F), and Cnp (G) mRNA in scAAV-infected brain tissue of WT and Plp1-Tg mice (n = 5 mice per group). Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
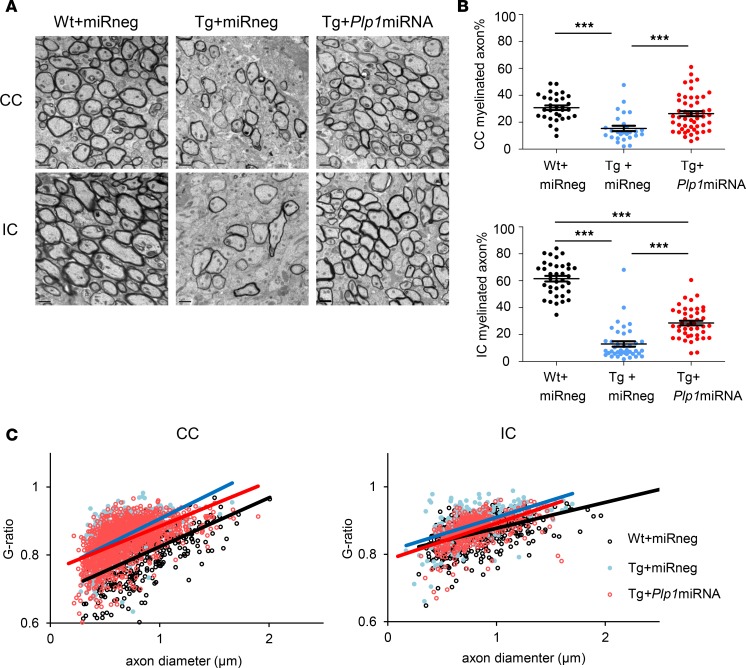
scAAV.CNP.Venus.Plp1miRNA treatment improved myelin structure in Plp1-Tg mice.
Myelin in the CNS of Plp1-Tg mice exhibits structural abnormality in a dose-dependent manner (8, 9). To investigate whether scAAV.CNP.Venus.Plp1miRNA can ameliorate such abnormalities in myelin structure, we examined the ultrastructure of myelin using transmission electron microscopy in the CC and IC (Figure 8A). Compared with the Wt+miRneg mice, fewer axons were myelinated in Tg+miRneg mice (Figure 8, A and B). However, scAAV.CNP.Venus.Plp1miRNA therapy significantly increased the proportion of myelinated axons in Plp1-Tg mice almost 2-fold (Figure 8, A and B). We also examined G-ratios, which represent the thickness of the myelin sheath with respect to the inner diameter of the myelinated fibers (Figure 8C). Tg+miRneg mice had significantly higher G-ratios (meaning thinner myelin) than Wt+miRneg mice, whereas G-ratios of larger-caliber axons were reduced in Tg+Plp1miRNA mice relative to Tg+miRneg mice (meaning thicker myelin) (Figure 8, A and C). Collectively, these data indicated that scAAV.CNP.Venus.Plp1miRNA treatment preserved myelin structure in Plp1-Tg mice.
Figure 8. scAAV.CNP.Venus.Plp1miRNA treatment improved myelin structure in Plp1-Tg mice.
(A) Electron microscopic images of the CC (top row) and IC (bottom row) from P25 Wt+miRneg mice (n = 2), Tg+miRneg mice (n = 2), and Tg+Plp1miRNA mice (n = 3). Scale bars: 1 μm. (B) Quantification of the percentage of myelinated axons in CC sections and IC sections. (C) G-ratios were calculated for each myelinated axon (>500 per group) to examine the thickness of myelin in the CC and IC. Graphs show plotted G-ratios against axonal diameters and approximating curves (CC: Wt+miRneg vs. Tg+miRneg, P < 0.001; Wt+miRneg vs. Tg+Plp1miRNA, P < 0.001; Tg+miRneg vs. Tg+Plp1miRNA, P < 0.01. IC: Wt+miRneg vs. Tg+miRneg, P < 0.001; Wt+miRneg vs. Tg+Plp1miRNA, P < 0.01; Tg+miRneg vs. Tg+Plp1miRNA, P < 0.001). Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test and MANOVA test. ***P < 0.001.
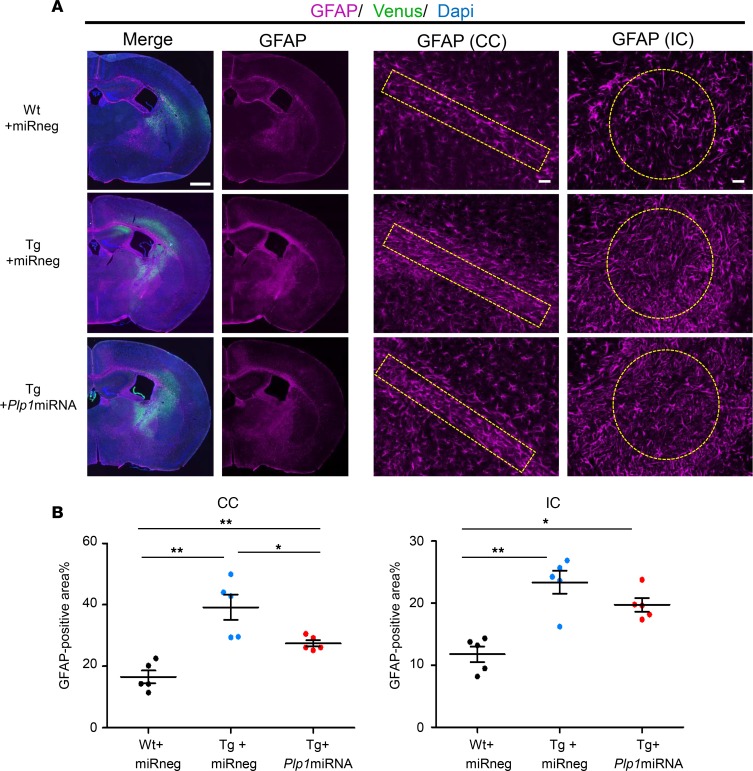
scAAV.CNP.Venus.Plp1miRNA treatment improved astrogliosis and microgliosis in Plp1-Tg mice.
Astrogliosis and inflammatory microglial activation are commonly observed phenomena in the lesions of neurodegenerative diseases. Recent studies suggest that these inflammatory reactions are not just a nonspecific tissue response, but also important modifiers of the disease phenotypes in PMD (28–30). We therefore determined the effect of scAAV.CNP.Venus.Plp1miRNA treatment on the response of astrocytes and microglia by GFAP and Iba1 immunostaining in the CC and IC. First, we confirmed that, in WT mice, scAAV.CNP.Venus.Plp1miRNA treatment itself did not affect the number of GFAP-positive astrocytes or Iba1-positive microglia (data not shown). As expected, Tg+miRneg mice showed enhanced astrogliosis in comparison with the Wt+miRneg mice, suggested by increased proportions of GFAP-positive areas both in the CC and IC (Figure 9, A and B). Tg+Plp1miRNA mice showed a significant reduction of GFAP-positive areas in the CC, but not a significant reduction in the IC.
Figure 9. scAAV.CNP.Venus.Plp1miRNA treatment improved astrogliosis in Plp1-Tg mice.
(A) Immunostaining for astrocyte marker GFAP in the coronal brain sections of P25 Wt+miRneg mice (top row), Tg+miRneg mice (middle row), and Tg+Plp1miRNA mice (bottom row). Low-magnification images (first and second columns, scale bar: 1 mm) and the high-magnification images (scale bar: 50 μm) of the CC (third column) and IC (fourth column) are shown. (B) Quantification of the percentage of GFAP-positive areas in CC sections and IC sections (n = 5 mice per group). Region of interest (ROI) was 700 μm × 100 μm in the CC and 15,1976 μm2 in the IC. Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test. *P < 0.05; **P < 0.01.
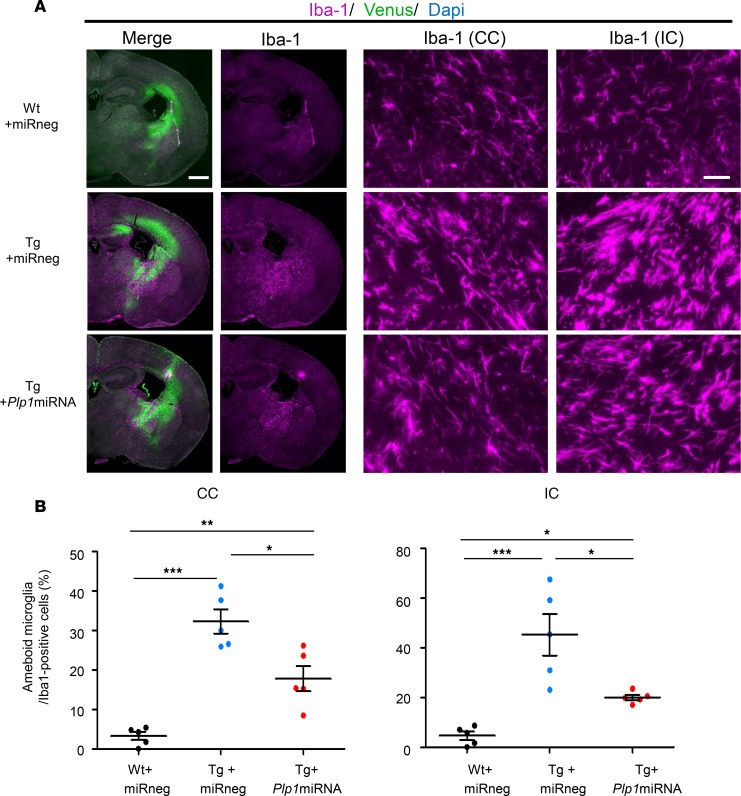
Regarding the microglia, we observed increased numbers of Iba1-positive microglia both in the CC and IC of Tg+miRneg mice in comparison with the WT mice (Figure 10A), and Tg+Plp1miRNA mice showed similarly increased numbers of microglia as in Tg+miRneg mice (data not shown). However, when we determined the proportions of activated microglia by measuring the size of each cell body, Tg+Plp1miRNA mice showed a significantly lower proportion of activated microglia than Tg+miRneg mice (Figure 10, A and B). These findings suggest that scAAV.CNP.Venus.Plp1miRNA treatment not only ameliorated oligodendrocyte pathology, but also suppressed reactive astrogliosis and microgliosis. It is unlikely that these changes are the direct consequence of AAV treatment on the astrocytes or microglia, but rather secondary to improved myelination and reduced oligodendrocyte cell death, because our AAV vectors are essentially silent in these non–oligodendrocyte lineage cells. Nevertheless, the therapeutic effect appeared to expand beyond myelination to include inflammatory tissue reactions.
Figure 10. scAAV.CNP.Venus.Plp1miRNA treatment improved microgliosis in Plp1-Tg mice.
(A) Immunostaining for microglia marker Iba1 in the brain sections from P25 Wt+miRneg mice (top row), Tg+miRneg mice (middle row), and Tg+Plp1miRNA mice (bottom row). Low-magnification images (first and second columns, scale bar: 1 mm) and high-magnification images (scale bar: 50 μm) of the CC (third column) and IC (fourth column) are shown. (B) The percentages of amoeboid microglia in total Iba1-positive cells were examined in CC sections and IC sections (n = 5 mice per group). Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
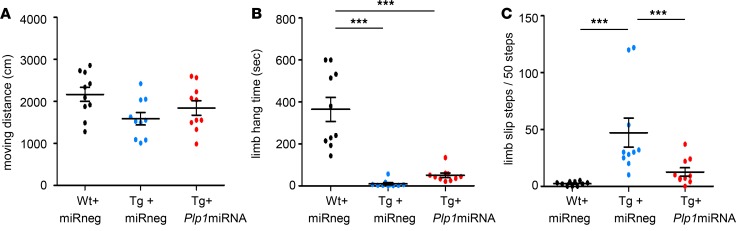
scAAV.CNP.Venus.Plp1miRNA treatment improved motor functions in Plp1-Tg mice.
Given the histological and molecular improvement observed in Tg+Plp1miRNA mice, we next determined if scAAV.CNP.Venus.Plp1miRNA treatment could also improve the motor function of Plp1-Tg mice. For behavioral analysis, Wt+miRneg, Tg+miRneg, and Tg+Plp1miRNA mice were subjected to the open field test, limb hang test, and limb slip test (23, 27). The locomotor activities examined in the open field test (Figure 11A) showed no difference in moving distance among these 3 groups. Limb hang tests (31) were used to examine whole-body force and muscle coordination (Figure 11B). Tg+Plp1miRNA mice were able to remain hanging on a bar significantly longer than Tg+miRneg mice (P = 0.008, 2-tailed Student’s t test); however, the large difference from Wt+miRneg mice diminished this statistical significance in 1-way ANOVA test. The limb slip test was previously reported as a sensitive examination to monitor the coordination of motor function in the CNP-deficient dysmyelinating mice (23, 27). Tg+Plp1miRNA mice had 4-fold fewer slips than Tg+miRneg mice (Figure 11C and Supplemental Videos 1–3). These results suggest that scAAV.CNP.Venus.Plp1-miRNA treatment also improved the motor abnormalities of Plp1-Tg mice.
Figure 11. scAAV.CNP.Venus.Plp1miRNA-treated Plp1-Tg mice show improved motor functions.
(A) The total moving distance in open field test, (B) the time hanging on a bar in limb hang test, and (C) the number of missteps in 50 steps of gait in limb slip test for P25 Wt+miRneg mice, Tg+miRneg mice, and Tg+Plp1miRNA mice (n = 10 mice per group) are shown. Statistical significance was determined using 1-way ANOVA with Bonferroni’s post hoc test. ***P < 0.001.
Discussion
In this study, we have developed a molecular system for targeted gene suppression therapy specifically controlled within the oligodendrocytes as a potential treatment for PMD caused by PLP1 duplication. A combination of a human CNP promoter, a Plp1-targeting artificial miRNA cassette, and an scAAV vector achieved widespread transduction with high oligodendrocyte specificity and sufficient gene suppression efficacy. We have also provided evidence for its significant therapeutic effects in Plp1-Tg mice. With these findings, we have established what we believe is a novel gene therapy platform for the most common form of the PLP1 mutations causing PMD, which may serve as a basis for the development of a clinically applicable molecular therapy for PMD.
The key concept of our therapeutic approach is straightforward. The primary molecular cause of PLP1 duplication is the overexpression of WT PLP1 transcripts and proteins. Therefore, we took advantage of RNAi to specifically lower the level of PLP1 in oligodendrocytes. We employed artificial miRNA technology in an scAAV delivery system, which provides a number of advantages in comparison with shRNA driven by RNA polymerase III–mediated promoters, such as U6, and conventional single-strand AAV (ssAAV). First, cell-type-specific expression can be achieved because a wide range of RNA polymerase II–driven promoters can be utilized. In this study, we originally cloned the human CNP promoter with a length of 1.8 kb, which is shorter than the previously reported mouse Cnp promoter (32–34) or Mbp promoter (22). With this size, the human CNP promoter can fit within the limit of the scAAV vector. More importantly, the human CNP promoter showed high promoter activity comparable to the CAG promoter in the oligodendrocyte-derived Oli-neu cell line (data not shown) and exhibited oligodendrocyte-specific gene expression in vivo. In addition, the human-derived sequence serves as a better source for future clinical applications in terms of origin of species. Second, artificial miRNA may cause fewer adverse effects than RNA polymerase III–driven shRNA. Application of shRNAs in vivo may cause serious cytotoxicity, which is associated with saturation of the endogenous components necessary for the miRNA pathway being overwhelmed by shRNA (35–38). The artificial miRNA–based approach shows little cytotoxic reactivity because it mimics the endogenous miRNA–expressing system (35). Third, it allows for greater freedom in designing the RNAi cassette to boost the level of interference. When a single copy of miRNA is not sufficient, multiple miRNAs can be tandemly concatenated to target the same or different sequences in a gene to enhance the silencing efficiency. Therefore, the degree of gene suppression can be controlled by designing the units. Fourth, scAAV vectors enable higher efficiency and faster expression than the conventional ssAAV expression vectors (39, 40). In comparison with our preliminary efforts with poor selectivity and low transduction efficiency in oligodendrocytes using the U6-driven shRNA system and CAG-driven miRNA system, as also reported elsewhere (40), our present platform shows significant advantages in tissue specificity of gene expression, safety, and efficacy of gene suppression and viral transduction.
With these features of our molecular system for targeted gene suppression therapy, we first validated the effectiveness of scAAV.CNP.Venus.miRNA in WT mice. The immunostaining for cell-type-specific markers demonstrated that the human CNP promoter in our AAV delivery system successfully drove oligodendrocyte-specific gene expression. Essentially, little expression was observed in neurons, astrocytes, and microglia. This is particularly important to avoid nonspecific adverse effects of miRNA in nontargeted cells. Second, a rapid and widespread distribution of scAAV-mediated transduction is verified by examining the expression of reporter protein Venus at 1 week after injection. Locally administered scAAV spread widely in the cerebrum, including the CC, fimbria, and most area of the CS and IC, in just 1 week. In clinical applications, the wide distribution of gene transduction is critical because PMD affects all oligodendrocytes in the CNS. Compared with a previous report using a similar oligodendrocyte-targeting ssAAV system (23), the usage of scAAV may have contributed to higher efficacy and faster transduction (39, 40). Third, artificial Plp1-miRNA effectively lowers Plp1 mRNA and protein expression. The suppression efficacy of 40%–50% both at mRNA and protein levels in vivo is ideal for the treatment of PMD caused by PLP1 duplication, which presumably doubles the expression in patients’ oligodendrocytes. Although additional optimization can be pursued, we conclude that our system is sufficiently established to lower PLP1 expression to a therapeutic level in a targeted, oligodendrocyte-specific manner.
We also provide evidence suggesting that our therapeutic system can significantly mitigate the phenotypic presentation of the Plp1-Tg mice. Notably, the cytosolic accumulation of Plp1, which is already present at P10 when the treatment is initiated, mostly disappeared at P25. Accumulated Plp1 appears to incorporate cholesterol to form raft aggregates, which potentially disturb lipid membrane trafficking, prevent late endosomes/lysosomes from recycling and degrading myelin membrane, interrupt myelination signal molecules, and impair the process of myelination (12). Because the cytoplasmic accumulation of Plp1 is one of the earliest pathological hallmarks before other phenotypes become apparent, it may have a fundamental role in the pathogenesis of Plp1 overexpression. Therefore, our findings that Plp1 suppression promptly alleviated the Plp1 cytoplasmic accumulation in Plp1-Tg mice suggest that this therapy may potentially serve as a definitive cure for PMD.
Histological and molecular analyses demonstrated that treatment with Plp1-miRNA significantly rescued the myelination. We observed increased mature oligodendrocyte number with decreased degeneration, increased expression of myelin proteins, MBP, MOG, and CNPase, and an ultrastructural recovery of myelin density and myelin sheath thickness. Furthermore, the associated global inflammatory changes in the white matter of Plp1-Tg mice, including astrogliosis and microglia activation, were effectively rescued. To our surprise, local AAV administration and Plp1 silencing in the restricted areas achieved significantly extended lifespan as well as improvement in behavioral phenotypes. This indicated that even the local recovery of myelination can improve the global phenotypes. These findings strongly suggest the potential of our artificial miRNA–harboring scAAV system as a promising molecular therapy for PMD.
Before future clinical application, there are several issues to be resolved to ensure the therapeutic efficacy and safety in humans. First, selection of a serotype with higher specificity for oligodendrocyte infection is preferred to reduce the risk for adverse effects on nontarget cells. Our current system exhibits oligodendrocyte-specific gene expression solely by the selectivity of the CNP promoter. However, AAV1/2 vector infection itself may occur in a nonselective fashion, and hence cryptic promoter activity in non–oligodendrocyte lineage cells potentially allows leaky expression of artificial miRNA. In fact, low level expression of CNP in astroglia and microglia has been reported (41). We have not completely excluded the possibility that our CNP-driven AAV has cryptic effects on cells other than oligodendrocytes, which could influence the survival of Tg mice. Optimization using serotypes that have higher tropism for oligodendrocytes such as AAV8 (40) or a synthetic novel capsid, olig001 (42), may further improve safety and cell-specific viral infection of our system. Second, development of delivery systems with broader distribution may lead to better therapeutic outcome. In our study, although a widespread oligodendrocyte-specific gene expression was achieved by local AAV administration, numerous oligodendrocytes outside the infected areas were still in the untreated state. Dystonia, the main cause of death in Plp1-Tg mice, may involve neuronal networks both in the basal ganglia and cerebellum (43), where few infected oligodendrocytes were found in our study. Use of AAV delivery systems enabling broader distribution, such as intraventricular injection or ultrasound treatment combined with microbubbles (44–48), may further improve the therapeutic effect of our AAV treatment system.
In conclusion, we developed an AAV platform harboring artificial miRNA under the control of an oligodendrocyte-specific promoter and demonstrated that correcting Plp1 overexpression ameliorated cytoplasmic accumulation of Plp1, preserved myelination, and improved disease phenotype in Plp1-Tg mice. We provided evidence for the proof of concept that AAV-mediated gene suppression therapy can possibly serve as a fundamental cure for PMD resulting from PLP1 duplication. Furthermore, the artificial miRNA–based platform may be applicable to the treatment of other genomic disorders caused by segmental genomic duplications, such as Charcot-Marie-Tooth disease type 1A.
Methods
Animals.
Jcl:B6C3F1 mice (WT) were purchased from CLEA Japan, Inc. Plp1-Tg mice (Plp1-Tg 4e/C57BL/6, gifted by T. Kagawa, National Institute of Physiological Sciences, Okazaki, Japan) were mated with Jcl:B6C3F1 mice over 10 generations to obtain a Plp1-Tg/B6C3 backcross. We housed mice individually with ad libitum food and water in an EBAC-S breeding apparatus (CL-5351, CLEA), where environmental conditions were controlled: temperature (22°C ± 1°C), humidity (60% ± 5%), and a 12-hour light/12-hour dark cycle (LD; lights on from 08:00 to 20:00 hours). The mice were from different litters and assigned to the different experimental groups randomly.
scAAV vector cloning.
The artificial miRNA sequence to target mouse Plp1 mRNA (NM_011123.3) was designed by BLOCK-iT RNAi Designer (Thermo Fisher Scientific). The negative control miRNA sequence (miRneg) was predicted not to target any known vertebrate gene (21). The engineered miRNA sequences were subcloned into the cloning site of the pcDNA6.2-GW/miR vector (Thermo Fisher Scientific), which was flanked on either side by sequences from murine miR-155 to allow proper processing of the engineered pre-miRNA sequence (19). Out of 6 candidate sequences obtained, 1 sequence listed below was selected based on the efficacy of Plp1 downregulation and availability of viral production. The Plp1miRNA sequence was 5′-ACTCCAAAGAAACACAATCCAGTTTTGGCCACTGACTGACTGGATTGTTTCTTTGGAGT-3′. The miRneg sequence was 5′-AAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′.
The engineered miRNA cassette was cut out and inserted into an scAAV backbone vector, pscw.CAG.Venus, which harbors a CAG promoter and fluorescent protein Venus cDNA. The artificial miRNA sequences were subcloned at the 3′ UTR region of Venus to generate pscw.CAG.Venus.Plp1miRNA and pscw.CAG.Venus.miRneg backbone vectors.
We then replaced the CAG promoter with the human CNP promoter. A pair of primers (Supplemental Table 1) with linkers for restriction enzymes SpeI or EcoRI was designed for PCR amplification of the proximal 1.8-kb promoter of the human CNP gene. The CAG promoter in the pscw.CAG.Venus.Plp1miRNA or pscw.CAG.Venus.miRneg vector was removed at the SpeI/EcoRI sites and replaced with the human CNP promoter to generate pscw.CNP.Venus.Plp1miRNA or pscw.CNP.Venus.miRneg vector plasmid.
AAV vector production and titration.
To produce chimeric AAV1/2 vectors, we cotransfected AAV-293 cells with the scAAV backbone plasmid (pscw.CNP.Venus.Plp1miRNA or pscw.CNP.Venus.miRneg), the serotype-specific AAV helper plasmids, p5E18RXC1 (49) and pAAV-RC (Agilent Technologies), encoding the rep and cap genes of AAV1 and AAV2, respectively, and the adenovirus helper plasmid pHelper (Agilent Technologies), using polyethyleneimine (PEI), as described elsewhere (50). Cells were harvested 72 hours after transfection and AAVs were purified using an AAVpro Purification Kit (Takara Bio Inc.). Vector genome titers were determined by quantitative PCR using the AAVpro Titration Kit (Takara Bio Inc.). We performed the AAV transduction experiments with 8 different AAV lots at different timing.
Direct brain injection of AAV.
We delivered the scAAV vector (scAAV.CNP.Venus.miRneg or scAAV.CNP.Venus.Plp1miRNA) into the CS and IC of either WT or Plp1-Tg mouse brains at P10.
We injected 1 μl of each scAAV vector (1.2 × 1012 genome copies/ml) into the CS and IC at the following coordinates: for the CS, 0 mm anterior-posterior (AP), +1.5 mm medial-lateral (ML), and –2 mm dorsal-ventral (DV) from the bregma; for the IC, +4.0 mm AP, +1.7 mm ML, and –2.5 mm DV from the lambda. For the quantification of viral distribution and effect of Plp1 expression silencing in WT mice, we injected scAAV exclusively in the right hemisphere. For the experimental treatment trials using WT and Plp1-Tg mice, 1 μl scAAV vector solution was injected into the CS and IC in both hemispheres. Following the injections, the pups were returned to their home cages and were analyzed after an appropriate number of days for subsequent studies. Because Plp1-Tg mice began to die at about P25, we perform behavioral tests and biochemical and histology analyses at P25 for comparative study.
Antibodies.
The following primary antibodies were used in this study: GFAP (clone 6F2, Dako/Agilent Technologies); NeuN protein (clone A60, Chemicon/MilliporeSigma), MBP (clone SMI99, BioLegend); Iba1 (clone EPR16588, Abcam); Gst-π (MBL312, Medical and Biological Laboratories); Olig2 (IBL18953, Immuno-Biological Laboratories); MOG (ab32760, Abcam); myelin-CNPase (clone SMI91, BioLegend); β-actin (ab8227, Abcam); and Plp1 (gift from M. Itoh, National Center of Neurology and Psychiatry, Tokyo, Japan).
Immunostaining.
Mice were anesthetized with isoflurane inhalant liquid and transcardially perfused with phosphate-buffered saline (PBS) followed by fresh 4% paraformaldehyde (n = 5 per group). We further fixed the harvested brain in 4% paraformaldehyde overnight at 4°C, cryoprotected in 30% sucrose, embedded in Tissue-Tek optimal cutting temperature (O.C.T.) compound (Sakura Finetek), and coronally cryosectioned at 20-μm thickness. The sections were treated with HistoVT One (Nacalai Tesque) and blocked in 5% goat serum in PBS with 0.05% Triton X-100 for 3 hours. We diluted primary and secondary antibodies in PBS with 3% goat serum and 0.05% Triton X-100. The following dilutions of primary antibodies were incubated overnight at 4°C: anti-GFAP 1:400, anti-NeuN 1:400, anti-MBP 1:500, anti-Iba1 1:500, anti–Gst-π 1:300, anti-Olig2 1:400, and anti-Plp1 1:500. Secondary antibody incubation was performed for 1 hour at room temperature (22°C–25°C) with 1:400 dilutions of Alexa Fluor 647–goat anti-mouse, Alexa Fluor 647–goat anti-rabbit, and Alexa Fluor 594–goat anti-rabbit (Thermo Fisher Scientific). Cell nuclei were visualized with 4′,6′-diamidino-2-phenylindole (DAPI; MillipporeSigma). Slides were mounted with ProLong Diamond mounting agent (Thermo Fisher Scientific) and examined under a KEYENCE BZ-X710 fluorescence microscope or a Zeiss LSM 710 confocal microscope (Carl Zeiss AG).
To assess the intracranial AAV vector distribution and AAV-mediated transgene expression, we measured the Venus-positive transduced area in the coronal sections (n = 5 mice, 32 sections were imaged per mouse) using the KS-analyzer (KEYENCE). To evaluate the population of Gst-π–positive cells and the protein expression levels of Plp1 and GFAP, the number of fluorescently stained cells and mean fluorescence intensity were quantified (region of interest [ROI] = 0.29 mm2 in the CC and ROI = 0.615 mm2 in the IC, 6 slices per mouse, n = 5 mice per group) using ImageJ 1.45s software (NIH).
To examine the proportion of activated microglia, we measured the minor axis of the Iba1-positive microglia cell body in Wt+miRneg mice, which is predominantly in the silent state (i.e., ramified form), using ImageJ 1.45s. We obtained a mean value of 4.41 ± 1.30 μm in the CC and 4.94 ± 1.49 μm in the IC (mean ± SD, from 219 cells and 250 cells, respectively). We defined the activated microglia, which show an enlarged cell body (i.e., amoeboid form), as the Iba1-positive cells whose minor axes were larger than the mean + 2 SD (7.00 μm in the CC and 7.93 μm in the IC). We measured the proportion of activated microglia against the total number of microglia in the scAAV-infected brain area from the Wt+miRneg, Tg+miRneg, and Tg+Plp1miRNA mice.
FACS.
To sort scAAV-infected Venus-positive cells, mice were anesthetized with isoflurane inhalant liquid and transcardially perfused with 20 ml PBS. We dissected the mouse brains and coarsely chopped them with microdissection scissors. One milliliter of Accutase (MilliporeSigma) was added to each brain, followed by incubation for 30 minutes at 37°C. We then added 2 ml of Hank’s balanced salt solution (HBSS) with 10% FBS to each sample and manually triturated the tissue with a transfer pipette. Triturated samples were transferred to 100-μm cell strainers and the suspensions were centrifuged. To purify cells from the myelin debris, we resuspended the pellet in 40% Percoll (GE Healthcare) in HBSS and centrifuged at 700 g for 25 minutes at room temperature. The myelin debris in the top layer was aspirated and cells in the pellet were resuspended in 1 ml DMEM/F-12 (Thermo Fisher Scientific) with 2% FBS. Samples were run on a FACSAria Fusion flow cytometer (BD Biosciences) using a 100-μm nozzle to sort the Venus-positive cells, which were then subjected to the following gene expression studies.
RT-qPCR.
To test the Plp1 gene expression exclusive to scAAV-infected oligodendrocytes of WT mice, we used sorted Venus-positive cells (brains from 3 mice were pooled into 1 sample, and 4 samples were prepared per group). In the treatment trials, we cut out the Venus-fluorescent areas from coronal sections of brain slices (n = 5 per group) under an MA16FA fluorescence stereo microscope (Leica Camera AG). We isolated total RNA from sorted Venus-positive cells or excised tissue using an RNeasy Mini Kit (Qiagen) and synthesized cDNA with a SuperScript reverse transcriptase kit (Thermo Fisher Scientific). We measured gene expression by RT-qPCR using LightCycler 480 SYBR Green I Master on a LightCycler 480 instrument (Roche). Gene-specific primer sequences are shown in Supplemental Table 1. Results were standardized to the housekeeping gene Actb. We calculated the gene expression levels relative to Wt+miRneg mice using 2–ΔΔCt based on the Livak and Schmittgen method (51). Results are reported as mean ± SEM.
Western blotting.
To assess the protein expression levels in the scAAV-infected region, we excised the Venus-positive areas from the mouse brains (n = 5 per group) under the MA16FA fluorescence stereo microscope because FACS could not efficiently collect the myelin fraction. We harvested the samples in RIPA buffer (Nacalai Tesque) with protease inhibitor and phosphatase inhibitor cocktails (Thermo Fisher Scientific), and then quantified protein levels using a Pierce BCA protein assay kit (Thermo Fisher Scientific). Eighty micrograms of total protein was separated by SDS-PAGE (Sigma-Aldrich, 4%–12% TruPAGE with Tris-glycine running buffer). We used Trans-Blot Turbo transfer packs (Bio-Rad) for protein transfer. Membranes were blocked with Blocking One solution (Nacalai Tesque) and then incubated overnight at 4°C with primary antibodies as follows: anti-MOG (1:500), anti-MBP (1:500), anti-CNPase (1:500), anti-Plp1 (1:1000), and anti–β-actin (1:1000). On the following day, we washed the membranes with 0.01% Tween 20 in PBS and incubated them for 1 hour with species-appropriate secondary antibodies conjugated to HRP: goat anti-rabbit HRP (1:6000; Jackson ImmunoResearch), goat anti-mouse HRP (1:6000; Jackson ImmunoResearch), and donkey anti-rat HRP (1:6000; Jackson ImmunoResearch). The membranes were then rewashed and incubated with ECL Prime Western blot detection reagent for 5 minutes (GE Healthcare), followed by visualization using an LAS-4000 lumino-image analyzer (GE Healthcare).
Toluidine blue staining and electron microscopic observation.
For morphometric analysis of degenerative oligodendrocytes and myelination, P25 Wt+miRneg, Tg+miRneg, and Tg+Plp1miRNA mice (n = 2–3 mice per group) were transcardially perfused with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.2) and further fixed overnight. We excised the AAV-infected corpus regions from 600-μm coronal sections under the fluorescence stereo microscope. The samples were treated with 1% osmium (OsO4) in cacodylate buffer for 60 minutes, and then stained with 3% uranyl acetate for 60 minutes. After serial dehydration, the tissues were embedded in Epon812 (TAAB).
To examine degenerative oligodendrocytes, we obtained transverse semi-thin sections (0.5 μm) stained with toluidine blue. The number of degenerative oligodendrocytes showing numerous vacuoles and highly condensed chromatin was determined (10–12 slices per region per mouse, n = 2–3 mice per group).
For the ultrastructural analyses, we cut ultrathin sections at 70 nm and observed them under a Tecnai Spirit electron microscope (Thermo Fisher Scientific-FEI). To examine the ratio of myelinated fibers, we counted all axons in each field and determined how many of them were myelinated. To examine the myelin sheath thickness, we calculated the G-ratio, which represents the relative thickness of the myelin sheath by measuring the ratio of inner and outer diameters of myelinated axons (n > 500 axons, from 2–3 mice per group).
Behavior analysis.
To assess the motor performance of the mice, we carried out the open field test, limb hang test, and limb slip test as previously described (27). Wt+miRneg, Tg+miRneg, and Tg+Plp1miRNA mice (n = 10–12 mice per group) were examined at P25. Before each test, mice were acclimated to the environment and instrument for 30 minutes. The motor activity was examined using the open field test. Each mouse was placed in the center of a 40 × 40 cm activity arena with 30-cm-high walls and allowed to explore for 10 minutes. Locomotion was measured as the total distance moved throughout the arena. After each experiment was recorded on a hard disk drive (HDD) video recorder, the data were transferred and converted into AVI format video files to analyze the moving distance of each mouse using EthoVision 3.0 software (Noldus Information Technology).
To assess whole-body force and muscle coordination, we used the limb hang test (52). Each mouse was allowed to grasp a 6-mm-diameter metal stick by their 4 paws, which was maintained horizontally 35 cm above a thick layer of soft bedding. The length of time until each mouse fell from the stick was recorded. After each fall, the mice were allowed to recover for 2 minutes. Each session consisted of 3 trials from which the times were averaged.
To examine the coordination behavior in CNS demyelination models, we carried out the limb slip test (23, 27). Mice were placed on a 40 × 40 cm metal grid with 1-cm spacing between bars (bar diameter: 3 mm) and the number of limbs slipping through the grid were counted as missteps. Each trial consisted of 50 steps. A video camera was used to film the mice to ensure accurate counts.
Statistics.
All values are expressed as mean ± SEM. For survival analysis, we used the Kaplan-Meier method combined with a generalized Wilcoxon test. For G-ratio analysis, we used multivariate analysis of variance (MANOVA). For bodyweight gain analysis, we used 2-way ANOVA. Other than these experiments, we used 1-way ANOVA with Bonferroni’s post hoc test and the 2-tailed Student’s t test. Statistical significance was defined as P < 0.05 in all comparisons. MANOVA was performed using R programming language (R Development Core Team, R Foundation for Statistical Computing). All other statistical examinations were carried out using GraphPad Prism.
Study approval.
All experimental protocols were conducted in accordance with the guidelines for animal care regulated by the animal committee of the National Institute of Neuroscience, National Center of Neurology and Psychiatry, Tokyo, Japan.
Author contributions
KI and HL conceived the study. TO, YG, and NI provided professional advice and instruction. HL, HO, KS, and KI acquired and validated the data. HL, SS, KS, HI, and YM executed the experiments. HL, SS, HI, and YM analyzed the data. HL and KI wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Tetsushi Kagawa and Kazuhiro Ikenaka (National Institute for Physiological Sciences) for providing Plp-Tg e4 mice, Masayuki Itoh (National Center of Neurology and Psychiatry) for providing anti-Plp1 antibody, Hirohiko Hojo (National Center of Neurology and Psychiatry) for helpful suggestions about designing miRNA, and Hitoshi Osaka (Jichi Medical University) for helpful advise and critical review of this manuscript. This study was supported in part by grants from the Japan Agency of Medical Research and Development, AMED, Practical Research Project for Rare/Intractable Diseases (16ek0109016h0003 and 19ek0109270h0003 to KI), and Grant-in-Aid for Scientific Research (KAKENHI) (16H05361 to KI and 16K19679 to HL).
Version 1. 05/16/2019
Electronic publication
Footnotes
Conflict of interest: Ken Inoue receives a research grant from Daiichi-Sankyo Company, Limited.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(10):e125052. https://doi.org/10.1172/jci.insight.125052.
Contributor Information
Heng Li, Email: yinzhidr@ncnp.go.jp.
Hironori Okada, Email: kanitamaokdhrnr@gmail.com.
Sadafumi Suzuki, Email: suzuki-sadafumi@ncnp.go.jp.
Kazuhisa Sakai, Email: kazusaka@ncnp.go.jp.
Hitomi Izumi, Email: izumi@ncnp.go.jp.
Yukiko Matsushima, Email: matsushima-yk@ncnp.go.jp.
Noritaka Ichinohe, Email: nichinohe@brain.riken.jp.
Yu-ichi Goto, Email: goto@ncnp.go.jp.
Takashi Okada, Email: t-okada@nms.ac.jp.
Ken Inoue, Email: kinoue@ncnp.go.jp.
References
- 1.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14(10):417–422. doi: 10.1016/S0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 2.Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6(1):1–16. doi: 10.1007/s10048-004-0207-y. [DOI] [PubMed] [Google Scholar]
- 3.Inoue K, et al. A duplicated PLP gene causing Pelizaeus-Merzbacher disease detected by comparative multiplex PCR. Am J Hum Genet. 1996;59(1):32–39. [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue K, et al. Proteolipid protein gene duplications causing Pelizaeus-Merzbacher disease: molecular mechanism and phenotypic manifestations. Ann Neurol. 1999;45(5):624–632. doi: 10.1002/1531-8249(199905)45:5<624::AID-ANA11>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Hudson LD, Puckett C, Berndt J, Chan J, Gencic S. Mutation of the proteolipid protein gene PLP in a human X chromosome-linked myelin disorder. Proc Natl Acad Sci USA. 1989;86(20):8128–8131. doi: 10.1073/pnas.86.20.8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trofatter JA, Dlouhy SR, DeMyer W, Conneally PM, Hodes ME. Pelizaeus-Merzbacher disease: tight linkage to proteolipid protein gene exon variant. Proc Natl Acad Sci USA. 1989;86(23):9427–9430. doi: 10.1073/pnas.86.23.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue K. Cellular pathology of Pelizaeus-Merzbacher disease involving chaperones associated with endoplasmic reticulum stress. Front Mol Biosci. 2017;4:7. doi: 10.3389/fmolb.2017.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Readhead C, Schneider A, Griffiths I, Nave KA. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12(3):583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 9.Kagawa T, et al. Glial cell degeneration and hypomyelination caused by overexpression of myelin proteolipid protein gene. Neuron. 1994;13(2):427–442. doi: 10.1016/0896-6273(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 10.Anderson TJ, et al. Late-onset neurodegeneration in mice with increased dosage of the proteolipid protein gene. J Comp Neurol. 1998;394(4):506–519. doi: 10.1002/(SICI)1096-9861(19980518)394:4<506::AID-CNE8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Inoue Y, Kagawa T, Matsumura Y, Ikenaka K, Mikoshiba K. Cell death of oligodendrocytes or demyelination induced by overexpression of proteolipid protein depending on expressed gene dosage. Neurosci Res. 1996;25(2):161–172. doi: 10.1016/0168-0102(96)01039-5. [DOI] [PubMed] [Google Scholar]
- 12.Simons M, et al. Overexpression of the myelin proteolipid protein leads to accumulation of cholesterol and proteolipid protein in endosomes/lysosomes: implications for Pelizaeus-Merzbacher disease. J Cell Biol. 2002;157(2):327–336. doi: 10.1083/jcb.200110138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simons M, Krämer EM, Thiele C, Stoffel W, Trotter J. Assembly of myelin by association of proteolipid protein with cholesterol- and galactosylceramide-rich membrane domains. J Cell Biol. 2000;151(1):143–154. doi: 10.1083/jcb.151.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saher G, et al. Therapy of Pelizaeus-Merzbacher disease in mice by feeding a cholesterol-enriched diet. Nat Med. 2012;18(7):1130–1135. doi: 10.1038/nm.2833. [DOI] [PubMed] [Google Scholar]
- 15.Inoue K, et al. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet. 2002;71(4):838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garbern JY, et al. Proteolipid protein is necessary in peripheral as well as central myelin. Neuron. 1997;19(1):205–218. doi: 10.1016/S0896-6273(00)80360-8. [DOI] [PubMed] [Google Scholar]
- 17.Hocquemiller M, Giersch L, Audrain M, Parker S, Cartier N. Adeno-associated virus-based gene therapy for CNS diseases. Hum Gene Ther. 2016;27(7):478–496. doi: 10.1089/hum.2016.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cearley CN, Vandenberghe LH, Parente MK, Carnish ER, Wilson JM, Wolfe JH. Expanded repertoire of AAV vector serotypes mediate unique patterns of transduction in mouse brain. Mol Ther. 2008;16(10):1710–1718. doi: 10.1038/mt.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung KH, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34(7):e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, Jiang X, Li W, Cao D, Shen K, Yang J. Inhibition of the long non-coding RNA MALAT1 suppresses tumorigenicity and induces apoptosis in the human ovarian cancer SKOV3 cell line. Oncol Lett. 2016;11(6):3686–3692. doi: 10.3892/ol.2016.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Jonquieres G, et al. Glial promoter selectivity following AAV-delivery to the immature brain. PLoS ONE. 2013;8(6):e65646. doi: 10.1371/journal.pone.0065646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgiou E, et al. Gene therapy targeting oligodendrocytes provides therapeutic benefit in a leukodystrophy model. Brain. 2017;140(3):599–616. doi: 10.1093/brain/aww351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawlor PA, Bland RJ, Mouravlev A, Young D, During MJ. Efficient gene delivery and selective transduction of glial cells in the mammalian brain by AAV serotypes isolated from nonhuman primates. Mol Ther. 2009;17(10):1692–1702. doi: 10.1038/mt.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geisler A, Fechner H. MicroRNA-regulated viral vectors for gene therapy. World J Exp Med. 2016;6(2):37–54. doi: 10.5493/wjem.v6.i2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gow A, Southwood CM, Lazzarini RA. Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus-Merzbacher disease. J Cell Biol. 1998;140(4):925–934. doi: 10.1083/jcb.140.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prukop T, et al. Progesterone antagonist therapy in a Pelizaeus-Merzbacher mouse model. Am J Hum Genet. 2014;94(4):533–546. doi: 10.1016/j.ajhg.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marteyn A, et al. Modulation of the innate immune response by human neural precursors prevails over oligodendrocyte progenitor remyelination to rescue a severe model of Pelizaeus-Merzbacher disease. Stem Cells. 2016;34(4):984–996. doi: 10.1002/stem.2263. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu T, et al. The balance between cathepsin C and cystatin F controls remyelination in the brain of Plp1-overexpressing mouse, a chronic demyelinating disease model. Glia. 2017;65(6):917–930. doi: 10.1002/glia.23134. [DOI] [PubMed] [Google Scholar]
- 30.Ip CW, et al. Immune cells contribute to myelin degeneration and axonopathic changes in mice overexpressing proteolipid protein in oligodendrocytes. J Neurosci. 2006;26(31):8206–8216. doi: 10.1523/JNEUROSCI.1921-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aartsma-Rus A, van Putten M. Assessing functional performance in the mdx mouse model. J Vis Exp. 2014;(85):e51303. doi: 10.3791/51303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gravel M, Di Polo A, Valera PB, Braun PE. Four-kilobase sequence of the mouse CNP gene directs spatial and temporal expression of lacZ in transgenic mice. J Neurosci Res. 1998;53(4):393–404. doi: 10.1002/(SICI)1097-4547(19980815)53:4<393::AID-JNR1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 33.Kagiava A, et al. Gene delivery targeted to oligodendrocytes using a lentiviral vector. J Gene Med. 2014;16(11-12):364–373. doi: 10.1002/jgm.2813. [DOI] [PubMed] [Google Scholar]
- 34.Sargiannidou I, et al. Connexin32 mutations cause loss of function in Schwann cells and oligodendrocytes leading to PNS and CNS myelination defects. J Neurosci. 2009;29(15):4736–4749. doi: 10.1523/JNEUROSCI.0325-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride JL, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci USA. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beer S, et al. Low-level shRNA cytotoxicity can contribute to MYC-induced hepatocellular carcinoma in adult mice. Mol Ther. 2010;18(1):161–170. doi: 10.1038/mt.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snøve O, Rossi JJ. Toxicity in mice expressing short hairpin RNAs gives new insight into RNAi. Genome Biol. 2006;7(8):231. doi: 10.1186/gb-2006-7-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 39.McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16(10):1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 40.Aschauer DF, Kreuz S, Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS ONE. 2013;8(9):e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L, Kan EM, Lu J, Wu C, Ling EA. Expression of 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase) and its roles in activated microglia in vivo and in vitro. J Neuroinflammation. 2014;11:148. doi: 10.1186/s12974-014-0148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell SK, et al. Characterization of a novel adeno-associated viral vector with preferential oligodendrocyte tropism. Gene Ther. 2016;23(11):807–814. doi: 10.1038/gt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neychev VK, Fan X, Mitev VI, Hess EJ, Jinnah HA. The basal ganglia and cerebellum interact in the expression of dystonic movement. Brain. 2008;131(Pt 9):2499–2509. doi: 10.1093/brain/awn168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Karakatsani ME, Fung C, Sun T, Acosta C, Konofagou E. Direct brain infusion can be enhanced with focused ultrasound and microbubbles. J Cereb Blood Flow Metab. 2017;37(2):706–714. doi: 10.1177/0271678X16637881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Meo I, Marchet S, Lamperti C, Zeviani M, Viscomi C. AAV9-based gene therapy partially ameliorates the clinical phenotype of a mouse model of Leigh syndrome. Gene Ther. 2017;24(10):661–667. doi: 10.1038/gt.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katz ML, et al. AAV gene transfer delays disease onset in a TPP1-deficient canine model of the late infantile form of Batten disease. Sci Transl Med. 2015;7(313):313ra180. doi: 10.1126/scitranslmed.aac6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dirren E, Aebischer J, Rochat C, Towne C, Schneider BL, Aebischer P. SOD1 silencing in motoneurons or glia rescues neuromuscular function in ALS mice. Ann Clin Transl Neurol. 2015;2(2):167–184. doi: 10.1002/acn3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hironaka K, et al. Enzyme replacement in the CSF to treat metachromatic leukodystrophy in mouse model using single intracerebroventricular injection of self-complementary AAV1 vector. Sci Rep. 2015;5:13104. doi: 10.1038/srep13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao W, Chirmule N, Berta SC, McCullough B, Gao G, Wilson JM. Gene therapy vectors based on adeno-associated virus type 1. J Virol. 1999;73(5):3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomono T, et al. Ultracentrifugation-free chromatography-mediated large-scale purification of recombinant adeno-associated virus serotype 1 (rAAV1) Mol Ther Methods Clin Dev. 2016;3:15058. doi: 10.1038/mtm.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Dorchies OM, et al. The anticancer drug tamoxifen counteracts the pathology in a mouse model of Duchenne muscular dystrophy. Am J Pathol. 2013;182(2):485–504. doi: 10.1016/j.ajpath.2012.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.