Abstract
Imaging research laboratories are rapidly creating machine learning systems that achieve expert human performance using open-source methods and tools. These artificial intelligence systems are being developed to improve medical image reconstruction, noise reduction, quality assurance, triage, segmentation, computer-aided detection, computer-aided classification, and radiogenomics. In August 2018, a meeting was held in Bethesda, Maryland, at the National Institutes of Health to discuss the current state of the art and knowledge gaps and to develop a roadmap for future research initiatives. Key research priorities include: 1, new image reconstruction methods that efficiently produce images suitable for human interpretation from source data; 2, automated image labeling and annotation methods, including information extraction from the imaging report, electronic phenotyping, and prospective structured image reporting; 3, new machine learning methods for clinical imaging data, such as tailored, pretrained model architectures, and federated machine learning methods; 4, machine learning methods that can explain the advice they provide to human users (so-called explainable artificial intelligence); and 5, validated methods for image de-identification and data sharing to facilitate wide availability of clinical imaging data sets. This research roadmap is intended to identify and prioritize these needs for academic research laboratories, funding agencies, professional societies, and industry.
© RSNA, 2019
Summary
This summary of the 2018 NIH/RSNA/ACR/The Academy Workshop on Artificial Intelligence in Medical Imaging provides a roadmap to identify and prioritize research needs for academic research laboratories, funding agencies, professional societies, and industry.
Key Points
■ New image reconstruction and enhancement methods are needed to produce images suitable for human interpretation from the source data produced by the imaging device.
■ Automated labeling methods are needed to rapidly produce training data for machine learning research by extracting information from narrative reports and clinical notes.
■ Novel machine learning algorithms are needed that are tailored for the complexity of clinical imaging data, which are often high resolution, 3D, 4D, multimodality, and multichannel.
■ Machine learning systems must be capable of explaining or illustrating the advice they provide to human users (so-called explainable artificial intelligence).
■ Aggregation methods for clinical imaging data are needed to produce the large volume of data necessary to train machine learning algorithms.
Introduction
Artificial intelligence (AI) concerns the development of methods that enable computers to behave in ways we consider intelligent when those same behaviors are exhibited by humans. AI is the most general term to define this field of inquiry and is broadly understood by scientists and lay public. Although some have expressed concerns about the development of an “artificial general intelligence” that could mimic the totality of human behavior, for the foreseeable future, AI systems will be “narrow,” typically constructed to assist humans with a specific task, such as driving a car, targeting an advertisement, or interpreting a mammogram.
A more specific term to describe the most popular form of AI is machine learning. Machine learning systems are constructed by feeding many positive and negative examples to an algorithm that modifies itself based on feedback from its response to these examples. Until recently, the most accurate machine learning methods for image analysis involved painstaking feature engineering, the manual development of software to preprocess images, segment anatomic structures, and detect or compute features suggested by an expert.
To classify the images, the extracted features were fed to a suitable machine learning algorithm, such as penalized regression (1), support vector machines (2), conditional random fields (3), or random forests (4). These machine learning methods were generally effective, but often required years of software development, and faced challenges of developing accurate feature extraction methods and selecting an appropriate machine learning algorithm.
Recent advances in artificial intelligence have replaced feature engineering with a more time-efficient process of machine learning from large sets of labeled training data using neural networks with many layers—sometimes called deep learning (5). Image classifiers that had taken years to develop now can be created in weeks or months.
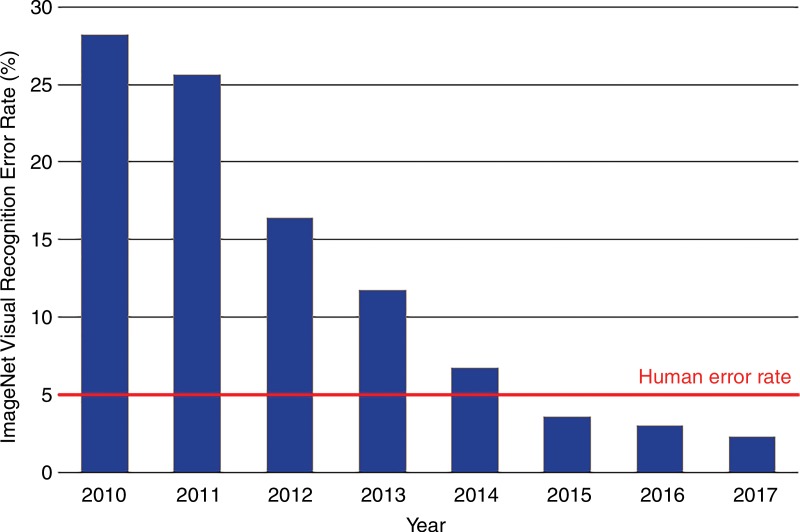
ImageNet, a database of over 14 million human-annotated nonmedical images has been instrumental to the success of these new systems (6). The classification error for the annual ImageNet large-scale visual recognition challenge has declined more than eightfold over the past 6 years to a rate of below 3% in 2017, which surpasses human performance (7,8) (Fig 1). This progress has been catalyzed by the rapid advances in computer hardware in the past decade.
Figure 1:
Error rates on the ImageNet Large-Scale Visual Recognition Challenge. Accuracy dramatically improved with the introduction of deep learning in 2012 and continued to improve thereafter. Humans perform with an error rate of approximately 5%.
The recent successes of these AI techniques in the analysis of nonmedical images has led to high interest and explosive growth in the use of deep learning in the analysis of clinical images and other medical data. These promising techniques create computer vision systems that perform some clinical image interpretation tasks at the level of expert physicians (9–12). The resulting computer vision systems have the potential to transform medical imaging, thereby reducing diagnostic errors, improving patient outcomes, enhancing efficiency, and reducing costs.
Diagnostic errors may cause patient harm, and they play a role in up to 10% of patient deaths (13). Between 3% and 6% of image interpretations rendered by radiologists contain clinically important errors (14–16). Inter- and intraobserver variability, another indicator of error, occurs at rates as high as 37%, depending on the imaging modality (17). Cardiologists and pathologists experience similar error rates (18,19).
In August 2018, the National Institutes of Health (NIH) assembled multiple relevant stakeholders at a public meeting. The goal was to assess the current state of the science of AI in medical imaging, to identify gaps in current data, knowledge, and science, and to provide a research and translation roadmap that maximizes the benefit to patients in the years to come. This report summarizes key priorities of that meeting for acceleration of foundational AI research.
Research Priorities in Machine Learning Research
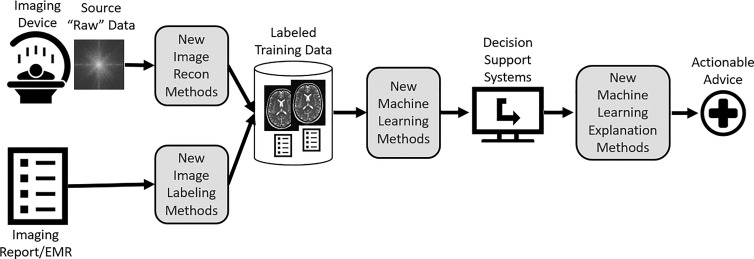
The ultimate purpose of AI research in medical imaging is to create tools that improve patient outcomes. AI tools typically take the form of imaging decision support systems that provide actionable advice to imaging professionals. The gaps in foundational machine learning research can be viewed along the path from the raw materials for machine learning to the production of decision support systems that provide actionable advice to imaging professionals (Fig 2).
Figure 2:
Diagram illustrates the use of images and narrative reports to produce decision support systems that provide actionable advice. There are several opportunities for AI in medical imaging research (shaded boxes). EMR = electronic medical record.
There are several opportunities for AI in medical imaging research from image acquisition device to actionable advice:
New image reconstruction and enhancement methods are needed to produce images suitable for human interpretation from the source data produced by the imaging device. These methods can produce high-quality images using smaller doses of intravenous contrast material, lower radiation dose, and shorter scan and reconstruction times.
Automated labeling and annotation methods are needed to rapidly produce training data for machine learning research. These labeling methods often use machine learning algorithms that process information from the imaging report or the electronic medical record.
Because most deep learning research has been conducted on photographs and videos of natural images, there is a need to develop novel machine learning algorithms trained for the complexity of clinical imaging data, which are often high resolution, 3D, 4D, multimodality, and multichannel.
Because these algorithms will be operating as a sophisticated clinical “autopilot,” in partnership with a human imaging expert, there is a need for research on machine learning methods that can explain or illustrate advice to human users.
Because privacy concerns are paramount when using clinical data, methods are needed to facilitate the aggregation of clinical imaging data for training of machine learning algorithms.
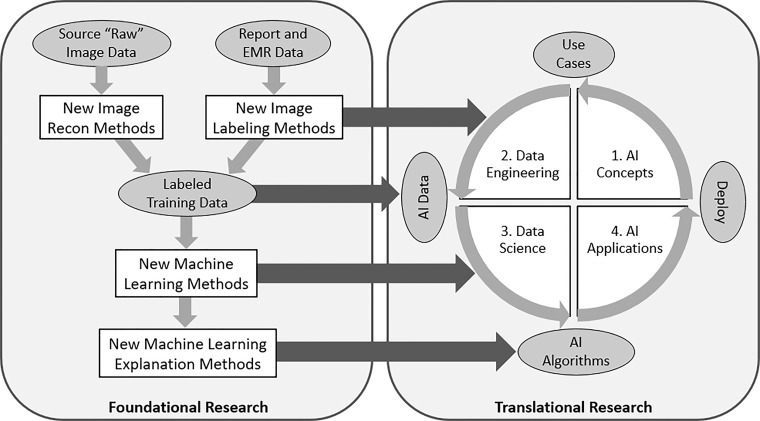
These priorities directly affect the translational research priorities discussed at the workshop. The relationship between foundational and translational research roadmaps is illustrated in Figure 3.
Figure 3:
Diagram shows how foundational and translational research activities are connected. Foundational research leads to new image reconstruction and labeling methods, new machine learning algorithms, and new explanation methods, each of which enhance the data sets, data engineering, and data science that lead to the successful deployment of AI applications in medical imaging. EMR = electronic medical record.
Data Availability
The first set of challenges for foundational AI research relates to data availability and sharing. Table 1 highlights the important opportunities in this area.
Table 1:
Research Opportunities in AI for Medical Imaging Related to Data Sharing and Data Availability
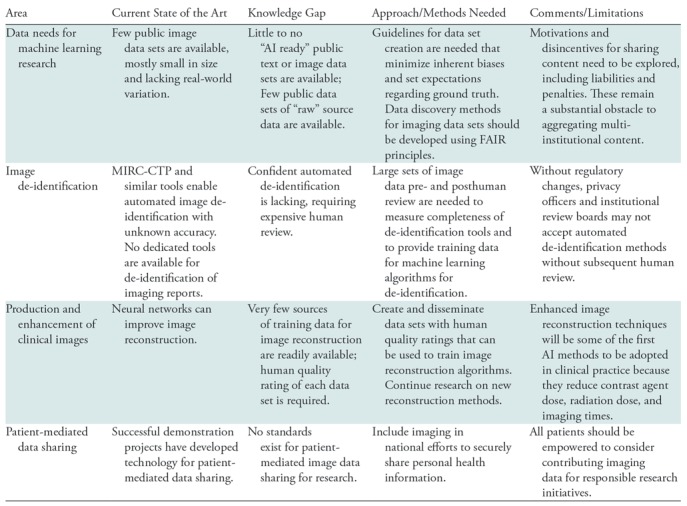
Note.—FAIR = findable, accessible, interoperable and reusable, MIRC-CTP = Medical Image Resource Center-Clinical Trials Processor.
Data Needs for Machine Learning Research
The first major barrier to progress in machine learning in medical imaging is the lack of standard and accessible imaging data for training of machine learning algorithms. The development of new AI methods requires high-quality, labeled, curated, publicly available data. While health care organizations worldwide control vast stores of data that could be exploited to train machine learning algorithms, most imaging data are not accessible for research purposes. Accessible imaging data are often unusable because they are not curated, organized, anonymized, or appropriately annotated, and rarely linked to a ground-truth diagnosis. Few well-curated and validated image data sets are available to the research and vendor community. To address these gaps, more effective methods are needed for data collection, de-identification, and management of images for research that use findable, accessible, interoperable and reusable (FAIR) principles for scientific data management and stewardship (20).
A small number of imaging data sets have been made public across imaging domains. For example, there are several data sets available for neuroimaging research (21–26). However, many public data sets are too small to support clinically meaningful machine learning experiments or consist mostly of healthy individuals or functional MRI data from patients with psychiatric diseases. Those that are available often were obtained from a single institution and do not reflect the variety of imaging devices and clinical scenarios that will be encountered in real-world settings. A few larger data sets have been constructed, with the potential to be useful for machine learning experiments. For example, The Cancer Imaging Archive hosts imaging data from multiple cancers; the largest data set, containing over 50 000 patient studies, comes from the National Lung Screening Trial (27) and is patient matched to clinical and genomics data in The Cancer Genome Atlas. The NIH has recently released large chest x-ray (28) and chest CT (29) data sets with weak annotations specifically for training machine learning models. A data set for detecting abnormalities on musculoskeletal radiographs is available (30), as is a data set containing both k-space and reconstructed images for knee MRI studies (31). A data set of nearly 600 000 chest radiographs with high-quality labels was recently released as a joint effort of two large research groups (32).
Researchers face significant challenges in finding and accessing the medical imaging data sets that are publicly available. Google recently announced a data set search tool, which provides pointers to data sets across a variety of web-based repositories (33). As more clinical imaging data sets become available, tools of this kind will become essential to find clinical imaging data sets.
Image De-identification
The automated de-identification of images is addressed today by open-source tools like the Medical Imaging Resource Center–Clinical Trials Processor (MIRC-CTP), which receives DICOM images and replaces the protected health information (PHI) they contain with de-identified data (34). While anecdotal evidence suggests these systems sometimes miss PHI in custom DICOM tags, training data to measure the accuracy of these de-identification systems are scarce because they contain PHI. Lack of validated standards and technology for the de-identification of images and reports reduces the willingness of individual sites to share data, for fear of a breach of PHI leading to litigation and federal penalties. Research is needed to measure and enhance the capabilities of current image and report de-identification methods.
Quality Rated Data for Reconstruction and Enhancement of Clinical Images
Clinical images produced by sophisticated imaging devices, such as CT, MRI, PET/SPECT, US, and optical scanners, are reconstructed from “raw” or source data produced by detectors. The resulting signals are indirect and imperfect evidence of anatomic, functional, cellular, and molecular features. The relationships between these tomographic data and underlying structures are generally nonlinear and complex. The optimal conversion of source data from these sensors into reconstructed tomographic images suitable for human interpretation or radiomics is an emerging area of extensive study. Deep learning methods can be highly effective in reconstructing images directly from source data (35). For example, these data-driven image reconstruction methods can perform superior MRI reconstruction, predict the image enhanced with a full contrast agent dose from a partial dose counterpart, or a high-quality image from a low-radiation-dose scan (36–39). The result may be shorter scan times and, because less expensive components are needed, lower cost imaging devices.
In some cases, training data for these methods are readily available. For example, the first serial counts from a PET scanner (ie, in “list mode”) are a realistic simulation of a low-dose study; the full study can serve as the reference standard. Likewise, a test-dose of contrast agent before a full dose study can serve as training data for machine learning. But in many other cases, we can only measure the synthetic images against radiologists’ perception of image quality, against basic measures of quality such as noise and spatial resolution, or against patient outcomes that are noisy and can take many years to occur. For this reason, data sets and benchmarks are needed for subjective image quality ratings that address the suitability of the images for specific clinical tasks. These resources would serve as training data for the production of many image reconstruction and enhancement methods and could also be used to produce deep learning algorithms that simulate human image quality ratings.
The source data needed for this form of research can be difficult to obtain. List-mode data, sinogram data, and k-space data are often kept in a proprietary format that impedes data sharing and can raise intellectual property concerns if shared, so industry cooperation often is required. The International Society for Magnetic Resonance in Medicine has proposed a standard for the exchange of MRI source data (40), but few vendors make MRI source data available in this format. The challenges of obtaining CT sinogram data are even more formidable. Professional societies and industry must work together to make this form of data more accessible to researchers.
Methods and Standards for Patient-mediated Data Sharing
In the past decade, we have overcome many technical challenges of cloud-based sharing of clinical images and reports between care teams at disparate facilities. But the logistical, operational, and regulatory challenges of record sharing between institutions for research requires substantial additional effort and resources. Attempts to aggregate data for research initiatives remain limited and do not scale effectively to meet the needs of AI research: large data sets, acquired from heterogeneous sources, with diverse patient representation. While there are national imperatives that encourage sharing of research data, there are barriers to data access and usability and challenges in combining data sets from multiple sources.
Patient-mediated data sharing has the potential to dramatically increase the number, type, and variety of available data for machine learning by breaking down institutional barriers to sharing. Patients today are more involved and engaged in their care and more recently have become actively engaged in advancing medical research. The RSNA Image Share Network demonstrated how patients can take ownership of their imaging examinations and exchange them at their discretion, first using Simple Object Access Protocol–based standards developed by Integrating the Healthcare Enterprise, then updated to incorporate Fast Health care Interoperability Resources and DICOMweb (41).
Despite these important enabling steps, there are no established mechanisms or standards for patients to share their medical data for research, and there is no common repository or exchange clearinghouse for patient data outside individual health care organizations. Initiatives such as the NIH-sponsored All of Us/Sync 4 Science program, and the nonprofit Count Me In collaborative, aim to advance personalized medicine and research (42). As these initiatives are expanded to educate patients about the importance of data sharing for research, patients should be encouraged to share not only their electronic medical record (EMR) information but also their images.
Image Labeling and Annotation
Most health care organizations maintain picture archiving and communication systems (PACS) that store millions of clinical imaging studies and their associated reports. But imaging studies stored in PACS are not suitable for most machine learning research because they contain no labels or annotations for machine learning. Accordingly, a second imperative AI research theme in medical imaging is the development of rapid labeling and annotation methods for clinical images. The opportunities in this area are summarized in Table 2.
Table 2:
Research Opportunities in AI for Medical Imaging Related to Image Labeling and Annotation
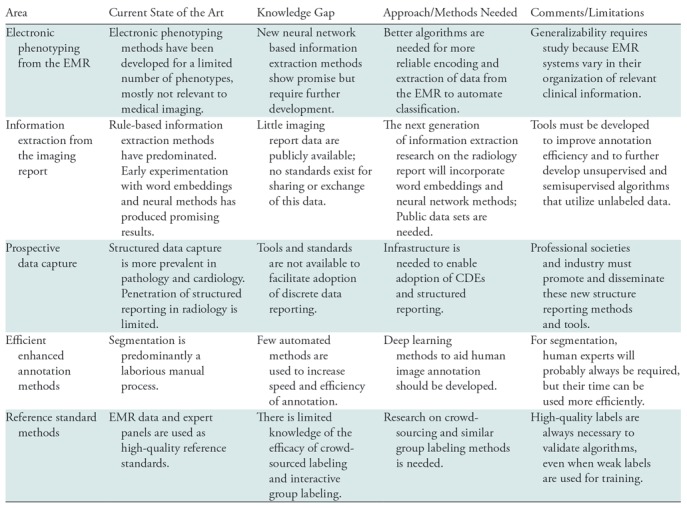
Note.—EMR = electronic medical record, CDE =common data element.
We define “labeling” as attaching a category, class, or tag to an entire image or imaging study. This form of labeling is particularly helpful in the development of machine learning systems that perform classification tasks, such as whether an imaging study shows the presence of tuberculosis or a lung nodule. We define “annotation” as furnishing information about a particular portion of an image—for example, whether a pixel is part of a tumor or not. Annotations are particularly useful training data for segmentation or detection tasks.
When using unstructured data to create labels from the EMR or from the imaging report, an important consideration is the degree of noise that can be tolerated in the training data. Both mathematical analysis and empirical results indicate that neural network models are quite robust to large amounts of noise (43–48). Thus, the resulting classifiers often achieve an F1 statistic (a measure of accuracy) in the 85%–90% range and can serve as noisy labels for training data (73).
Electronic Phenotyping from the Electronic Medical Record
The EMR is a rich source of information about the patient, including disease states that can serve as image labels. For example, the EMR documents an oncologic condition through a pathologic tissue diagnosis, or it confirms a major medical condition like liver disease with clear biochemical test results. In those instances, the EMR can provide labels for diagnostic images. Methods to extract labels from the EMR are often called “electronic phenotyping” because they identify patients with a defined disease, clinical condition, or outcome based on the contents of the EMR (49–51).
Electronic phenotyping methods typically have relied on rule-based definitions (47,52). Research networks, such as the Electronic Medical Records and Genomics network (53,54), create and validate electronic phenotypes at multiple institutions and make them available online in PheKB, the Phenotype Knowledge-Base (55). An alternative is the use of machine learning to train classifiers from patient records labeled as having the phenotype or not (44,45,56,57). For example, regression-based phenotype models for rheumatoid arthritis learned from expert labeled data can identify cases with a positive predictive value of 94% (58). Supplementing diagnosis codes and medications with images and terms extracted from clinical narratives has been shown to improve phenotyping classifiers (59,60). However, to date only a few dozen phenotyping algorithms have been developed, only a fraction of which are relevant to medical imaging (61). Electronic phenotyping research must be scaled up to incorporate the phenotypes that can serve as ground truth for the training of computer vision algorithms.
Information Extraction from the Radiology Report
Almost all clinical imaging studies are accompanied by a report that describes the imaging findings, produced by an expert physician. When the abnormality is clearly visible on the images, such as the presence of a lung nodule, the imaging report can be a rich source of image labels for training of deep learning systems.
To date, information extraction methods have been applied to detect specific report features or to extract labels from reports of a particular imaging modality and body region. The earliest and most widely studied radiology information extraction system is the Medical Language Extraction and Encoding System, which uses a custom controlled vocabulary and grammatical rules to convert chest radiography reports into a structured format (62,63). General toolkits have been applied to extract concepts from medical narrative (64–67). However, the performance of these systems decreases when applied to data from multiple organizations (68) or to more complex narrative reports from CT and MRI head images (69).
More recent work on information extraction from the radiology report focuses on specific elements of the report to improve accuracy for specific use cases. Rule-based natural language processing methods can extract critical results (70), and notification statements (71). The Lexicon Mediated Entropy Reduction system extracts and classifies phrases with important findings and recommendations from radiology reports (72–74). However, these techniques are not scalable for image classification, because they require manual annotation of a training set for each new class. Weak supervision to create inferred generative models is a special case of automated rule-based labeling that improves efficiency (75) but still requires new rule sets for each class. A simple model for the information in diagnostic imaging reports that generalizes across imaging modalities and body regions can improve scalability of weak labeling (76).
Recent research has repeatedly shown that unsupervised training of recurrent neural networks, or RNNs, on a large unlabeled collection of narrative reports yields superior performance to previous machine learning methods, which relied on hand-engineered features and specialized knowledge sources (77,78). The latest vector-based methods, such as the Word2Vec (79), and GloVe (80), rely on representations of words that encode not only information about the word, but also the context in which the word occurs. These methods typically achieve improved accuracy relative to conventional machine learning methods (81,82). These methods have not yet been used extensively to extract information from diagnostic imaging reports or the EMR, but early research suggests they will have substantial value to generate an efficient, automated, scalable method to extract labels (83–85).
Most information extraction methods currently used in medical imaging are based on supervised machine learning methods. Further research into semisupervised methods that utilize both labeled and unlabeled data can help reduce the annotation burden, currently a bottleneck to training robust models (86). Semisupervised and unsupervised learning with generative adversarial networks, or GANs, also shows promise (87).
Prospective Discrete Data Capture and Structured Reporting
Image labels that are consistently and prospectively applied by experts are the most valuable because they are highly accurate. To obviate the need for subsequent retrospective labeling of vast amounts of data, clinical images could be labeled or annotated as part of the routine clinical workflow using structured reporting (88). Structured reporting and associated standard interpretation criteria can reduce interobserver variability.
Standards are being developed to facilitate this approach. The ACR-RSNA common data elements (CDEs) initiative directly supports labeling and annotation during clinical work (89). The creation of a canonical set of structured imaging observations and responses represents a significant step forward in improving discovery of report annotations with high accuracy. CDEs enable structured annotated image and text data to be continuously updated, shared, and fully integrated for use in patient care and within a learning health care system.
Use of Machine Learning for Efficient Image Annotation
For image segmentation or detection tasks, large numbers of experts must create training and validation data by labeling the images and annotating the structures of interest. Research is needed on newer tools that reduce the annotation burden on human experts. For example, some algorithms can semiautomatically trace structures on images, so that a human annotator need only modify machine-generated traces, rather than generate each annotation de novo (90). It is possible to train deep learning methods in a semisupervised manner with minimally annotated data sets to get reasonable approximations of structures, thereby iteratively reducing the human effort in tracing structures (91).
Reference Standard
To definitively measure the accuracy of machine learning algorithms, a smaller data set (typically containing hundreds of cases) with high-quality labels is required. Often this reference standard is produced from manual chart review or tissue sampling. But many conditions on diagnostic images, such as pneumonia, congestive heart failure, or the presence of a lung nodule on a screening study, are only visible to the imaging professional reviewing the images. In these cases, a panel of clinical experts must serve as the reference standard by reviewing comprehensive longitudinal clinical data sets that include imaging, reporting, and pathology.
But variability, even between experts, is well documented. Bias and variability are inherent when manual annotations are acquired and can lead to label noise. Acquiring annotations from multiple experts for each case is currently considered best practice (10,12) yet is achieved infrequently due to the cost. Selecting an appropriate method to synthesize the judgment of many observers has been an active research area for many years (92). Fundamental methods to reduce interrater variability that can learn from noisy, biased labels need to be further developed. For example, the availability of “crowd-sourced” image labeling platforms and methods provide new opportunities to create valid image labels (93).
Machine Learning Challenges Unique to Clinical Imaging Data
Clinical images present unique challenges to AI researchers. The challenges in this area are highlighted in Table 3.
Table 3:
Research Opportunities Related to the Unique Challenges of Clinical Imaging Data
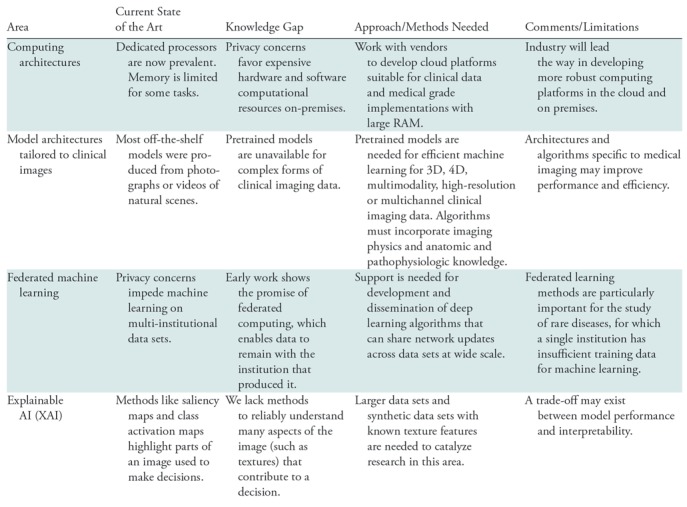
Computing Architectures
Computing power has increased by orders of magnitude over the past decade, enabling rapid progress in deep learning and computer vision. Computational power for deep learning methods is typically supplied by graphical processing units (GPUs) or specially designed chips like tensor processing units (94). These high-performance computers can be implemented locally (“on-premises”) or in the cloud. Cloud computing is typically more cost-effective when bursts of high-performance computing are needed, while sustained computing is typically more cost-effective on dedicated machines in local data centers. Cloud computing poses challenges because it entails transferring clinical data to a cloud platform, which may raise privacy concerns.
Deep learning methods often require multiple GPUs to get results in reasonable time, requiring special software systems to route data and allocate computing across a network of processors. For cases with large image files (such as 3D segmentation algorithms), memory can also be a limitation. While some software libraries can spread computation across GPUs, methods for opportunistic allocation of computing and data across GPUs require further development.
Model Architectures Tailored to Clinical Imaging
Most off-the-shelf machine learning models were constructed for ImageNet, which is a data set of 256 × 256 color photographs of natural scenes (7). Pretrained weights for these images enhance the accuracy of machine learning classifiers, even for some clinical imaging tasks (95). However, these models can incorporate neither the knowledge of the physics of clinical image acquisition, nor the anatomy and pathology of human physiology and disease. Therefore, these pretrained models do not work effectively for many forms of clinical data, which are often high resolution, multimodality, multichannel, and 3D and 4D. Work is needed to develop a library of deep learning models, including pretrained weights, for these more complex forms of clinical imaging data. New architectures also are needed to perform image segmentation, reconstruction, and enhancement on more complex clinical data sets. Developing deep learning techniques that can incorporate such knowledge has the potential to improve the efficiency of training (eg, by using more realistic augmentation schemes) and to attain more biologically plausible results.
Federated Learning Methods
Research outside medicine typically strives to aggregate massive data sets from multiple sources, such as the ImageNet database. Such methods are less feasible for clinical data because they must meet the requirements of privacy officers and institutional review boards from each contributing institution. Due to privacy and data sharing limitations, most machine learning models to date have been generated using data from single institutions. The performance of such models may be limited due to sample size (in the case of rare diseases) or due to the lack of diverse training data, and therefore may not generalize well to other institutions. Federated learning methods are being developed whereby network weights and parameters, rather than the training data, are transferred between sites, enabling learning across multi-institutional data while preserving the privacy (96,97). More research is needed to develop and refine these new methods.
Explainable Artificial Intelligence
An important impediment to clinical adoption (and possibly regulatory approval) of AI algorithms is a lack of understanding of how the algorithm made its decision. Trained deep learning networks have been described as “black boxes.” Deep learning models are susceptible to bias (98) and subject to adversarial attack (99). These challenges are not unique to medical imaging; similar concerns have arisen in finance, defense, and self-driving cars. As a result, there are increasing efforts to develop explainable artificial intelligence (XAI) methods.
XAI methods such as attention or saliency maps can draw attention to the locations in an image that contributed to the decision for a particular case (100). These methods also can filter certain frequencies or textures to understand their contribution to a decision (100). Another important benefit of XAI methods are their potential to increase our understanding of a disease and to improve imaging devices. If a network can predict cancer survival and the saliency maps indicate a risk factor outside the visible signal abnormality, then microscopic invasion may be an indicator of tumor progression. If a certain texture frequency is critical to identifying a label or annotation, one may be able to optimize an imaging device to enhance (or avoid suppressing) that frequency.
Conclusion
Machine learning algorithms will transform clinical imaging practice over the next decade. Most imaging research laboratories are now employing machine learning methods to solve computer vision problems. Yet machine learning research is still in its early stages. We have outlined several key research themes and described a roadmap to accelerate advances in foundational machine learning research for medical imaging. To produce generalizable algorithms, rather than just those that serve communities with a wealth of AI researchers, we describe innovations that would help to produce more publicly available, validated, and reusable data sets against which to evaluate new algorithms and techniques. To be useful for machine learning, these data sets require methods to rapidly create labeled or annotated imaging data. Finally, novel pretrained model architectures, tailored for clinical imaging data, must be developed, along with methods for federated training that reduce the need for data exchange between institutions. Standards bodies, professional societies, government agencies, and private industry must work together to accomplish these goals in service of patients, who are sure to benefit from the innovative imaging technologies that will result.
Acknowledgments
Acknowledgments
The authors thank all the presenters and participants at the NIH Workshop on Artificial Intelligence in Medical Imaging. We also are indebted to Maryellen Giger, Paul Kinahan, and Cynthia McCullough for their comments on earlier versions of the manuscript.
Disclosures of Conflicts of Interest: C.P.L. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: Founder, shareholder, board member, received consulting fees and travel reimbursement, Montage Healthcare Solutions; shareholder and advisory board member for Nines.ai, whiterabbit.ai, GalileoCDS, Inc, Bunker Hill, Inc. Other relationships: disclosed no relevant relationships. B.A. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: disclosed no relevant relationships. Other relationships: Chief Medical Officer Of The ACR Data Science Institute. B.J.E. disclosed no relevant relationships. J.K.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: personal fees, INFOTECH, Soft. Other relationships: disclosed no relevant relationships. K.B. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employee of GE Healthcare. Other relationships: disclosed no relevant relationships. T.S.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: Board membership and reimbursement of travel expenses, SIIM, AUR, PRRS, PRS; grants/grants pending, ACRIN, Beryl Institute, Siemens Healthineers, SIIM; Payment for lectures including service on speakers bureaus, Osler Institute. Other relationships: disclosed no relevant relationships. A.E.F. disclosed no relevant relationships. M.P.L. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: consultancy, Nines Inc; stock/stock options, Nines Inc. Other relationships: disclosed no relevant relationships. D.S.M. Activities related to the present article: grant and support for travel to meetings, NIH/RSNA; RSNA Image Sharecontract from NIBIB sponsored development of solutions described in this article regarding patient directed contribution of imaging exams. Activities not related to the present article: Board membership and stock options, Nines, Inc, and Maverick AI; general radiology IT advisory board, Bayer; PACS advisory board, GE. Other relationships: disclosed no relevant relationships. J.D.R. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: Pending research grant applications with RSNA and ASNR. Other relationships: disclosed no relevant relationships. G.W. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: support for travel to meetings, NIH; patents pending, AI related imaging IPs filed by RPI; research license to GE, Hologic. Other relationships: disclosed no relevant relationships. K.K. disclosed no relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- CDE
- common data element
- EMR
- electronic medical record
- GPU
- graphical processing unit
- PACS
- picture archiving and communication system
- PHI
- protected health information
- XAI
- explainable artificial intelligence
References
- 1.Tibshirani R. Regression Shrinkage and Selection via the Lasso. J R Stat Soc Series B Stat Methodol 1996;58(1):267–288. [Google Scholar]
- 2.Suykens JAK, Vandewalle J. Least Squares Support Vector Machine Classifiers. Neural Process Lett 1999;9(3):293–300. [Google Scholar]
- 3.Lafferty J, McCallum A, Pereira FCN. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. https://repository.upenn.edu/cis_papers/159. Published 2001.
- 4.Breiman L. Random forests. Mach Learn 2001;45(1):5–32. [Google Scholar]
- 5.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 6.Socher R. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE, 2009; 248–255. [Google Scholar]
- 7.Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis 2015;115(3):211–252. [Google Scholar]
- 8.Karpathy A. What I learned from competing against a ConvNet on ImageNet. http://karpathy.github.io/2014/09/02/what-i-learned-from-competing-against-a-convnet-on-imagenet/. Published 2014.
- 9.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542(7639):115–118 [Published correction appears in Nature 2017;546(7660):686.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316(22):2402–2410. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Gadepalli K, Norouzi M, et al. Detecting Cancer Metastases on Gigapixel Pathology Images. http://arxiv.org/abs/1703.02442. Published 2017.
- 12.Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 2018;15(11):e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Academy of Medicine . Improving Diagnosis in Health Care. Washington, DC: The National Academies Press, 2015. [PubMed] [Google Scholar]
- 14.Borgstede JP, Lewis RS, Bhargavan M, Sunshine JH. RADPEER quality assurance program: a multifacility study of interpretive disagreement rates. J Am Coll Radiol 2004;1(1):59–65. [DOI] [PubMed] [Google Scholar]
- 15.Berlin L. Accuracy of diagnostic procedures: has it improved over the past five decades? AJR Am J Roentgenol 2007;188(5):1173–1178. [DOI] [PubMed] [Google Scholar]
- 16.Waite S, Scott J, Gale B, Fuchs T, Kolla S, Reede D. Interpretive error in radiology. AJR Am J Roentgenol 2017;208(4):739–749. [DOI] [PubMed] [Google Scholar]
- 17.Elmore JG, Wells CK, Lee CH, Howard DH, Feinstein AR. Variability in radiologists’ interpretations of mammograms. N Engl J Med 1994;331(22):1493–1499. [DOI] [PubMed] [Google Scholar]
- 18.Saraf RP, Suresh P, Maheshwari S, Shah SS. Pediatric echocardiograms performed at primary centers: Diagnostic errors and missing links! Ann Pediatr Cardiol 2015;8(1):20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson SL, Frederick PD, Pepe MS, et al. Diagnostic reproducibility: what happens when the same pathologist interprets the same breast biopsy specimen at two points in time? Ann Surg Oncol 2017;24(5):1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson MD, Dumontier M, IJJ Aalbersberg, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Essen DC, Smith SM, Barch DM, et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 2013;80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mennes M, Biswal BB, Castellanos FX, Milham MP. Making data sharing work: the FCP/INDI experience. Neuroimage 2013;82:683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jack CR, Jr, Bernstein MA, Fox NC, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27(4):685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Martino A, Yan CG, Li Q, et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 2014;19(6):659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4:170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prior FW, Clark K, Commean P, et al. TCIA: An information resource to enable open science. Conf Proc IEEE Eng Med Biol Soc 2013;2013:1282–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. http://arxiv.org/abs/1705.02315. Published 2017. Accessed DATE.
- 29.Yan K, Wang X, Lu L, Summers RM. DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging (Bellingham) 2018;5(3):036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajpurkar P, Irvin J, Bagul A, et al. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs. arXiv [physics.med-ph]. http://arxiv.org/abs/1712.06957. Published 2017.
- 31.Zbontar J, Knoll F, Sriram A, et al. fastMRI: An Open Dataset and Benchmarks for Accelerated MRI. arXiv [cs.CV]. http://arxiv.org/abs/1811.08839. Published 2018.
- 32.Irvin J, Rajpurkar P, Ko M, et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. arXiv [cs.CV]. http://arxiv.org/abs/1901.07031. Published 2019.
- 33.Noy N. Making it easier to discover datasets. https://www.blog.google/products/search/making-it-easier-discover-datasets/. Accessed January 1, 2019.
- 34.MIRC CTP Wiki . http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor. Accessed September 23, 2018.
- 35.Wang G, Ye JC, Mueller K, Fessler JA. image reconstruction is a new frontier of machine learning. IEEE Trans Med Imaging 2018;37(6):1289–1296. [DOI] [PubMed] [Google Scholar]
- 36.Chen KT, Gong E, de Carvalho Macruz FB, et al. Ultra-low-dose 18F-florbetaben amyloid pet imaging using deep learning with multi-contrast MRI inputs. Radiology 2019;290(3):649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong E, Pauly JM, Wintermark M, Zaharchuk G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Imaging 2018;48(2):330–340. [DOI] [PubMed] [Google Scholar]
- 38.Yang Q, Yan P, Zhang Y, et al. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE Trans Med Imaging 2018;37(6):1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature 2018;555(7697):487–492. [DOI] [PubMed] [Google Scholar]
- 40.Inati SJ, Naegele JD, Zwart NR, et al. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn Reson Med 2017;77(1):411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendelson DS, Erickson BJ, Choy G. Image sharing: evolving solutions in the age of interoperability. J Am Coll Radiol 2014;11(12,12 Pt B):1260–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon HU. General Bounds on the Number of Examples Needed for Learning Probabilistic Concepts. J Comput Syst Sci 1996;52(2):239–254. [Google Scholar]
- 44.Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016;23(6):1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halpern Y, Horng S, Choi Y, Sontag D. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc 2016;23(4):731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banda JM, Halpern Y, Sontag D, Shah NH. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc 2017;2017:48–57. [PMC free article] [PubMed] [Google Scholar]
- 47.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013;20(e1):e147–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rolnick D, Veit A, Belongie S, Shavit N. Deep Learning is Robust to Massive Label Noise. arXiv [cs.LG]. http://arxiv.org/abs/1705.10694. Published 2017.
- 49.Rasmussen LV, Thompson WK, Pacheco JA, et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms. J Biomed Inform 2014;51:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richesson RL, Sun J, Pathak J, Kho AN, Denny JC. Clinical phenotyping in selected national networks: demonstrating the need for high-throughput, portable, and computational methods. Artif Intell Med 2016;71:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 2014;21(2):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Overby CL, Pathak J, Gottesman O, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc 2013;20(e2):e243–e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011;3(79):79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016;23(6):1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mosley JD, Witte JS, Larkin EK, et al. Identifying genetically driven clinical phenotypes using linear mixed models. Nat Commun 2016;7(1):11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peissig PL, Santos Costa V, Caldwell MD, et al. Relational machine learning for electronic health record-driven phenotyping. J Biomed Inform 2014;52:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62(8):1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei WQ, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc 2016;23(e1):e20–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiley LK, Moretz JD, Denny JC, Peterson JF, Bush WS. Phenotyping Adverse Drug Reactions: Statin-Related Myotoxicity. AMIA Jt Summits Transl Sci Proc 2015;2015:466–470. [PMC free article] [PubMed] [Google Scholar]
- 61.Phe KB. A knowledge base for discovering phenotypes from electronic medical records. https://phekb.org/. Accessed January 2, 2019.
- 62.Friedman C, Alderson PO, Austin JHM, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1(2):161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hripcsak G, Austin JHM, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002;224(1):157–163. [DOI] [PubMed] [Google Scholar]
- 64.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17(5):507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 67.Manning CD, Surdeanu M, Bauer J, Finkel J, Bethard SJ, McClosky D. The Stanford CoreNLP Natural Language Processing Toolkit. Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics. Baltimore, Md: Association for Computational Linguistics, 2014; 55–60. [Google Scholar]
- 68.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med 1998;37(1):1–7. [PubMed] [Google Scholar]
- 69.Elkins JS, Friedman C, Boden-Albala B, Sacco RL, Hripcsak G. Coding neuroradiology reports for the Northern Manhattan Stroke Study: a comparison of natural language processing and manual review. Comput Biomed Res 2000;33(1):1–10. [DOI] [PubMed] [Google Scholar]
- 70.Lakhani P, Kim W, Langlotz CP. Automated detection of critical results in radiology reports. J Digit Imaging 2012;25(1):30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lakhani P, Langlotz CP. Automated detection of radiology reports that document non-routine communication of critical or significant results. J Digit Imaging 2010;23(6):647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dreyer KJ, Kalra MK, Maher MM, et al. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study. Radiology 2005;234(2):323–329. [DOI] [PubMed] [Google Scholar]
- 73.Sistrom CL, Dreyer KJ, Dang PP, et al. Recommendations for additional imaging in radiology reports: multifactorial analysis of 5.9 million examinations. Radiology 2009;253(2):453–461. [DOI] [PubMed] [Google Scholar]
- 74.Yetisgen-Yildiz M, Gunn ML, Xia F, Payne TH. A text processing pipeline to extract recommendations from radiology reports. J Biomed Inform 2013;46(2):354–362. [DOI] [PubMed] [Google Scholar]
- 75.Ratner AJ, Bach SH, Ehrenberg HR, Ré C. Snorkel: Fast Training Set Generation for Information Extraction. Proceedings of the 2017 ACM International Conference on Management of Data. New York, NY: ACM, 2017; 1683–1686. [Google Scholar]
- 76.Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artif Intell Med 2016;66:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lipton ZC, Berkowitz J, Elkan C. A Critical Review of Recurrent Neural Networks for Sequence Learning. http://arxiv.org/abs/1506.00019. Published 2015.
- 78.Sutskever I, Martens J, Hinton GE. Generating text with recurrent neural networks. Proceedings of the 28th International Conference on Machine Learning (ICML-11), 2011; 1017–1024. [Google Scholar]
- 79.Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. http://arxiv.org/abs/1301.3781. Published 2013.
- 80.Pennington J, Socher R, Manning C. Glove: Global Vectors for Word Representation. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). 2014. [Google Scholar]
- 81.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural Architectures for Named Entity Recognition. http://arxiv.org/abs/1603.01360. Published 2016.
- 82.Dernoncourt F, Lee JY, Szolovits P. NeuroNER: an easy-to-use program for named-entity recognition based on neural networks. http://arxiv.org/abs/1705.05487. Published 2017.
- 83.Chen MC, Ball RL, Yang L, et al. Deep Learning to Classify Radiology Free-Text Reports. Radiology 2018;286(3):845–852. [DOI] [PubMed] [Google Scholar]
- 84.Banerjee I, Chen MC, Lungren MP, Rubin DL. Radiology report annotation using intelligent word embeddings: Applied to multi-institutional chest CT cohort. J Biomed Inform 2018;77:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banerjee I, Ling Y, Chen MC, et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif Intell Med 2018 Nov 23 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kingma DP, Rezende DJ, Mohamed S, Welling M. Semi-Supervised Learning with Deep Generative Models. arXiv [cs.LG]. http://arxiv.org/abs/1406.5298. Published 2014.
- 87.Schlegl T, Seeböck P, Waldstein SM, Schmidt-Erfurth U, Langs G. Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery. arXiv [cs.CV]. http://arxiv.org/abs/1703.05921. Published 2017. [DOI] [PubMed]
- 88.Kahn CE, Jr, Langlotz CP, Burnside ES, et al. Toward best practices in radiology reporting. Radiology 2009;252(3):852–856. [DOI] [PubMed] [Google Scholar]
- 89.Rubin DL, Kahn CE, Jr. Common Data Elements in Radiology. Radiology 2017;283(3):837–844. [DOI] [PubMed] [Google Scholar]
- 90.Hoogi A, Beaulieu CF, Cunha GM, et al. Adaptive local window for level set segmentation of CT and MRI liver lesions. Med Image Anal 2017;37:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weston AD, Korfiatis P, Kline TL, et al. Automated Abdominal Segmentation of CT Scans for Body Composition Analysis Using Deep Learning. Radiology 2019;290(3):669–679. [DOI] [PubMed] [Google Scholar]
- 92.Revesz G, Kundel HL, Bonitatibus M. The effect of verification on the assessment of imaging techniques 1983. Invest Radiol 1990;25(4):461–464. [DOI] [PubMed] [Google Scholar]
- 93.Krishna R, Zhu Y, Groth O, et al. Visual Genome: Connecting Language and Vision Using Crowdsourced Dense Image Annotations. Int J Comput Vis 2017;123(1):32–73. [Google Scholar]
- 94.Jouppi NP, Young C, Patil N, et al. In-datacenter performance analysis of a tensor processing unit. 2017 ACM/IEEE 44th Annual International Symposium on Computer Architecture (ISCA), 2017; 1–12. [Google Scholar]
- 95.Dunnmon JA, Yi D, Langlotz CP, Ré C, Rubin DL, Lungren MP. Assessment of Convolutional Neural Networks for Automated Classification of Chest Radiographs. Radiology 2019;290(2):537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheller MJ, Anthony Reina G, Edwards B, Martin J, Bakas S. Multi-Institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation. arXiv [cs.LG]. http://arxiv.org/abs/1810.04304. Published 2018. [DOI] [PMC free article] [PubMed]
- 97.Chang K, Balachandar N, Lam C, et al. Distributed deep learning networks among institutions for medical imaging. J Am Med Inform Assoc 2018;25(8):945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Char DS, Shah NH, Magnus D. Implementing Machine Learning in Health Care - Addressing Ethical Challenges. N Engl J Med 2018;378(11):981–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Finlayson SG, Chung HW, Kohane IS, Beam AL. Adversarial Attacks Against Medical Deep Learning Systems. arXiv [cs.CR]. http://arxiv.org/abs/1804.05296. Published 2018.
- 100.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016; 2921–2929. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The first set of challenges for foundational AI research relates to data availability and sharing. Table 1 highlights the important opportunities in this area.
Table 1:
Research Opportunities in AI for Medical Imaging Related to Data Sharing and Data Availability

Note.—FAIR = findable, accessible, interoperable and reusable, MIRC-CTP = Medical Image Resource Center-Clinical Trials Processor.
Data Needs for Machine Learning Research
The first major barrier to progress in machine learning in medical imaging is the lack of standard and accessible imaging data for training of machine learning algorithms. The development of new AI methods requires high-quality, labeled, curated, publicly available data. While health care organizations worldwide control vast stores of data that could be exploited to train machine learning algorithms, most imaging data are not accessible for research purposes. Accessible imaging data are often unusable because they are not curated, organized, anonymized, or appropriately annotated, and rarely linked to a ground-truth diagnosis. Few well-curated and validated image data sets are available to the research and vendor community. To address these gaps, more effective methods are needed for data collection, de-identification, and management of images for research that use findable, accessible, interoperable and reusable (FAIR) principles for scientific data management and stewardship (20).
A small number of imaging data sets have been made public across imaging domains. For example, there are several data sets available for neuroimaging research (21–26). However, many public data sets are too small to support clinically meaningful machine learning experiments or consist mostly of healthy individuals or functional MRI data from patients with psychiatric diseases. Those that are available often were obtained from a single institution and do not reflect the variety of imaging devices and clinical scenarios that will be encountered in real-world settings. A few larger data sets have been constructed, with the potential to be useful for machine learning experiments. For example, The Cancer Imaging Archive hosts imaging data from multiple cancers; the largest data set, containing over 50 000 patient studies, comes from the National Lung Screening Trial (27) and is patient matched to clinical and genomics data in The Cancer Genome Atlas. The NIH has recently released large chest x-ray (28) and chest CT (29) data sets with weak annotations specifically for training machine learning models. A data set for detecting abnormalities on musculoskeletal radiographs is available (30), as is a data set containing both k-space and reconstructed images for knee MRI studies (31). A data set of nearly 600 000 chest radiographs with high-quality labels was recently released as a joint effort of two large research groups (32).
Researchers face significant challenges in finding and accessing the medical imaging data sets that are publicly available. Google recently announced a data set search tool, which provides pointers to data sets across a variety of web-based repositories (33). As more clinical imaging data sets become available, tools of this kind will become essential to find clinical imaging data sets.
Image De-identification
The automated de-identification of images is addressed today by open-source tools like the Medical Imaging Resource Center–Clinical Trials Processor (MIRC-CTP), which receives DICOM images and replaces the protected health information (PHI) they contain with de-identified data (34). While anecdotal evidence suggests these systems sometimes miss PHI in custom DICOM tags, training data to measure the accuracy of these de-identification systems are scarce because they contain PHI. Lack of validated standards and technology for the de-identification of images and reports reduces the willingness of individual sites to share data, for fear of a breach of PHI leading to litigation and federal penalties. Research is needed to measure and enhance the capabilities of current image and report de-identification methods.
Quality Rated Data for Reconstruction and Enhancement of Clinical Images
Clinical images produced by sophisticated imaging devices, such as CT, MRI, PET/SPECT, US, and optical scanners, are reconstructed from “raw” or source data produced by detectors. The resulting signals are indirect and imperfect evidence of anatomic, functional, cellular, and molecular features. The relationships between these tomographic data and underlying structures are generally nonlinear and complex. The optimal conversion of source data from these sensors into reconstructed tomographic images suitable for human interpretation or radiomics is an emerging area of extensive study. Deep learning methods can be highly effective in reconstructing images directly from source data (35). For example, these data-driven image reconstruction methods can perform superior MRI reconstruction, predict the image enhanced with a full contrast agent dose from a partial dose counterpart, or a high-quality image from a low-radiation-dose scan (36–39). The result may be shorter scan times and, because less expensive components are needed, lower cost imaging devices.
In some cases, training data for these methods are readily available. For example, the first serial counts from a PET scanner (ie, in “list mode”) are a realistic simulation of a low-dose study; the full study can serve as the reference standard. Likewise, a test-dose of contrast agent before a full dose study can serve as training data for machine learning. But in many other cases, we can only measure the synthetic images against radiologists’ perception of image quality, against basic measures of quality such as noise and spatial resolution, or against patient outcomes that are noisy and can take many years to occur. For this reason, data sets and benchmarks are needed for subjective image quality ratings that address the suitability of the images for specific clinical tasks. These resources would serve as training data for the production of many image reconstruction and enhancement methods and could also be used to produce deep learning algorithms that simulate human image quality ratings.
The source data needed for this form of research can be difficult to obtain. List-mode data, sinogram data, and k-space data are often kept in a proprietary format that impedes data sharing and can raise intellectual property concerns if shared, so industry cooperation often is required. The International Society for Magnetic Resonance in Medicine has proposed a standard for the exchange of MRI source data (40), but few vendors make MRI source data available in this format. The challenges of obtaining CT sinogram data are even more formidable. Professional societies and industry must work together to make this form of data more accessible to researchers.
Methods and Standards for Patient-mediated Data Sharing
In the past decade, we have overcome many technical challenges of cloud-based sharing of clinical images and reports between care teams at disparate facilities. But the logistical, operational, and regulatory challenges of record sharing between institutions for research requires substantial additional effort and resources. Attempts to aggregate data for research initiatives remain limited and do not scale effectively to meet the needs of AI research: large data sets, acquired from heterogeneous sources, with diverse patient representation. While there are national imperatives that encourage sharing of research data, there are barriers to data access and usability and challenges in combining data sets from multiple sources.
Patient-mediated data sharing has the potential to dramatically increase the number, type, and variety of available data for machine learning by breaking down institutional barriers to sharing. Patients today are more involved and engaged in their care and more recently have become actively engaged in advancing medical research. The RSNA Image Share Network demonstrated how patients can take ownership of their imaging examinations and exchange them at their discretion, first using Simple Object Access Protocol–based standards developed by Integrating the Healthcare Enterprise, then updated to incorporate Fast Health care Interoperability Resources and DICOMweb (41).
Despite these important enabling steps, there are no established mechanisms or standards for patients to share their medical data for research, and there is no common repository or exchange clearinghouse for patient data outside individual health care organizations. Initiatives such as the NIH-sponsored All of Us/Sync 4 Science program, and the nonprofit Count Me In collaborative, aim to advance personalized medicine and research (42). As these initiatives are expanded to educate patients about the importance of data sharing for research, patients should be encouraged to share not only their electronic medical record (EMR) information but also their images.